Abstract
In this work we have studied the intracellular localization properties of the Gag and Env proteins of Moloney murine leukemia virus (MLV) and human immunodeficiency virus (HIV) in dorsal root ganglion (DRG) neurons of rat. These neurons form thick bundles of axons, which facilitates protein localization studies by immunofluorescence analyses. When such neuron cultures were infected with recombinant Semliki Forest virus particles carrying the gag genes of either retrovirus, the expressed Gag proteins were localized to both the somatic and the axonal regions of the DRG neurons. In contrast, the Env proteins were confined only to the somatic region. When the Gag and Env proteins were coexpressed, the Gag proteins were also excluded from the axons. This effect of the Env proteins was shown to be dependent on the concentration of the Gag proteins in the neuron and also to be specific for homologous pairs of retrovirus proteins. Therefore, the results suggest that there are specific interactions between the Env and the Gag proteins of MLV and HIV in the DRG neurons.
All enveloped viruses are equipped with transmembrane proteins, called envelope (Env) proteins or spikes, on their surface. These proteins enable the particles to bind to host cell receptors and, further, to penetrate into the cell cytoplasm by a virus membrane-host membrane fusion event. The most reasonable explanation of how the envelope proteins become incorporated into the viral membrane is that there are specific interactions between the internal (cytoplasmic) core or capsid structure of the virus and the cytoplasmic domains (tails) of the viral membrane proteins. If these interactions drive the budding process of the virus, only entry-competent particles which contain spike proteins will be produced. While this budding model has been verified for several alphaviruses (e.g., Semliki Forest virus [SFV], Sindbis virus, and Ross River virus) (5, 33, 36) and hepadnaviruses (e.g., hepatitis B virus) (1a, 2, 9), it does not hold true for retroviruses. Several studies have shown that intracellular expression of the cytoplasmic Gag precursor protein alone results in its membrane binding, core formation, and budding into the medium of the host cells (7, 13, 15, 26, 35). These results have confirmed some early reports on the existence of helper-dependent retrovirus particles that lack the Env proteins (25, 28). This ability of the Gag precursor protein raises an important question about how Env proteins are incorporated into the retroviral envelope. One possibility is that the Env proteins are incorporated by specific Env-Gag interactions that are functionally uncoupled from the budding reaction. According to another model, there is no interaction between the Env and Gag proteins at all, but the Env proteins end up in the particle passively after being localized to that region of the cell membrane where the Gag-driven budding takes place. Therefore, a key question concerning the incorporation mechanism of Env is whether there is an Env-Gag interaction or not.
Several recent studies suggest that such an interaction exists, at least in the case of lentiviruses. First, Owens et al. (23) showed that human immunodeficiency virus (HIV) Env will restrict HIV Gag budding to the basolateral (BL) plasma membrane (PM) domain of the polarized epithelial cell line MDCK. When Gag was expressed separately, budding occurred from both the BL and the apical PM domains. The most reasonable explanation for this phenomenon is that Gag proteins interact with Env proteins, which are known to be targeted to the BL PM. Second, studies with both HIV and simian immunodeficiency virus have shown that certain mutations in the NH2-terminal domain (MA) of Gag will block incorporation of Env into the viral envelope during budding (10, 14). However, Env proteins with tail deletions were not blocked. These data suggested that Env proteins with intact tails can enter the envelope only by interactions with Gag but that if the tail has been deleted, then Env can be incorporated unspecifically. Third, Cosson (6) used an in vitro assay to demonstrate a specific binding between HIV MA and the tail of HIV Env. Thus, it is very likely that HIV incorporates the Env proteins into its envelope by an interaction with the Gag proteins during budding. In the case of other retroviruses, the existence of an Env-Gag interaction is still an open question.
In the present work we have studied this interaction in Moloney murine leukemia virus (MLV) and HIV by monitoring the localization properties of their Gag and Env proteins in primary neuron cultures from rat dorsal root ganglia (DRG). The corresponding retroviral genes were introduced into DRG neurons by infection with recombinant SFV particles. We show that the Gag proteins localize to both the somatic and axonal regions of the DRG cells when expressed separately. However, when coexpressed with the homologous Env protein, but not with the heterologous one, the Gag protein localization becomes restricted to the somatic region of the cell, the domain that is used by the retroviral Env proteins. Thus, these results suggest that the Env and Gag proteins of MLV and HIV can interact with each other in a specific way in the DRG neurons.
MATERIALS AND METHODS
Primary cell culture.
DRG neurons were obtained from embryos of pregnant Sprague-Dawley rats (B&K, Stockholm, Sweden) taken on day 15 to 18 of gestation. The cultures were prepared as described by Sotelo et al. (31). Briefly, DRG were dissociated by several passages through a constricted Pasteur pipette, and the cells were seeded on collagen-coated (Collagen Corporation, Palo Alto, Calif.) glass coverslips (G. Menzel, Braunschweig, Germany) attached to the bottom of sterile plastic petri dishes (Costar, Cambridge, Mass.). The culture medium consisted of minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 10% horse serum, 2% chicken embryo extract, gentamicin sulfate (15 μg/ml), and l-glutamine (200 mM) (all obtained from Gibco, Paisley, Scotland) and glucose (6 mg/ml) and nerve growth factor (1 ng/ml) (Sigma Chemical Co., St. Louis, Mo.). The medium was changed three times a week. The cultures were exposed for 48 h to a cytostatic agent, cytosine arabinoside (3 μg/ml) (Sigma Chemical Co.), after 4 to 5 days to inhibit proliferation of nonneuronal cells. For further incubation the bovine serum was omitted from the medium. The primary DRG cultures consisted of differentiated neurons and a population of nonneuronal cells, predominantly fibroblasts and Schwann cells. Neurons constituted approximately 30 to 50% of all cells in such cultures. The choices of medium supplements, inclusion or exclusion of neurotrophic factors, and adhesive properties of the underlying substrate were based on observation of the survival and differentiation of the sensory neurons. The proportion, presence, or absence of the various substances was found to be critical for an optimal microenvironment.
Hippocampal cells were prepared from rat embryos after 18 days of gestation, generally as described before (27). The hippocampi were dissected, trypsinized (0.1% trypsin [Gibco] for 15 min at 37°C), and dissociated by several passages through a constricted Pasteur pipette. Cell suspensions were seeded on glass coverslips coated with poly-l-lysine hydrobromide (Sigma Chemical Co.) and placed in petri dishes. The culture medium, Dulbecco’s MEM-Nutrient Mix F12 (Gibco) and 10% fetal bovine serum, was supplemented with glucose (1.2 mg/ml), 20 nM progesterone, 100 μM putrescine, and 30 nM selenium dioxide (all from Sigma Chemical Co.) and bovine insulin (5 μg/ml) and human transferrin (100 μg/ml) (both from Gibco). Penicillin and streptomycin (Gibco) were added to final concentrations of 20 U/ml and 20 μg/ml, respectively.
After 14 days in culture, DRG and hippocampal cells were incubated with recombinant SFV diluted in MEM (total volume, 600 μl/petri dish). After 1 h of incubation at 37°C, culture medium was added for a total of 2 ml/petri dish. The cultures were sampled at 6 and 16 h postinfection, which were found to be optimal infection times as estimated from the protein expression level and preservation of neurons. Morphological cytotoxic effects became evident after 24 h, when neuronal perikarya swelled, processes retracted, and, finally, the cells detached from the coverslips.
