Abstract
Retroviruses are produced as immature particles containing structural polyproteins, which are subsequently cleaved by the viral proteinase (PR). Extracellular maturation leads to condensation of the spherical core to a capsid shell formed by the capsid (CA) protein, which encases the genomic RNA complexed with nucleocapsid (NC) proteins. CA and NC are separated by a short spacer peptide (spacer peptide 1 [SP1]) on the human immunodeficiency virus type 1 (HIV-1) Gag polyprotein and released by sequential PR-mediated cleavages. To assess the role of individual cleavages in maturation, we constructed point mutations abolishing cleavage at these sites, either alone or in combination. When all three sites between CA and NC were mutated, immature particles containing stable CA-NC were observed, with no apparent effect on other cleavages. Delayed maturation with irregular morphology of the ribonucleoprotein core was observed when cleavage of SP1 from NC was prevented. Blocking the release of SP1 from CA, on the other hand, yielded normal condensation of the ribonucleoprotein core but prevented capsid condensation. A thin, electron-dense layer near the viral membrane was observed in this case, and mutant capsids were significantly less stable against detergent treatment than wild-type HIV-1. We suggest that HIV maturation is a sequential process controlled by the rate of cleavage at individual sites. Initial rapid cleavage at the C terminus of SP1 releases the RNA-binding NC protein and leads to condensation of the ribonucleoprotein core. Subsequently, CA is separated from the membrane by cleavage between the matrix protein and CA, and late release of SP1 from CA is required for capsid condensation.
The retroviral Gag polyprotein and its proteolytic cleavage products are the major structural components of the virion, and Gag is sufficient to direct the assembly and release of virus-like particles in the absence of any other viral protein (17; reviewed in references 7 and 64). For human immunodeficiency virus (HIV) and other lentiviruses and type C retroviruses, the accumulation of Gag proteins at the plasma membrane leads to the assembly of immature virions that bud from the cell surface. Immature particles contain a spherical protein shell closely apposed to the lipid membrane (12, 15, 42). Within the nascent virion, the Pr55gag polyprotein of HIV is cleaved by the viral proteinase (PR) into the matrix (MA), capsid (CA), nucleocapsid (NC), and C-terminal p6 domain (for nomenclature, see reference 38). Gag processing induces a dramatic reorganization of the internal virion structure, termed maturation (61). In mature HIV particles, MA lines the inner surface of the membrane, while CA forms the conical capsid shell which encases the genomic RNA complexed with NC (16). Cleavage and maturation are not required for particle formation but are essential for infectivity (31). Mature cores are readily dissociated by detergent-mediated removal of the viral membrane, while immature cores are significantly more stable (44, 51, 53, 58), suggesting that maturation is important for uncoating. However, the relevance of individual cleavages and the precise functions of the cleavage products in the entry process are currently not known.
Specific functions have been assigned to individual Gag domains. The myristoylated MA domain directs intracellular targeting of polyproteins (36), NC is tightly bound to the genomic RNA and is important for RNA packaging and reverse transcription (2), and the C-terminal p6 domain is involved in particle release (19) and mediates the incorporation of Vpr (45). CA plays essential roles early and late in infection. The C-terminal segment of CA appears to contain an important assembly domain (8, 62, 65) and is required for Gag oligomerization (11, 65) and CA dimerization (14). Mutations in the N-terminal part of CA did not block the assembly or release of virions but nevertheless abolished viral infectivity (8, 51, 63).
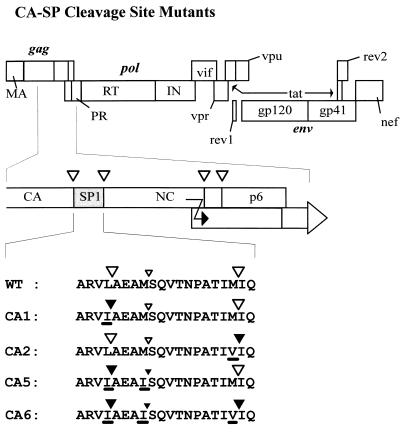
Sequence analysis of proteins from mature HIV type 1 (HIV-1) particles as well as in vitro processing experiments showed that CA and NC as well as NC and p6 are separated on the polyprotein by short spacer peptides of 14 and 10 amino acids, respectively (spacer peptide 1 [SP1] and SP2, respectively) (Fig. 1) (25, 26, 40). These spacer peptides are released by PR-mediated cleavages at their N and C termini during maturation. Similar spacer peptides separating CA and NC have been observed for simian (24), bovine (59), and feline (9) immunodeficiency viruses, for equine infectious anemia virus (27), and for avian retroviruses (6, 47).
FIG. 1.
Schematic representation of various mutants. At the top, the coding region of the HIV-1 genome is shown, with the different open reading frames depicted as boxes. The 3′-terminal part of the CA coding region and the NC coding region of the gag reading frame, including SP1, are expanded in the middle. Cleavage sites for HIV-1 PR are indicated as open triangles. The signal for translational frameshifting is also marked (black arrow). At the bottom, the amino acid sequence of wild-type (WT) SP1 and flanking cleavage sites as well as the altered sequences of the various mutant constructs are shown. The name of each construct is indicated on the left. Mutated amino acids are underlined. Large open triangles indicate cleavage sites that are processed by HIV-1 PR; large closed triangles represent sequences that are not cleaved by HIV-1 PR. The smaller triangles correspond to a cryptic PR cleavage site within SP1 which is cleaved much more slowly.
