Abstract
The E2 spike glycoprotein of Semliki Forest virus is produced as a p62 precursor protein, which is cleaved by host proteases to its mature form, E2. Cleavage is not necessary for particle formation or release but is necessary for infectivity. Previous results had shown that phenotypic revertants of cleavage-deficient p62 mutants are generated, and here we show that these may contain second-site suppressor mutations in the vicinity of the cleavage site. These hot-spot sites were mutated to abolish the generation of such suppressor mutations; however, secondary mutations in another distant domain of the E2 protein appeared instead, all of which still caused cleavage-deficient mutations. Such mutants grew very poorly and were inefficient in virus entry and release. The mutated sites define domains of the spike protein which probably interact to regulate its structure and function. Because of their highly attenuated phenotype and the lower probability of reversion, the new mutations close to the cleavage site were used to make new helper vectors for packaging of recombinant RNA into infectious particles, thus increasing further the biosafety of the vector system based on the Semliki Forest virus replicon.
Incorporation of transmembrane spike glycoproteins into viral envelopes is a prerequisite for particle infectivity. However, since the spike proteins contain fusion peptides, which are not to be exposed and utilized before the virion encounters a new host cell, many viruses initially synthesize their spike proteins in a fusogenically inactive precursor form. On their way from the endoplasmic reticulum to the cell surface, these precursors mature by cleavage (10, 56). This cleavage is performed by host cell furin-like proteases in the trans-Golgi or post-Golgi compartment (46, 48) and occurs either at motifs of basic di-, tri-, or tetrabasic residues or, more commonly, after (R/K)X(R/K)R or RXXR sequences (43). The sequence RXXXXR may also be used for cleavage (35).
The alphaviruses (family Togaviridae) are enveloped viruses harboring a nonsegmented positive-strand RNA genome (42, 45). The virion has a T = 4 icosahedral symmetry and is composed of 240 copies of the capsid monomer encapsidating a single molecule of the positive-strand RNA. The nucleocapsid (NC) is surrounded by an envelope containing 240 copies of the transmembrane glycoproteins E2 and E1, which, in complexes of three E2E1 heterodimers, form the viral spikes (4, 11). The spike proteins are synthesized as a polyprotein precursor on the endoplasmic reticulum membrane, which is cleaved to p62 (precursor of E2) and E1 by signal peptidase cleavage (26). The proteins heterodimerize in the ER (58), after which they are transported to the cell surface by the exocytic pathway. During this transport, p62 is cleaved to E2 and E3 by host furin-like proteases (8, 54). The E3 fragments of Semliki Forest virus (SFV) and Sindbis virus (SIN) are 66 and 64 amino acids long, respectively. In SFV, E3 remains part of the mature virion (13), whereas it is shed from the spike in SIN (55).
Cleavage of the SFV p62 precursor protein occurs after the sequence RHRR, but it can be abolished by mutation of this motif to either RHRL (29, 40) or SHQL (2). Such mutations do not hamper virus assembly and release but are severely impaired in virus binding and entry into new cells. Similar observations, although not distinguishing the exact defective event, have been made for SIN (9, 19, 54) and Venezuelan equine encephalitis (VEE) virus (5) mutants. Infectivity can, however, be restored by distortion of the spike structure (18, 30, 39, 51, 52), by in vitro protease cleavage (21, 29), or by low-pH treatment (40).
A series of expression vectors based on the SFV replicon were previously constructed, which allow cloning of foreign sequences as part of the SFV replicon replacing the subgenomic RNA portion of the genome. For packaging of such recombinant RNAs into infectious virus particles, a helper vector was made in which the replicase region was deleted but which carried the structural genes (27). Although such helper RNA, when cotransfected with recombinant RNA into cells is not packaged into particles (due to lack of packaging signal), small amounts of wild-type SFV, which stem from amplification of vector-helper recombinants in the culture, could be recovered. To prohibit the amplification of these recombinants, a new helper was created which harbored mutations (SHQL) at the p62 furin cleavage site, thus rendering particles noninfectious unless activated by chymotrypsin treatment in vitro. While the SHQL mutations did not reduce the frequency of recombination itself, they efficiently prohibited the spread of wild-type virus to the extent that no such particles have been found to date. The SHQL mutation was also tested in the context of the full-length cDNA clone of SFV and turned out to be severely attenuated (2).
In our previous study, we found that transfection of full-length SFV RNA harboring the cleavage-deficient SHQL mutation resulted in production of phenotypically revertant virus that appeared as microplaques with a frequency close to 10−6 (for unactivated virus) (2). In this study, we have further characterized such revertants and found that they were second-site escape mutations accumulated in the vicinity of the furin cleavage site. A similar observation was made earlier when an RHRL mutant was tested in mice and found to be severely attenuated but not fully avirulent (14), suggesting the presence of infectious revertants. We reasoned, therefore, that it should be possible to engineer less leaky variants by mutating the hot spots, thus abolishing or at least severely reducing the possibility of new suppressor mutations. Such constructs, if tight, would be useful in creating an even safer helper construct for packaging of recombinant RNA into SFV particles (2, 27). We show here that the new mutants indeed remained cleavage deficient; however, other mutations located at a significant distance from the original cleavage site still appeared, albeit at low frequency.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Baby hamster kidney (BHK-21) cells were cultured in either Glasgow minimal essential medium (MEM) supplemented with 5% fetal calf serum, 10% tryptose phosphate broth, 2 mM glutamine, and 20 mM HEPES (Life Technologies, Inc., Paisley, Scotland) or MEM with Earl’s salts supplemented with 0.2% bovine serum albumin, 2 mM glutamine, and 20 mM HEPES. SFV was derived from the pSFV4 infectious clone plasmid (28). Packaging of in vitro-produced viral RNA into infectious particles has been described previously (2, 27). Antibodies were used as mouse ascites preparations. The anti-E1 monoclonal antibody K-22/98 was a kind gift from A. Salmi (Department of Virology, University of Turku, Turku, Finland). It recognizes E1 in both free form and heterodimeric form with p62/E2 (1).
