Abstract
The spread of antimicrobial resistant Campylobacter strains, linked to antimicrobials use and abuse in humans and food animals, has become a global public health problem. In this study, we determine the prevalence of antimicrobial resistance (AMR) in human Campylobacter isolates (n = 820) collected in Piedmont, Italy, from March 2020 to July 2023. The strains underwent susceptibility testing to determine the minimal inhibitory concentration for erythromycin, ciprofloxacin, gentamicin, streptomycin, and tetracycline: 80.1% of the strains showed resistance to at least one antibiotic. The highest prevalence of AMR was noted for ciprofloxacin and tetracycline (72.1% and 52.9%, respectively) and the lowest for erythromycin and aminoglycosides (streptomycin/gentamicin) (3.2% and 5.4%, respectively). The prevalence of co-resistance against fluoroquinolones and tetracyclines was 41.1%. The prevalence of multidrug resistant strains was 5.7%. Our data support evidence that AMR in human Campylobacter strains is common, particularly against ciprofloxacin and tetracycline, two medically important antimicrobials for humans.
Keywords: antimicrobial susceptibility testing, Campylobacter jejuni, Campylobacter coli, foodborne pathogens, human strains, occurrence
1. Introduction
Campylobacter species are Gram-negative, spiral, or curve-shaped bacteria. These microaerophilic, non-fermentative, non-spore-forming, mobile [1] microorganisms are one of the most common causes of diarrhea worldwide. Many Campylobacter species are zoonotic pathogens, associated with a range of gastrointestinal diseases in humans termed campylobacteriosis [2]. Campylobacteriosis is the most commonly reported foodborne zoonosis in Europe (127,840 cases, EU notification rate of 41.1 per 100,000 of population), with a 2.1% increase in the EU notification rate compared with 2020 [3]. Active surveillance through the U.S. Foodborne Diseases Active Surveillance Network (FoodNet) reports about 20 cases of campylobacteriosis per 100,000 people diagnosed each year [4]. Since many more cases probably go undiagnosed or unreported, the CDC estimates that 1.5 million people in the United States become ill from Campylobacter infection every year [4].
Campylobacter infection is associated with gastrointestinal symptoms and inflammation of the gastrointestinal tract involving the small intestine. Symptoms are generally diarrhea. Extra-gastrointestinal complications include reactive arthritis, bacteremia, septicemia, endocarditis, and meningitis [5]. Most Campylobacter infections are mild and self-limiting and require only supportive therapy, while appropriate antibiotic treatment is indicated for severe or prolonged infections and in immunocompromised patients [6]. Among Campylobacter species, C. jejuni and C. coli are the most prevalent causative agents of gastroenteritis. Most infections in humans are correlated with food handling and the consumption of contaminated food (e.g., meat, unpasteurized milk, fruits, vegetables, or water) [7]. Poultry, domestic and wild animals are the primary reservoirs of Campylobacter species. The consumption of raw or undercooked poultry meat is the main risk factor for human campylobacteriosis [8,9,10].
The prevention and control of Campylobacter colonization in food-producing animals, such as poultry flocks, for example, involve public health strategies to reduce the incidence of campylobacteriosis in humans. Within the national surveillance networks for enteric pathogens in human medicine (Enter-NET), the regional reference center for Salmonella typing (Centro di Riferimento per la Tipizzazione delle Salmonelle, CeRTiS) is involved in the identification and characterization of pathogens, including Campylobacter. CeRTiS performs analysis of samples in official microbiological controls, investigates antimicrobial resistance (AMR) profiles, and provides support to agencies in risk analysis and epidemiological outbreak investigations. AMR surveillance of strains of human origin is carried out on a panel of molecules established by the European Centre for Disease Prevention and Control (ECDC) [11].
The antimicrobial resistance of Campylobacter species has increased worldwide [12] largely due to antimicrobial overuse in humans and in food-producing animals. Certain antimicrobials have been used extensively to treat Campylobacter infection in humans. For example, fluoroquinolones and macrolides are the drugs of choice to treat campylobacteriosis and tetracyclines and gentamicin to treat systemic infection in some cases [13]. High rates of AMR to ciprofloxacin, tetracycline, and erythromycin have been observed in C. coli and C. jejuni from human samples and food-producing animals in Europe [14].
The development of AMR has serious implications for treating Campylobacter infection in humans, but antimicrobial susceptibility testing can help guide appropriate therapy and monitor trends in AMR. With the present study, we wanted to determine the occurrence of phenotypic AMR of human Campylobacter species isolated in Piedmont in the period 2020–2023.