BHK-21 cell culture.
Baby hamster kidney (BHK-21) cells, obtained from the American Type Culture Collection, were grown in Glasgow MEM (BHK-21) and supplemented with 10% tryptose phosphate broth, 5% fetal bovine serum, 20 mM HEPES, and 2 mM glutamine (all obtained from Gibco). Cells were washed once with phosphate-buffered saline (PBS) (without Ca2+ and Mg2+) (Gibco), trypsinized with 1× trypsin-EDTA (0.5 and 0.2 mg/ml, respectively) (Gibco) in PBS (without Ca2+ and Mg2+), and subcultivated 1:5 every second day. Cells were incubated at 37°C in 5% CO2–95% O2.
Plasmids.
High-expression-level plasmids were (i) SFV-C-hTR, which encodes the human transferrin receptor (hTR) (19); (ii) SFV-C-hTRΔ2, which encodes a variant hTR in which most of its cytoplasmic domain is deleted (32); (iii) SFV-C-gagMLV, which encodes the Gag precursor protein of MLV (32); and (iv) SFV-C-gagHIV, which encodes the Gag precursor of HIV (16a). Low-expression-level plasmids were (i) SFV-1-gagMLV, which encodes the Gag precursor of MLV (32); (ii) SFV-1-envMLV, which encodes the Env precursor of MLV (32); (iii) SFV-1-gagHIV, which encodes the Gag precursor of HIV (16a); and (iv) SFV-1-envHIV, which encodes the Env precursor of HIV (24). In addition, the SFV Helper 1 plasmid was used (19).
Antibodies.
The following primary antibodies were used: (i) anti-HIV Env antibody P4/D10, a monoclonal antibody (MAb) which recognizes GP120 (kindly provided by B. Wahren, Karolinska Institute, Stockholm, Sweden); (ii) anti-HIV Gag antibody RL4.72.1.1, a MAb which recognizes p24/p55 (Aalton, Dublin, Ireland); (iii) anti-MLV antibody HC185, a polyclonal pig antiserum which recognizes whole MLV (Quality Biotech Inc.); (iv) anti-MLV Gag, a rabbit antiserum which recognizes p30 (kindly provided by G. Smith, Institute for Molecular Virology, GSF, Münich, Germany); (v) anti-MLV Env MAb 500, a MAb which recognizes gp70 (kindly provided by B. W. Chesebro, Rocky Mountain Laboratories, National Institutes of Health, Hamilton, Mont.); (vi) anti-hTR antibody OKT9, a MAb which recognize hTR (prepared by T. Ebel by using the corresponding hybridoma cell line, which was obtained from the American Type Culture Collection); and (vii) anti-MAP2, a MAb which recognizes the microtubule-associated protein 2 (MAP2) (Sigma Chemical Co.).
Secondary antibodies were (i) fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse F(ab)2 fragment of immunoglobulin (IgG) (Dakopatts A/S, Copenhagen, Denmark), (ii) FITC-conjugated swine anti-rabbit IgG (Dakopatts), and (iii) tetramethylrhodamine isothiocyanate-conjugated rabbit anti-mouse IgG (Dakopatts).
Preparation of recombinant SFV.
Expression plasmids were used for transcription of recombinant SFV RNA in vitro as described before (19). Recombinant SFV particles were produced by cotransfecting 107 BHK-21 cells with recombinant SFV and Helper 1 RNA, also as described before (19). Yields of recombinant virus varied between 107 and 108 infectious particles per ml. Recombinant viruses were stored in small aliquots at −80°C.
Immunofluorescence analyses.
Cells grown on glass coverslips were washed twice with Dulbecco’s PBS (PBS with Mg2+ and Ca2+) (Gibco) and then fixed either in cold methanol (−20°C) for 5 to 6 min or in 4% formaldehyde at room temperature for 30 min. For immunolabelling with the MAbs, the cells were preincubated with 2% normal rabbit serum (Dakopatts) and 0.3% Triton X-100 (Eastman Kodak Company, Rochester, N.Y.) for 5 min at room temperature. The cultures were then incubated for 1 h at room temperature with one of the MAbs diluted in 2% normal rabbit serum and 0.3% Triton X-100. After being rinsed in Dulbecco’s PBS, the cells were incubated with FITC-conjugated rabbit anti-mouse F(ab)2 fragment of IgG (Dakopatts) diluted 1:15 in Dulbecco’s PBS with 2% normal rat serum (Dakopatts) for 30 min at 37°C. After being rinsed in distilled water, the cultures were mounted in glycerol. For labelling with the rabbit anti-MLV Gag hyperimmune serum, the cells were preincubated with 2% normal swine serum (Dakopatts) and 0.3% Triton X-100 for 5 min at room temperature followed by the anti-MLV Gag antiserum diluted in 2% normal swine serum and 0.3% Triton X-100 for 1 h at room temperature. After being rinsed, the cells were incubated with FITC-conjugated swine anti-rabbit IgG (Dakopatts) diluted 1:15 in Dulbecco’s PBS containing 2% normal rat serum for 30 min at 37°C. For double labelling the cells were first incubated with the anti-MLV Gag antiserum, washed, and then incubated with the anti-HIV Env MAb diluted in 2% normal horse serum (Vector Laboratories, Burlingame, Calif.). After being rinsed, the cells were incubated with FITC-conjugated swine anti-rabbit IgG, followed by incubation with tetramethylrhodamine isothiocyanate-conjugated rabbit anti-mouse IgG (Dakopatts) diluted 1:30 in 2% normal rat serum for 30 min at 37°C, and mounted.
Metabolic labelling of cells and preparation of cell lysates.
Neuron and BHK-21 cell cultures were used for labelling with [35S]methionine 6 h after infection. Culture media were replaced with methionine-free MEM (Gibco). After 30 min at 37°C, media were replaced with 500 μl of the same methionine-free medium containing 50 μCi of [35S]methionine (Amersham International plc, Bucks, England), and cells were incubated at 37°C for 30 min. After the 30-min pulse, cells were washed twice with MEM (Gibco) containing a 10-fold excess of cold methionine (Gibco) and then incubated in the same medium for 15 or 120 min at 37°C (chase). After the 120-min chase, media were collected and clarified by centrifugation in an Eppendorf centrifuge for 5 min at 5,000 rpm and 4°C. After pulse-chasing of HIV-infected cells, the medium was removed and cells were washed with cold PBS and lysed with 300 μl of Nonidet P-40 (NP-40) lysis buffer. This contained 1% NP-40, 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 10 μg of phenylmethylsulfonyl fluoride per ml, and 20 mM N-ethylmaleimide. Nuclei were removed from cell lysates by centrifugation in an Eppendorf centrifuge for 5 min at 6,000 rpm and 4°C. After pulse-chasing of MLV-infected cells, the medium was removed and cells were washed with PBS and lysed with 300 μl of sodium dodecyl sulfate (SDS) lysis buffer. This contained 1% SDS, 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 2 mM EDTA, 10 μg of phenylmethylsulfonylfluoride per ml, and 20 mM N-ethylmaleimide. SDS lysis was done at room temperature. The SDS lysate was passed five times through a 20 G1 1/2 9 by 40 needle, heated to 95°C for 2 min, and centrifuged in an Eppendorf centrifuge for 2 min at 14,000 rpm. The supernatant was transferred to a fresh tube.
