Abstract
A 40-amino-acid sequence located in the nonstructural 5A (NS5A) protein of hepatitis C virus genotype 1b (HCV-1b) was recently suggested to be the interferon sensitivity-determining region (ISDR), because HCV-1b strains with an ISDR amino acid sequence identical to that of the prototype strain HCV-J were found to be resistant to alpha interferon (IFN-α) whereas strains with amino acid substitutions were found to be sensitive (N. Enomoto, I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato, J. Clin. Invest. 96:224–230, 1995; N. Enomoto, I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato, N. Engl. J. Med. 334:77–81, 1996). We used single-strand conformation polymorphism (SSCP) analysis, combined with cloning and sequencing strategies, to characterize NS5A quasispecies in HCV-1b-infected patients and determine the relationships between pre- and posttreatment NS5A quasispecies mutations and the IFN-α sensitivity of HCV-1b. The serine residues involved in phosphorylation of NS5A protein were highly conserved both in the various patients and in quasispecies in a given patient, suggesting that phosphorylation is important in NS5A protein function. A hot spot for amino acid substitutions was found at positions 2217 to 2218; it could be the result of either strong selection pressure or tolerance to these amino acid replacements. The proportion of synonymous mutations was significantly higher than the proportion of nonsynonymous mutations, suggesting that genetic variability in the region studied was the result of high mutation rates and viral replication kinetics rather than of positive selection. Sustained HCV RNA clearance was associated with low viral load and low nucleotide sequence entropy, suggesting (i) that the replication kinetics when treatment is started plays a critical role in HCV-1b sensitivity to IFN-α and (ii) that HCV-1b resistance to IFN-α could be conferred by numerous and/or related mutations that could be patient specific and located at different positions throughout the viral genome and could allow escape variants to be selected by IFN-α-stimulated immune responses. No NS5A sequence appeared to be intrinsically resistant or sensitive to IFN-α, but the HCV-J sequence was significantly more frequent in nonresponder quasispecies than in sustained virological responder quasispecies, suggesting that the balance between NS5A quasispecies sequences in infected patients could have a subtle regulatory influence on HCV replication.
Hepatitis C virus (HCV) is a small enveloped RNA virus which causes chronic liver disease, including chronic active hepatitis in up to 80% of infected individuals, as well as cirrhosis and hepatocellular carcinoma (1). The estimated mean prevalence of infection in industrialized countries is 1 to 2%. Many patients have been infected through blood transfusions or intravenous drug use, but the cause of infection remains unknown in about 20% of cases (63). No antiviral drugs targeting specific HCV proteins have been developed, although in vitro models are now available to screen such drugs (28, 35, 54). The only approved drug is alpha interferon (IFN-α), whose antiviral effect results from both direct nonspecific inhibition of viral replication and modulation of the immune response to viral epitopes (2, 51, 70).
In a recent meta-analysis of controlled trials of the effect of IFN-α in chronic hepatitis C (60), the rate of sustained biochemical responses (defined as sustained normalization of serum alanine aminotransferase [ALT] activity after treatment withdrawal) to a standard regimen of IFN-α (3 MU subcutaneously 3 times a week for 6 months) was 22%. Recent data suggest that about 80% of patients with a sustained biochemical response achieve a sustained virological response, defined as sustained HCV RNA clearance (4). Several independent predictors of a sustained virological response to IFN have been identified in multivariate analyses. They include age, the presence of cirrhosis on liver biopsy, the titer of anti-HCV core immunoglobulin M, and virological parameters such as viral load, the HCV genotype, and the genetic heterogeneity of hypervariable region 1 (HVR1) of the HCV genome (9, 22, 43, 46, 49, 53, 58, 66).
The independent predictive influence of virological parameters on sustained HCV clearance in patients receiving IFN-α therapy raises the question whether intrinsic characteristics of HCV strains confer sensitivity or resistance to IFN-α. Recently, Enomoto et al. (15, 16) analyzed the NS5A gene of HCV genotype 1b (HCV-1b) strains by direct sequencing. They found a 40-amino-acid stretch in the NS5A protein that they designated the interferon sensitivity-determining region (ISDR), because HCV-1b strains with an amino acid sequence identical to that of the prototype HCV-1b strain HCV-J, isolated in 1990 in Japan by Kato et al. (33), were resistant to IFN-α while strains with amino acid substitutions were sensitive to IFN-α (15). The authors therefore suggested that sequence analysis of the ISDR before therapy might be used to predict the response to IFN-α (15, 16). Using a similar approach based on direct sequencing of the NS5A gene, several authors confirmed the results of Enomoto et al. (3, 40, 47, 48) while others found frequent amino acid substitutions in the NS5A gene of nonresponders to IFN-α and concluded that the response was not related to the number of amino acid substitutions in the putative ISDR (26, 34, 67, 76).
HCV, like other RNA viruses (5, 6, 10–12, 14, 29, 59, 71, 74), exists within its hosts as pools of genetically distinct but closely related variants referred to as quasispecies (44). This probably confers a significant survival advantage, since the simultaneous presence of multiple variant genomes and the high rate at which new variants are generated allow rapid selection of mutants better suited to new environmental conditions (44). The genetic heterogeneity of the HCV quasispecies population results from a high RNA-dependent RNA polymerase error rate (with misincorporation frequencies averaging about 10−4 to 10−5 per base site) and the apparent absence of any error correction or proofreading mechanism (6). Most mutant viral particles cannot replicate, but the remainder can transmit the new genetic information to their progeny. The “fittest” infectious particles are selected on the basis of their replication capacities and especially by the selective pressure exerted by their interactions with host cell proteins and the immune response, which targets regions encoding both cytotoxic and neutralizing epitopes (12, 13, 17, 37, 38, 72, 73).
The quasispecies distribution of HCV genomes in infected individuals implies that direct sequencing of any genomic region gives access only to dominant or consensus viral sequences. Thus, direct sequencing is not suited to addressing the issue of IFN-α sensitivity- and resistance-associated sequences within any coding region of the HCV genome, including the NS5A region. In this study, we used single-strand conformation polymorphism (SSCP) analysis, combined with cloning and sequencing strategies, to characterize NS5A quasispecies and their nucleotide and amino acid sequences in HCV-1b-infected patients who responded or failed to respond to IFN-α therapy. Using these techniques, we determined the relationships between pre- and posttreatment NS5A quasispecies mutations and HCV-1b sensitivity to IFN-α therapy. Hypotheses on the mechanisms underlying HCV-1b resistance to IFN-α are given.
MATERIALS AND METHODS
Patients and samples.
We studied 45 consecutive patients who had chronic hepatitis C related to HCV-1b infection and who were eligible for IFN-α therapy. The genetic heterogeneity of the NS5A gene was studied before treatment in all 45 patients by means of reverse transcriptase-PCR-SSCP. SSCP analysis was initially developed to detect sequence differences in single-stranded DNA by nondenaturing polyacrylamide gel electrophoresis (52, 65). In this technique, the target sequences are amplified by PCR. The products are denatured by heating in the presence of formamide to obtain single-stranded DNA and are analyzed by nondenaturing polyacrylamide gel electrophoresis. The mobility of the separated strands depends on their sequence-specific three-dimensional conformation. Therefore, heterogeneous mixtures of mutant genomes, such as viral quasispecies, can be separated into different bands by PCR-SSCP analysis (52, 65).
The patients were treated with 3 MU of IFN-α2a (Roferon-A; Roche Laboratories, Basel, Switzerland) subcutaneously 3 times a week for at least 3 months. At month 3 of therapy, patients with elevated ALT levels were considered nonresponders while patients with normal ALT activity received a further 3 months of treatment with the same dose (i.e., until month 6). The ALT activity in serum was used as a biochemical index of the response to IFN-α. A sustained biochemical response was defined as normal ALT activity at month 12, i.e., 6 months after treatment withdrawal. All patients with a sustained biochemical response at month 12 underwent HCV RNA determination by highly sensitive nested PCR. Patients who were PCR negative at this time were defined as sustained virological responders.