2. Materials and Methods
2.1. Sample Collection and Campylobacter Isolation
Isolates were collected via laboratory passive surveillance from March 2020 to July 2023. The Campylobacter strains originated from biological samples collected from symptomatic humans. The strains were first isolated using selective enrichment in Preston broth for 24 h at 42 °C in microaerobic atmosphere (approximately 5% O2, 10% CO2, 85% N2) at clinical laboratories throughout Piedmont. The isolates were then sent to the CeRTiS where they were subcultured on Columbia blood agar (Becton Dickinson, Franklin Lakes, NJ, USA) at 37 °C for 24 h and identified by MALDI TOF/TOF mass spectrometry (Bruker Daltonics GmbH, Bremen, Germany).
2.2. Antimicrobial Susceptibility Testing
Phenotypic testing based on the determination of minimum inhibitory concentrations (MICs) to erythromycin (1–128 µg/mL), ciprofloxacin (0.12–16 µg/mL), gentamicin (0.12–16 µg/mL), streptomycin (0.25–16 µg/mL), and tetracycline (0.5–64 µg/mL) was performed using a commercial microdilution tool (Sensititre Campylobacter plate–EUCAMP2, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. We applied the MIC interpretive resistance standards defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and EFSA [15,16] to define isolates of C. jejuni resistant to erythromycin > 4 µg/mL, ciprofloxacin > 0.5 µg/mL, tetracycline > 2 µg/mL, streptomycin > 4 µg/mL or gentamicin > 2 µg/mL. C. coli was defined resistant when the MIC equated to erythromycin was > 8 µg/mL, ciprofloxacin > 0.5 µg/mL, tetracycline > 2 µg/mL, streptomycin > 4 µg/mL or gentamicin > 2 µg/mL. The MIC ranges, MIC50 and MIC90, were calculated separately for each species; MIC50 and MIC90 represented the antibiotic concentrations (µg/mL) at which 50% and 90% of the Campylobacter isolates could be inhibited, respectively. Multidrug resistance (MDR) was defined as resistance to at least three antimicrobial classes.
2.3. Statistical Analysis
Prevalence of AMR was calculated as the percentage of microbial strains exhibiting resistance to at least one antibiotic, together with corresponding exact 95% confidence intervals (95% CI), for each Campylobacter species and by antibiotic. Differences in AMR prevalence were evaluated using a Chi-squared test, while the cumulate number of AMR per isolate was modelled with Poisson regression, considering the Campylobacter species as a covariate. The prevalence ratio (PR) was used to express the results of the model. Data analysis was performed using STATA 17 [17] and the statistical significance level was set at 5%.
3. Results
Between 2020 and 2023, the regional laboratory surveillance system identified 820 cases of Campylobacter infection in humans. The leading causative agents were C. jejuni and C. coli, with C. jejuni accounting for 87.7% of cases. The strains were collected from patients with gastrointestinal symptoms (n = 802), septicemia (n = 17) or urinary tract infection (n = 1).
AMR was frequent: 80.1% of the strains were resistant to at least one antibiotic. AMR was highest against ciprofloxacin and tetracycline (prevalence of 72.1% and 52.9%, respectively) and lowest against erythromycin (3.2%) and aminoglycosides (streptomycin/gentamicin) (5.4%). The distribution of MICs, MIC50 and MIC90 of the Campylobacter isolates against five common antibiotics are shown in Table 1.
Table 1.
Distribution of MICs, MIC50 and MIC90 of human Campylobacter species against five common antibiotics.