Analyses of virus particles.
Virus particles were harvested from clarified 120-min chase medium by pelleting them through a 10% sucrose cushion in a Beckman JA 18.1 rotor at 17,000 rpm for 2 h at 4°C. Pelleted particles were taken up into SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, heated for 5 min at 95°C, and analyzed by SDS-PAGE.
Immunoprecipitation.
MLV Gag and Env precursors were immunoprecipitated from SDS cell lysates with the anti-MLV antiserum. HIV precursors were immunoprecipitated from NP-40 cell lysates with anti-HIV Env and anti-HIV Gag antibodies. Aliquots of SDS lysates (100 μl) were diluted with 900 μl of NET buffer, which contained 1% NP-40, 50 mM Tris-HCl (pH 8.0), 400 mM NaCl, 5 mM EDTA (pH 8.0), and 0.02% NaN3, and mixed with the antibody and 40 μl of protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden) (50% [vol/vol] in 10 mM Tris-HCl, pH 7.5). The samples were then rotated end over end for 16 h at 4°C. Pellets were washed twice with a solution containing 0.2% NP-40, 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2 mM EDTA; twice with a solution containing 0.2% NP-40, 10 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 2 mM EDTA; and finally once with a solution containing 10 mM Tris-HCl (pH 7.5). The immunoprecipitation in NP-40 cell lysates was done as previously described (34).
SDS-PAGE.
Immunoprecipitates and pelleted virus particles were taken up into 40 μl of SDS-gel sample buffer, which contained 200 mM Tris-HCl (pH 8.8), 20% glycerol, 5 mM EDTA, 0.02% bromphenol blue, 1 mM methionine, 4% SDS, and 50 mM dithiothreitol. The sample mixture was heated for 5 min at 95°C before being analyzed on a 10 or 15% gel. After electrophoresis, gels were processed for autoradiography (30). Quantitation of radioactivity of the protein in gel bands was done with a Fuji phosphorimager (type FUJIX BAS 2000 TR). For calculation of protein ratios, the PSL values of corresponding proteins were normalized for the methionine content of respective protein.
Electron microscopic analyses.
Hippocampal and DRG cultures were infected with SFV-C-gagHIV or SFV-C-gagMLV recombinant virus particles. At 6 and 16 h postinfection the cultures were fixed in 1% glutaraldehyde–PBS, postfixed in OsO4, and embedded in LX112 (Ladd, Burlington, Vt.). For orientation, semithin sections stained with toluidine blue were used. For electron microscopy, ultrathin sections were cut and stained with uranyl acetate and lead citrate.
RESULTS
Analyses of protein targeting in dorsal root ganglia neurons with recombinant SFV.
It has recently been shown that rat hippocampus neurons can be infected with recombinant SFV carrying a foreign gene and that these cells retain their typical morphological features for several hours (21, 22). This has facilitated gene expression experiments with these cells, for instance, for the purpose of studying the mechanisms of axonal and dendritic protein transport. However, analyses of the axonal distribution of a protein in hippocampus neurons are sometimes difficult because the latter extensions have such small diameters. Therefore, we investigated whether DRG cultures could be used for similar studies (31). The DRG neuron differs considerably in morphology from the hippocampus neuron (8, 17). DRG neurons form very thick bundles of axons, while dendritic extensions are completely lacking. A typical DRG neuron is shown in Fig. 1a. The compact cell body (the somatic region) and the thick bundle of axons are clearly visible. We first analyzed the distribution of the endogenous MAP2 in these neurons by using immunofluorescence after membrane permeabilization. This marker protein is known to be present in free and tubulin-associated forms in the somatodendritic domain of hippocampus neurons but to be excluded from their axon extensions (3, 20). The staining of the cells in our DRG culture showed a corresponding polarity: the cell body was positive, whereas the axon bundle was negative (Fig. 1b). For comparison, MAP2 staining in a hippocampus neuron is also shown (Fig. 1c). In this case, clear somatic and dendritic staining was observed.
FIG. 1.
Immunofluorescence analyses of MAP2 in nerve cells. DRG neurons (a and b) and a hippocampus neuron (c) were stained with anti-MAP2 antibodies and examined by fluorescence (b and c) and Nomarski (a) microscopy. Arrow, axon bundle of the DRG neuron. Magnification, ×375.
Next we studied the distribution of hTR in DRG neurons. This marker protein was expressed by infecting the neurons with SFV-C-hTR recombinant particles. Rat TR has earlier been shown to be confined to the dendrites of hippocampus neurons (4). Consistent with these earlier findings, staining of the infected DRG cultures with an hTR-specific MAb (which does not react with the endogenous receptor) showed clear staining of the cell body, whereas the thick axon bundle remained unstained (Fig. 2a and b). It should be noted that this restricted localization of hTR was maintained although the SFV-C-based vector is known to express very high levels of this receptor molecule (29). As a control we also stained hippocampus neurons and observed strong somatodendritic staining (Fig. 2e). The distribution of the hTR between the internal (endosomal) and surface pools was not examined. However, we know that a fraction of hTR is also stained in nonpermeabilized hippocampus and DRG neurons (data not shown).
FIG. 2.
Immunofluorescence analyses of hTR in nerve cells. DRG neurons (a to d) and a hippocampus neuron (e) were infected with SFV-C-hTR (a, b, and e) or SFV-C-hTRΔ2 (c and d), incubated for 6 h, and stained with anti-hTR antibodies that do not react with rat TR. Panels b and d are Nomarski images of the stained cells in panels a and c, respectively. Note the restricted localization of hTR to the soma of the DRG neuron in panel a, whereas the corresponding cytoplasmic tail deletion variant localizes to the axon bundle as well (c). Magnification, ×375.
In order to test the importance of the cytoplasmic tail of hTR for the localization behavior of TR, we infected the DRG neurons with a vector expressing a cytoplasmic tail deletion variant of hTR (SFV-C-hTRΔ2). This corresponds to a deletion variant which has been used before to show that the tail contains a BL targeting signal of this protein in epithelial cells (18). The results of this localization analyses are exemplified by the immunofluorescence staining in Fig. 2c. It is evident not only that the hTRΔ2 is present in the somatic region but also that strong staining is seen in the axon bundle. This finding was consistent for all infected and stained DRG neurons of the culture. These results suggest that there is somatic targeting information in the cytoplasmic tail of hTR. We conclude that the DRG cultures can be used with advantage to study signal-mediated protein targeting in neurons.
SFV recombinants expressing retrovirus proteins.
