Abstract
Introduction
Genetic mutations and amplifications found in hepatocellular carcinoma (HCC) have a potentially prognostic impact. The aim of this study was to investigate the prognostic value of mutations and amplifications in HCC from patients that were liver resected.
Methods
Patients liver resected for HCC at Copenhagen University Hospital Rigshospitalet between May 2014 and January 2018 were included. DNA from freshly frozen tumour tissue was investigated with TruSight Oncology 500. Mutations and amplifications were correlated with disease-free survival and overall survival using multivariate Cox regression to assess the effect on prognosis.
Results
Of the 51 patients included, 88% were male and the median age was 69 years. Most patients had a single tumour (84%) with no vascular invasion (67%) in a non-cirrhotic liver (76% with fibrosis, 24% with cirrhosis). The median follow-up was 37 months. Patients with a MYC amplification (8%) were significantly younger than the remaining patients. Furthermore, they had a significantly shorter overall survival (15 months (95% CI: 0.0–31.6) vs. 59 months (95% CI: 34.4–83.6), p = < 0.001) and disease-free survival (8 months (95% CI: 4.6–11.4) vs. 19 months (95% CI: 12.3–25.7), p = 0.03). However, only overall survival remained statistically significant in the adjusted analysis. Furthermore, all patients with an ARID1A mutation (6%) had microvascular invasion and significantly larger tumours than the patients without ARID1A mutation.
Conclusion
MYC amplifications had a prognostic influence on survival, whereas ARID1A gene mutations were correlated with microvascular invasion. These may serve as prognostic biomarkers and should be validated in large, independent cohort.
Keywords: Hepatocellular carcinoma, HCC, MYC amplification, ARID1A, Biomarker, Prognosis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer in the world and a leading cause of cancer-related mortality worldwide [1]. Chronic hepatitis B and C virus infections are common in HCC patients, especially in Asia and Africa, and are drivers of HCC development and progression. Other important aetiologies, especially in Western countries, include non-alcoholic steatohepatitis and chronic alcohol abuse leading to liver cirrhosis [2].
HCC is an aggressive cancer with a high mortality rate in advanced stages. However, in early stages, patients may benefit from curative treatments, such as liver resection, ablation, or liver transplantation [2–4]. The decision of a treatment strategy is primarily based on tumour burden, liver function, and patient performance status [2]. Resection retains sufficient liver function and is typically the first choice for HCC patients with a single tumour. However, as many as 70% of the resected patients develop recurrence within five years [2, 5].
Key drivers in the malignant development of HCC are a number of genetic alterations (often (in)activating mutations or gene amplifications). Among the genes frequently altered in HCC patients are TERT, TP53, MYC amplification, CTNNB1, and ARID1A, which occur in up to 60%, 48%, 33%, 30%, and 17% of cases, respectively [6–8]. Several genetic alterations in these genes have been associated with a negative impact on survival. In previous studies, MYC amplification was found to be associated with larger, more aggressive tumours in patients who are often younger. ARID1A was associated with carcinogenesis and metastasis in already established HCC tumours [7, 9]. Notably, few prognostic biomarkers are available today for a personalized HCC treatment approach allowing to stratify patients in the groups that would benefit the most from surgical treatment and other groups where less aggressive treatment would be preferable [4].
Most previous studies on genetic alterations in HCC have been conducted on patients of Asian, American, or Southern European descent with a high prevalence of cirrhosis as well as chronic viral hepatitis C infection [7, 10–14]. The association between clinically significant genetic alterations and prognosis have yet to be described in a population with a low prevalence of cirrhosis and viral hepatitis undergoing resection for HCC.
The aim of this study was to investigate genetic alterations as prognostic markers in a Danish population of patients with HCC with a low prevalence of cirrhosis and viral hepatitis C undergoing liver resection.
Materials and methods
This study was a retrospective cohort study including all patients that were liver resected for HCC at Copenhagen University Hospital Rigshospitalet, Denmark, between May 2014 and January 2018, with available freshly frozen tumour tissue stored in The Danish Cancer Biobank. The study was approved by the regional ethics committee (journal-nr.: H-18,015,944).