Viral genome sequencing by reverse transcriptase PCR (RT-PCR).
Plaque purification, virus amplification, RNA extraction, and sequencing were done as previously described (57). The oligonucleotides used for first-strand synthesis were 5′-gttatatgccattgcttaccc-3′ (nucleotides [nt] 11237 to 11257) and 5′-cggtagcttcggacctgaccgc-3′ (nt 8525 to 8546).
Construction of mutants.
Site-directed mutagenesis was performed by the method of Kunkel et al. (25) or by PCR (36). Briefly, the EcoRI fragment (nt 1301 to 2656) from plasmid pSFV-Helper 1 (27) modified around the p62 cleavage site as described previously (2) was cloned into the phagemid vector pBK (Stratagene, La Jolla, Calif.) by standard procedures (41). Single-stranded template was prepared as specified by the manufacturer and used for site-directed mutagenesis as described previously (26). The mutagenized EcoRI fragments were recloned back to the original pSFV-Helper 1 plasmid. The same mutations were generated in pSFV4 (28) with the unique BglII (nt 6715) and NsiI (nt 8927) restriction endonuclease sites. Mutants SHQL I244 and SHQL K244 were generated by the previously mentioned PCR mutagenesis protocol. The primers used were 5′-tactggcaaagtgccaccg-3′ (nt 8721 to 8739) and 5′-ttttgccgtggatgacggtt-3′ (nt 9237 to 9257) as the 5′ and 3′ primers, respectively. The mutagenic primers were 5′-ttcgtcccga(a/t)agccgacgaa-3′ (nt 9140 to 9160). The PCR products were cloned into pSFV4 via the unique Sse8337I (nt 8749) and BssHII (nt 9227) restriction sites. All the constructs were sequenced to verify the presence of the mutations by cycle sequencing by using the Dyedeoxy terminator method as specified by Applied Biosystems and run on an automated DNA sequencer (ABI 373A; Applied Biosystems).
Metabolic labelling.
[35S]methionine-labelled wild-type and mutant SFV were prepared as previously described (31). Briefly, in vitro-transcribed viral RNA was used to transfect BHK-21 cells by electroporation as previously described (28). At 6 to 7 h postelectroporation, the medium was replaced with [35S]methionine-containing MEM supplemented with 1% fetal calf serum at 200 μCi/ml (>1,000 Ci/mmol; Amersham) and incubation was continued overnight. The supernatant was clarified from cell debris by low-speed centrifugation, and virus was pelleted through a 20% (wt/wt) sucrose cushion in TNE buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.5 mM EDTA) in an SW28 Beckman ultracentrifuge rotor for 90 min at 25,000 rpm. The viral pellet was resuspended in TNE buffer and kept at −80°C. For pulse-chase assays, transfected BHK-21 cells were labelled 8 to 9 h posttransfection.
Packaging of recombinant viral genomes.
Packaging of recombinant viral genomes with the SFV-helper viruses constructed in this work was performed as previously described (27).
Binding and internalization assays.
Binding and internalization of [35S]methionine-labelled SFV on BHK-21 cells were performed as previously described (31, 53). Briefly, 80% confluent monolayers were incubated on ice with virus at a multiplicity of infection of 10 to 20, in MEM-bovine serum albumin medium for 1 to 2 h with continuous shaking. Unbound virus was removed by washing the cells twice with cold medium. Binding was assayed, after solubilization in 1% Nonidet P-40-containing buffer, by scintillation counting (1214 Rackbeta counter; LKB Wallac). The input amount was always in good agreement with the sum of the bound and washed viral levels. Internalization was initiated by incubation at 37°C for the required time intervals. Uninternalized virus was removed by treatment with proteinase K (0.5 mg/ml in phosphate-buffered saline).
Immunoprecipitation analysis.
Cell lysates were incubated with the antibody for 3 h at 4°C with continuous shaking. Immune complexes were precipitated with protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden), analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by fluorography. When necessary, protein levels were quantified by phosphoimager analysis on a FUJI BAS2000 instrument.
Stability of the heterodimer.
The buffers used to generate the different pH values were as previously described (40). Purified viral particles were incubated in the respective buffer for 10 min on ice. After immunoprecipitation with the anti-E1 monoclonal antibody, protein samples were analyzed by SDS-PAGE.
RESULTS
Isolation of infectious revertants of the SHQL isolate.