Deletion of spacer peptides in Rous sarcoma virus (RSV) (6, 47) or HIV-1 (34, 48) led to the production of noninfectious virus particles. For RSV, extracellular virions appeared morphologically normal but exhibited significantly reduced stability of the mature core (6, 47) and size heterogeneity (36a). Mutant HIV-1 particles, on the other hand, were morphologically aberrant and heterogeneous in size (34), indicating an important function of SP1 in ordered particle assembly. Size heterogeneity was also observed with a linker insertion within SP1 (51), and a dramatic alteration of the internal core structure was seen with a nonconservative amino acid substitution in this region (20). Furthermore, C-terminal deletions of Gag affecting SP1 were shown to abolish the formation of regular virus-like particles following expression of the mutant Gag proteins by recombinant baculoviruses (17, 29). Mutation of the N- and C-terminal cleavage sites of HIV-1 SP1 led to severely reduced or completely abolished viral infectivity (34), but a detailed analysis of the morphological phenotype of these mutant virions was not reported.
Individual cleavage sites on the HIV Gag and Gag-Pol polyproteins are processed at different rates which can be reproduced in in vitro reactions (10, 32, 35, 48, 60). Sequential processing of the Gag polyprotein results in discrete intermediates appearing transiently before the final products. Such intermediates may be important for virion morphogenesis or maturation but are incompatible with the structure of the mature virion. The initial cleavage occurs at the C terminus of SP1 and separates an N-terminal MA-CA-SP1 intermediate and a C-terminal NC-SP2-p6 intermediate (Fig. 1). Subsequent cleavages separating MA from CA-SP1 and NC-SP2 from p6 occur at an approximately 10-fold-lower rate (48). Cleavage of SP1 from the C terminus of CA is a late event, and C-terminally extended CA species can be observed in virions or virus-like particles with reduced PR activity (39, 53). Cleavage at the CA-SP1 site occurred at a 400-fold-lower rate than that at the SP1-NC site in an in vitro processing reaction (48).
To investigate the role of individual cleavages in the CA-NC region, we introduced point mutations abolishing cleavage at these sites, either alone or in combination. We report here that the separation of CA from NC is required for condensation of the immature core. Cleavage of SP1 from the C terminus of CA, which appears to be the last event in the processing cascade, is not needed for condensation of the ribonucleoprotein (RNP) core but is essential for condensation of the capsid shell. These results indicate that HIV maturation is a sequential process which is kinetically controlled by the rate of proteolytic processing at individual cleavage sites.
MATERIALS AND METHODS
Cells, transfections, and infections.
COS-7 cells were maintained in Dulbecco modified minimal Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine. For transfections, cells were seeded in 10-cm culture dishes and transfected with 15 μg of plasmid DNA by a modified calcium phosphate method (5). To normalize for transfection efficiency, 5 μg of an expression vector containing the lacZ gene under the control of the cytomegalovirus immediate-early promoter-enhancer was cotransfected, and cell extracts were normalized for β-galactosidase activity. Cells and culture media were harvested 72 h after transfection. HIV-1-permissive MT-4 (23) and C8166 (54) cells were maintained in RPMI 1640 with the supplements listed above and infected with culture medium from transfected cells which had been filtered through a 0.45-μm-pore-size filter. Productive infection was scored by indirect immunofluorescence with serum from an HIV-positive person and by an enzyme-linked immunosorbent assay (ELISA) for CA antigen.
Expression plasmids.
All mutations were made with a subclone of the full-length infectious provirus pNL4-3 (1) containing the PstI-ApaI fragment (nucleotides [nt] 1415 to 2006 of pNL4-3) in a pBluescript vector (Stratagene) (pBS-HIVP/A). Site-directed mutagenesis was performed with a single-stranded DNA template by the method of Kunkel (37). The CA1 mutation, encoding a Leu-to-Ile substitution in the P1 position (nomenclature according to reference 56) of the first cleavage site at the C-terminal end of CA (Fig. 1), and the CA2 mutation, encoding a Met-to-Val substitution in the P1 position of the second cleavage site (Fig. 1), have been described elsewhere (34). The CA5 mutation encodes the same Leu-to-Ile substitution as CA1 and in addition a Met-to-Ile substitution (nt 1888 to 1891) four codons downstream in the P1 position of a cryptic cleavage site within the spacer peptide (Fig. 1). Like CA1 (34), CA5 contains a substitution of nt 1869 and 1870 from AA to TC, which does not change the coding sequence but which creates a new XhoI site at nt 1869 to 1873 of pNL4-3. Mutation CA6, combining all three cleavage site substitutions, was created by PCR with plasmid pBS-HIVP/A containing the CA2 mutation as a template, the CA5 primer (5′GGCCATAAAGCTCGAGTTATCGCTGAAGCAATCAGCCAGGTAACCAATCCAGCTACC3′) as the forward primer, and an oligonucleotide complementary to the p6 region at the 3′ end of gag as the reverse primer. The PCR product was subsequently cleaved with XhoI (see above) and ApaI, and the resulting fragment (corresponding to nt 1869 to 2006 of pNL4-3) was inserted into plasmid pBS-HIVP/A containing the CA1 mutation and cleaved with XhoI and ApaI as well. The entire HIV-specific region was sequenced in all mutant plasmids, and the respective SphI-ApaI fragments (nt 1443 to 2006 of pNL4-3) were cloned into pNL4-3.
Purification and detergent treatment of virus particles.
Extracellular particles were collected from cleared media (10 min at 400 × g followed by filtration through a 0.45-μm-pore-size filter) by centrifugation through a 2-ml cushion of 20% (wt/wt) sucrose in phosphate-buffered saline (PBS) at 130,000 × g for 90 min at 4°C. Pellets were resuspended in PBS and analyzed by gel electrophoresis and Western blotting.
For detergent treatment of virus particles, cleared culture medium from transfected COS-7 cells was divided into two aliquots, and one was treated with 0.5% Triton X-100 for 10 min at 37°C. Detergent-treated and untreated samples were subsequently layered over a cushion of 20% (wt/wt) sucrose in PBS and centrifuged at 130,000 × g for 90 min at 4°C. Alternatively, cleared culture medium from transfected cells was centrifuged through preformed step gradients containing a cushion of 20% (wt/wt) sucrose in PBS and a 1-ml layer of 10% sucrose in PBS with or without 0.5% Triton X-100. Centrifugation was performed at 130,000 × g for 120 min at 4°C. The pellets from both sets of experiments were resuspended in PBS and analyzed by polyacrylamide gel electrophoresis and Western blotting.