The collected tumour tissue was investigated for genomic changes through next generation sequencing with the TruSight Oncology (TSO) 500 Assay of 523 genes for DNA variants. DNA was extracted from fresh frozen tumour samples using AllPrep DNA/RNA/protein Extraction Kit (Qiagen) according to manufacturer’s instructions. Library preparation was done using TSO500 solid kit from Illumina according to Illumina’s reference guide. Libraries were sequencing using 2 × 150 bp paired-end sequencing on Illumina NovaSeq6000 platform. Reads were aligned to the human reference genome (hg19/GRCh37) and gene variants and amplifications were called using the TSO500 Local App pipeline for data processing (Illumina). Further filtering and identification of cancer-associated mutations (variants classified as pathogenic or likely pathogenic) was performed using Qiagen Clinical Insight (QCI) software from Qiagen.
The following characteristics of the patients, pathology data, and follow-up-data were retrieved from electronic medical records: patient characteristics (age, sex, comorbidities including cirrhosis, and medical history), tumour characteristics (tumour size, number of tumours, microvascular invasion, and resection margins), and follow-up including recurrence, disease-free survival (defined as the time from date of liver resection to the date of detected recurrence), and overall survival (defined as the time from date of liver resection to the date of death). Tumour-stage of HCC was determined according to the AJCC 7th edition criteria [15]. A free margin in the pathological specimen was defined as 1 mm or more between tumour and resection margin. To determine fibrosis/cirrhosis in non-tumour liver tissue, a METAVIR score was evaluated by a specialized pathologist [16].
Patients were preoperatively assessed radiologically and clinically using the Barcelona Clinic Liver Cancer staging and discussed at multidisciplinary team conference. The diagnosis was confirmed in approximately one third of the cases by a preoperative needle biopsy. This is usually an option in non-cirrhotic patients with suspected HCC. Liver function was assessed using the Child-Pugh score. Portal hypertension was assessed with imaging (e.g. collaterals, splenomegaly, re-canalization of the umbilical vein, and ascites) and thrombocyte count. In cases with suspicion of impaired liver function or cases with large resections, ICG-clearance was performed. Patients with impaired liver function were generally not considered for resection and was treated according to the Barcelona Clinic Liver Cancer staging system [17].
As part of clinical practice, patients were followed with abdominal Computed Tomography-scans 3, 6, 9, 12, 18, 24, 36, 48, and 60 months after resection to identify potential recurrence.
Statistical analyses
The patients were divided into groups according to the genetic alterations detected by the TSO 500 assay. Association between groups of interest and prognosis (disease-free survival and overall survival) was determined by Kaplan Meier statistics reported as median survival with 95% confidence intervals (CI). Log-rank test was used to compare the survival of the different groups. In multivariate Cox-regressions, the association was adjusted for possible confounders. Of the possible confounders (male sex, age, size of HCC, number of tumours, cirrhosis, microvascular invasion, and resection margins), variables significantly associated with the outcome in univariate analyses were included in the multivariate analysis. Moreover, the association between the groups and markers of tumour biology (microvascular invasion, tumour grade, and size) was investigated with Fisher’s exact test. Continuous variables are reported as median with range or interquartile range. The Mann-Whitney Test was used to compare the median age of the patients as well as median size of the largest tumour in patients with and without MYC amplification as well as mutations affecting ARID1A. The reverse Kaplan-Meier method was used to determine the median follow-up. Follow-up was defined as the median time from the date of liver resection to either event (death) or last follow-up (03.11.2022).
An a priori sample size calculation was based on an earlier study in patients with HCC, where a 5-Gene Score was associated with inferior five-year disease-free survival (78% vs. 33%) [18]. Based on an alpha of 5% and a power of 80%, we needed 34 patients in the study to be able to detect a difference of this magnitude (G-power, version 3.1.9.3.).