| C. jejuni | C. coli | |||||
|---|---|---|---|---|---|---|
| Antibiotic | MIC Range (µg/mL) | MIC50 (µg/mL) |
MIC90 (µg/mL) |
MIC Range (µg/mL) | MIC50 (µg/mL) |
MIC90 (µg/mL) |
| Erythromicin | 1–128 | ≤1 | 2 | 1–128 | ≤1 | 128 |
| Ciprofloxacin | 0.12–16 | 4 | 8 | 0.12–16 | 4 | 8 |
| Tetracycline | 0.5–64 | 4 | 32 | 0.5–64 | 32 | 64 |
| Streptomycin | 0.25–16 | 0.5 | 1 | 0.25–16 | 1 | 16 |
| Gentamicin | 0.12–16 | ≤0.125 | 0.25 | 0.12–16 | 0.25 | 1 |
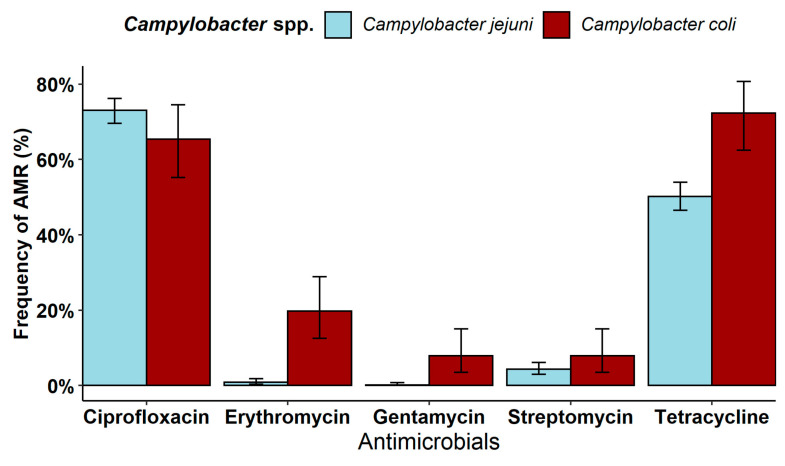
The frequency of AMR observed in C. jejuni strains (79.6%; 95% CI 76.4–82.4) was comparable to that observed in C. coli strains (84.2; 95% CI 75.6–90.7; Chi-squared test, p > 0.05). However, differences in the occurrence of AMR against single antibiotics were observed, especially against erythromycin and tetracycline for which C. coli displayed higher AMR levels than those observed in C. jejuni isolates (Figure 1). In addition, differences were noted in AMR against gentamycin and streptomycin: the prevalence in C. coli (7.9%; 95% CI 3.5–15.0) was higher than that observed against gentamycin in C. jejuni (0.1%; 95% CI 0.004–0.8; p < 0.001), while no differences were noted between the two Campylobacter species against streptomycin (7.9% in C. coli vs. 4.3% in C. jejuni, p > 0.05).
Figure 1.
Antimicrobial resistance (95% confidence interval) of Campylobacter spp. against antibiotics tested.
C. coli strains were more likely than C. jejuni strains to exhibit co-occurring AMR against two or more antimicrobial classes (PR 1.32; 95% CI 1.15–1.51). The most common co-resistance pattern was resistance against two antimicrobial classes, recorded for 41.1% (n = 337) of Campylobacter strains. Within this pattern, co-resistance against fluoroquinolones and tetracyclines was the most prevalent combination (Table 2). The profiles involving macrolides with tetracyclines and aminoglycosides and fluoroquinolones with aminoglycosides were found only in C. coli (n = 3). Furthermore, we noted a prevalence of 5.7% for MDR strains circulating in the study area and most commonly involving fluoroquinolones. Concurrent resistance against all four antimicrobial classes was identified only in C. coli strains, which accounted for 10.6% of the total number of resistant strains identified.
Table 2.
Co-resistance profiles of human Campylobacter species.
| Co-Resistance | Campylobacter Species | N° of Strains | Resistance Profile | N° of Strains | % of Strains |
|---|---|---|---|---|---|
| Resistant to two antimicrobial classes |
C. jejuni | 327 | Fluoroquinolones Tetracyclines | 291 | 89.9 |
| C. coli | 36 | 11.0 | |||
| C. jejuni | 4 | Aminoglycosides Tetracyclines | 2 | 50.0 | |
| C. coli | 2 | 50.0 | |||
| C. jejuni | 3 | Fluoroquinolones Macrolides | 1 | 33.3 | |
| C. coli | 2 | 66.7 | |||
| C. jejuni | 1 | Macrolides Tetracyclines | 0 | 0 | |
| C. coli | 1 | 100 | |||
| C. jejuni | 1 | Aminoglycosides Macrolides | 0 | 0 | |
| C. coli | 1 | 100 | |||
| C. jejuni | 1 | Fluoroquinolones Aminoglycosides | 0 | 0 | |
| C. coli | 1 | 100 | |||
| Resistant to three antimicrobial classes |
C. jejuni | 26 | Fluoroquinolones Aminoglycosides Tetracyclines | 25 | 96.2 |
| C. coli | 1 | 3.8 | |||
| C. jejuni | 13 | Fluoroquinolones Macrolides Tetracyclines | 1 | 7.7 | |
| C. coli | 12 | 92.3 | |||
| C. jejuni | 1 | Fluoroquinolones Macrolides Aminoglycosides | 1 | 100 | |
| C. coli | 0 | 0 | |||
| Resistant to four antimicrobial classes |
C. jejuni | 7 | Fluoroquinolones Macrolides Aminoglycosides Tetracyclines | 0 | 0 |
| C. coli | 7 | 100 |
4. Discussion
Since 2005, Campylobacter has become the most commonly reported cause of bacterial food-borne illness in the European Union [10]. Because it is a zoonotic pathogen, it is exposed to antibiotics for both human and veterinary medicine. For example, fluoroquinolones, which are critically important antimicrobials (CIAs), and tetracycline have been used over the past 50 years to promote growth and to treat infection in poultry [13]. Since campylobacteriosis is generally a self-limiting illness, it is not usually treated. In contrast, macrolides and fluoroquinolones are the antibiotics of choice in the treatment of severe or persistent illness [18]. Antibiotic resistance of Campylobacter to these classes of antibiotics, especially fluoroquinolones, continues to increase. For these reasons, Campylobacter has been identified as a public health threat by both the World Health Organization (WHO) and the U.S. CDC [19]. The increase in Campylobacter strains resistant to common antibiotics highlights the need for improved surveillance and data sharing.