Analysis of expression products.
HIV-1 antigens were analyzed by a quantitative ELISA for the viral CA protein (33). For protein analysis, cell or particle extracts were separated on sodium dodecyl sulfate (SDS)–17.5% polyacrylamide gels (200:1 ratio of acrylamide to N,N-methylenebisacrylamide) and stained with Coomassie blue or silver stain (28). For immunoblot analysis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) by electroblotting. Membranes were blocked with 10% lowfat dry milk in 0.1 M Tris-HCl (pH 7.5)–0.15 M NaCl (TBS) for 1 h and subsequently reacted with rabbit polyclonal antiserum against HIV-1 CA or reverse transcriptase (39). The antisera were diluted 1:2,000 in TBS containing 5% lowfat dry milk and 0.025% Tween 20. Incubation was done overnight at room temperature with shaking. After three washes with TBS containing 0.05% Tween 20 and blocking for 30 min with TBS containing 5% lowfat dry milk, peroxidase-conjugated antibody against rabbit IgG (Jackson Immunochemicals Inc.; dilution, 1:10,000) was used as a secondary antibody; incubation was done for 90 min at room temperature. Immune complexes were visualized by enhanced chemiluminescence (ECL system; Amersham) according to the manufacturer’s instructions.
Electron microscopy.
At 60 h after transfection, cells were fixed in situ for 20 min with cold 2.5% glutaraldehyde in 100 mM PIPES [piperazine-1,4-bis(2-ethanesulfonic acid); pH 6.9]. Fixed cells were scraped from the plate with a rubber policeman, collected by low-speed centrifugation at 4°C, washed three times in cold PIPES, and postfixed with 1% osmium tetroxide in PIPES for 30 min at 4°C. After extensive washing in PIPES, cells were embedded in agarose (low gelling temperature; Sigma) and further treated with 1% tannic acid for 10 min. Finally, agar blocks were washed in water, stained with 1% uranyl acetate for 30 min in the cold, dehydrated in ethanol, and embedded in ERL resin (57). Silver-grey sections were stained with lead citrate and uranyl acetate. Sections were examined in a Philips CM120 electron microscope at 60 kV.
RESULTS
Construction of CA cleavage site mutations and analysis of protein expression and processing.
In a previous study, we had shown that the presence of SP1 is essential for the regular assembly of immature HIV-1 particles and that cleavages on both sides of SP1 are important for viral infectivity (34). To analyze the role of sequential cleavages of Gag and to determine whether the release of SP1 from the C terminus of CA is important for capsid maturation, we constructed additional point mutations in this region. In each case, the amino acid in the P1 position of the respective cleavage site was altered into a residue with a β-branched side chain, since HIV PR will not cleave such sequences (48–50, 52). Mutants CA1 and CA2 (Fig. 1) were described previously (34) and caused the loss of cleavage at either the N terminus (mutant CA1) or the C terminus (mutant CA2) of SP1. Blocking the N-terminal SP1 cleavage site had revealed an additional cryptic site which was processed by HIV-1 PR in mutant particles and yielded a CA species extended by 4 amino acids (34). We therefore constructed a Met-Ile substitution in the P1 position of this cryptic site, in addition to blocking the N-terminal SP1 cleavage site (mutant CA5; Fig. 1). Finally, a combination of all three mutations was constructed (mutant CA6; Fig. 1) to completely block the separation of CA and NC. All mutations were cloned into the infectious proviral clone pNL4-3, and polyprotein synthesis and processing as well as particle release and infectivity were analyzed following transient transfection of the respective plasmids into COS-7 cells.
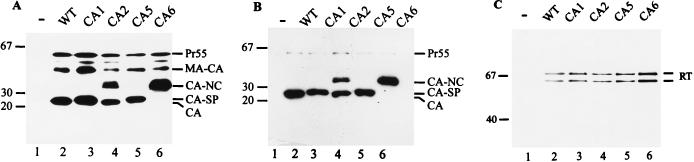
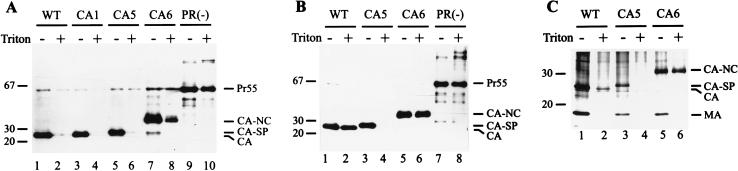
The expression of HIV-1-specific proteins was analyzed 72 h after transfection. Western blot analysis of transfected cells with antiserum against CA revealed comparable amounts of immunoreactive material in wild-type- and mutant-transfected cells. Major immunoreactive products in pNL4-3 (wild-type)-transfected cells corresponded to the Gag polyprotein Pr55gag, the intermediate cleavage product MA-CA, and the cleaved CA protein (Fig. 2A, lane 2). A similar pattern was seen with the CA1 mutant (Fig. 2A, lane 3), while the CA2 mutant yielded an additional product, CA-NC, which had been reported previously (Fig. 2A, lane 4) (34). Thus, blocking the cleavage site between SP1 and NC significantly reduced cleavage that would create the N terminus of SP1. The CA-NC product was the predominant immunoreactive product in the case of the CA6 mutation, and no completely processed CA protein was observed with this mutation (Fig. 2A, lane 6). The CA5 mutation yielded an extended CA protein, corresponding to CA-SP1 (Fig. 2A, lane 5), which migrated slightly slower on SDS gels than CA.
FIG. 2.