We used SPSS, version 23 (IBM Corp, Armonk, NY, USA) to conduct the statistical analysis. The significance level was adjusted to the number of analyses of gene groups using the Bonferroni correction setting the significance level to 0.005.
Results
Patient characteristics
In total, 117 patients were liver resected for HCC at Copenhagen University Hospital Rigshospitalet, Copenhagen between May 2014 and January 2018. We identified 54 patients with tumour tissue as well as non-tumorous adjacent tissue stored as freshly frozen tissue in the Danish cancer biobank. Patients with freshly frozen tissue were primarily from the later part of the period where storage of freshly frozen tissue was part of the standard treatment. Of these 54 patients, one was excluded due to liver transplantation as the surgical choice of treatment. Another patient died 41 days after resection due to liver failure, probably caused by the surgery. Unfortunately, the patients’ genetic characteristics were not determined as the patient was excluded prior to analyses. Lastly, one patient appeared twice in the biobank, leaving 51 patients available for analyses. The median follow-up was 37 months (31–41).
Patient characteristics are shown in Table 1. Most patients were male (88%), had a median age of 69 years, a pT1 stage tumour, a median tumour size of 45 mm (largest tumour), and a single tumour without microvascular invasion which were resected with free resection margins. The majority had fibrosis (METAVIR stage 1–3) at time of the resection. However, 12 patients (24%) had cirrhosis (METAVIR stage 4). Complications occurred in 5 (9.8%) of the 51 patients including two cases of fascial dehiscence (3.9%), two cases of bile leakage (3.9%), and one case of pulmonary embolism (2%). Furthermore, 42 patients (82.4%) had recurrence during the study period with a median time to recurrence of 14 months.
Table 1.
Patient characteristics
| All patients, n = 51 | Patients w MYC, n = 4 | Patients w/o MYC, n = 47 | |||
|---|---|---|---|---|---|
| Sex, n (%) | |||||
| Women | 6 (12) | 0 | 6 | ||
| Men | 45 (88) | 4 | 41 | ||
| Age, years, median (range) | 69 (23–88) | 58 (50–69) | 80 (23–88) | ||
| Liver disease, n (%) | |||||
|
Alcoholic cirrhosis Hepatitis C virus Hemochromatosis Non-alcoholic steatohepatitis No known liver disease |
8 (16) 10 (20) 2 (4) 1 (2) 30 (59) |
2 1 0 0 1 |
6 9 2 1 29 |
||
| Stage, n (%) | |||||
| pT1 | 24 (47) | 1 | 23 | ||
| pT1b | 4 (8) | 2 | 2 | ||
| pT2 | 9 (18) | 0 | 9 | ||
| pT2a | 1 (2) | 0 | 1 | ||
| pT3 | 1 (2) | 0 | 1 | ||
| pT3a | 8 (16) | 1 | 7 | ||
| pT3b | 3 (6) | 0 | 3 | ||
| pT4 | 1 (2) | 0 | 1 | ||
| Number of tumours, n (%) | |||||
| 1 | 43 (84) | 4 | 39 | ||
| 2 | 6 (12) | 0 | 6 | ||
| 3 | 2 (4) | 0 | 2 | ||
| Free resection margins, n (%) | |||||
| Yes | 41 (80) | 2 | 39 | ||
| No | 10 (20) | 2 | 8 | ||
| Diameter of the largest tumour, mm, median (range) | 45 (17–250) | 75 (30–85) | 45 (17–250) | ||
| METAVIR, n (%) | |||||
| Stage 1–3 | 39 (76) | 3 | 36 | ||
| Stage 4 (cirrhosis) | 12 (24) | 1 | 11 | ||
| Vascular invasion, n (%) | |||||
| Yes | 17 (33) | 0 | 17 | ||
| No | 34 (67) | 4 | 30 | ||
| MELD, median (IQR)* | 28.9 (27.3–29.7) | NA | NA | ||
| BCLC | |||||
|
Very early stage (0) Early stage (A) Intermediate stage (B) |
3 40 8 |
0 4 0 |
3 36 8 |
* 1 of 4 patients with a MYC amplification and 22 of 51 patients in total had a MELD-score available. These are therefore displayed as “NA”.