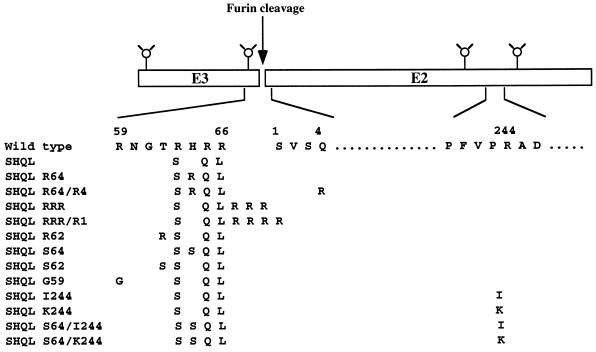
The p62 spike glycoprotein of SFV is normally cleaved after the RHRR motif (Fig. 1). Previous characterization of the noncleavable SHQL mutant, which as such is noninfectious, had shown that replicative virus was generated at a frequency of about 10−6 but that plaques of such virus were quite small (2). To analyze the genotype of such revertants, a plaque assay was used by plating virus particles onto BHK cells at a multiplicity of infection of 5. Plaques were scored after 48 h, and individual plaques were picked and further passaged on BHK cells for the production of small virus stocks. To determine whether the p62 precursor protein of these isolates had converted to a cleavable form, infected BHK cells were pulse-labelled and the p62/E2 and E1 spike proteins produced were analyzed by SDS-PAGE. Figure 2A shows the profiles of two such isolates, both of which produce an uncleaved p62 protein. To determine the genotype in the E3/E2 cleavage region, the RNA from 20 isolates was prepared and sequenced by RT-PCR. Most of the revertants (14/20) harbored a single base substitution, resulting in an H-to-R mutation at position 64 of E3 (mutant SHQL R64) (Fig. 1). One isolate was a double mutant, with an H-to-R mutation at position 64 of E3 and a Q-to-R mutation at position 4 of E2 (mutant SHQL R64/R4), and two isolates had the T-62 residue in E3 changed to an R (mutant SHQL R62). One isolate had three R residues inserted at the cleavage site between E3 and E2 (mutant SHQL RRR), and two isolates had the same insertion plus the S residue at position 1 of E2 changed to R (mutant SHQL RRR/R1), creating a string of four R residues.
FIG. 1.
Schematic presentation of the mutations in the E3/E2 SFV glycoprotein.
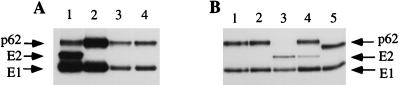
FIG. 2.
Characterization of SFV isolates by SDS-PAGE. BHK-21 cells transfected with SFV RNA were pulse-labelled for 15 min and chased for 3 h. Cell lysates were immunoprecipitated with anti-E2 and anti-E1 polyclonal antibodies. (A) Lanes: 1, wild type; 2, SHQL; 3 and 4, two independent revertant isolates, which later turned out to be of type SHQL R64. (B) Reconstructed mutants with the SHQL background. Lanes: 1, SHQL R64; 2, SHQL R64/R4; 3, SHQL RRR; 4, SHQL RRR/R1; 5, SHQL R62.
To determine the phenotype of these mutants in a clean background, the mutations were reconstructed into the parental pSFV4-SHQL clone. Full-length viral RNA was produced in vitro by SP6 transcription and subsequently transfected into BHK-21 cells, and the metabolically labelled proteins were analyzed by SDS-PAGE (Fig. 2B). The SHQL R64 and SHQL R64/R4 mutations did not result in p62 cleavage (Fig. 2B, lanes 1 and 2), while the presence of the mutation leading to a string of either three or four R residues (SHQL RRR and SHQL RRR/R1) resulted in full and partial p62 cleavage, respectively (Fig. 2B, lane 3 and 4). The SHQL R62 mutation did not result in p62 cleavage but produced a slightly smaller p62 protein (Fig. 2B, lane 5). This was probably due to loss of a sugar group, since the mutation disrupted the motif N-60/T-62 in E3, a site known to become glycosylated (12).
New variants of SHQL.
Since most of the infectious revertants represented single base substitutions leading to single amino acid changes, we attempted to create less leaky SHQL variants by introducing secondary mutations that would diminish the probability of such conversions by requiring at least two simultaneous base changes. We also wanted to avoid the possibility that combinations of mutations in the region would allow furin-type enzymes to cleave the protein. The furin family of proteases cleave at either RXXR or RXXXXR motifs, and such motifs had been created by the SHQL R64 mutation in combination with wild-type residue R59 or by the SHQL R62 mutation in combination with wild-type residue R59. The reason why these particular mutants were not cleaved could be that the site was not accessible for the enzyme, but there was a risk that new mutations in this region might change the conformation of the protein and thus allow cleavage. For these reasons, we chose to make three new variants of SHQL, one in which H64 was changed to S (SHQL S64), one in which T62 was changed to S (SHQL S62), and one in which R59 was changed to G (SHQL G59). The S and G substitutions were chosen because other alphaviruses have such residues in the corresponding positions (45).
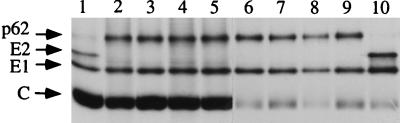
RNA from the new clones was produced in vitro and transfected into BHK-21 cells which were then metabolically labelled, and the lysates were analyzed directly by SDS-PAGE (Fig. 3). All new variants produced an uncleaved p62 protein with the same apparent size as p62 from the SHQL variant. To determine whether the mutations had any effect on virus assembly and release from cells, transfected cells were labelled for 30 min and chased for 3 h to determine the kinetics of virus release (Fig. 3). Quantitation showed that all the variants released about 30% of the labelled material into the growth medium at the end of the 3-h chase, which was the same amount produced by both wild-type and SHQL virus, suggesting that the new mutations had not affected the virus assembly process.