Western blot analysis of gag and pol gene products after transient transfection. COS-7 cells transfected with pNL4-3 or derivatives and media were harvested 72 h after transfection. Lysates of transfected cells (A) or viral particles collected by centrifugation through a sucrose cushion (B and C) were resolved by SDS-polyacrylamide gel electrophoresis, and Western blots were reacted with polyclonal antiserum against CA (A and B) or RT (C) and tested with chemiluminescence. The wild-type (WT) and mutant proviral constructs are indicated above each lane. Molecular mass standards (in kilodaltons) are indicated on the left. HIV-specific precursor proteins, intermediate processing products, and cleaved proteins are indicated on the right.
The reduced mobility of the extended CA species was more obvious on immunoblot analysis of particle fractions showing CA (wild type; Fig. 2B, lane 2), CA extended by 4 amino acids (mutant CA1; Fig. 2B, lane 3), and CA-SP1 extended by 14 amino acids (mutant CA5; Fig. 2B, lane 5). These results indicate that the introduced mutations abolished cleavage at the respective sites. Accordingly, CA-NC was the predominant immunoreactive species in particles derived from pNL43-CA6-transfected cells (Fig. 2B, lane 6). CA-NC was also detected in CA2-derived particles (Fig. 2B, lane 4). The predominant immunoreactive species in this case was completely processed CA (Fig. 2B, lane 4), indicating that N-terminal cleavage of SP1 occurred at a reduced rate but was not blocked by the downstream mutation. Only minor differences in the relative amounts of extracellular particle-associated antigen were observed for wild-type HIV-1 and the four mutants, indicating that the cleavage site mutations did not affect Gag polyprotein assembly or particle release. Consistently, the signal for the CA-NC product in the case of mutant CA6 appeared slightly more intense than that for the other CA products, a result which is most likely due to an increased stability of the immature CA6-derived particles (see below). To analyze processing at other sites of the Gag-Pol polyprotein, we subjected particle fractions from wild-type- and mutant-transfected cells to Western blot analysis using an antiserum against reverse transcriptase for detection. All samples contained predominantly the completely processed heterodimer of reverse transcriptase (p66-p51), and no significant alterations in Pol domain processing were observed (Fig. 2C). Similar results were obtained when an antiserum against integrase was used (data not shown).
To assay for the infectivity of mutant viruses, HIV-1-permissive MT-4 and C8166 cells were infected with cleared culture medium from wild-type- and mutant-transfected COS-7 cells. No infection was detected by indirect immunofluorescence or an ELISA with undiluted or diluted medium from three independent transfections with the proviral constructs pNL43-CA2, pNL43-CA5, and pNL43-CA6 during an observation period of up to 28 days. Coculturing of these cells with fresh MT-4 cells also did not lead to infection, and no reversion was observed. In contrast, medium from wild-type-transfected cells contained infectious HIV-1 particles with a titer of approximately 105 to 106 per ml, and infection was always detectable after 2 to 5 days (data not shown). As reported previously, considerably delayed infectivity and reduced titers were observed with the CA1 mutant (34).
Effect of cleavage site mutations on particle morphology and maturation.
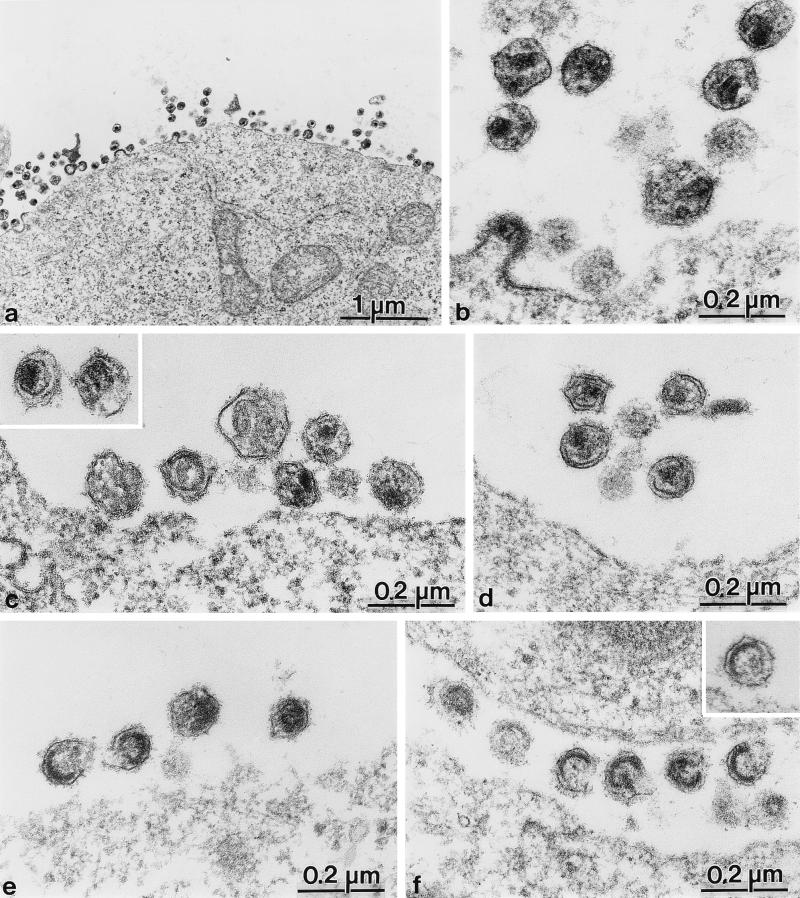
To analyze the morphogenesis and morphology of mutant viruses, we performed thin-section electron microscopic analysis of COS-7 cells transfected with pNL4-3 and derivatives. Cells transfected with the wild-type proviral construct showed many particles of immature and mature morphology in the vicinity of the cell surface as well as some budding structures (Fig. 3a). Extracellular virions of regular size and of a morphology typical of mature HIV-1 were readily observed (Fig. 3b and 4, lowest panel on left). Mature wild-type particles contained an electron-dense internal structure corresponding to an RNP complex consisting of the genomic RNA and the NC protein surrounded by a thin electron-dense capsid shell. This capsid structure may appear conically shaped or circular, depending on the plane of the section (Fig. 3b).