Thirteen patients were selected for sequence analysis of the NS5A gene. They comprised five patients with a sustained virological response, defined by normal ALT activity and negative HCV RNA PCR 6 months after IFN-α withdrawal (sustained HCV clearance) and eight nonresponders randomly selected from the initial 45 patients, all of whom had elevated ALT levels and detectable HCV viremia at treatment withdrawal and 6 months later. For the epidemiological, histological, and virological features of these 13 patients, see Table 1.
TABLE 1.
Epidemiological, clinical, and histological features of the 13 patients studied and characteristics of the NS5A quasispecies before treatment
| Patient | Sex | Age (yr) | Liver histologya | Response to IFN-αb | Viral load (log10 copies/ml)c | Nd | Normalized entropy
|
Mean genetic distance (SEM) within samplese | Type of mutational changef
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleo- tide | Amino acid | Mean (SEM) synonymous | Mean (SEM) nonsynonymous | ||||||||
| 1 | Female | 37 | CAH | SVR | 5.5 | 30 | 0.156 | 0.114 | 0.0715 (0.0279) | 0.2022 (0.0668) | 0.0373 (0.0143)* |
| 2 | Male | 32 | CAH | SVR | 5.9 | 31 | 0.265 | 0.246 | 0.0135 (0.0017) | 0.0000 (0.0000) | 0.0183 (0.0084) |
| 3 | Male | 53 | CAH | SVR | 3.9 | 48 | 0.127 | 0.085 | 0.0115 (0.0012) | 0.0101 (0.0102) | 0.0116 (0.0069) |
| 4 | Male | 30 | CAH | SVR | 4.2 | 48 | 0.219 | 0.000 | 0.0110 (0.0012) | 0.0362 (0.0216) | 0.0032 (0.0032) |
| 5 | Female | 38 | CAH | SVR | 4.7 | 36 | 0.455 | 0.436 | 0.0608 (0.0107) | 0.1220 (0.0401) | 0.0287 (0.0103)* |
| 6 | Male | 44 | CAH | NR | 5.3 | 30 | 0.236 | 0.180 | 0.0391 (0.0109) | 0.1279 (0.0539) | 0.0131 (0.0093)* |
| 7 | Female | 69 | CAH | NR | 5.4 | 30 | 0.695 | 0.466 | 0.0656 (0.0039) | 0.2007 (0.0562) | 0.0264 (0.0090)** |
| 8 | Male | 32 | CAH | NR | 6.2 | 35 | 0.439 | 0.085 | 0.0380 (0.0061) | 0.1340 (0.0444) | 0.0112 (0.0056)** |
| 9 | Male | 55 | CAH | NR | 5.6 | 40 | 0.424 | 0.240 | 0.0262 (0.0028) | 0.0846 (0.0295) | 0.0090 (0.0054)* |
| 10 | Male | 67 | CAH +cirrhosis | NR | 5.8 | 36 | 0.761 | 0.473 | 0.0643 (0.0021) | 0.2135 (0.0508) | 0.0239 (0.0088)*** |
| 11 | Female | 63 | CAH | NR | 5.7 | 34 | 0.750 | 0.331 | 0.0528 (0.0032) | 0.2053 (0.0590) | 0.0131 (0.0067)** |
| 12 | Male | 28 | CAH | NR | 5.6 | 48 | 0.652 | 0.226 | 0.0371 (0.0023) | 0.1158 (0.0378) | 0.0116 (0.0070)** |
| 13 | Female | 64 | CAH | NR | 6.1 | 36 | 0.551 | 0.291 | 0.0595 (0.0047) | 0.2073 (0.0532) | 0.0200 (0.0076)**** |
CAH, chronic active hepatitis.
SVR, sustained virological response, characterized by normal ALT and negative HCV RNA PCR during treatment and 6 months after; NR, nonresponse, characterized by the lack of effect of IFN-α on ALT and HCV RNA.
Viral load was measured by the Amplicor HCV Monitor assay (Roche Molecular Systems, Branchburg, N.J.).
Number of independent clones analyzed before treatment.
The within- and between-sample genetic distances within NS5A quasispecies were calculated by using the DNADIST program in the PHYLIP package version 3.752 (20).
The proportions of synonymous mutations per synonymous site and of nonsynonymous mutations per nonsynonymous site were calculated by means of the MEGA program (39). P values lower than 0.05 were considered to indicate significant differences. The proportion of synonymous substitutions per potential synonymous site was compared to the proportion of nonsynonymous mutations per potential nonsynonymous site by using a t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; P ≥ 0.05 was considered not significant).
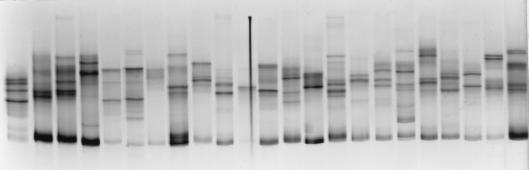
A 185-bp fragment of the NS5A gene was amplified by PCR in the pretreatment samples from all 13 patients. PCR products were cloned into the pTAg vector, and 30 to 48 clones per patient were analyzed by SSCP to determine the frequency of each quasispecies variant (clonal frequency analysis [24]). Indeed, clones with the same sequence gave identical patterns in this technique while clones with different sequences gave consistently different patterns (Fig. 1). In all instances, a second SSCP analysis was performed in the putative order of frequency deduced from the first analysis to better discriminate between clones with close but nonidentical SSCP patterns (Fig. 1). One to three (when available) clones were then sequenced for each SSCP pattern. The same procedure was used at month 12 (end of follow-up) with four nonresponders.
FIG. 1.
Clonal frequency analysis by SSCP. A 185-bp fragment in the NS5A gene of HCV was amplified by PCR, and the PCR product was cloned into the pTAg vector. Thirty clones were isolated, clonal DNA was amplified by PCR, and the PCR products were analyzed by SSCP. Each clone theoretically gives two bands in SSCP, corresponding to the two strands of double-stranded PCR products, which migrate to positions depending strictly on their nucleotide sequence. At the low temperature used for migration (3°C), minor conformations of DNA strands can generate weak additional bands which also migrate to specific positions. A first SSCP round was performed in random order. The second SSCP round, in which the clones are in the order of frequency deduced from the first analysis, is shown. Each lane corresponds to a different NS5A clone. After clonal frequency analysis, one to three (when available) clones per SSCP pattern were sequenced.
HCV RNA quantification.
HCV RNA was quantified before therapy by means of a commercial noncompetitive PCR-based assay (Amplicor HCV Monitor; Roche Molecular Systems, Branchburg, N.J.) as specified by the manufacturer (55). The results were expressed in log10 genome copies per milliliter.
5′ untranslated region PCR.
A sustained virological response was defined by a negative 5′ untranslated region PCR in patients with normal ALT activity 6 months after IFN-α withdrawal. A previously described nested reverse transcriptase PCR technique was used (56).
NS5A PCR.
RNA was first extracted from 50 μl of serum with RNAzol (RNA-B; Bioprobe Systems, Montreuil-sous-Bois, France) and chloroform. After alignment of European HCV genotype 1b sequences, conserved primers were designed (34) to amplify a 185-bp fragment containing, downstream, the portion of the so-called ISDR in which mutations had been reported in responders (15, 16) and, upstream, three serine residues at amino acid positions 2197, 2201, and 2204, which were recently reported to be mandatory for hyperphosphorylation of the NS5A gene product (68). RNA was reverse transcribed at 42°C for 90 min with 100 pmol of random hexamers in the presence of 8 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). Seminested PCR was performed. The first round used 5 pmol of upstream primer HCPr 378 (5′GTGCTCACTTCCATGCTCACCG; nucleotides 6837 to 6858 in the HCV-J sequence) and downstream primer HCPr 382 (5′GTTTCCGCCCATCTCCTGCCGC3′; nucleotides 7028 to 7049) (34). The second round used upstream primer HCPr 379 (5′CCCACATTACAGCAGAGACGGC3′; nucleotides 6865 to 6886) and downstream primer HCPr 382 (34). The two rounds of PCR were performed with 2.5 U of Taq DNA polymerase (Pharmacia Biotech, Uppsala, Sweden) under the same conditions, i.e., a 5-min denaturation at 94°C followed by 35 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s) and then by a final PCR extension at 72°C for 5 min. Amplified products were analyzed by electrophoresis through a 3% NuSieve agarose gel (FMC, Rockland, Maine) and staining with ethidium bromide.