Resistance to the empirical drugs erythromycin, ciprofloxacin, and tetracyclines has been reported for human clinical strains in many countries worldwide: a previous study in Quebec (Canada) showed a 50% tetracycline resistance among C. coli isolates and 39% among C. jejuni isolates [20] and another study in South Africa recorded resistance to ciprofloxacin and erythromycin in 33.3% and 38.9% of C. coli and 20% and 31.5% of C. jejuni, respectively [21].
Our data show high resistance to ciprofloxacin (72.1%) for the period between 2020 and 2023 in Piedmont. These findings are shared by previous reports for Italy: high levels of resistance in C. jejuni and C. coli to ciprofloxacin (76% and 70%, respectively) from human samples [13,22]. These rates are consistent also with those reported for the EU between 2019 and 2021, where ciprofloxacin resistance in Campylobacter isolated from human samples was high to extremely high (range, 22.2% to 100% for C. jejuni and for C. coli). Very high levels of resistance, higher for C. coli than for C. jejuni, were observed for ciprofloxacin also in isolates from food-producing animals (range, 41.7% to 80.4%) [14,23]. Moreover, the high resistance to tetracycline (52.9%) we observed is consistent with data for the EU: 45.3% in C. jejuni and 70.3% in C. coli from human isolates. The resistance to tetracycline ranged from high to extremely high (43.3–90.5%) also in food-producing animals [14]. Antimicrobial resistance to aminoglycosides (gentamicin and streptomycin) was less frequent (5.4%), roughly similar to previous studies where the resistance to gentamicin and streptomycin was low (0.7% and 2.4% on average, respectively) in C. jejuni and C. coli isolates from human samples [13,14]. Conversely, we observed low rates of resistance to erythromycin (3.2%), another CIA, supporting previous data that indicated low resistance in C. jejuni from human and animal samples [12,13,14]. However, higher rates have been reported for C. coli isolates from human samples (8.5%), similar to our findings, and animals (range, 4.4% to 35.7%) [14].
The high rate of co-occurrence of resistance to ciprofloxacin and tetracycline in C. jejuni (89.9%) raises concerns for public health. Lower percentages were reported by a Campylobacter spp. surveillance study performed in the 2013–2016 period by the Enter-Net Italia network where the same co-occurrence of resistance was observed in 48% of C. jejuni and 41% of C. coli (total n = 647) [22]. The higher resistance we observed could be due to the many intensive poultry farms operating in our area. Indeed, Campylobacter spp. isolates from slaughtered poultry in northern Italy demonstrated high resistance to quinolones, tetracycline, and macrolides [22]. This high prevalence of co-resistance underscores the importance of monitoring for resistant Campylobacter spp. strains in food-producing animals and the transmission to humans through the food chain.
Regarding multidrug resistance (MDR), co-resistance to ciprofloxacin, tetracycline, and aminoglycosides was markedly higher in C. jejuni (96.2%) than in C. coli, In contrast, the highest rate of co-resistance to ciprofloxacin, tetracycline, and erythromycin (92.3%) was noted for C. coli although the number of isolates was not high; lower rates of MDR to these three antimicrobials were previously reported for C. coli of human origin (29%) [22].