Six different SFV recombinants were used to express the MLV and HIV Gag and Env proteins. SFV-C-gagMLV was used for high-level expression of the MLV Gag protein, whereas SFV-1-gagMLV and SFV-1-envMLV were used to express the MLV proteins at a comparatively lower level. SFV-C-gagHIV, SFV-1-gagHIV, and SFV-1-envHIV were the corresponding vectors for the expression of HIV proteins. It should be noted that the infectivity and RNA replication of SFV-1 and SFV-C vectors are equal; only the mRNA translatability differs. The SFV-C-based vectors contain a translation-enhancing RNA segment in the SFV capsid (C)-coding part of the subgenomic mRNA which is used for heterologous gene expression in this vector (29). We also constructed SFV-C-env vectors for high-level expression of the Env proteins. However, high-level synthesis of the MLV and HIV Env proteins was associated with severe folding problems in the endoplasmic reticulum, and these vectors therefore could not be used (1).
Synthesis of Gag and Env proteins of MLV and HIV in DRG neurons.
DRG cultures were infected with the various SFV-1-based recombinant viruses and analyzed for protein expression by SDS-PAGE. The results are shown in Fig. 3. The cells were labelled with [35S]methionine for 30 min and then chased as indicated before the preparation of cell and medium samples. Figure 3a, lanes 5 to 7, shows that 65-kDa Gag precursors of MLV are produced in the SFV-1-gagMLV-infected ganglion neurons and also are released as particles into the medium. This is very similar to their behavior in BHK cell cultures (Fig. 3a, lanes 2 to 4), which has been reported before (32). Note the efficient shutdown of host protein synthesis when the SFV expression system is used. This facilitates quantitative and qualitative analyses of the expressed proteins directly from cell extracts. Note also that much less protein is expressed in DRG than in BHK cultures. The major reason is that there are fewer cells in the DRG culture. Figure 3b, lanes 5 and 6, shows the synthesis of the Env precursor protein of MLV in SFV-1-envMLV-infected cells. This is the same size (80 kDa) as the corresponding product in BHK cells (Fig. 3b, lanes 2 and 3). After the 120-min chase, the transmembrane cleavage product Pr15E can be detected. There is also some degradation of the Env precursors in the DRG neurons during the 120-min chase.
FIG. 3.
SDS-gel analyses of retroviral proteins expressed in DRG neurons and BHK-21 cells. DRG neuron and BHK-21 cell cultures were infected with recombinant SFV carrying the genes for Gag and Env precursors, incubated, pulse-labelled, chased (15 and 120 min), and processed (without immunoprecipitation if not indicated) for analyses on 10% gels. SFV-infected cells were used as a control. Pelletable material from medium samples was also analyzed. L and M, cell lysate and medium samples. (a, b, c, and d) Analyses of MLV Gag precursor, MLV Env precursor, HIV Gag precursor, and HIV Env precursor, respectively. The MLV-specific Pr65gag, Pr80env, and Pr15E protein bands and the HIV-specific Pr55gag and Pr160env protein bands, as well as the SFV-specific E1, E2, and C protein bands, are indicated.
Figure 3c and d show corresponding analyses of cells infected with SFV-1-gagHIV and SFV-1-envHIV. It is evident that the Gag and Env products of the nerve cells (Fig. 3c and d, lanes 5 and 6) migrated similarly to the 55-kDa Gag and the 160-kDa Env precursors that were produced in infected BHK cell cultures (Fig. 3c and d, lanes 2 and 3), which has been previously observed (16a, 24). In the present experiment, no Gag particles could be detected in the medium. The release of such particles is much less efficient in the case of HIV than with MLV (16a). We conclude that authentic MLV and HIV proteins were produced in the infected DRG cultures.
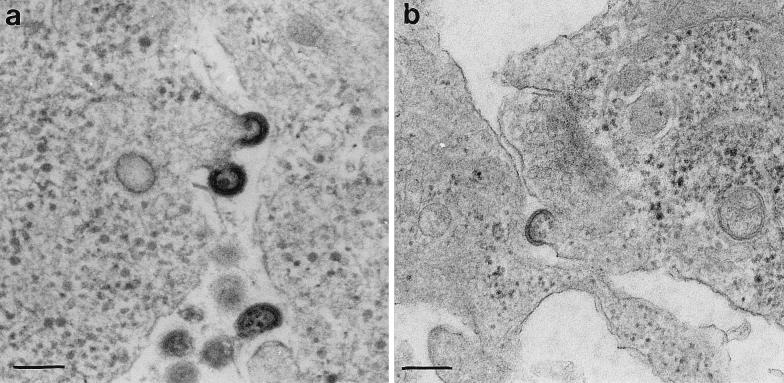
Morphological analyses of Gag particles in SFV-1-gagMLV- and SFV-1-gagHIV-infected nerve cells.
An electron micrograph of an SFV-1-gagMLV-infected DRG neuron is shown in Fig. 4a. Several free and budding MLV Gag particles are present. Most particles were spherical, with a diameter of approximately 130 nm. In Fig. 5a to c, infected hippocampus neurons are shown for comparison. In these cells, budding and accumulation of MLV Gag particles were also found at or near synaptic membranes (Fig. 5b and c). Corresponding analyses of SFV-1-gagHIV-infected DRG and hippocampus neurons are shown in Fig. 4b and 5d. Note the difference between HIV and MLV Gag particle morphology: the MLV particle appear to have a double-layered structure below the viral envelope, whereas the HIV particle has only a single layer. These are typical features for these two different retroviruses (11, 12).
FIG. 4.
Electron microscopic analyses of Gag particle budding in DRG neurons. Cultures with DRG neurons were infected with recombinant SFV carrying the genes for MLV Gag (a) and HIV Gag (b), incubated for 6 h, and processed for electron microscopic analyses. Bars, 200 nm.
FIG. 5.
Electron microscopic analyses of Gag particle budding in hippocampus neurons. Cultures with hippocampus neurons were infected with recombinant SFV carrying the genes for MLV Gag (a to c) and HIV Gag (d), incubated for 16 h, and processed for electron microscopic analyses. In panels b and c, MLV Gag particles are shown to bud at dendrite-like processes. Note the morphological difference between the membrane-associated layers of the MLV and HIV particles. Bars, 200 nm.
Localization of Env and Gag precursors in neurons.
The distribution of the MLV and HIV proteins in the infected neurons was studied by immunofluorescence analyses of permeabilized cells. We first compared the staining patterns of separately expressed Gag and Env proteins in infected hippocampus and DRG neurons. Figure 6 shows photomicrographs of infected hippocampus neurons. The Gag and Env proteins of both retroviruses are localized in the entire somatodendritic region of the hippocampus neurons. The thin axon extensions of these cells are not easily detected, but two possible ones which are positive for HIV and MLV Gag precursors, respectively, are indicated in Fig. 6b and d. These are thin nontapering extensions lacking branches at acute angles. Figures 7 and 8a (upper panels) show corresponding micrographs of infected DRG neurons. In these, the Gag precursors of both retroviruses are shown to be distributed in the cell body as well as in the thick, clearly visible axon bundle. Figure 8b shows a larger view of a DRG culture that has been infected with SFV-1-gagHIV and stained for the HIV Gag protein. However, it should be noted that there was an apparent difference between the two kinds of Gag proteins in their efficiency in reaching the axons. While MLV Gag proteins entered axons in virtually all DRG neurons which were infected with SFV-1-gagMLV, the HIV Gag proteins entered the axon extensions in only about 65% of the SFV-1-gagHIV-infected neurons (Table 1). In addition, some infected ganglion neurons showed HIV Gag protein staining only in the proximal part of the axon bundle. In contrast to the Gag proteins, the Env proteins of both viruses were found to be restricted to the cell body. This is shown for MLV Env in Fig. 7 (upper panel) and for HIV Env in Fig. 8a (upper panel). It should be noted that these analyses do not indicate how much of the Env precursors is present at the cell surface. A major fraction of the Env precursors might actually reside in intracellular membranes of the cell body. However, this possibility does not interfere with our main conclusions from the results of the coexpression experiments described below.