Cloning.
PCR products were purified with the Sephaglas BandPrep kit (Pharmacia Biotech) as specified by the manufacturer. Purified products were quantified by ethidium bromide staining with DNA standards as controls; 50 ng was directly ligated into 50 ng of pTAg vector (LigATor cloning kit; R&D Systems, Abingdon, United Kingdom). Transformation of recombinant plasmid DNA into Escherichia coli competent cells was performed as specified by the manufacturer (R&D Systems), and transformants were grown on ampicillin-tetracycline plates. Cloned DNA was reamplified by the same NS5A-specific PCR procedure as that described above and used for SSCP analysis.
SSCP analysis.
Amplified products were extracted from the agarose gel and purified with the Sephaglas BandPrep kit (Pharmacia Biotech) as specified by the manufacturer. Purified PCR products were eluted in 20 ml of dilution buffer. SSCP analysis was performed with the PCR fragment analysis kit (Pharmacia Biotech). An average of 50 ng of amplified DNA recovered from each serum sample was diluted in 4.5 μl of sterile distilled water plus 4.5 μl of a solution containing 10 mM NaOH and 2 mM EDTA, and bromophenol blue was added. The samples were denatured for 10 min at 100°C and chilled on ice immediately. Then 8-μl portions of the denatured samples were loaded into the wells of a discontinuous polyacrylamide gel (CleanGel; Pharmacia Biotech) which had been rehydrated to 0.5-mm thickness with a buffer specially designed for DNA separation (pH 7.3). Horizontal electrophoresis was performed in a Multiphor II electrophoresis apparatus (Pharmacia Biotech) at 3°C and 100 V for 20 min (penetration) and 600 V for 70 min (migration and stacking). The gel was then subjected to a rapid, sensitive silver-staining procedure with a silver-staining DNA kit (Pharmacia Biotech), a procedure that can detect 0.5 to 2 ng of DNA. After electrophoresis, the gel was fixed for 30 min at room temperature in 10% acetic acid, washed, and incubated for 20 min in 200 ml of a solution containing 0.1% (wt/vol) AgNO3 and 0.1% formaldehyde. The gel was rinsed, placed in 200 ml of a solution containing 2.5% Na2CO3, 0.1% formaldehyde, and 0.002% sodium thiosulfate, and slowly agitated until staining became visible. The reaction was stopped by incubation for 20 min in 40 mM EDTA, and the stain was fixed by incubation for 20 min at room temperature in a solution containing 10% glycerol.
Sequencing.
After cloning and PCR amplification of the clones, the PCR products were amplified by the dye termination method in an ABI 377 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction (Applied Biosystems) as specified by the manufacturer. The sequencing primers were HCPr 379 and HCPr 382.
Calculation of genetic diversity and genetic distances.
We determined the genetic diversity of HCV strains in the region of interest by calculating entropy, which is defined in terms of the probabilities of the different sequences or clusters of sequences that can appear at a given time point. This measure, also known as Shannon entropy (74), is calculated as S = −∑i (pi ln pi), where pi is the frequency of each sequence in the viral quasispecies. The normalized entropy, Sn, was calculated as Sn = S/ln N, where N is the total number of sequences analyzed. Sn theoretically varies from 0 (no diversity) to 1 (maximum diversity). Normalized entropy was calculated at both the nucleotide and amino acid levels on the basis of the first 30 clones isolated before treatment in the 13 patients.
Nucleotide sequences were aligned with the CLUSTAL W program version 1.5 (69). Distances between pairs of sequences were calculated by using the DNADIST module in the PHYLIP package version 3.572 (20). The calculation was based on a Kimura two-parameter distance matrix with a transition-to-transversion ratio of 2.0. The mean and standard error of the mean (SEM) within-sample genetic distances were calculated for the quasispecies in each of the 13 patients before treatment. The numbers of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site, respectively, were calculated with the Jukes-Cantor correction for multiple substitutions, using the MEGA program (31, 39). In the four nonresponders studied at month 12, the mean and SEM of the between-sample genetic distances were calculated on the basis of distances between pairs of pre- and posttreatment sequences, as were the numbers of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site. Statistical comparisons were made with a t-test.
Construction of phylogenetic trees.
The PHYLIP program version 3.572 (20) was used to construct phylogenetic trees by means of the neighbor-joining method (64) with a sequence matrix determined by the two-parameter method of Kimura. Bootstrap support was determined by 100 resamplings of the sequences (19).
RESULTS
Quasispecies distribution of HCV in the NS5A gene.
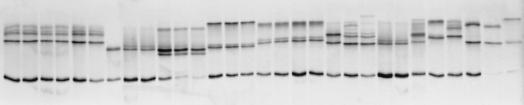
A 185-bp NS5A PCR product was generated from the pretreatment samples from 45 HCV-1b-infected patients. This fragment included, upstream, the three serine residues mandatory for hyperphosphorylation of the NS5A protein (68) and, downstream, the region containing the putative IFN-α sensitivity-determining sequences (15, 16). The PCR products were initially analyzed by SSCP. As shown in Fig. 2, sera were characterized by specific SSCP patterns, with two to eight bands (mean ± SD, 3.5 ± 1.5) migrating to different positions in the gel. This suggested that different mixtures of NS5A sequences were circulating in the patients, i.e., that the NS5A gene of HCV-1b had a quasispecies distribution.
FIG. 2.
SSCP analysis of NS5A PCR products obtained from pretreatment samples from HCV-1b-infected patients. One patient’s sample is analyzed per lane. The SSCP patterns were different from one patient to the next and were characterized by the presence of two to eight bands (mean, 3.5 ± 1.5) migrating to different positions in the gel. These patterns suggested that the patients harbored mixes of different sequences in the region of interest, i.e., that this region had a quasispecies distribution.
The quasispecies distribution of the region of interest was confirmed by cloning NS5A PCR products obtained from 13 patients before therapy into the pTAg vector. A total of 482 pretreatment NS5A clones (i.e., 30 to 48 clones per patient) were generated and analyzed. For each sample, the clonal frequency was calculated by SSCP and the clones were sequenced. This procedure showed in all instances that the patients harbored complex mixtures of genetically distinct but closely related variants (Fig. 3). The genetic diversity of the region of interest was estimated by calculating normalized nucleotide sequence entropy in the 13 samples on the basis of the first 30 clones isolated. The mean entropy in the 13 patients was 0.441 ± 0.063 (range, 0.127 to 0.761). The degree of diversification of each quasispecies was evaluated as the average genetic distance within the quasispecies (within-sample genetic distance). The mean within-sample genetic distance in the 13 patients was 0.0408 ± 0.0063 (range, 0.0110 to 0.0656). The results are given for each patient in Table 1. The nucleotide sequence entropy and the average within-sample genetic distance were significantly related to each other (r = 0.575, P < 0.04) but were not related to the viral load measured at the same time point (Table 1).
FIG. 3.
Alignment of nucleotide sequences of independent HCV NS5A clones from two patients, characteristic of a quasispecies distribution in the region of interest. Relative frequencies determined by SSCP are shown on the right (the nucleotide sequence between codons 2195 and 2240 of the HCV-1b sequence is shown).