HR, hazard ratio; CI, confidence interval; METAVIR, Meta-analysis of Histological Data in Viral Hepatitis; MELD, Model for End-Stage Liver Disease; BCLC, Barcelona Clinic Liver Cancer
DNA sequencing
The ten most frequently occurring genomic alterations within the study group are shown in Table 2. Mutations affecting TERT, CTNNB1, and TP53 were the most frequent genetic alterations and were observed in 51%, 31%, and 31% of the patients, respectively. The most frequent amplification affected the CCDN1 gene, appearing in 10% of the patients. MYC amplification occurred in 8% of the patients.
Table 2.
Frequently occurring mutations and amplifications
| Frequent genomic mutations and amplifications | |
|---|---|
| TERT, n (%) | 26 (51) |
| CTNNB1, n (%) | 16 (31) |
| TP53, n (%) | 16 (31) |
| ATM, n (%) | 5 (10) |
| FGF3, n (%) | 5 (10) |
| FGF19, n (%) | 5 (10) |
| FGF4, n (%) | 5 (10) |
| ARID1A, n (%) | 3 (6) |
| CCND1 amplification, n (%) | 5 (10) |
| MYC amplification, n (%) | 4 (8) |
Of note, the median age of patients with MYC amplification was significantly lower (58 years (50–69)) than the remaining patients (69 years (23–88), p = 0.018). The median diameter of the largest tumour was 75 mm in patients with MYC amplification and 45 mm in the remaining patients (p = 0.754).
Survival analysis
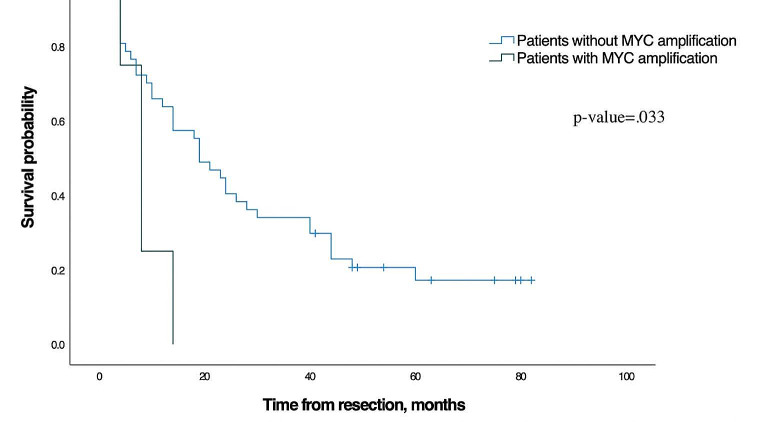
The median overall survival of all the patients in this study was 56 months (95% CI: 39.2–72.8). For all patients without MYC amplification the median overall survival was 59 months (95% CI: 34.4–83.6) compared to 15 months in patients with MYC amplification (95% CI: 0.0–31.6), p = < 0.001, Fig. 1). Among possible confounders (male sex, age, size of HCC, number of tumours, cirrhosis, microvascular invasion, and resection margins) only male sex and age were significantly associated with overall survival in the univariate Cox-regression (Table 3). Thus, these variables were included in the multivariate analysis with MYC amplification. MYC amplification remained independently associated with inferior overall survival when adjusted for male sex and age (p = < 0.001, HR: 8.6, 95% CI: 2.5–30.3), and thus remained significant when corrected for multiple comparison with Bonferroni correction. Furthermore, male sex and age were independently associated with inferior overall survival (Table 4).
Fig. 1.
Overall survival for patients without MYC amplification and for patients with MYC amplification
Table 3.