FIG. 6.
Immunofluorescence analyses of MLV and HIV Gag and Env precursor proteins in hippocampus neurons. Cultures with hippocampus neurons were infected with recombinant SFV carrying the genes for HIV Env (a), HIV Gag (b), MLV Env (c), and MLV Gag (panel d). The cultures were incubated for 6 h and stained with anti-HIV Env (a), anti-HIV Gag (b), anti-MLV Env (c), and anti-MLV Gag (d). Arrows, putative axon extensions. Magnification, ×375.
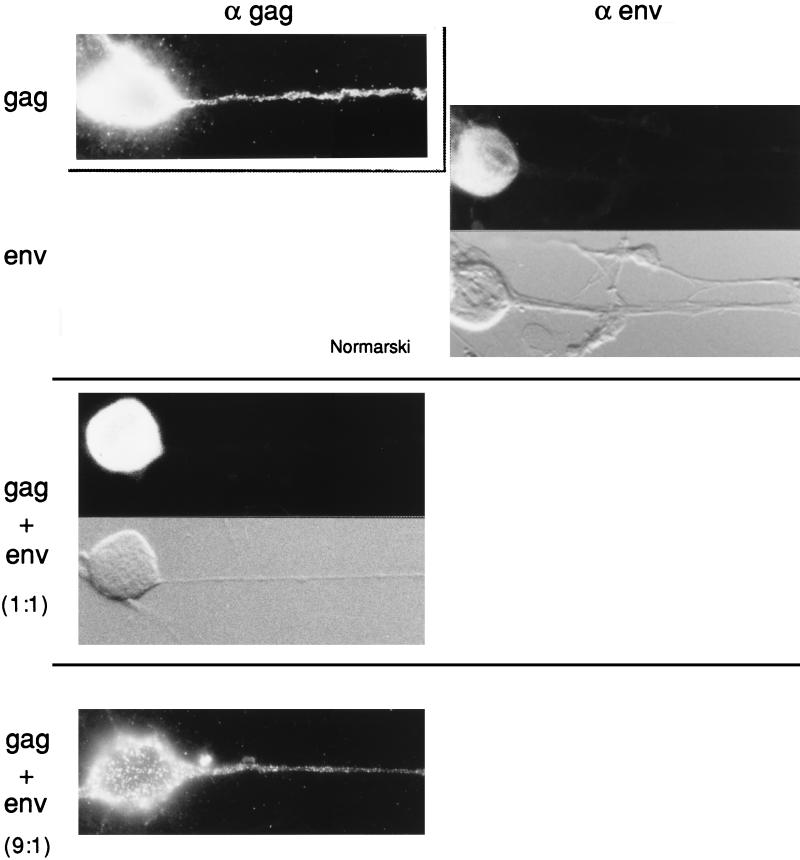
FIG. 7.
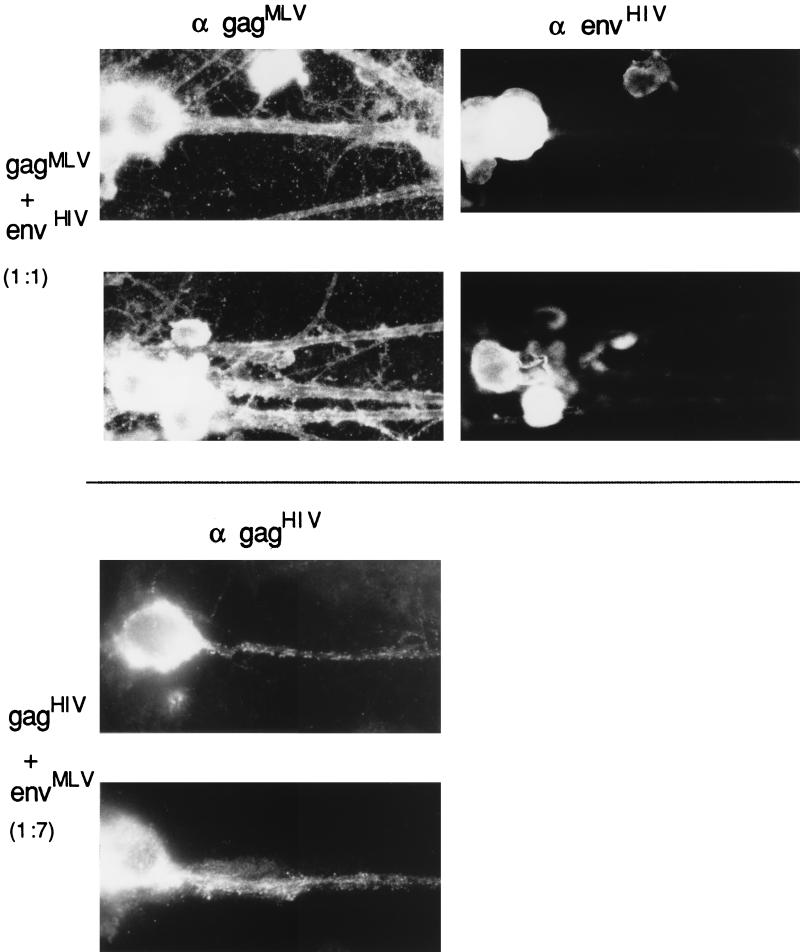
Immunofluorescence analyses of MLV Gag and Env precursor proteins in DRG neurons. Several parallel DRG cultures were infected with recombinant SFV carrying the genes for MLV Gag and Env either separately or together, as indicated to the left. After incubation, the cultures were stained with anti-MLV Gag (α gag) or anti-MLV Env (α env) antibodies, as indicated at the top. The ratios given to the left indicate the ratio of Gag to Env proteins as determined by quantitation of radioactive bands in gel analyses of samples from parallel cultures. Nomarski views are also indicated. Magnification, ×380.