Figure 4 shows the amino acid sequences found before therapy in the NS5A quasispecies from the 13 patients and their relative frequencies. The mean normalized amino acid sequence entropy was 0.244 ± 0.042 (range, 0.000 to 0.473). The amino acid sequence entropy was significantly related to the nucleotide sequence entropy (r = 0.755, P < 0.003) but not to the average within-sample genetic distance within the quasispecies or to the viral load. The types of mutational changes were then examined. As shown in Table 1, the proportion of synonymous substitutions was significantly higher than the proportion of nonsynonymous substitutions in all the patients except for three, who had very low entropy. This suggested that quasispecies mutations in the region of interest were due mainly to random genetic drift. Most of the nonsynonymous mutations resulted in amino acid substitutions at positions 2217 to 2218 (Fig. 4), suggesting either that, in contrast to the rest of the region of interest, this short amino acid stretch was subjected to strong selection pressure or that amino acid replacements encoded by genomic molecules selected for other traits were tolerated at these positions. Other amino acid substitutions were rare and seemed to occur at random positions. The three serine residues at positions 2197, 2201, and 2204, recently suggested to be important for hyperphosphorylation of the NS5A protein (68), were highly conserved among the patients studied and within the quasispecies in each patient. The other serine residues probably involved in phosphorylation of the NS5A protein (61) were also well conserved, except for the serine at position 2200, which was often replaced by a threonine, and the serine at position 2210, which was sometimes replaced by a proline.
FIG. 4.
Amino acid composition (amino acid positions 2195 to 2240) of NS5A protein of quasispecies from 13 patients. The alignment of amino acid sequences was deduced from the nucleotide sequences of 30 to 48 independent clones obtained before IFN-α treatment in 13 patients and 30 to 48 independent clones obtained 6 months after IFN-α withdrawal in 4 of them (patients 10 to 13). Relative frequencies of the variants are shown on the right, as determined by clonal frequency analysis by means of SSCP. Patients 1 to 5 were sustained virological responders to 3 MU of IFN-α three times a week for 6 months; i.e., they cleared HCV during treatment and were still nonviremic 6 months later. Patients 6 to 13 were nonresponders. The HCV-J sequence, reported in 1990 by Kato et al. (33), is given for comparison at the top, and the variants with a sequence identical to HCV-J are identified by an asterisk. The three serine residues mandatory for hyperphosphorylation of the product of the NS5A gene (68) are underlined. Amino acid positions 2217 to 2218, at which nonsynonymous mutations were frequently found, are identified by +.
Relationship between NS5A and sensitivity to IFN-α.
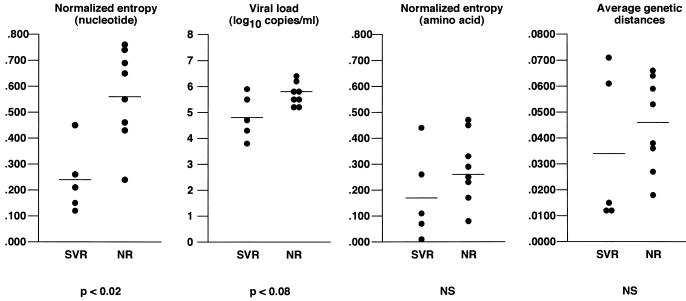
We then compared nucleotide sequence entropy, amino acid sequence entropy, the average within-sample genetic distances, and viral load between the sustained virological responders and nonresponders. As shown in Fig. 5, the genetic diversity of the region of interest, as evaluated by normalized nucleotide sequence entropy, was significantly lower in the sustained virological responders than in the nonresponders (0.244 ± 0.058 and 0.564 ± 0.066, respectively; P < 0.02). Moreover, only one sustained virological responder had a nucleotide sequence entropy higher than 0.300, while only one nonresponder had a nucleotide sequence entropy lower than 0.400 (Table 1). There was also a strong tendency toward a lower viral load in sustained responders than in nonresponders (mean log10 viral load, 4.84 ± 0.38 and 5.38 ± 0.19, respectively; P < 0.08) (Fig. 5; Table 1). In contrast, neither the amino acid sequence entropy nor the average within-sample genetic distance was related to the response to IFN therapy (Fig. 5).
FIG. 5.
Nucleotide sequence entropy, amino acid sequence entropy, average genetic distances within the quasispecies, and viral load in the five patients with a sustained virological response (SVR) to IFN-α therapy and in the eight nonresponders (NR). NS, not significantly different.
No specific NS5A nucleotide or amino acid sequence of pretreatment isolates appeared to be associated with sustained HCV clearance (Fig. 4). Similarly, the nonresponders harbored various NS5A nucleotide and amino acid sequences. However, the HCV-J sequence was significantly more frequent in the nonresponders than in the sustained responders. Indeed, it was found in all eight nonresponders, in whom it represented 16.7 to 94.3% of the quasispecies variants (Fig. 4). Nevertheless, the HCV-J sequence was the dominant sequence in only five of these eight patients. In contrast, only one of the five patients with a sustained virological response harbored the HCV-J sequence within his quasispecies (patient 5, in whom it represented 8.3% of the quasispecies variants). Another responder (patient 1) had a sequence with only one mutation relative to HCV-J, representing 3.3% of the quasispecies variants. In patient 5, the HCV-J sequence was cleared after treatment, together with the other variants, and the patient was still nonviremic 3 years after IFN-α withdrawal, showing that the HCV-J sequence is not intrinsically resistant to IFN-α treatment. Finally, both the nonresponders and the sustained responders harbored quasispecies variants with one to seven mutations relative to the HCV-J sequence, showing that these mutations are not characteristic of sensitive strains (Fig. 4).
Evolution of NS5A quasispecies during and after IFN-α treatment.
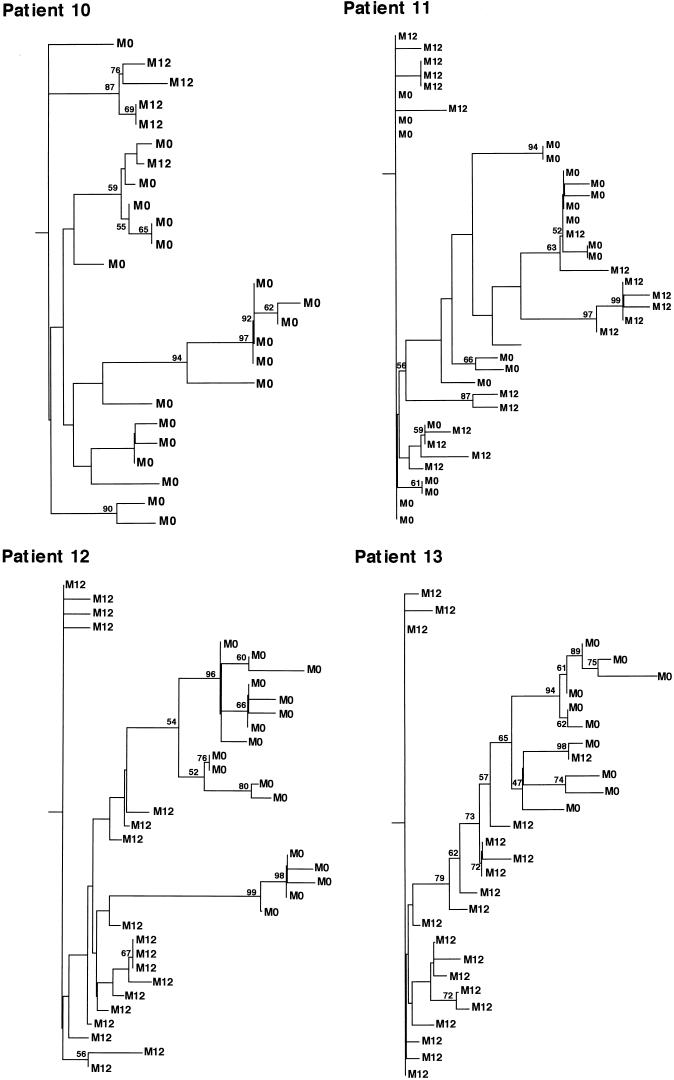
NS5A PCR products were generated from sera sampled 12 months after initiation of therapy, i.e., at the end of follow-up, to determine the evolutionary pattern of NS5A quasispecies during this period. PCR products were cloned into the pTAg vector, and 36 to 48 clones per patient were studied by SSCP for clonal frequency (Table 2). One to three clones (when available) per SSCP pattern were sequenced and compared with pretreatment sequences. As shown in Table 2, the average between-sample (posttreatment versus pretreatment) genetic distances were significantly higher than the average pretreatment within-sample genetic distances, indicating that changes in sequences occurred and that these changes were evolutionary. In addition, when pre- and posttreatment sequences were compared, the proportion of synonymous mutations was always significantly higher than the proportion of nonsynonymous mutations (Table 2). Phylogenetic analysis was used (see Materials and Methods) to study the evolution and viral diversification of NS5A sequences over time. As shown in Fig. 6, variants isolated at treatment initiation and variants isolated 12 months later showed distinctive clustering with the sampling time, compatible with shifts in the population of viruses. This suggested that a true evolutionary process occurred during the study period. Intermingling of sequences was rarely observed, possibly suggesting that pretreatment variants survived.