FIG. 3.
Secondary mutations in the SHQL background. Transfected BHK cells were pulse-labelled for 15 min and chased for 3 h. Cell lysates were loaded directly on the gel (lanes 1 to 5). Virus was quantitatively sedimented from the medium by centrifugation through a 20% sucrose cushion (lanes 6 to 10). Lanes: 1 and 10, wild type; 2 and 6, SHQL; 3 and 7, SHQL S64; 4 and 8, SHQL S62; 5 and 9, SHQL G59.
To test whether the new variants reverted to the replicative form, virus stocks were prepared and used to infect BHK cells at a multiplicity of infection of 5. Indeed, after 48 h, plaques could be seen; however, they were significantly smaller than the revertant plaques obtained after plating the SHQL virus, and they appeared with a frequency which was 1 order of magnitude lower than that determined previously for SHQL. The SHQL S62 variant did not give any revertants, while variant SHQL S64 gave two and SHQL G59 gave one. The three plaques were isolated, and stocks were prepared from which the RNA was isolated for RT-PCR sequencing all across E3 and 90 bases into E2. In no case could we identify additional mutations in this region, which in every case had retained their original genotype. The two independent revertant isolates of variant SHQL S64 were chosen and sequenced across the complete structural gene region (nt 8079 to 11240). Single amino acid changes were identified at position 244 of E2, which had been changed from R to I in one case and to K in the other (Fig. 1). To assess the effect of the I244 and K244 mutations in the absence of the S64 mutation, they were transferred to the SHQL background. Analysis showed that they did not allow p62 cleavage to any extent, but virus production appeared to be significantly reduced compared to either wild-type or SHQL virus (Fig. 4; Table 1).
FIG. 4.
Production of SFV isolates. BHK-21 cells were transfected with RNA, pulse-labelled for 15 min, and chased for 3 h. Cell lysates were loaded directly on the gel (lanes 1 to 6). Virus particles were quantitatively sedimented through a 20% sucrose cushion before loaded on the gel (lanes 7 to 12). Lanes: 1 and 7, wild type; 2 and 8, SHQL; 3 and 9, SHQL R64; 4 and 10, SHQL R64/R4; 5 and 11, SHQL I244; 6 and 12, SHQL K244.
TABLE 1.
Characteristics of SFV isolates
| Isolate | Stock titer (PFU/ml)a | Specific infectivityb | Binding to cellsc | Internalizationd |
|---|---|---|---|---|
| Wild type | 3.0 × 108 | 100 | 72.9 | 71.25 |
| SHQL | 3.3 × 108 | 0 | 8.45 | 4.09 |
| SHQL R64 | 4.0 × 105 | 0.26 | 8.04 | 5.96 |
| SHQL R64/R4 | 1.6 × 106 | 0.55 | 11.73 | 9.54 |
| SHQL I244 | 2.0 × 104 | 0.037 | 26.44 | 7.27 |
| SHQL K244 | 2.0 × 104 | 0.030 | 16.12 | 7.0 |
Virus produced from transfection of 107 BHK cells during a 24-h incubation.
The infectivity of an arbitrarily chosen amount (PFU) was correlated with the protein content of the viral particles as measured by gel electrophoresis. Wild type was given a value of 100.
Percentage of input virus (mean of three experiments [SD = 10%]).
Percentage of bound virus (mean of three experiments [SD = 10%]).
Characterization of the revertant isolates.
Since all revertants except those with R insertions gave rise to infectious virus in the absence of p62 cleavage, it was of interest to analyze the stability of the spike structure and the infectivity of the virus particles. Radiolabelled viral particles of the wild type, SHQL R64, SHQL R64/R4, SHQL K244, and SHQL I244 were produced after continuous labelling during growth on BHK cells. During this production, the SHQL R64 and SHQL R64/R4 variants produced virus as much as the wild type or SHQL, while production of SHQL K244 and SHQL I244 virus was reduced by a factor of 3 (Fig. 4). The specific infectivity was estimated for each isolate (Table 1). All the mutants had reduced infectivity; mutants SHQL K244 and SHQL I244 were the least infectious, being 4 orders of magnitude less infectious than the wild type (Table 1).
To analyze whether the mutations had any effect on binding of the virus particle to the cell, radiolabelled virus was loaded on BHK-21 cells at 0°C (to inhibit endocytosis) for 60 min at a multiplicity of infection of 10 or 20 (the same result was obtained for both), after which the cells were extensively washed with ice-cold medium. The amount of virus bound was then determined by cell solubilization and scintillation counting. While 70% of the input wild-type virus bound to the cells, only 9% of the SHQL virus bound. The revertants showed a slight increase in binding, with SHQL I244 having a threefold increase compared to SHQL (Table 1).
Internalization of the viral particles was also monitored. About 70% of wild-type particles were internalized during a 10- to 20-min interval, while only 4% of the SHQL particles were internalized. Moderate increases in uptake efficiency were observed for the mutants with respect to SHQL, with the exception of SHQL R64/R4 mutant, which showed a twofold increase (Table 1).