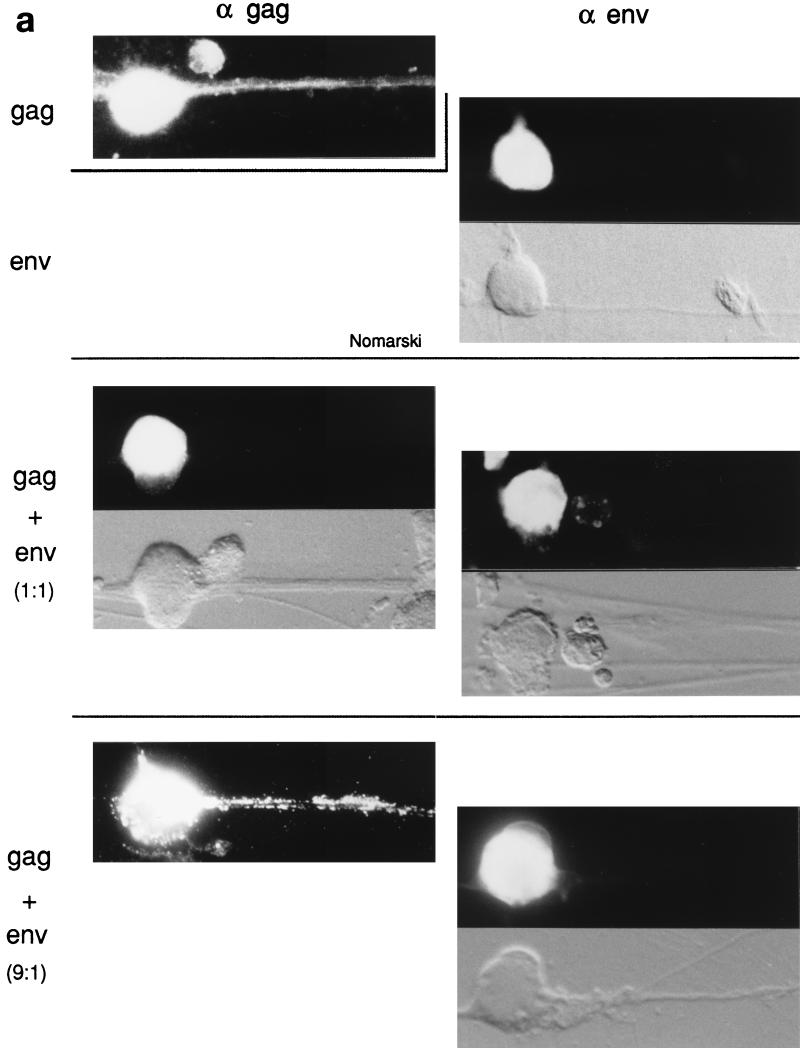
FIG. 8.
Immunofluorescence analyses of HIV Gag and Env precursors in DRG neurons. (a) Analyses were done as described in the legend to Fig. 7 for the corresponding MLV proteins. α gag, staining with anti-HIV Gag antiserum; α env, staining with anti-HIV Env antibodies. Magnification, ×380. (b) A larger view of a DRG culture that has been infected with SFV-1-gagHIV and stained with anti-HIV Gag antibodies. Magnification, ×225. (c) Left panel, immunofluorescence analysis of HIV Env in a nonpermealized DRG neuron; right panel, corresponding Nomarski view.
TABLE 1.
MLV and HIV Gag protein distributions in DRG neurons when expressed separately or together with their homologous envelope precursor
| Recombinant SFV used for infection | Gag/Env ratio in cellsa | % of positive cells with Gag in both soma and axonb |
|---|---|---|
| SFV-1-gagMLV | NAc | 98 |
| SFV-C-gagMLV | NA | 98 |
| SFV-1-gagMLV plus SFV-1-envMLV | 1:1 | 5 |
| SFV-C-gagMLV plus SFV-1-envMLV | 9:1 | 95 |
| SFV-1-gagHIV | NA | 65 |
| SFV-C-gagHIV | NA | 70 |
| SFV-1-gagHIV plus SFV-1-envHIV | 1:1 | 5 |
| SFV-C-gagHIV plus SFV-1-envHIV | 9:1 | 65 |
Determined from quantitation of radioactive proteins in SDS gels.
In each experiment, 50 to 60% of DRG neurons were transfected. Analyses were then performed with 100 to 350 of the transfected neurons. Staining was with anti-MLV Gag antiserum or with anti-HIV Gag antibodies.
NA, not applicable.
Coexpression of homologous Env and Gag proteins restricts Gag protein localization to the somatic region of DRG neurons.
Analyses of the Gag precursor distribution in DRG neurons coinfected with SFV-1-gagMLV and SFV-1-envMLV revealed a location of the Gag protein which was restricted to the somatic region of the neuron (Fig. 7, middle panel). Of 200 Gag-positive cells, 95% showed this distribution (Table 1). Although double-immunofluorescence analyses were not possible in this experiment, the very frequent soma-specific Gag localization observed in the coinfected cells suggests an efficient coexpression of the two viral proteins in the neurons.
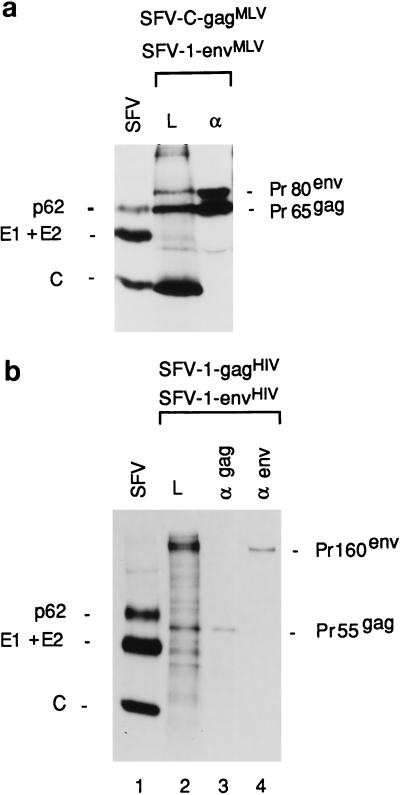
Thus, these results indicate that the MLV Env proteins, which are inserted into the membranes of the somatic part of the DRG, can interact with the Gag proteins and thereby restrict the latter to the somatic domain of the neuron. If this model is correct, then one would expect that the overexpression of Gag relative to Env proteins should allow the Gag proteins to enter into the axons. We tested this by using the SFV-C-gagMLV vector, which expresses about 10 times more Gag protein than SFV-1-gagMLV (29). When ganglion neurons were coinfected with SFV-1-envMLV and SFV-C-gagMLV recombinant viruses, the immunofluorescence analyses showed that the Gag proteins reached the axon in 95% of Gag-positive cells (Fig. 7 [lower panel] and Table 1). The ratio of the Env and Gag protein concentrations in the neurons was estimated by SDS-PAGE analyses of corresponding proteins in the lysate of another ganglion cell culture which had been coinfected in parallel and then pulse-labelled (Fig. 9a). After quantitation of the radioactivities in Gag and Env protein bands and normalization of these values for the number of methionine residues for the respective proteins, we found that there were about nine times more Gag than Env proteins in the cell sample (Table 1). This should be compared with an approximately 1:1 ratio of Gag to Env proteins that we found in those cultures which had been coinfected with the two SFV-1 vectors and which showed the soma-specific Gag protein distribution (Table 1).
FIG. 9.
SDS-gel analyses of retroviral proteins coexpressed in DRG neurons and BHK-21 cells. (a) Analyses of MLV, Gag, and Env proteins coexpressed in same DRG culture at a 9:1 molar ratio. Lane L, lysate sample; lane α, sample obtained by immunoprecipitation of the lysate with anti-MLV serum. (b) Analyses of HIV Gag and Env proteins coexpressed in the same DRG culture at a 1:1 molar ratio. Lane α gag, sample obtained by immunoprecipitation of the lysate with anti-HIV Gag antibodies; lane α env, sample obtained by immunoprecipitation of the lysate with anti-HIV Env antibodies.