TABLE 2.
Effect of IFN-α therapy on NS5A quasispecies in four patients tested before treatment and at the end of follow-up
| Patient | N prea | Mean genetic distance (SEM) within sampleb | N postc | Mean genetic distance (SEM) between sampled | Type of mutational changese
|
|
|---|---|---|---|---|---|---|
| Mean (SEM) synonymous | Mean (SEM) nonsynonymous | |||||
| 10 | 36 | 0.0643 (0.0021) | 37 | 0.0944 (0.0131)* | 0.2435 (0.0086) | 0.0216 (0.0016)*** |
| 11 | 34 | 0.0528 (0.0032) | 42 | 0.0610 (0.0007)* | 0.1621 (0.0051) | 0.0136 (0.0005)*** |
| 12 | 48 | 0.0371 (0.0023) | 48 | 0.0450 (0.0011)** | 0.2192 (0.0029) | 0.0172 (0.0007)*** |
| 13 | 36 | 0.0595 (0.0047) | 36 | 0.0936 (0.0018)*** | 0.3336 (0.0074) | 0.0356 (0.0021)*** |
Number of independent clones analyzed before treatment.
Quasispecies average within-sample genetic distances calculated before therapy.
Number of independent clones analyzed at the end of follow-up.
The between-sample mean genetic distance was compared to the pretreatment within-sample mean genetic distance by using a t test. See footnote e for an explanation of asterisks.
The proportion of synonymous substitutions per potential synonymous site was compared to the proportion of nonsynonymous mutations per nonsynonymous site among pre- and posttreatment samples by using a t test (*, P < 0.05; **, P < 0.01; ***, P < 0.0001).
FIG. 6.
Phylogenetic trees of the NS5A region within the four subjects studied before IFN-α treatment and 12 months later. The phylogenetic reconstructions shown are neighbor-joining trees with bootstrap proportions of greater than 50 of 100 bootstrap replicates shown at appropriate branch points. M0 indicates a variant isolated before treatment, and M12 indicates a variant isolated 12 months later.
When pre- and posttreatment NS5A amino acid sequences were compared (Fig. 4), it indeed appeared that most of the variants found after treatment were not detected before treatment whereas most pretreatment variants were no longer found after treatment. Most amino acid changes occurred at positions 2217 to 2218, again suggesting that this short stretch is subjected to strong selection pressure that could be enhanced by IFN-α or that it tolerates amino acid replacements. The HCV-J sequence was present in the quasispecies from the four patients before treatment and was the dominant sequence in three of the patients. This sequence was still found 12 months later in all four patients but was the dominant sequence in only one. Sequences with 1 to 6 amino acid substitutions relative to HCV-J were present in significant proportions in the four patients before and after treatment, indicating that NS5A sequences with mutations relative to HCV-J are not intrinsically sensitive to IFN-α treatment. Finally, it is noteworthy that the serine residues involved in NS5A protein phosphorylation (61, 68) remained highly conserved among the quasispecies variants from the four patients after treatment.
DISCUSSION
This study shows that the NS5A gene of HCV genotype 1b has a quasispecies distribution, which is a characteristic of other regions of the HCV genome, including the 5′ untranslated region, E2, NS2, and NS3 (18, 44, 45, 73). Nucleotide and amino acid sequence analysis of quasispecies variants provides valuable information on how a virus behaves in a constant or changing environment, provided that a sufficient number of clones are analyzed per time point and per patient, i.e., more than 20 (12, 23). We analyzed 30 to 48 clones from each of 13 patients with chronic hepatitis C related to HCV-1b. Calculation of normalized nucleotide sequence entropy showed a high genetic diversity of the region of interest. The three serine residues at positions 2197, 2201, and 2204 showed remarkable conservation, both within quasispecies in a given patient and among the various patients, and also before and after treatment, as did (although to a slightly lesser extent) the other serine residues involved in phosphorylation of the NS5A protein (68). This finding, together with the recent observation that seryl phosphorylation of NS5A protein is a feature conserved among divergent HCV isolates (61), suggests that phosphorylation of NS5A protein, a protein thought to be integrated into the polymerase complex during replication (27, 75), is important for its function (which is unknown). In contrast, amino acid substitutions were frequently observed at positions 2217 to 2218, both within quasispecies in a given patient and among the various patients. This suggested either tolerance to amino acid replacements at these positions or strong selection pressure acting on this short amino acid stretch. Although an HLA B38-restricted cytotoxic epitope including these two amino acids was recently identified in the liver biopsy specimen of an HCV-infected patient (74a), this region does not seem to be a major target for cytotoxic responses. Therefore, selection might instead be due to interactions with host cellular proteins.
In contrast to HVR1, another highly diverse region of the HCV genome subjected to strong selection pressure by neutralizing antibodies (17, 62, 73), the proportion of synonymous mutations was significantly higher than the proportion of nonsynonymous mutations in the portion of the NS5A gene studied here. Entropy measures the repertoire size of viral quasispecies, which results from the accumulation of nucleotide substitutions over time under the variable influence of three parameters: (i) viral production, i.e., replication rates; (ii) rates of misincorporation of viral RNA-dependent RNA polymerases; and (iii) selection of the fittest newly produced variants (13, 14). The significantly higher proportion of synonymous mutations suggests that the overall genetic diversity of the region studied, as measured by nucleotide sequence entropy, reflects a combination of high mutation rates, viral replication kinetics, and limitations to acceptance of amino acid substitutions rather than the positive selection of viral variants (74).
An important finding in this study was that pretreatment nucleotide sequence entropy was significantly lower in the patients who eliminated HCV than in the nonresponders (Fig. 5). The predictive value of low pretreatment entropy for a sustained virological response cannot be deduced from our data, because the eight nonresponders were randomly selected from a larger group of patients. In contrast, all five sustained virological responders were studied. Our results therefore show that patients with a high pretreatment entropy in the region of interest are very unlikely to eradicate HCV when given the classical IFN-α regimen. The viral load was also lower in sustained responders than in nonresponders, and the difference was close to statistical significance. It is noteworthy that in a multivariate analysis of the series of patients from whom the present study group was extracted, a low viral load emerged as an independent predictor of a sustained virological response to IFN-α, in keeping with numerous reports (8, 25, 42, 46, 49, 58).