FIG. 3.
Electron micrographs showing thin sections of COS-7 cells transfected with pNL4-3 and mutant derivatives at 72 h after transfection. (a and b) Overview (a) and higher-magnification view (b) of wild-type-transfected cells and extracellular particles. (c and inset) Mutant pNL43-CA1. (d) Mutant pNL43-CA5. (e) Mutant pNL43-CA2. (f and inset) Mutant pNL43-CA6.
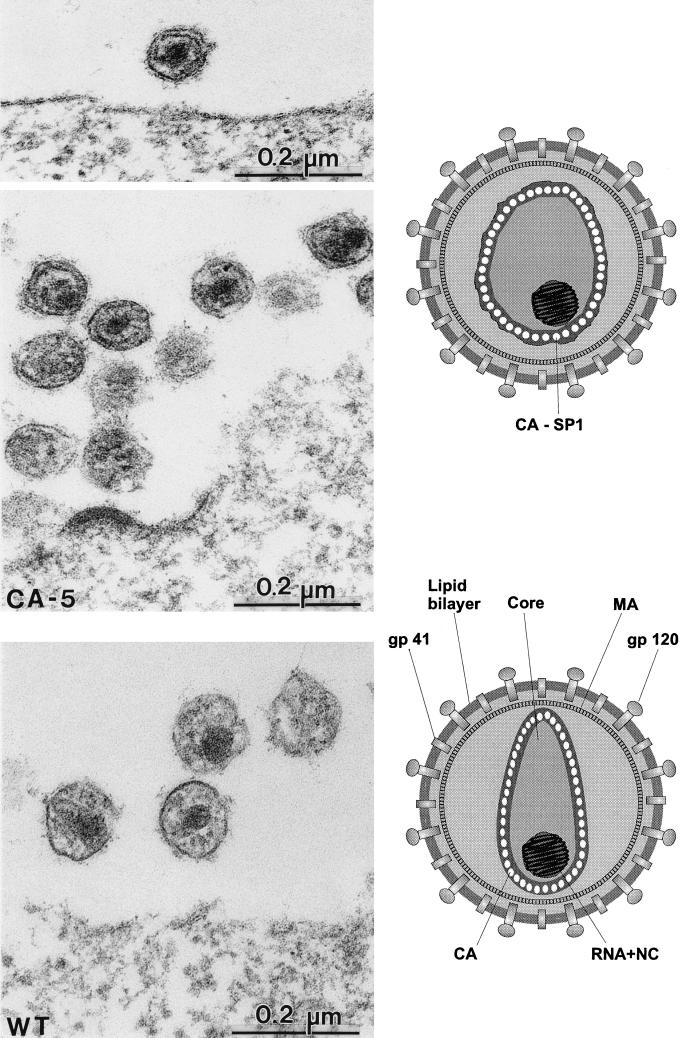
FIG. 4.
Comparative analysis of particle morphology of HIV-1 wild-type (WT) and mutant CA5 particles. Electron micrographs of WT particles show an electron-dense centrally located nucleocapsid surrounded by a conical capsid shell (best seen in the particle at the left). For the CA5 mutant, particles are of a similar size and also contain a centrally located electron-dense nucleocapsid but lack the conical capsid structure. Instead, a thin electron-dense layer is observed in the vicinity of the viral membrane but clearly separated from it. Schematic representations of the morphologies of the different particles are shown on the right.
Different morphologies were observed for mutant viruses (Fig. 3). Extracellular particles derived from CA6-transfected cells containing uncleaved CA-NC were of regular size but showed a membrane-apposed electron-dense structure with an electron-lucent interior (Fig. 3f). No condensed RNP core and no mature capsid structure were observed. Electron-dense rings were not completely closed, and many particles exhibited a crescent-shaped structure. Morphologically similar particles containing immature core structures were also observed for the CA2 mutation (Fig. 3e, particles at the left). Other CA2-derived particles exhibited more diffuse electron-dense material inside the viral membrane (Fig. 3e, particles at the right) but neither a defined RNP core nor a typical condensed capsid structure.
Mutations affecting the release of SP1 from the C terminus of CA resulted in a different phenotype. Formation of the electron-dense RNP core was observed in these cases, but condensation of the capsid shell was blocked (Fig. 3c and d and 4). This phenotype was more obvious for the CA5 mutant (Fig. 3d and 4, upper panels), which showed normal budding structures (Fig. 4) and normal amounts of regularly sized extracellular particles. All particles contained a tightly condensed internal RNP core with a size and morphology similar to those of wild-type particles (Fig. 4, compare upper panels with lower panel). However, CA5-derived particles never contained a condensed capsid shell. Instead, a thin electron-dense layer in the vicinity of the viral membrane but clearly separated from it was observed (Fig. 3d and 4, upper panels). Most likely, this electron-dense layer corresponded to a homomultimer of the unprocessed CA-SP1 intermediate, which was blocked in the process of capsid maturation (Fig. 4, upper schematic drawing). A less severe phenotype was observed for the CA1 mutant containing a CA protein extended by only 4 amino acids. Again, regularly sized particles with a condensed internal RNP structure but no regular capsid structure were observed (Fig. 3c and inset). Capsid condensation appeared to be less affected than that of the CA5 mutant, and the thin electron-dense layer was separated further from the membrane (Fig. 3c, left particle in inset). As in the case of the CA5 mutant, no conically shaped or round capsid structures were detected.
Effect of cleavage site mutations on core stability.
Immature retroviral cores have been shown to be stable against detergent treatment, while mature capsids are readily solubilized in Triton X-100 (44, 51, 53, 58). In order to analyze alterations in mutant capsid stabilities, we collected wild-type and mutant particles and subjected them to brief detergent treatment prior to centrifugation. Figure 5A shows that detergent-stripped capsids from PR-defective HIV-1 particles (33) containing only uncleaved Gag polyprotein were stable (lanes 9 and 10), while wild-type capsids were almost completely solubilized (lanes 1 and 2). Detergent-stripped capsids from CA6-derived particles were much less stable than those from PR-defective virions (Fig. 5A, compare lanes 8 and 10), although the morphological appearances of these particles were similar. Detergent treatment of CA1- and CA5-derived particles completely solubilized the capsid structures (Fig. 5A, lanes 3 to 6), as for wild-type particles.