When DRG neurons were coinfected with SFV-1-gagHIV and SFV-1-envHIV recombinant viruses and analyzed for Gag and Env protein distribution by immunofluorescence, we observed the same Gag distribution as described above for MLV proteins. With this procedure the Gag precursors were localized in the somatic region and not in the axons as was the case when the Gag protein had been separately expressed (Fig. 8a, middle panel). Upon examination of several hundred Gag-positive cells, we found only 5% of the neurons with Gag proteins in the axon extension (Table 1). The Env protein-directed effect seemed to be dependent on the concentration of the HIV precursor molecules, because when we used the SFV-C-gagHIV vector to overexpress the Gag protein, the latter was found in the axon extensions (Fig. 8a, lower panel). In this case about 65% of the Gag-positive cells displayed Gag proteins in the axons (Table 1). Quantitation of Env and Gag proteins from SDS gel analyses of labelled lysates from SFV-1-gagHIV/SFV-1-envHIV- and SFV-C-gagHIV/SFV-1-envHIV-infected neurons and subsequent calculation of relative concentrations of Gag and Env proteins showed that these were about 1:1 and 9:1, respectively (Fig. 9b; Table 1). Therefore, we conclude that the exclusion of Gag localization in axons depends on the relative concentrations of the Gag and Env proteins in the neuron. This clearly supports the hypothesis of direct binding of the Gag protein to the Env protein.
Coexpression of heterologous Gag-Env protein pairs cannot restrict Gag expression to the somatic region of DRG neurons.
In order to analyze the specificity of the proposed Gag-Env interaction, we performed a series of coexpression experiments using vector combinations which directed the expression of heterologous pairs of Gag and Env proteins. We first analyzed whether the HIV Env protein could influence the distribution of the MLV Gag protein by using DRG neurons coinfected with SFV-1-envHIV and SFV-1-gagMLV recombinant viruses. As previous expression studies using the homologous combinations have demonstrated that the Env protein-directed somatodendritic restriction in Gag localization requires a Gag-to-Env precursor concentration ratio of close to 1 or lower, we tried to vary the ratio of the recombinant particles in the mixtures used for the infection of the neuron cultures accordingly. This was somewhat difficult with the combination of SFV-1-gagMLV and SFV-1-envHIV, since the HIV Env protein was expressed at a considerably lower level than the corresponding MLV protein. However, reasonable precursor ratios were produced (Table 2). By using the Gag-specific polyclonal antibody and the Env-specific MAb, it was possible to make double stainings and study the Env and Gag protein distribution specifically in neurons that were coexpressing the two proteins. The results showed that the MLV Gag proteins reached the axon extensions in all double-stained cells, whereas the HIV Env proteins were confined to the somatic region only (Table 2). Representative stainings are shown in Fig. 10 (upper panel).
TABLE 2.
MLV and HIV Gag protein distributions in DRG neurons when coexpressed with a heterologous Env protein
| Combination of recombinant SFV | Ratio of recombinant particlesa | Gag/Env ratio in cellsb | Coexpression frequency (%)c | % Coexpressing cells with Gag in soma and axon |
|---|---|---|---|---|
| SFV-1-gagMLV plus SFV-1-envHIV | 1:15 | 1:4 | 24 | 100 |
| 5:15 | 3:1 | 46 | 100 | |
| 10:15 | 1.6:1 | 44 | 100 | |
| 10:10 | NDd | 61 | 100 | |
| SFV-1-gagHIV plus SFV-1-envMLV | 20:1 | 1:4 | NAe | 70 |
| 15:5 | 1:12 | NA | 62 | |
| 10:5 | 1:7 | NA | 60 |
Multiplicity of infection determined from titration on BHK-21 cells.
Determined from quantitation of radioactive proteins in SDS gels.
Stainings were with anti-MLV Gag antisera and anti-HIV antibodies for SFV-1-gagMLV plus SFV-1-envHIV and with anti-HIV Gag antibodies for SFV-1-gagHIV plus SFV-1-envMLV.
ND, not determined.
NA, not applicable.
FIG. 10.
Immunofluorescence analyses of heterologous retrovirus precursor combinations in DRG neurons. (Top) Analyses with cells infected with recombinant SFV carrying genes for MLV Gag and HIV Env. The Gag/Env protein ratio was 1:1. Anti-MLV Gag antiserum and anti-HIV Env antibodies were used for staining. (Bottom) DRG cells infected with recombinant SFV carrying the genes for HIV Gag and MLV Env. The Gag/Env protein ratio was 1:7. Staining was with anti-HIV Gag antibodies. Magnification, ×370.
Similar studies were also done with several combinations of SFV-1-gagHIV and SFV-1-envMLV recombinant viruses (Table 2). Unfortunately, we could not use double stainings in these experiments. However, as the MLV Env protein expression was always severalfold higher than that of HIV Gag protein, we assume that a substantial fraction of the HIV Gag-positive neurons were also expressing the MLV Env protein (Table 2). The staining with the anti-HIV Gag antibody showed that 60 to 70% of the Gag-positive cells displayed Gag proteins in both the soma and axon (Fig. 10 [lower panel]; Table 2). This is about the same frequency that we observed in the experiments in which the HIV Gag protein was expressed separately (Table 1). Thus, these results indicate that the MLV Env protein was not able to influence the distribution of the HIV Gag protein in the DRG neurons. We conclude from these experiments that the Env protein is able to restrict only its homologous Gag protein to the somatic region of the neuron. This suggests that the Gag protein-Env protein interaction causing this effect is specific for homologous proteins.
DISCUSSION
We have used an SFV vector-DRG cell system to study the Env-Gag protein interaction of two retroviruses, MLV and HIV. Our major finding is that the Env proteins of these viruses are able to restrict the distribution of the homologous Gag proteins to the somatic region of DRG neurons. If expressed separately, the Gag proteins of both viruses will also distribute into the axon bundles of the neurons. This Env protein-directed restriction of Gag protein distribution was shown to be dependent on the relative concentrations of the two proteins in the neuron. If the Gag protein was expressed in excess over the Env protein, the Gag protein was able to enter into the axon. These results support a mechanism for Env protein-directed Gag localization that is based on an interaction between the two viral proteins. Furthermore, we showed that the Gag protein localization was not restricted by heterologous Env proteins. The latter result indicates that the observed effect is based on a specific interaction between homologous Gag and Env proteins in DRG neurons. Thus, our results confirm earlier data on the existence of a Gag-Env interaction in HIV and, most importantly, suggest that such an interaction also takes place in MLV. This supports a model for Env incorporation into MLV particles that is based on an Env-Gag interaction. Previously, the sole support for an Env-Gag interaction in MLV has been the observation that MLV variants with certain tail mutations in the Env protein were defective in Env incorporation into virions (15, 16). However, in those studies it remained unclear whether the mutant Env proteins were actually defective in Gag binding or whether they simply were unable to reach the Gag budding sites at the PM.