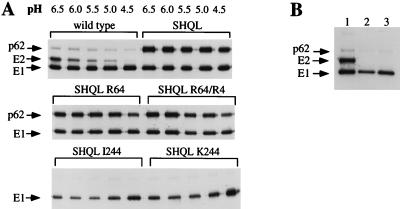
During the infectious process, the spike heterodimer (E2E1) of wild-type virus is dissociated in the increasingly acidic milieu of the developing endosome. This dissociation is important for release of the E1 protein from E2 to allow a trimeric E1 to catalyze the fusion of the viral and endosomal membrane (52, 53). Accordingly, to analyze the effect of the reversions on the stability of the spike heterodimer, radiolabelled virus preparations were incubated on ice for 10 min in buffer of decreasing pH. Dissociation of the spike complex was monitored by immunoprecipitation from Nonidet P-40-lysed virus with an anti-E1 monoclonal antibody, which coprecipitates an intact heterodimer at neutral pH (1). The wild-type spike heterodimer started to dissociate at pH 6.0, and dissociation was completed at pH 4.5, while the SHQL mutant spike heterodimers did not dissociate even at pH 4.5, due to the uncleaved p62 protein (Fig. 5A). The SHQL R64 and SHQL R64/R4 variants also displayed resistance to low-pH-induced dissociation. However, for mutants SHQL I244 and SHQL K244, the p62-E1 heterodimeric complex had already dissociated at pH 7.4 (Fig. 5B).
FIG. 5.
(A) Dissociation of the heterodimer by pH values ranging from 6.5 to 4.5. (B) Dissociation of the heterodimer at pH 7.4. Lanes: 1, wild type; 2, SHQL I244; 3, SHQL K244.
New helpers for recombinant particle production.
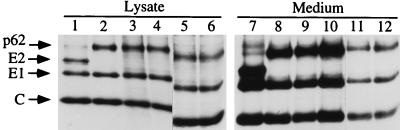
Since the new SHQL variants appeared to result in significantly fewer revertants and since those found grew very poorly, it was of value to test whether these new mutations would work in the context of the helper vector. The SHQL mutation has been extensively used for production of recombinant SFV particles in a cotransfection setup where only recombinant but not helper RNAs are packaged into forming virions (2, 27). To test this, the mutations S64, S62, and G59 were cloned into the helper 2 vector (2), which originally carried the SHQL mutation alone. RNA was produced in vitro and cotransfected with recombinant vector RNA encoding LacZ into BHK-21 cells for particle production. At 9 h posttransfection, the cells were labelled for 15 min and chased for 3 h, and the labelled products both in the lysate and in the growth medium were analyzed by SDS-PAGE (Fig. 6). It was found that all new helper constructs readily expressed the uncleaved p62 protein, E1, and capsid protein. When the efficiency of particle production was assessed, it was found that the new helpers were as efficient as the original SHQL helper in producing recombinant virus.
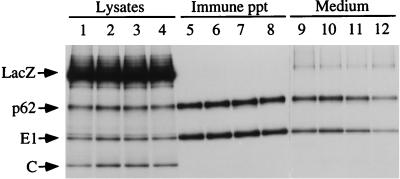
FIG. 6.
New SFV SHQL variants as helpers. Recombinant RNA encoding LacZ and helper RNAs were cotransfected into BHK-21 cells, pulse-labelled for 15 min, and chased for 3 h. Lysates were either loaded directly on the gel (lanes 1 to 4) or immunoprecipitated with anti-E1 and anti-E2 antibodies (lanes 5 to 8). Packaged virus was pelleted from the growth medium by ultracentrifugation through a 20% sucrose cushion (lanes 9 to 12). Lanes: 1, 5, and 9, SHQL helper (original helper 2); 2, 6, and 10, helper SHQL S64; 3, 7, and 11, helper SHQL S62; 4, 8, and 12, helper SHQL G59. The C protein of purified virus often smears in SDS-PAGE and is not a good indicator of quantitation (lanes 9 to 12). ppt, precipitate.
DISCUSSION
Mutations at the p62 cleavage site can inhibit the cleavage of p62 to E3 and E2 glycoproteins, rendering the virus essentially noninfectious. Preliminary tests on the reversion frequency of the SHQL mutation concluded that second-site mutations, rather than true revertants, contributed to the observed revertant phenotypes. In this work, we identified such mutational hot spots by isolating revertants and sequencing the genes coding for the structural envelope glycoproteins. None of the mutations restored the original cleavage-site motif at the same position. Instead, we found second site-mutations that mapped in the vicinity of the p62 cleavage site (SHQL RRR and SHQL RRR/R1), creating alternative substrates for endoproteases (43). There is no mechanistic explanation for the way in which these insertions might be generated, although such phenomena, related to the replication of RNA viruses, have been observed (37).
We also found second-site suppressor mutations which rendered the virus infectious without affecting the cleavage site (SHQL R64 and SHQL R64/R4). These mutants retained the uncleaved phenotype, suggesting that p62 cleavage per se is not an absolute requirement for infectivity. Similar observations have been made for the SIN isolate, where creation of a glycosylation site abolishing cleavage did not affect virus replication (7). However, it appears that the replication-proficient phenotype was due to the particular genetic background of the S.A.AR86 SIN strain used in these studies (38, 39), since the same mutations in the AR339 background were lethal (19, 54).
Since most of the sequenced revertants were R64 or R64/R4, we chose to study them in more detail as part of the parental SHQL background. Pulse-labelling experiments on cells transfected with the mutant viral RNAs showed that virus assembly and release were not affected by the mutants but that infectivity was significantly lower than for wild-type virus. Cell binding assays with the mutant viruses suggested no major difference between SHQL and either of the mutants; however, R64/R4 appeared to enter BHK-21 cells more efficiently. Both mutations had partially restored infectivity profiles and stable spike structures as measured by resistance to low-pH dissociation. The R64/R4 double mutant was clearly more infectious, suggesting that the addition of an extra charged R residue may have resulted in enhanced structural changes in the domain. Taken together, these results suggest that distortion of the spike structure by mutation in the vicinity of the cleavage-site domain can allow replication by changing the spike structure, thereby enhancing the binding of the cellular receptor and/or enhancing the entry into cells.