As recently reported, it can be assumed that the viral load and infected-cell numbers are in steady state before treatment, with a minimal estimated production of the order of 1010 to 1011 virions per day (41, 77), and a maximal estimated half-life of free virions of 0.3 day (41). In this model, the viral load is a good estimate of the level of virus production (41), while our findings indicate that nucleotide sequence entropy in the region of the NS5A gene we studied is dependent largely on viral replication kinetics. Our results therefore suggest that replication kinetics when treatment is started is a major determinant of the response of HCV-1b to 3 MU of IFN-α three times a week. On the other hand, the fact that no major selection pressure appeared to be exerted on the region of the NS5A gene studied (except possibly at amino acid positions 2217 to 2218) suggests that its genetic diversity reflects the overall diversity of the HCV strain whereas other regions subjected to strong pressure, such as HVR1, might be less representative. Since only patients with low nucleotide sequence entropy, i.e., a small HCV quasispecies repertoire, can have sustained HCV RNA clearance, HCV resistance to therapy is unlikely to be related to a small number of specific mutations on the genome, which could be achieved by rapid viral replication starting from a small quasispecies repertoire. The resistance of HCV to IFN-α would instead depend on the presence of complex (i.e., numerous and/or related) mutations, which could be achieved only by starting from a large quasispecies repertoire. Indeed, in quasispecies with low entropy, there is a low probability that all the mutations needed for resistance will be present before treatment or will occur after the initiation of therapy, given the initial effect of IFN-α on virus production (41). In contrast, high pretreatment entropy is associated with a higher probability that the resistance mutations are present, even in minor poorly fit variants, and that these variants can be selected (7).
IFN-α acts on HCV through two phenomena: (i) it causes direct inhibition of HCV replication through activation of the expression of various proteins, such as 2′,5′-oligoadenylate synthetase, Mx protein, or double-stranded RNA-activated protein kinase (PKR), that act as nonspecific antiviral agents in infected cells (2, 51); and (ii) it is a multifunctional immunomodulatory cytokine which induces a number of immunological changes, including expression of class I and class II major histocompatibility complex antigens, activation of cytotoxic T cells and macrophages, and complex interactions with the cytokine cascade (51, 70). Early loss of HCV RNA in serum is necessary for long-term eradication of HCV. This results from the direct antiviral effect of IFN-α (41) and could be largely dependent on the viral kinetics at the time treatment is started. Indeed, the antiviral effect of a single injection of IFN-α was recently shown to be dose dependent and short-lived (less than 24 h) (41). It is therefore possible that primary HCV resistance is due mainly to a mismatch between the recommended IFN-α (3 MU three times per week) and the viral kinetics, which would strongly support the use of higher IFN-α doses and more frequent administration at the outset (41). It must be stressed, however, that in spite of prolonged IFN-α treatment, sustained HCV eradication is obtained in a relatively small proportion of the patients who initially clear HCV RNA, owing to subsequent breakthroughs and relapses (58, 60). It is therefore possible that quasispecies variants against which the host has mounted an immune response are eliminated during IFN-α treatment but that the remaining variants bear mutations giving a selective advantage to escape immune system elimination (36, 50). Such mutations would be numerous, possibly related, and located at different positions throughout the viral genome corresponding to the various epitopes involved in immune system-mediated viral clearance, a hypothesis supported by our results. If so, the mutations conferring IFN-α resistance on HCV-1b would differ among patients, being strictly dependent on the state of the immune system relative to HCV epitopes during therapy. The better efficacy of longer maintenance IFN-α therapy on sustained HCV clearance (30, 60) could then be explained by the fact the IFN-α stimulated immune system has more time to mount an efficient response to escape variants selected during the first months of therapy.
Altogether, these findings suggest that the resistance of HCV-1b to IFN-α is mediated through viral replication kinetics and complex nonspecific mutations related to the host immune response, rather than through specific sequences in the NS5A gene of HCV-1b. Indeed, the HCV-J sequence does not appear to be intrinsically resistant to IFN-α, and mutant sequences are not intrinsically sensitive to IFN-α, while their presence does not appear to confer sensitivity. Nevertheless, the HCV-J sequence was consistently found more often in the nonresponders than in the sustained virological responders. This could explain why the HCV-J sequence or sequences with few mutations relative to HCV-J were more frequently found in nonresponders than in sustained responders in studies based on direct sequencing, which gives access only to dominant or consensus sequences (15, 16, 40, 47, 48).
The presence of the HCV-J sequence in all the nonresponders and in the only sustained virological responder with high entropy suggests that NS5A quasispecies mutations could play a role in the resistance of HCV-1b to IFN-α by influencing viral kinetics and, as a consequence, genomic entropy. Indeed, NS5A protein seems to be integrated into the polymerase complex during replication (27) and could therefore play a critical role in the regulation of viral replication. Recent studies have suggested that the amino acid sequences in the region we studied could be important in the structure and function of the protein. First, the NS5A gene product was recently shown to repress, in vitro, the IFN-α-induced PKR, a mediator of the antiviral effect of IFN-α which directly inhibits the protein synthesis initiation factor eIF2-α (21). Interestingly, the HCV-J sequence bound PKR and inhibited the phosphorylation of eIF2-α, while certain forms with as few as two mutations lost their ability to bind PKR and to inhibit eIF2-α phosphorylation (33a). The relative proportion of these two functionally distinct sets of sequences at a given time point in a given patient might participate in the regulation of HCV replication rates in infected cells, in which antiviral conditions exist naturally through endogenous IFN-α production (57). It could also play a role in IFN-α resistance by influencing replication kinetics at the time treatment is started and/or by at least partly inhibiting the antiviral effect of IFN-α through the interaction with PKR and possibly other mediators of IFN-α action. Second, it has recently been found, in vitro and in cultured cells, that NS5A protein is phosphorylated in the region we studied by an associated cellular serine/threonine kinase that differed from PKR (61). The action of this kinase seems to be highly dependent on the amino acid sequences found immediately upstream and downstream of the phosphate acceptor sites, namely, serines and, to a lesser extent, threonines (61). In this respect, it must be noted that amino acid 2217 in the HCV-J sequence is a threonine which follows another threonine and that the kinase would prefer to phosphorylate serine or threonine residues followed by acidic amino acids (61), which is the case in the region immediately following amino acids 2217 to 2218 in the HCV-J sequence. It is therefore possible that quasispecies mutations regulate the rate of phosphorylation of the protein. It is also possible that, as reported for dengue virus type 2 NS5 protein, the phosphorylation state of NS5A protein correlates with its subcellular location (32). Since replication seems to occur on cytoplasmic membranes surrounding the nucleus and since PKR is mostly a cytoplasmic enzyme, the coexistence of unequally phosphorylated NS5A variants in infected cells might conceivably participate in the regulation of viral replication during chronic infection and might therefore play a role in the resistance of HCV-1b to IFN-α therapy. Strong selection pressures exerted on amino acid positions 2217 to 2218 would be consistent with the involvement of these two amino acids in the interaction with host proteins and/or in the phosphorylation processes. Both in vitro and in vivo structure-function studies are now needed to determine the relative biological activities of NS5A quasispecies variants isolated from individual patients.
The results of the analysis of NS5A quasispecies at the end of the follow-up period in four nonresponders were compatible with a true evolutionary process characterized by shifts in the populations of viruses. No control group of untreated patients was studied, but it has recently been shown that the IFN-α regimen used in this study is associated with significantly higher rates of accumulation of mutations in various regions of the HCV genome than during the natural progression of the disease in untreated patients (22a). Again, most amino acid changes occurred at positions 2217 to 2218, suggesting that viral variants were selected or that substitutions at these positions were tolerated. It must be noted, however, that the posttreatment sequences were different from one patient to the next, indicating that no specific NS5A amino acid sequence is resistant to IFN-α and reinforcing our hypothesis that resistant variants could be specific to each patient. In contrast, nucleotide substitutions not involving amino acids 2217 to 2218 were synonymous in most cases, suggesting that the observed changes reflected enhanced replication kinetics (74). This is consistent with our recent observation of replication rebound during and after treatment (breakthrough and relapse, respectively) in patients who do not clear HCV (58a). It is possible that this rebound is dependent on changes in regions of the HCV genome that are more directly involved both in replication and in elimination by the immune system. In this respect, we recently observed profound changes in the composition of HCV quasispecies in the HVR1, the target of neutralizing responses, during and after IFN-α treatment (58a). These changes could at least partly account for the changes in viral replication kinetics caused by treatment.