Univariate Cox regression analysis
| DFS | OS | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | p-value | HR | 95% CI | p-value |
| TERT | 1.78 | 0.96–3.32 | 0.07 | 1.76 | 0.88–3.52 | 0.10 |
| CTNNB1 | 1.17 | 0.61–2.25 | 0.64 | 1.35 | 0.67–2.72 | 0.40 |
| TP53 | 0.72 | 0.36–1.44 | 0.36 | 0.68 | 0.32–1.43 | 0.31 |
| ATM | 0.76 | 0.27–2.12 | 0.59 | 0.67 | 0.23–1.93 | 0.46 |
| FGF3 | 1.23 | 0.44–3.47 | 0.69 | 0.94 | 0.33–2.70 | 0.91 |
| FGF19 | 1.23 | 0.44–3.47 | 0.69 | 0.94 | 0.33–2.70 | 0.91 |
| FGF4 | 1.23 | 0.44–3.47 | 0.69 | 0.94 | 0.33–2.70 | 0.91 |
| ARID1A | 0.79 | 0.19–3.27 | 0.74 | 0.65 | 0.15–2.76 | 0.56 |
| CCND1 amplification | 1.23 | 0.44–3.47 | 0.69 | 0.94 | 0.33–2.70 | 0.91 |
| MYC amplification | 3.29 | 1.10–9.83 | 0.03 | 5.66 | 1.80-17.84 | 0.003 |
| Male sex | 14.22 | 1.91-105.97 | 0.01 | 11.10 | 1.42–86.74 | 0.02 |
| Age | 1.02 | 0.99–1.05 | 0.23 | 1.05 | 1.00-1.09 | 0.04 |
| Number of tumours | 1.39 | 0.80–2.40 | 0.24 | 1.31 | 0.72–2.40 | 0.38 |
| Free resection margins | 0.73 | 0.33–1.58 | 0.41 | 0.66 | 0.29–1.54 | 0.34 |
| Diameter of the largest tumour | 0.99 | 0.99-1.00 | 0.68 | 1.00 | 0.99-1.00 | 0.91 |
| METAVIR | 1.24 | 0.60–2.53 | 0.56 | 1.69 | 0.80–3.55 | 0.17 |
| Microvascular invasion | 1.34 | 0.70–2.57 | 0.38 | 1.05 | 0.51–2.12 | 0.90 |
HR, hazard ratio; CI, confidence interval; DFS, disease-free survival; OS, overall survival; METAVIR, Meta-analysis of Histological Data in Viral Hepatitis; Bold, statistically significant association (before correction)
Table 4.
Multivariate Cox regression analysis with overall survival
| HR (95% CI) | p-value | |
|---|---|---|
| MYC amplification | 8.6 (2.5–30.3) | < 0.001 |
| Age | 1.1 (1.0–1.1) | 0.013 |
| Male sex | 5.7 (1.2–26.8) | 0.029 |
HR, hazard ratio; CI, confidence interval
The disease-free survival was shorter in patients with MYC amplification (8 months (95% CI: 4.6–11.4)) compared with the remaining patients (19 months (95% CI: 12.3–25.7), p = 0.03, Fig. 2). Among possible confounders (male sex, age, size of HCC, number of tumours, cirrhosis, microvascular invasion, and resection margins) only male sex was significantly associated with disease-free survival in the univariate Cox-regression and, thus, included in the multivariate analysis (Tables 3 and 5). MYC amplification was not associated with microvascular invasion, tumour grade, or size.
Fig. 2.
Disease-free survival for patients without MYC amplification and for patients with MYC amplification
Table 5.
Multivariate Cox regression analysis with disease-free survival
| HR (95% CI) | p-value | |
|---|---|---|
| MYC amplification | 2.8 (0.9–8.4) | 0.064 |
| Male sex | 13.8 (1.8–102.8) | 0.011 |
HR, hazard ratio; CI, confidence interval
The remaining mutations and amplifications listed in Table 2 were not associated with overall or disease-free survival.