In our attempt to create a less leaky variant of SHQL, we engineered the viral genome around the p62 cleavage site, giving rise to mutants S64, S62, and G59. We showed that the mutations affected neither the production of the structural glycoprotein nor the virus assembly or release. While the reversion frequency of the new engineered forms to the infectious phenotype was now significantly lower than for SHQL, it was still possible to isolate infectious mutants. They harbored second-site mutations that map at position 244 of E2. Interestingly, in VEE virus, second-site suppressor mutations to a noncleavable E3/E2 precursor (PE2 in VEE virus) harbored a mutation at neighboring position 243 of E2 (5), which corresponds to position 242 of SFV. While the authors did not attempt to elucidate the infectious phenotype by assaying for the binding and internalization steps, our data clearly show a substantial enhancement in binding of the mutant to BHK-21 cells compared to that of SHQL. At the same time, the mutant heterodimer was more labile to low-pH dissociation.
Other studies have shown that SIN second-site suppressors to a noncleavable PE2 glycoprotein precursor harbored mutations in E2 at position 169 (His to Leu), 216 (Gly to Glu) and 239 (His to Lys), among others (19). Furthermore, a single amino acid substitution at position 162 of the SFV E2 (Glu to Lys) showed a destabilizing effect on the E2/E1 heterodimer and on virus release (14).
The suppressor mutations at position 244 of the SFV E2 protein are within a domain which has been predicted to be a major conformational epitope recognized by protective antibodies (16, 17, 44). The same epitope has been found in other alphaviruses (15, 22, 49). This region is located between two N-linked glycosylation sites (residues 201 and 262), suggesting that indeed this charged domain is presented on the surface of the E2 molecule, where it has a complex folding and where the epitopes have been identified as nonlinear (23). The domain is believed to be the target in neutralization and to function in virus binding and/or entry, and several mutations within this domain have conferred a rapid penetration phenotype (33, 50).
Mutations at position 4 of E2 may also confer a rapid-penetration phenotype (6, 24), consistent with the R4 mutation in this study. Interestingly, competition assays with antipeptide antibodies have indicated a spatial overlap with the E2 amino terminus and region close to the 244 domain (18, 20, 22), suggesting that the rapid-penetration phenotype of R4 may be an indirect effect that occurs by uncovering the 244 domain to enhance virus binding. There are several observations of mutations in other regions of E2 which confer a rapid-penetration phenotype in response to a change in conformation (47), which may be achieved either by the mutation directly (3, 39) or after binding of the virus to its receptor on the cell surface (34).
The cleavage site of p62 would be expected to be exposed on the surface of the protein, since it is recognized by furin-like protease in the wild-type protein. This is supported by the fact that uncleaved p62 can be cleaved at this site by protease in vitro. The charged character of the cleavage domain also supports the notion that the domain is exposed, and the site is also permissive for insertion of foreign peptides and epitopes (9, 32). The domain around residue 240 in SIN also appears to be exposed, since short peptides can be inserted into this site as well (9).
The results of this study, taken together with previously obtained data, suggest that the domains covering residues 1 and 244 of the E2 protein are exposed on the surface and that they may interact in the quaternary structure of the protein. Mutation at either site can distort this structure. The high lability of the heterodimer for the SHQL I244 and SHQL K244 mutants, even at pH values of 6.5 and 7.4, is particularly illustrative in this respect. A change in the E2 structure may lead to exposure of domains which enhance the binding of the virus to its receptor and, through decreasing the association of E2 and E1, may increase the uptake of bound virus into cells by allowing the formation of the E1 trimer structure needed for fusion of the viral and endosomal membranes (51–53).
The noncleaved phenotype of the SHQL variant of SFV makes it a perfect candidate for a helper-based packaging expression system for the production of conditionally infectious recombinant particles (2). The new mutations described in this study (R62, S64, and S62) are far less prone for mutations, which occur at very low frequency, are highly deficient in replication. As demonstrated here, these mutations can be used in the context of the original SHQL mutations to create new helper vectors, which further enhance the biosafety of the SFV packaging system.
ACKNOWLEDGMENTS
This work was supported by the Swedish Medical Research Council, the Swedish Council for Engineering Sciences, the EU Biotechnology Programme, and the EU EVA Programme.