5. Conclusions
The present study provides evidence that AMR is common among human Campylobacter strains isolated in the study area, particularly against ciprofloxacin and tetracycline. The further emergence of Campylobacter resistance may be prevented through the implementation of good antimicrobial stewardship at the farm level, since Campylobacter can easily reach the consumer via the food production chain and pose a serious public health risk. Moreover, clinicians should consider optimal treatment before beginning empiric treatment and use antibiotics judiciously for the protection of their patients and the community.
Acknowledgments
We are thankful to CeRTiS Clinical Laboratories Group (Zaccaria T., Laboratorio di Microbiologia AOU Città della Salute e della Scienza Torino; Mussino S., Laboratorio Analisi Humanitas Gradenigo Torino; Del Re S., Laboratorio di Microbiologia e Virologia PO Amedeo di Savoia Torino; Amarù G., Laboratorio Analisi Unificato Rivoli—Pinerolo ASL TO3 Rivoli-TO; Li Vigni N., Laboratorio Analisi e Microbiologia Ospedale Civico di Ivrea-TO; Brossa S. Laboratorio analisi Ospedale di Ciriè-TO; Barreca P., Ravarino D. Laboratorio analisi Ospedale di Chivasso-TO; Allocco A., Laboratorio analisi chimico-cliniche e microbiologiche ASL TO5 ospedali riuniti Carmagnola, Chieri, Moncalieri-TO; Leli C., Laboratorio di Microbiologia Ospedale Civile Santi Antonio e Biagio e Cesare Arrigo Alessandria; Salerno A., Laboratorio Analisi e Microbiologia Ospedale Civile SS Antonio e Margherita Tortona-AL; Concialdi E., Laboratorio Analisi chimiche cliniche e microbiologiche Ospedale Cardinal Massaia Asti; Piana F., Laboratorio Analisi Chimico Cliniche e microbiologia AO S. Croce e Carle Cuneo; Vinai E., Laboratorio Microbiologia Ospedale Regina Montis Regalis Mondovì-CN; Squillario P., Laboratorio Analisi e Microbiologia Nuovo Ospedale degli Infermi Biella; Canale C., Laboratorio Analisi Chimico Cliniche e Microbiologiche, Ospedale Castelli Verbania-VCO; Caffiero G., Laboratorio Analisi e Microbiologia Ospedale S. Andrea Vercelli).
Author Contributions
Conceptualization, M.P. and C.T.; methodology, C.T. and M.P.; data curation, M.P., A.G.-V., D.M.B. and C.M.; writing—original draft preparation, M.P., C.T. and A.G.-V.; writing—review and editing, M.P., C.T., A.G.-V., C.M. and L.D.; supervision, M.P., D.M.B. and L.D.; project administration, M.P., C.M., D.M.B. and L.D.; CeRTiS Clinical Laboratories Group provided Campylobacter strains. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Campylobacter strains, isolated from patient samples, examined for this study were submitted to the CeRTiS for surveillance within the framework of Directive 2003/99/EC on the monitoring of zoonoses and zoonotic agents. In addition, information on and consent to medical intervention were obtained from patients examined by medical doctors, in accordance with current Italian legislation. All data on humans were treated in an anonymized manner and were used solely for the purposes of scientific research.
Informed Consent Statement
Patient consent was revoked because the samples were anonymous and personal data were not employed.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Aydin F., Abay S., Kayman T., Karakaya E., Mustak H.K., Mustak I.B., Bilgen N., Goncuoglu M., Duzler A., Guran O., et al. Campylobacter anatolicus sp. nov., a novel member of the genus Campylobacter isolated from feces of Anatolian Ground Squirrel (Spermophilus xanthoprymnus) in Turkey. Syst. Appl. Microbiol. 2021;44:126265. doi: 10.1016/j.syapm.2021.126265. [DOI] [PubMed] [Google Scholar]
- 2.Kaakoush N.O., Natalia Castaño-Rodríguez N., Mitchell H.M., Man S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015;28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA. ECDC The European Union One Health 2021 Zoonoses Report. EFSA J. 2022;20:7666. doi: 10.2903/j.efsa.2022.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC Campylobacter (Campylobacteriosis) [(accessed on 16 February 2023)];2023 Available online: https://www.cdc.gov/campylobacter/faq.html#:~:text=The%20Foodborne%20Diseases%20Active%20Surveillance,million%20U.S.%20residents%20every%20year.