ACKNOWLEDGMENTS
This work was supported by a grant from the French Ministry of Health (Programme Hospitalier de Recherche Clinique 1996, contract AOM96-136) and a grant from Roche Products (Neuilly-sur-Seine, France).
We thank Lieven Stuyver for designing NS5A-specific primers for PCR; Anne Bastie, Jean-Michel Métreau, Ariane Mallat, Jean-Philippe Mavier, Gilles Duverlie, and Laurent Castéra for providing patient samples; Isabelle Da Silva, Jocelyne Rémiré, and Françoise Darthuy for excellent technical assistance; and Jean-Marc Josse, Carole Dupont, and the Management Staff of Pharmacia Biotech, Orsay, France, for their support in this study. We are also grateful to Marie-France Saint-Marc Girardin (Produits Roche) for her help. We are indebted to Margaret J. Koziel for critical review of the manuscript.
REFERENCES
- 1.Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 2.Baron S, Tyring S K, Fleischmann W R, Coppenhaver D H, Niesel D W, Klimpel G R, Stanton G J, Hughes T K. The interferons. Mechanisms of action and clinical applications. JAMA. 1991;266:1375–1383. doi: 10.1001/jama.266.10.1375. [DOI] [PubMed] [Google Scholar]
- 3.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 4.Chemello L, Cavalletto L, Casarin C, Bonetti P, Bernardinello E, Pontisso P, Donada C, Belussi F, Martinelli S, Alberti A the TriVeneto Viral Hepatitis Group. Persistent hepatitis C viremia predicts late relapse after sustained response to interferon-α in chronic hepatitis C. Ann Intern Med. 1996;124:1058–1060. doi: 10.7326/0003-4819-124-12-199606150-00005. [DOI] [PubMed] [Google Scholar]
- 5.Chumakov K M. PCR engineering of viral quasispecies: a new method to preserve and manipulate genetic diversity of RNA virus populations. J Virol. 1996;70:7331–7334. doi: 10.1128/jvi.70.10.7331-7334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke D K, Duarte E A, Elena S F, Moya A, Domingo E, Holland J. The red queen reigns in the kingdom of RNA viruses. Proc Natl Acad Sci USA. 1994;91:4821–4824. doi: 10.1073/pnas.91.11.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 8.Conjeevaram H S, Everhart J E, Hoofnagle J H. Predictors of a sustained beneficial response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1326–1329. doi: 10.1016/0270-9139(95)90646-0. [DOI] [PubMed] [Google Scholar]
- 9.Davis G L. Prediction of response to interferon treatment of chronic hepatitis C. J Hepatol. 1994;21:1–3. doi: 10.1016/s0168-8278(94)80128-2. [DOI] [PubMed] [Google Scholar]
- 10.Delwart E L, Sheppard H W, Walker B D, Goodsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dockter J, Evans C F, Tishon A, Oldstone M B A. Competitive selection in vivo by a cell for one variant over another: implications for RNA virus quasispecies in vivo. J Virol. 1996;70:1799–1803. doi: 10.1128/jvi.70.3.1799-1803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingo E. Biological significance of viral quasispecies. Viral Hepatitis Rev. 1996;2:247–261. [Google Scholar]
- 13.Duarte E A, Novella I S, Weaver S C, Domingo E, Wain-Hobson S, Clarke D K, Moya A, Elena S F, de la Torre J C, Holland J J. RNA virus quasispecies: significance for viral disease and epidemiology. Infect Agents Dis. 1994;3:201–214. [PubMed] [Google Scholar]
- 14.Eigen M, Biebricher C. Role of genome variation in virus evolution. In: Domingo E, Holland J, Ahlquist P, editors. RNA genetics. 3. Variability of RNA genomes. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 211–245. [Google Scholar]
- 15.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 17.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feller J A, Grace K, Clarke B E, Lau J Y N. Variations in the translational efficiency of naturally occurring hepatitis C virus (HCV) variants. Implication for the design of antiviral therapy. Hepatology. 1996;24:263A. [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP (phylogeny inference package): version 3.57. Distributed by the author. Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- 21.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Peralta R P, Qian K, She J Y, Davis G L, Ohno T, Mizokami M, Lau J Y N. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J Med Virol. 1996;49:242–247. doi: 10.1002/(SICI)1096-9071(199607)49:3<242::AID-JMV14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 22a.Gretch, D. R. Personal communication.
- 23.Gretch D R, Polyak S J. Groupe Français d’Etudes Moléculaires des Hépatites (GEMHEP) (ed.), Hepatitis C virus: genetic heterogeneity and viral load. Paris, France: John Libbey Eurotext; 1997. The quasispecies nature of hepatitis C virus: research methods and biological implications; pp. 57–72. [Google Scholar]
- 24.Gretch D R, Polyak S J, Wilson J J, Carithers R L, Perkins J D, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara H, Hayashi N, Mita E, Takehara T, Kasahara A, Fusamoto H, Kamada T. Quantitative analysis of hepatitis C virus RNA in serum during interferon alfa therapy. Gastroenterology. 1993;104:877–883. doi: 10.1016/0016-5085(93)91025-d. [DOI] [PubMed] [Google Scholar]
- 26.Halimi G, Halfon P, Gerolami V, Castets F, Khiri H, Bourliere M, Gauthier A P, Cartouzou G. Mutations in NS5A region and interferon response in hepatitis C-1b-infected patients. Hepatology. 1996;24:162A. [Google Scholar]
- 27.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirowatari Y, Hijikata M, Shimotohno K. A novel method for analysis of viral proteinase activity encoded by hepatitis C virus in cultured cells. Anal Biochem. 1995;225:113–120. doi: 10.1006/abio.1995.1116. [DOI] [PubMed] [Google Scholar]
- 29.Holland J J, De La Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 30.Jouët P, Roudot-Thoraval F, Dhumeaux D, Métreau J M le Groupe Français pour l’Etude du Traitement des Hépatites Chroniques NANB/C. Comparative efficacy of interferon alfa in cirrhotic and noncirrhotic patients with non-A, non-B, C hepatitis. Gastroenterology. 1994;106:686–690. doi: 10.1016/0016-5085(94)90703-x. [DOI] [PubMed] [Google Scholar]
- 31.Jukes T H, Cantor T R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 32.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 33.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Katze, M. G. Personal communication.