REFERENCES
- 1.Barth B U, Garoff H. The nucleocapsid-binding spike subunit E2 of Semliki Forest virus requires complex formation with the E1 subunit for activity. J Virol. 1997;71:7857–7865. doi: 10.1128/jvi.71.10.7857-7865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund P, Sjöberg M, Garoff H, Atkins G J, Sheahan B J, Liljeström P. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Nat Biotechnol. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 3.Boggs W M, Hahn C S, Strauss E G, Strauss J H, Griffin D E. Low pH-dependent Sindbis virus-induced fusion of BHK cells: differences between strains correlate with amino acid changes in the E1 glycoprotein. Virology. 1989;169:485–488. doi: 10.1016/0042-6822(89)90178-5. [DOI] [PubMed] [Google Scholar]
- 4.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis N L, Brown K W, Greenwald G F, Zajac A J, Zacny V L, Smith J F, Johnston R E. Attenuated mutants of Venezuelan equine encephalitis virus containing lethal mutations in the PE2 cleavage signal combined with a second-site suppressor mutation in E1. Virology. 1995;212:102–110. doi: 10.1006/viro.1995.1458. [DOI] [PubMed] [Google Scholar]
- 6.Davis N L, Powell N, Greenwald G F, Willis L V, Johnson B J B, Smith J F, Johnston R E. Attenuating mutations in the E2 glycoprotein gene of Venezuelan equine encephalitis virus: construction of single and multiple mutants in a full-length cDNA clone. Virology. 1991;183:20–31. doi: 10.1016/0042-6822(91)90114-q. [DOI] [PubMed] [Google Scholar]
- 7.Davis N L, Willis L V, Smith J F, Johnston R E. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology. 1989;171:189–204. doi: 10.1016/0042-6822(89)90526-6. [DOI] [PubMed] [Google Scholar]
- 8.de Curtis I, Simons K. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc Natl Acad Sci USA. 1988;85:8052–8056. doi: 10.1073/pnas.85.21.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubuisson J, Rice C M. Sindbis virus attachment: isolation and characterization of mutants with impaired binding to vertebrate cells. J Virol. 1993;67:3363–3374. doi: 10.1128/jvi.67.6.3363-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 11.Fuller S D, Berriman J A, Butcher S J, Gowen B E. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell. 1995;81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 12.Garoff H, Huylebroeck D, Robinson A, Tillman U, Liljeström P. The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J Cell Biol. 1990;111:867–876. doi: 10.1083/jcb.111.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garoff H, Simons K, Renkonen O. Isolation and characterization of the membrane proteins of Semliki Forest virus. Virology. 1974;61:493–504. doi: 10.1016/0042-6822(74)90285-2. [DOI] [PubMed] [Google Scholar]
- 14.Glasgow G M, Sheahan B J, Atkins G J, Wahlberg J M, Salminen A, Liljeström P. Two mutations in the envelope glycoprotein E2 of Semliki Forest virus affecting the maturation and entry patterns of the virus alter pathogenicity for mice. Virology. 1991;185:741–748. doi: 10.1016/0042-6822(91)90545-m. [DOI] [PubMed] [Google Scholar]
- 15.Grosfeld H, Lustig S, Gozes Y, Velan B, Cohen S, Leitner M, Lachmi B, Katz D, Olshevski U, Shafferman A. Divergent envelope E2 alphavirus sequences spanning amino acids 297 to 352 induce in mice virus-specific protective immunity and antibodies with complement-mediated cytolytic activity. J Virol. 1992;66:1084–1090. doi: 10.1128/jvi.66.2.1084-1090.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosfeld H, Velan B, Leitner M, Cohen S, Lustig S, Lachmi B E, Shafferman A. Semliki Forest virus E2 envelope epitopes induce a nonneutralizing humoral response which protects mice against lethal challenge. J Virol. 1989;63:3416–3422. doi: 10.1128/jvi.63.8.3416-3422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosfeld H, Velan B, Leitner M, Lustig S, Lachi B E, Cohen S, Shafferman A. Delineation of protective epitopes on the E2-envelope glycoprotein of Semliki Forest virus. Vaccine. 1991;9:451–456. doi: 10.1016/0264-410x(91)90134-r. [DOI] [PubMed] [Google Scholar]
- 18.Heidner H W, Johnston R E. The amino-terminal residue of Sindbis virus glycoprotein E2 influences virus maturation, specific infectivity for BHK cells, and virulence in mice. J Virol. 1994;68:8064–8070. doi: 10.1128/jvi.68.12.8064-8070.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidner H W, McKnight K L, Davis N L, Johnston R E. Lethality of PE2 incorporation into Sindbis virus can be suppressed by second-site mutations in E3 and E2. J Virol. 1994;68:2683–2692. doi: 10.1128/jvi.68.4.2683-2692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt A R, Johnson A J, Roehrig J T. Synthetic peptides of Venezuelan equine encephalitis virus E2 glycoprotein. Virology. 1990;179:701–711. doi: 10.1016/0042-6822(90)90137-g. [DOI] [PubMed] [Google Scholar]
- 21.Jain S K, DeCandido S, Kielian M. Processing of the p62 envelope precursor protein of Semliki Forest virus. J Biol Chem. 1991;266:5756–5761. [PubMed] [Google Scholar]
- 22.Johnson A J, Hunt A R, Roehrig J T. Synthetic peptides of Venezuelan equine encephalomyelitis virus E2 glycoprotein. III. Identification of a protective peptide derived from the carboxy-terminal extramembranal one-third of the protein. Virology. 1991;185:840–842. doi: 10.1016/0042-6822(91)90555-p. [DOI] [PubMed] [Google Scholar]
- 23.Kerr P J, Fitzgerald S, Tregear G W, Dalgarno L, Weir R C. Characterization of a major neutralization domain of Ross river virus using anti-viral and anti-peptide antibodies. Virology. 1992;187:338–342. doi: 10.1016/0042-6822(92)90324-i. [DOI] [PubMed] [Google Scholar]