FIG. 5.
Analysis of detergent stability of wild-type and mutant HIV particles. (A) Virus particles collected from the medium of transfected COS-7 cells were treated with 0.5% Triton X-100 for 10 min or left untreated, and both samples were subsequently centrifuged through a sucrose cushion. (B and C) In a second experiment, virus particles were layered on a sucrose step gradient containing a layer of 10% sucrose with or without 0.5% Triton X-100 on top of a 20% sucrose layer as described in Materials and Methods. Pellet fractions were resolved by SDS-polyacrylamide gel electrophoresis followed by silver staining (C) or Western blot analysis with polyclonal antiserum against CA and chemiluminescence detection (A and B). The wild-type (WT) proviral construct and the respective mutants are indicated above each lane. The presence or absence of 0.5% Triton X-100 is indicated by + or −, respectively. Molecular mass standards (in kilodaltons) are indicated on the left. HIV-specific precursor proteins and cleavage products are identified on the right.
To detect more subtle alterations in capsid stability, we centrifuged wild-type and mutant particles through a layer of sucrose either containing or lacking detergent. This treatment results in a brief contact time for particles and detergent and should therefore be less destabilizing. Approximately 20 to 30% of CA protein (as detected by an ELISA) remained as a stable complex after centrifugation of wild-type HIV-1 through a detergent layer (Fig. 5B and C, lanes 1 and 2). The membrane-associated MA protein, which is not part of the inner capsid structure, was completely solubilized under these conditions (Fig. 5C, lane 2). Centrifugation through a detergent layer abolished viral infectivity, also indicating that viral membranes were effectively stripped. Centrifugation of either PR-defective particles (Fig. 5B, lanes 7 and 8) or CA6-derived particles (Fig. 5B and C, lanes 5 and 6) through a detergent layer did not solubilize the stripped capsids. The levels of CA antigen were not reduced in the pellet fraction relative to lipid-containing particles (Fig. 5B), while MA was completely solubilized (Fig. 5C, lane 6). For the CA5 mutant, however, no CA-reactive material was observed in the pellet fraction after detergent treatment, indicating that mutant capsids were completely solubilized and were significantly less stable than wild-type capsids.
DISCUSSION
In this study, we have shown that sequential processing of cleavage sites separating the CA and NC domains on the HIV-1 Gag polyprotein is required for ordered maturation of the viral core and for the generation of infectious virus. When the two regular sites and a cryptic site within the intervening SP1 region were mutated, no condensation of the viral core was observed (mutant CA6). Extracellular particles exhibited crescent-shaped electron-dense structures near the viral membrane, similar to those in PR-defective HIV-1 mutants (46) and PR inhibitor-treated virions (30, 55). The same immature core morphology was also found for a small number of wild-type particles, indicating that the crescent-shaped core represents a normal intermediate in HIV-1 morphogenesis (43). Analysis of proteins from mutant CA6-derived particles revealed a stable CA-NC product, whereas all other cleavages of the Gag and Gag-Pol polyproteins proceeded normally. This CA-NC product was also detected with mutant CA2, albeit at a much lower level, and extracellular particles in this case appeared to have a delay or reduction in core maturation. These results suggest that the separation of NC from CA is necessary for condensation of the electron-dense RNP core. Some condensation occurs when N-terminally extended NC is released, but the core structure appears less tightly organized (mutant CA2) than that of the wild-type virus. If CA-NC is not cleaved at all, the stable intermediate cleavage products are likely to remain attached to the viral nucleic acid through NC-mediated interactions. The long genomic RNA may link these proteins together and maintain a rigid shell that cannot undergo ordered maturation. Such a restriction would also be consistent with the suggested organizing role of RNA in the assembly of the spherical core shell (12).
Recently, the in vitro assembly of helically organized hollow cylinders which are likely to be related to mature viral capsids was reported, both for RSV and for HIV-1 (3, 21). In contrast to the situation with virion maturation, purified CA-NC proteins readily formed tubular structures in vitro in an RNA-dependent manner (3, 21). Morphologically indistinguishable structures were observed when HIV-1 CA with or without the adjacent NC domain was used, and no difference was observed for CA proteins containing or lacking SP1 (21). Spherical particles were found when N-terminally extended forms of RSV CA containing the p10 domain of RSV (4) or of HIV-1 CA containing part of the MA domain of HIV (22) were used for in vitro assembly. These experiments showed that CA-NC can form helically organized nucleocapsid structures when synthesized and assembled without flanking domains. The production of CA-NC by proteolytic cleavage from the Gag polyprotein, on the other hand, is not sufficient for condensation of the spherical immature core to the conical mature core, and the separation of CA from NC is required in this case. It is likely that the release of NC and genomic RNA from the membrane-attached MA-CA intermediate is an essential first step in virion maturation which induces condensation of the RNP core. This cleavage also relieves the CA domain from RNA-induced restrictions, permitting subsequent condensation of the capsid shell. The separation of CA from NC is not required if the upstream MA domain is absent and if tubular particles are produced in the first place (in the in vitro assembly system). The requirement for the rapid release of the NC-RNA complex may also explain why the SP1-NC cleavage site is processed approximately 10-fold faster than other sites on the Gag polyprotein.