- 5.Igwaran A., Okoh A.I. Human campylobacteriosis: A public health concern of global importance. Heliyon. 2019;5:e02814. doi: 10.1016/j.heliyon.2019.e02814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker C.R., Painset A., Swift C., Jenkins C., Godbole G., Maiden M.C.J., Dallman T.J. Microevolution of Campylobacter jejuni during long-term infection in an immunocompromised host. Sci. Rep. 2020;10:10109. doi: 10.1038/s41598-020-66771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansarifar E., Riahi S.M., Tasara T., Sadighara P., Zeinali T. Campylobacter prevalence from food, animals, human and environmental samples in Iran: A systematic review and meta-analysis. BMC Microbiol. 2023;23:126. doi: 10.1186/s12866-023-02879-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucarelli C., Dionisi A.M., Trezzi L., Farina C., Passera M., Kärki T., Luzzi I. Molecular and epidemiological analysis of a Campylobacter jejuni outbreak in Northern Italy in November 2013. Foodborne Pathog. Dis. 2016;13:490–494. doi: 10.1089/fpd.2015.2104. [DOI] [PubMed] [Google Scholar]
- 9.Manfreda G., Parisi A., De Cesare A., Mion D., Piva S., Zanoni R.G. Typing of Campylobacter jejuni isolated from turkey by genotypic methods, antimicrobial susceptibility, and virulence gene patterns: A retrospective study. Foodborne Pathog. Dis. 2016;13:93–100. doi: 10.1089/fpd.2015.2048. [DOI] [PubMed] [Google Scholar]
- 10.Conesa A., Garofolo G., Di Pasquale A., Camma C. Monitoring AMR in Campylobacter jejuni from Italy in the last 10 years (2011–2021): Microbiological and WGS data risk assessment. EFSA J. 2022;20((Suppl. 1)):e200406. doi: 10.2903/j.efsa.2022.e200406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Centre for Disease Prevention and Control EU Protocol for Harmonised Monitoring of Antimicrobial Resistance in Human Salmonella and Campylobacter Isolates—June 2016. 2016.
- 12.Marotta F., Janowicz A., Romantini R., Di Marcantonio L., Di Timoteo F., Romualdi T., Zilli K., Barco L., D’Incau M., Mangone I., et al. Genomic and antimicrobial surveillance of Campylobacter population in Italian poultry. Foods. 2023;12:2919. doi: 10.3390/foods12152919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marotta F., Garofolo G., di Marcantonio L., DiSerafino G., Neri D., Romantini R., Sacchini L., Alessiani A., Di Donato G., Nuvoloni R., et al. Antimicrobial resistance genotypes and phenotypes of Campylobacter jejuni isolated in Italy from humans, birds from wild and urban habitats, and poultry. PLoS ONE. 2019;14:e0223804. doi: 10.1371/journal.pone.0223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.EFSA. ECDC The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023;21:7867. doi: 10.2903/j.efsa.2023.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020.
- 16.Aerts M., Battisti A., Hendriksen R., Kempf I., Teale C., Tenhagen B.A., Veldman K., Wasyl D., Guerra B., Liebana E., et al. EFSA (European Food Safety Authority), 2019. Scientific report on the technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 2019;17:5709. doi: 10.2903/j.efsa.2019.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.StataCorp LLC . Stata Statistical Software, Release 16. StataCorp LLC; Collage Station, TX, USA: 2019. [Google Scholar]
- 18.Yang Y., Feye K.M., Shi Z., Pavlidis H.O., Kogut M., JAshworth A., Ricke S.C. A historical review on antibiotic resistance of foodborne Campylobacter. [(accessed on 17 February 2024)];Front. Microbiol. 2019 26:1509. doi: 10.3389/fmicb.2019.01509. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6676416/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. WHO pathogens priority list working group. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 20.Guévremont E., Nadeau É., Sirois M., Quessy S. Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Can. J. Vet. Res. 2006;70:81–86. [PMC free article] [PubMed] [Google Scholar]
- 21.Shobo C.O., Bester L.A., Baijnath S., Somboro A.M., Peer A.K.C., Essack S.Y. Antibiotic resistance profiles of Campylobacter species in the South Africa private health care sector. J. Infect. Dev. Ctries. 2016;10:1214–1221. doi: 10.3855/jidc.8165. [DOI] [PubMed] [Google Scholar]
- 22.García-Fernández A., Dionisi A.M., Arena S., Iglesias-Torrens Y., Carattoli A., Luzzi I. Human campylobacteriosis in Italy: Emergence of multi-drug resistance to ciprofloxacin, tetracycline, and erythromycin. Front. Microbiol. 2018;9:1906. doi: 10.3389/fmicb.2018.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.EFSA. ECDC The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2022;20:7209. doi: 10.2903/j.efsa.2022.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are contained within the article.