- 24.Kerr P J, Weir R C, Dalgarno L. Ross River virus variants selected during passage in chick embryo fibroblasts: serological, genetic, and biological changes. Virology. 1993;193:446–449. doi: 10.1006/viro.1993.1143. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 26.Liljeström P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Nat Biotechnol. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 28.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobigs M, Garoff H. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J Virol. 1990;64:1233–1240. doi: 10.1128/jvi.64.3.1233-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobigs M, Zhao H X, Garoff H. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J Virol. 1990;64:4346–4355. doi: 10.1128/jvi.64.9.4346-4355.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loewy A, Smyth J, von Bonsdorff C H, Liljeström P, Schlesinger M J. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J Virol. 1995;69:469–475. doi: 10.1128/jvi.69.1.469-475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.London S D, Schmaljohn A L, Dalrymple J M, Rice C M. Infectious enveloped RNA virus antigenic chimeras. Proc Natl Acad Sci USA. 1992;89:207–211. doi: 10.1073/pnas.89.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meek A D J, Faragher S G, Weir R C, Dalgarno L. Genetic and phenotypic studies on Ross River virus variants of enhanced virulence selected during mouse passage. Virology. 1989;172:399–407. doi: 10.1016/0042-6822(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 34.Meyer W J, Johnston R E. Structural rearrangement of infecting Sindbis virions at the cell surface: mapping of newly accessible epitopes. J Virol. 1993;67:5117–5125. doi: 10.1128/jvi.67.9.5117-5125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama K, Watanabe T, Nakagawa T, Kim W-S, Nagahama M, Hosaka M, Hatsuzawa K, Kondoh-Hashiba K, Murakami K. Consensus sequence for precursor processing at mono-arginyl sites. Evidence for the involvement of a Kex2-like endoprotease in precursor cleavages at both dibasic and mono-arginyl sites. J Biol Chem. 1992;267:16335–16340. [PubMed] [Google Scholar]
- 36.Picard V, Ersdal-Badju E, Lu A, Bock S C. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994;22:2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilipenko E V, Gmyl A P, Agol V I. A model for rearrangements in RNA genomes. Nucleic Acids Res. 1995;23:1870–1875. doi: 10.1093/nar/23.11.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Presley J F, Polo J M, Johnston R E, Brown D T. Proteolytic processing of the Sindbis virus membrane protein precursor PE2 is nonessential for growth in vertebrate cells but is required for efficient growth in invertebrate cells. J Virol. 1991;65:1905–1909. doi: 10.1128/jvi.65.4.1905-1909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell D L, Dalrymple J M, Johnston R E. Sindbis virus mutations which coordinately affect glycoprotein processing, penetration, and virulence in mice. J Virol. 1989;63:1619–1629. doi: 10.1128/jvi.63.4.1619-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 825–841. [Google Scholar]
- 43.Smeekens S P. Processing of protein precursors by a novel family of subtilisin-related mammalian endoproteases. Nat Biotechnol. 1993;11:182–186. doi: 10.1038/nbt0293-182. [DOI] [PubMed] [Google Scholar]
- 44.Snijders A, Benaissa-Trouw B J, Oosterlaken T A, Puijk W C, Posthumus W P, Meloen R H, Boere W A, Oosting J D, Kraaijeveld C A, Snippe H. Identification of linear epitopes on Semliki Forest virus E2 membrane protein and their effectiveness as a synthetic peptide vaccine. J Gen Virol. 1991;72:557–565. doi: 10.1099/0022-1317-72-3-557. [DOI] [PubMed] [Google Scholar]
- 45.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270:28397–28401. doi: 10.1074/jbc.270.47.28397. [DOI] [PubMed] [Google Scholar]
- 47.Tucker P C, Lee S H, Bui N, Martinie D, Griffin D E. Amino acid changes in the Sindbis virus E2 glycoprotein that increase neurovirulence improve entry into neuroblastoma cells. J Virol. 1997;71:6106–6112. doi: 10.1128/jvi.71.8.6106-6112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks M S, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vrati S, Fernon C A, Dalgarno L, Weir R C. Location of a major antigenic site involved in Ross River virus neutralization. Virology. 1988;162:346–353. doi: 10.1016/0042-6822(88)90474-6. [DOI] [PubMed] [Google Scholar]
- 50.Vrati S, Kerr P J, Weir R C, Dalgarno L. Entry kinetics and mouse virulence of Ross River virus mutants altered in neutralization epitopes. J Virol. 1996;70:1745–1750. doi: 10.1128/jvi.70.3.1745-1750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahlberg J M, Boere W A, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahlberg J M, Bron R, Wilschut J, Garoff H. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J Virol. 1992;66:7309–7318. doi: 10.1128/jvi.66.12.7309-7318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahlberg J M, Garoff H. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J Cell Biol. 1992;116:339–348. doi: 10.1083/jcb.116.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson D G, Moehring J M, Moehring T J. A mutant CHO-K1 strain with resistance to Pseudomonas exotoxin A and alphaviruses fails to cleave Sindbis virus glycoprotein PE2. J Virol. 1991;65:2332–2339. doi: 10.1128/jvi.65.5.2332-2339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welch W J, Sefton B M. Two small virus-specific polypeptides are produced during infection with Sindbis virus. J Virol. 1979;29:1186–1195. doi: 10.1128/jvi.29.3.1186-1195.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 57.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziemiecki A, Garoff H, Simons K. Formation of the Semliki Forest virus membrane glycoprotein complexes in the infected cell. J Gen Virol. 1980;50:111–123. doi: 10.1099/0022-1317-50-1-111. [DOI] [PubMed] [Google Scholar]