In a study of sequential Gag polyprotein processing in vitro, Pettit et al. (48) reported that mutation of the SP1-NC cleavage site accelerated upstream cleavage at the CA-SP1 site 20-fold, presumably by blocking the formation of a CA-SP1 intermediate. In our experiments, however, reduced processing at the CA-SP1 site was observed for a downstream cleavage site mutant (CA2) and led to an increase in CA-NC levels in both transfected cells and virus particles and to an increased number of immature viruses. A possible explanation for this discrepancy is that the CA-SP1 site is not easily accessible to PR in the assembled immature core shell, while this restriction does not apply when Gag is translated in vitro and remains in a largely monomeric form. Conceivably, SP1 may also slow cleavage at the C terminus of CA during virion maturation, but it appears unlikely that the main function of this peptide is to inhibit its own removal from CA.
Cleavage at the MA-CA site is needed for the release of CA from the membrane-bound MA protein and is therefore a prerequisite for condensation of the capsid shell. Accordingly, mutation of the MA-CA site gave rise to noninfectious particles containing an electron-dense RNP core but no conical capsid shell and a thickening of the viral membrane, most likely due to the additional presence of CA (20). Structure determination of a fragment of HIV-1 CA (18, 41) revealed an N-terminal β-hairpin stabilized by hydrogen bonding involving the amino group of proline 1 of CA, which is only exposed after proteolytic cleavage (13, 18). This hairpin is part of a larger CA-CA interaction domain, and it has been proposed that a conformational change in this domain is necessary for condensation of the conical capsid shell (18). In agreement with this hypothesis, mutations in the N-terminal segment of CA yielded morphologically aberrant particles which did not exhibit regular core structures (8, 51), while mutations in the C-terminal dimerization domain severely reduced or abolished particle assembly (8, 62, 65). Our results show, however, that the release of NC and cleavage at the MA-CA site, which allow the separation of CA-SP1 from the membrane and the formation of the N-terminal hairpin structure, are not sufficient for capsid maturation. Electron microscopy of mutant CA5-derived particles revealed a regular electron-dense RNP core and a thin layer in the vicinity of the viral membrane, most likely corresponding to uncleaved CA-SP1. In contrast to the situation for a mutant in which the MA-CA cleavage site was blocked (20), this layer was clearly separated from the membrane, indicating that condensation of the capsid shell had been initiated but was arrested at an intermediate stage. It is interesting to note that the morphology of this mutant and especially of mutant CA1 bears some resemblance to the morphology of wild-type RSV, an avian type C retrovirus (6).
Cleavage between CA and SP1 appears to be the last event in the proteolytic cascade, and the CA-SP1 product trapped in the case of mutant CA5 probably corresponds to a regular maturation intermediate, although we cannot rule out a direct effect of the point mutation on capsid condensation. Conceivably, the transient presence of SP1 retains CA in an immature conformation until rearrangement of the N-terminal interaction domain has been completed and subsequent cleavage of SP1 relieves the final restriction. SP1 may exert its inhibitory function either by directly interlocking C-terminal domains of adjacent CA molecules or by keeping the C-terminal domains of CA in a conformation not compatible with the condensed capsid structure. In the latter case, sequential cleavages at the N and C termini of CA may trigger sequential conformational changes of the two-domain CA molecule (18), eventually leading to morphological maturation. The three-dimensional structure of the C-terminal fragment of CA was determined recently, revealing the primary dimer interface (14). However, the last 11 residues of CA were not ordered in this structure, and it is therefore not possible to draw any conclusions regarding the potential mechanism of SP1-mediated inhibition of maturation.
Detergent-stripped cores from CA5-derived particles were significantly less stable than those from wild-type particles, which were in turn much less stable than those from PR-defective particles. A similar observation was made for RSV: detergent treatment of extracellular particles led to a complete loss of C-terminally extended CA species, while a significant proportion of completely cleaved CA remained in particulate form (6, 47) and immature cores were largely stable against detergent (58). These results indicate that a stable assembly product (the immature spherical core) proceeds through a very unstable intermediate to the mature core of increased stability, and this stabilization appears to be required for early steps in viral infection. Conceivably, the capsid shell does not immediately dissociate upon fusion of viral and cellular membranes but may need to remain intact in the early phase of entry, and a capsid structure of intermediate stability would therefore be essential for early replication.
Based on this study and previous results summarized in this paper, we suggest that maturation of the HIV-1 core is an ordered process involving sequential steps which are kinetically controlled by the rate of processing at individual cleavage sites on the Gag polyprotein. Initial rapid cleavage at the C terminus of SP1 releases the RNA-binding NC protein and leads to condensation of the RNP core. Subsequently, CA is separated from the membrane by cleavage between MA and CA, leading to an altered CA-CA interface, while late (maturation) cleavage of SP1 from the C terminus of CA is required for final capsid condensation. It is possible, therefore, that changing the rate of processing at individual cleavage sites could disrupt the regular maturation process even if the specific mutations do not alter protein folding or interaction domains.
ACKNOWLEDGMENTS
We are grateful to J. Konvalinka for suggestions and to B. Müller, J. Konvalinka, and R. Welker for critically reading the manuscript. We thank C. Huckhagel, I. Ellhof, and A.-M. Heuser for expert technical assistance and for photography.