Markers of tumour biology
We found a larger tumour size in patients with microvascular invasion (median 85 mm) compared to patients without (median 41 mm, p = 0.03). All patients with a mutation in the ARID1A gene had microvascular invasion compared with only 29.2% of patients without the mutation (p = 0.03). However, ARID1A was not associated with disease-free survival, overall survival, tumour grade, or size.
The remaining mutations and amplifications listed in Table 2 were not significantly associated with tumour grade, size, or microvascular invasion. Furthermore, a correlation between aetiology and mutations could not be determined.
Discussion
In patients that were liver resected for HCC, we showed that MYC amplification was associated with a reduced overall survival. The association with overall survival remained significant when adjusted for male sex and age. The study also demonstrated that mutations in the gene ARID1A were associated with microvascular invasion in the tumour, and that patients with microvascular invasion had significantly larger tumours than patients without.
The association between the MYC amplification and a poorer prognosis has previously been shown in American, Italian, and Japanese studies [7, 10, 11]. All three studies found that MYC amplification was more frequent among HCC patients with high-grade tumours and found a higher prevalence of MYC amplification (19%, 33%, and 50%) compared with the 8% in our study. The reason for the variation in the frequency of MYC amplification may be due to different aetiologies for HCC between the studies. Only 24% of the patients included in the present study had cirrhosis at time of resection which is considerably lower than the 78.5% of the patients in the Italian study [7]. Moreover, the prevalence of hepatitis C virus in our study population was only 20% compared to 67% in the Japanese study [11]. In accordance with our results, the amplification was primarily found in younger patients (age < 50 years) with larger tumours in the Japanese study.
The clinical significance of mutations in ARID1A in HCC is still debated. One study reported that a loss of ARID1A was associated with a poorer prognosis and that ARID1A may exhibit a tumour suppressive role. The same study found that ARID1A is not carcinogenic alone but rather accelerates the carcinogenic process when associated with other oncogenes such as an amplification of MYC [8]. Another study showed that ARID1A had a context dependent role: overexpression of ARID1A was linked to the initiation of HCC, whereas a loss of ARID1A was linked to further metastasizing of a primary HCC [9]. We found that a mutation in the gene was associated with vascular invasion which is known to be a predictor of a poor prognosis [19]. However, ARID1A was not in itself prognostic in our study, which may be a result of the limited sample size and/or different aetiologies.
The present study adds the current evidence in the field by investigating the impact of MYC amplification and ARID1A on prognosis in a population of patients with a low prevalence of cirrhosis and viral hepatitis undergoing liver resection for HCC. Moreover, we evaluated HCC using the TSO 500 assay, which includes a wide range of oncogenes selected due to previously findings of prognostic significance. Our results corroborate with findings of previous studies, which emphasises the apparent clinical significance of MYC amplification. The patients in this study represent a homogenous group with long follow-up and high event rates. We conceived this work as an explorative study, using a limited number of available patient samples with detailed follow-up. This, however, is a limitation of this study. We were unable to adjust for potential confounders known to impact survival in a multivariate analysis, thus only show an association between MYC amplification and survival. Given the small sample size it is important to interpret these findings with caution and acknowledge the reduced generalizability. Therefore, future studies with a larger sample size are warranted, which could underline these results and possibly detect further relevant associations using the TSO 500 approach. This will enable adjustment for potential confounders and strengthen the results.
Few, biomarkers are used clinically in the treatment of HCC today. With a high recurrence rate as shown in the present study, biomarkers for personalized treatment are needed. Preoperative MYC amplification may have a strong prognostic impact but cannot be determined without a preoperative needle biopsy from the tumour. A preoperative needle biopsy bears a certain risk of tumour seeding to surrounding organs (2.7%) and structures and risk of bleeding [20]. Hence, we consider it of interest to further evaluate prospectively whether the knowledge of candidate biomarker status, such as MYC amplification, outweighs the potential negative effects risked by taking a preoperative tumour biopsy.
In conclusion, we showed that MYC amplification was associated with a poorer prognosis. Furthermore, we showed that a mutation in ARID1A was associated with vascular invasion. After validation in a prospective setup, the findings of the present study have the potential to affect surgical treatment strategy for patients with HCC.