- 34.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209–2248 amino acid sequence do not predict the response to recombinant interferon alfa therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 35.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O’Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–365. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 36.Koup R A. Virus escape from CTL recognition. J Exp Med. 1994;180:779–782. doi: 10.1084/jem.180.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koziel M J, Dudley D, Afdhal N, Choo Q L, Houghton M, Ralston R, Walker B D. Hepatitis C virus (HCV)-specific cytotoxic T-lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522–7532. doi: 10.1128/jvi.67.12.7522-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koziel M J, Dudley D, Afdhal N, Grakoui A, Rice C M, Choo Q L, Houghton M, Walker B D. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokines release. J Clin Invest. 1995;96:2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis for microcomputers. Comput Appl Biosci. 1993;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 40.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-β therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 41.Lam N P, Neumann A U, Gretch D R, Wiley T E, Perelson A S, Layden T J. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon alfa. Hepatology. 1997;26:226–231. doi: 10.1002/hep.510260130. [DOI] [PubMed] [Google Scholar]
- 42.Magrin S, Craxi A, Fabiano C, Simonetti R G, Fiorentino G, Marino L, Diquattro O, di Marco V, Loiacono O, Volpes R, Almasio P, Urdea M S, Neuwald P, Sanchez-Pescador R, Detmer J, Wilber J C, Pagliaro L. Hepatitis C viremia in chronic liver disease: relationship to interferon-α or corticosteroid treatment. Hepatology. 1994;19:273–279. [PubMed] [Google Scholar]
- 43.Mahaney K, Tedeschi V, Maertens G, Di Bisceglie A M, Vergalla J, Hoofnagle J H, Sallie R. Genotypic analysis of hepatitis C virus in American patients. Hepatology. 1994;20:1405–1411. doi: 10.1002/hep.1840200605. [DOI] [PubMed] [Google Scholar]
- 44.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martell M, Esteban J I, Quer J, Vargas V, Esteban R, Guardia J, Gomez J. Dynamic behavior of hepatitis C virus quasispecies in patients undergoing orthotopic liver transplantation. J Virol. 1994;68:3425–3436. doi: 10.1128/jvi.68.5.3425-3436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, Benhamou J P, Erlinger S. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 47.Murakami T, Enomoto N, Asahina Y, Izumi N, Marumo F, Sato C. Mutations in NS5A region and response to interferon in HCV genotype 2 infection. Hepatology. 1996;24:158A. doi: 10.1002/hep.510300405. [DOI] [PubMed] [Google Scholar]
- 48.Niiyama G, Kimura T, Mitani K, Tsugeno H, Sengoku N, Kinoyama S, Ito T, Kinoshita M, Yamada G. Mutations in the nonstructural protein 5A gene in chronic hepatitis C virus 1b infection patients with interferon treatment. Hepatology. 1996;24:164A. [Google Scholar]
- 49.Nousbaum J B, Pol S, Nalpas B, Landais P, Berthelot P, Bréchot C the Collaborative Study Group. Hepatitis C virus type 1b (II) infection in France and Italy. Ann Intern Med. 1995;122:161–168. doi: 10.7326/0003-4819-122-3-199502010-00001. [DOI] [PubMed] [Google Scholar]
- 50.Nowak M A, May R M, Phillips R E, Rowland-Jones S, Lalloo D G, McAdam S, Klenerman P, Koppe B, Sigmund K, Bangham C R M, McMichael A J. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature. 1995;375:606–611. doi: 10.1038/375606a0. [DOI] [PubMed] [Google Scholar]
- 51.O’Connell J F. Mechanisms of action of interferon: potential role in hepatitis C. Viral Hepatitis Rev. 1997;3:121–128. [Google Scholar]
- 52.Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 53.Orito E, Mizokami M, Suzuki K, Ohba K I, Ohno T, Mori M, Hayashi K, Kato K, Iino S, Lau J Y N. Loss of serum HCV RNA at week 4 of interferon-α therapy is associated with more favorable long-term response in patients with chronic hepatitis C. J Med Virol. 1995;46:109–115. doi: 10.1002/jmv.1890460205. [DOI] [PubMed] [Google Scholar]
- 54.Overton H, McMillan D, Gillespie F, Mills J. Recombinant baculovirus-expressed NS3 proteinase of hepatitis C virus shows activity in cell-based and in vitro assays. J Gen Virol. 1995;76:3009–3019. doi: 10.1099/0022-1317-76-12-3009. [DOI] [PubMed] [Google Scholar]
- 55.Pawlotsky J M. Measuring hepatitis C viremia in clinical samples: can we trust the assays? Hepatology. 1997;26:1–4. doi: 10.1002/hep.510260131. [DOI] [PubMed] [Google Scholar]
- 56.Pawlotsky J M, Fleury A, Choukroun V, Deforges L, Roudot-Thoraval F, Aumont P, Duval J, Dhumeaux D. Significance of highly positive c22-3 “indeterminate” second-generation hepatitis C virus (HCV) recombinant immunoblot assay (RIBA) and resolution by third-generation HCV RIBA. J Clin Microbiol. 1994;32:1357–1359. doi: 10.1128/jcm.32.5.1357-1359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawlotsky J M, Hovanessian A G, Roudot-Thoraval F, Lebon P, Robert N, Bouvier M, Babany G, Duval J, Dhumeaux D. Activity of the interferon-induced 2′-5′-oligoadenylate synthetase in patients with chronic hepatitis C. J Interferon Cytokines Res. 1995;15:857–862. doi: 10.1089/jir.1995.15.857. [DOI] [PubMed] [Google Scholar]
- 58.Pawlotsky J M, Roudot-Thoraval F, Bastie A, Darthuy F, Rémiré J, Métreau J M, Zafrani E S, Duval J, Dhumeaux D. Factors affecting treatment responses to interferon-α in chronic hepatitis C. J Infect Dis. 1996;174:1–7. doi: 10.1093/infdis/174.1.1. [DOI] [PubMed] [Google Scholar]
- 58a.Pawlotsky, J. M., et al. Unpublished data.
- 59.Plyusnin A, Cheng Y, Lehvaslaiho H, Vaheri A. Quasispecies in wild-type Tula hantavirus populations. J Virol. 1996;70:9060–9063. doi: 10.1128/jvi.70.12.9060-9063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poynard T, Leroy V, Cohard M, Thevenot T, Mathurin P, Opolon P, Zarski J P. Meta-analysis of interferon randomized trials in the treatment of viral hepatitis C: effects of dose and duration. Hepatology. 1996;24:778–789. doi: 10.1002/hep.510240405. [DOI] [PubMed] [Google Scholar]
- 61.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y N, Choo Q L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorometric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roudot-Thoraval F, Bastie A, Pawlotsky J M, Dhumeaux D the Study Group for the Prevalence and the Epidemiology of Hepatitis C Virus. Epidemiological factors affecting the severity of hepatitis C virus-related liver disease: a French survey of 6664 patients. Hepatology. 1997;26:485–490. doi: 10.1002/hep.510260233. [DOI] [PubMed] [Google Scholar]
- 64.Saitou N, Mei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 65.Sekiya T. Detection of mutant sequences by single-strand conformation polymorphism analysis. Mutat Res. 1993;288:79–83. doi: 10.1016/0027-5107(93)90209-x. [DOI] [PubMed] [Google Scholar]
- 66.Serfaty L, Giral P, Loria A, Andréani T, Legendre C, Poupon R. Factors predictive of the response to interferon in patients with chronic hepatitis C. J Hepatol. 1994;21:12–17. doi: 10.1016/s0168-8278(94)80130-4. [DOI] [PubMed] [Google Scholar]
- 67.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Bréchot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 68.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tilg H. New insights into the mechanisms of interferon alfa: an immunoregulatory and anti-inflammatory cytokine. Gastroenterology. 1997;112:1017–1021. doi: 10.1053/gast.1997.v112.pm9041265. [DOI] [PubMed] [Google Scholar]
- 71.Wain-Hobson S. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr Top Microbiol Immunol. 1992;176:181–193. doi: 10.1007/978-3-642-77011-1_12. [DOI] [PubMed] [Google Scholar]
- 72.Weiner A, Erickson A L, Kansopon J, Crawford K, Muchmore E, Hughes A L, Houghton M, Walker C M. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA. 1995;92:2755–2759. doi: 10.1073/pnas.92.7.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P J, Miyamura T, McHutchison J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptative evolution of human immunodeficiency virus type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 74a.Wong, D., M. J. Koziel, and B. D. Walker. Personal communication.
- 75.Yamashita T, Shirota Y, Kaneko S, Kobayashi K, Murakami S. Abstracts of the 4th International Meeting on Hepatitis C Virus and Related Viruses. 1997. RNA-dependent RNA polymerase activity of purified bacterial recombinant hepatitis C virus (HCV) NS5B and its binding activity to HCV NS5A; p. 35. [Google Scholar]
- 76.Zeuzem S, Lee J H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 77.Zeuzem S, Schmidt J M, Lee J H, Rüster B, Roth W K. Effect of interferon alfa on the dynamics of hepatitis C virus turnover in vivo. Hepatology. 1996;23:366–371. doi: 10.1002/hep.510230225. [DOI] [PubMed] [Google Scholar]