It will still be important to corroborate our findings in DRG neurons with direct in vitro binding studies. Further, the Env-Gag interactions of other retroviruses should also be studied. The most suitable approach is probably to use both in vitro binding and polarized cell assays. A full understanding of the Gag-Env interaction in the lentiviruses, MLV, and other retroviruses will, however, require a detailed structural analysis of the Env tail-Gag complexes.
ACKNOWLEDGMENTS
This study was supported by grants from the Swedish Natural Science Research Council (no. B-AA/BU 09353-311), the Cancer Foundation (no. 96 4165), the Human Capital and Mobility Network (no. CHRX-CT 94-0496), the Stanley Foundation Research Awards Program, and the Swedish Medical Research Council (no. 4480).
We thank Ingrid Jusinsky (Clinical Research Center, Huddinge, Sweden) for excellent preparation of the electron microscopy material, Ingrid Sigurdson for typing, and Mathilda Sjöberg for critical reading of the manuscript.
REFERENCES
- 1.Andersson, H., M. Ekström, and H. Garoff. Unpublished observations.
- 1a.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruss V, Vieluf K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J Virol. 1995;69:6652–6657. doi: 10.1128/jvi.69.11.6652-6657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caceres A, Banker G A, Steward O, Binder L, Payne M. MAP2 is localized to dendrites of hippocampal neurons which develop in culture. Dev Brain Res. 1984;13:314–318. doi: 10.1016/0165-3806(84)90167-6. [DOI] [PubMed] [Google Scholar]
- 4.Cameron P L, Südhof T C, Jahn R, De Camilli P. Colocalization of synaptophysin with transferrin receptors: implications for synaptic vesicle biogenesis. J Cell Biol. 1991;115:151–164. doi: 10.1083/jcb.115.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 7.Delchambre M, Gheysen D, Thines D, Thiriart C, Jacobs E, Verdin E, Horth M, Burny A, Bex F. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 1989;8:2653–2660. doi: 10.1002/j.1460-2075.1989.tb08405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dotti C G, Simons K. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- 9.Dyson M R, Murray K. Selection of peptide inhibitors of interactions involved in complex protein assemblies: association of the core and surface antigens of hepatitis B virus. Proc Natl Acad Sci USA. 1995;92:2194–2198. doi: 10.1073/pnas.92.6.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed E O, Martin M A. Virus incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 12.Gelderblom H R, Bauer P G, Özel M, Höglund P, Niedrig M, Renz H, Morath B, Lundquist P, Nilsson Å, Mattow J, Grund C, Pauli G. Morphogenesis and morphology of human immunodeficiency virus. In: Aloia R C, et al., editors. Membrane interactions of HIV. New York, N.Y: Wiley-Liss, Inc.; 1992. pp. 33–54. [Google Scholar]
- 13.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 14.González S A, Burny A, Affranchino J L. Identification of domains in the simian immunodeficiency virus matrix protein essential for assembly and envelope glycoprotein incorporation. J Virol. 1996;70:6384–6389. doi: 10.1128/jvi.70.9.6384-6389.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granowitz C, Colicelli J, Goff S P. Analysis of mutations in the envelope gene of Moloney murine leukemia virus: separation of infectivity from superinfection resistance. Virology. 1991;183:545–554. doi: 10.1016/0042-6822(91)90983-i. [DOI] [PubMed] [Google Scholar]
- 16.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Hewson, R., and H. Garoff. Unpublished data.
- 17.Inczedy-Marcsek M, Hsu L, Lindner E. Vitro Cell. Dev. Biol. 29A:661–670. 1993. An analysis of dorsal root ganglia differentiation using three tissue culture systems. [DOI] [PubMed] [Google Scholar]
- 18.Kundu A, Nayak D P. Analysis of the signals for polarized transport of influenza virus (A/WSN/33) neuraminidase and human transferrin receptor, type II transmembrane proteins. J Virol. 1994;68:1812–1818. doi: 10.1128/jvi.68.3.1812-1818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 20.Matus A, Bernhardt R, Bodmer R, Alaimo D. Microtubule-associated protein 2 and tubulin are differently distributed in the dendrites of developing neurons. Neuroscience. 1986;17:371–389. doi: 10.1016/0306-4522(86)90253-8. [DOI] [PubMed] [Google Scholar]
- 21.Olkkonen V M, Dupree P, Simons K, Liljeström P, Garoff H. Expression of exogenous proteins in mammalian cells with the Semliki Forest virus vector. Methods Cell Biol. 1994;43:43–53. doi: 10.1016/s0091-679x(08)60597-x. [DOI] [PubMed] [Google Scholar]
- 22.Olkkonen V M, Liljeström P, Garoff H, Simons K, Dotti C G. Expression of heterologous proteins in cultured rat hippocampal neurons using the Semliki Forest virus vector. J Neurosci Res. 1993;35:445–451. doi: 10.1002/jnr.490350412. [DOI] [PubMed] [Google Scholar]
- 23.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul N, Marsh M, McKeating J A, Schulz T F, Liljeström P, Garoff H, Weiss R A. Expression of HIV-1 envelope glycoprotein by Semliki Forest virus. AIDS Res Hum Retroviruses. 1993;9:963–970. doi: 10.1089/aid.1993.9.963. [DOI] [PubMed] [Google Scholar]
- 25.Pinter A, deHarven E. Protein composition of a defective murine sarcoma virus particle possessing the enveloped type-A morphology. Virology. 1979;99:103–110. doi: 10.1016/0042-6822(79)90041-2. [DOI] [PubMed] [Google Scholar]
- 26.Rhee S S, Hui H, Hunter E. Preassembled capsids of type D retroviruses contain a signal sufficient for targeting specifically to the plasma membrane. J Virol. 1990;64:3844–3852. doi: 10.1128/jvi.64.8.3844-3852.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothman S. Synaptic release of excitatory amino acid neurotransmitter mediates anoxic neuronal death. J Neurosci. 1984;4:1884–1891. doi: 10.1523/JNEUROSCI.04-07-01884.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheele C M, Hanafusa H. Proteins of helper-dependent RSV. Virology. 1971;45:401–410. doi: 10.1016/0042-6822(71)90341-2. [DOI] [PubMed] [Google Scholar]
- 29.Sjöberg M, Suomalainen M, Garoff H. A significantly improved Semliki Forest virus expression system based on translation enhancer segments from the viral capsid gene. Bio/Technology. 1994;12:1127–1131. doi: 10.1038/nbt1194-1127. [DOI] [PubMed] [Google Scholar]
- 30.Smyth J, Suomalainen M, Garoff H. Efficient multiplication of a Semliki Forest virus chimera containing Sindbis virus spikes. J Virol. 1997;71:818–823. doi: 10.1128/jvi.71.1.818-823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sotelo S, Gibbs C J, Gajdusek D C, Toh B H, Wurth M. Method for preparing cultures of neurons: cytochemical and immunochemical studies. Proc Natl Acad Sci USA. 1980;77:653–657. doi: 10.1073/pnas.77.1.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suomalainen M, Garoff H. Incorporation of homologous and heterologous proteins into the envelope of Moloney murine leukemia virus. J Virol. 1994;68:4879–4889. doi: 10.1128/jvi.68.8.4879-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahlberg J M, Boere W A, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to mildly acidic pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C-H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]