This work was supported in part by a grant from the German Ministry for Education and Research to H.-G.K.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Parent L J. Dynamic interactions of the Gag polyprotein. Curr Top Microbiol Immunol. 1996;214:65–94. doi: 10.1007/978-3-642-80145-7_3. [DOI] [PubMed] [Google Scholar]
- 8.Dorfman T, Bukovsky A, Öhagen A, Höglund S, Göttlinger H. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder J H, Schnölzer M, Hasselkus-Light C S, Henson M, Lerner D A, Phillips T R, Wagaman P C, Kent S B H. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J Virol. 1993;67:1869–1876. doi: 10.1128/jvi.67.4.1869-1876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison III C A, Loeb D D, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 11.Franke E K, Yuan H E H, Bossolt K L, Goff S P, Luban J. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J Virol. 1994;68:5300–5305. doi: 10.1128/jvi.68.8.5300-5305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller S D, Wilk T, Gowen B E, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 13.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 14.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;273:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 15.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 16.Gelderblom H R, Hausmann E H S, Özel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 17.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 18.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 19.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation on infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross I, Hohenberg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 22.Gross, I., H. Hohenberg, C. Huckhagel, and H.-G. Kräusslich. N-terminal extension of the human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 23.Harada S, Koyanagi Y, Yamamoto N. Infection of HTLV-III/LAV in HTLV-I carrying MT-2 and MT-4 cells and application in a plaque assay. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- 24.Henderson L E, Benveniste R E, Sowder R, Copeland T D, Schultz A M, Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson L E, Copeland T D, Sowder R C, Schultz A M, Oroszlan S. Analysis of proteins and peptides from sucrose banded HTLV-III. In: Bolognesi D, editor. Human retroviruses, cancer, and AIDS: approaches to prevention and therapy. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 135–147. [Google Scholar]
- 27.Henderson L E, Sowder R C, Smythers G W, Oroszlan S. Chemical and immunological characterization of equine infectious anemia virus gag-encoded proteins. J Virol. 1987;61:1116–1124. doi: 10.1128/jvi.61.4.1116-1124.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103–112. [Google Scholar]
- 29.Jowett J B M, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan A H, Zack J A, Knigge M, Paul D A, Kempf D J, Norbeck D W, Swanstrom R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67:4050–4055. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohl N, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A F, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konvalinka J, Heuser A-M, Hruskova-Heidingsfeldova O, Vogt V M, Sedlacek J, Strop P, Kräusslich H-G. Proteolytic processing of particle-associated retroviral polyproteins by homologous and heterologous viral proteinases. Eur J Biochem. 1994;228:191–198. doi: 10.1111/j.1432-1033.1995.tb20249.x. [DOI] [PubMed] [Google Scholar]
- 33.Konvalinka J, Litterst M A, Welker R, Kottler H, Rippmann F, Heuser A-M, Kräusslich H-G. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J Virol. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kräusslich H-G, Schneider H, Zybarth G, Carter C A, Wimmer E. Processing of in vitro-synthesized Gag precursor proteins of human immunodeficiency virus (HIV) type 1 by HIV proteinase generated in Escherichia coli. J Virol. 1988;62:4393–4397. doi: 10.1128/jvi.62.11.4393-4397.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kräusslich H-G, Welker R. Intracellular transport of retroviral capsid components. Curr Top Microbiol Immunol. 1996;214:25–63. doi: 10.1007/978-3-642-80145-7_2. [DOI] [PubMed] [Google Scholar]
- 36a.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mergener K, Fäcke M, Welker R, Brinkmann V, Gelderblom H R, Kräusslich H-G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 40.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 42.Nermut M V, Hockley D J. Comparative morphology and structural classification of retroviruses. Curr Top Microbiol Immunol. 1996;214:1–24. doi: 10.1007/978-3-642-80145-7_1. [DOI] [PubMed] [Google Scholar]
- 43.Öhagen A, Luftig R B, Reicin A S, Yin L, Ikuta K, Kimura T, Goff S P, Höglund S. The morphology of the immature HIV-1 virion. Virology. 1997;228:112–114. doi: 10.1006/viro.1996.8362. [DOI] [PubMed] [Google Scholar]
- 44.Park J, Morrow C D. Mutations in the protease gene of human immunodeficiency virus type 1 affect release and stability of virus particles. Virology. 1993;194:843–850. doi: 10.1006/viro.1993.1328. [DOI] [PubMed] [Google Scholar]
- 45.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng C, Ho B K, Chang T W, Chang N T. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepinsky R B, Papayannopoulos I A, Chow E P, Krishna N K, Craven R C, Vogt V M. Differential proteolytic processing leads to multiple forms of the CA protein in avian sarcoma and leukemia viruses. J Virol. 1995;69:6430–6438. doi: 10.1128/jvi.69.10.6430-6438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison III C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 50.Poorman R A, Tomasselli A G, Heinrikson R L, Kézdy F J. A cumulative specificity model for proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J Biol Chem. 1991;266:14554–14561. [PubMed] [Google Scholar]
- 51.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards A D, Phylip L H, Farmerie W G, Scarborough P E, Alvarez A, Dunn B M, Hirel P-H, Konvalinka J, Strop P, Pavlickova L, Kostka V, Kay J. Sensitive, soluble chromogenic substrates for HIV-1 proteinase. J Biol Chem. 1990;265:7733–7736. [PubMed] [Google Scholar]
- 53.Rosé J R, Babe L M, Craik C S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salahuddin S Z, Markham P D, Wong-Staal F, Franchini G, Kalyanaraman V S, Gallo R C. Restricted expression of human T-cell leukemia-lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology. 1983;129:51–64. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 55.Schätzl H, Gelderblom H R, Nitschko H, von der Helm K. Analysis of non-infectious HIV particles produced in presence of HIV proteinase inhibitor. Arch Virol. 1991;120:71–81. doi: 10.1007/BF01310950. [DOI] [PubMed] [Google Scholar]
- 56.Schechter I, Berger A. On the size of the active sites in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 57.Spurr A R. A low viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–34. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 58.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tobin G J, Sowder II R C, Fabris D, Hu M Y, Battles J K, Fenselau C, Henderson L E, Gonda M A. Amino acid sequence analysis of the proteolytic cleavage products of the bovine immunodeficiency virus Gag precursor polypeptide. J Virol. 1994;68:7620–7627. doi: 10.1128/jvi.68.11.7620-7627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tritch R J, Cheng Y-S E, Yin F H, Erickson-Viitanen S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J Virol. 1991;65:922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogt V M. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–132. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 62.Von Poblotzki A, Wagner R, Niedrig M, Wanner G, Wolf H, Modrow S. Identification of a region in the Pr55gag polyprotein essential for HIV-1 particle formation. Virology. 1993;193:981–985. doi: 10.1006/viro.1993.1210. [DOI] [PubMed] [Google Scholar]
- 63.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 65.Zhang W-H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]