Author contributions
Sophie Bull Nordkild: acquisition of data, data analysis and interpretation of results, writing the first draft, critical revision, final approval. Hans-Christian Pommergaard: conception and design, interpretation of results, critical revision, final approval. Susanne Dam Poulsen: interpretation of results, critical revision, final approval. Jeanett Klubien: data analysis and interpretation of results, critical revision, final approval. Günes Delal Akdag: data analysis and interpretation of results, critical revision, final approval. Lise Barlebo Ahlborn: data analysis and interpretation of results, critical revision, final approval. Christina Westmose Yde: data analysis and interpretation of results, critical revision, final approval. Gro Linno Willemoe: data analysis and interpretation of results, critical revision, final approval. Jan-Michael Kugler: data analysis and interpretation of results, critical revision, final approval.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Open access funding provided by National Hospital
Data availability
The Danish Data Protection Agency does not allow open access to the data included in this study. However, reasonable requests for additional analyses on the dataset can be made to the corresponding author.
Declarations
Ethics approval
The study was approved by the regional ethics committee (journal-nr.: H-18015944).
Informed consent
Informed consent has been obtained by all participants.
Competing interests
HC Pommergaard: Research grants from Danish Cancer Society, Svend Andersen Fund, Harboe Fund, Dagmar Marshalls Fund, and The Danish National Center for Circulating Tumor DNA Guided Cancer Treatment.SD Nielsen: Research grant from Novo Nordic Foundation and participation in advisory board for Gilead, Merck, and GSK.SB Nordkild, LB Ahlborn, CW Yde, JM Kugler, J Klubien, GD Akdag, and GL Willemoe: no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.F. J et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. https://gco.iarc.fr/today (accessed. 01.03.2023)
- 2.Llovet JM et al (p. 6, Jan 21 2021) Hepatocellular carcinoma. Nat Rev Dis Primers 7(1). 10.1038/s41572-020-00240-3 [DOI] [PubMed]
- 3.McGlynn KA, Petrick JL, El-Serag HB (Jan 2021) Epidemiol Hepatocellular Carcinoma Hepatol 73:4–13. 10.1002/hep.31288 [DOI] [PMC free article] [PubMed]
- 4.Pommergaard HC et al (Apr 2021) Peroxisome proliferator-activated receptor activity correlates with poor survival in patients resected for hepatocellular carcinoma. J Hepatobiliary Pancreat Sci 28(4):327–335. 10.1002/jhbp.745 [DOI] [PubMed]
- 5.Roayaie S et al (2013) Resection of hepatocellular cancer Hepatology, vol. 57, no. 4, pp. 1426-35, Apr 10.1002/hep.25832 [DOI] [PMC free article] [PubMed]
- 6.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM (2015) Genetic Landscape and Biomarkers of Hepatocellular Carcinoma, Gastroenterology, vol. 149, no. 5, pp. 1226–1239 e4, Oct 10.1053/j.gastro.2015.05.061 [DOI] [PubMed]
- 7.Pedica F et al (2013) A re-emerging marker for prognosis in hepatocellular carcinoma: the add-value of fishing c-myc gene for early relapse. PLoS ONE 8(7):e68203. 10.1371/journal.pone.0068203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y et al Loss of ARID1A promotes Hepatocellular Carcinoma Progression via Up-regulation of MYC Transcription. J Clin Transl Hepatol, 9, 4, pp. 528–536, Aug 28 2021, 10.14218/JCTH.2021.00111 [DOI] [PMC free article] [PubMed]
- 9.Sun X et al (2017) Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer, Cancer Cell, vol. 32, no. 5, pp. 574–589 e6, Nov 13 10.1016/j.ccell.2017.10.007 [DOI] [PMC free article] [PubMed]
- 10.Abou-Elella A, Gramlich T, Fritsch C, Gansler T c-myc amplification in hepatocellular carcinoma predicts unfavorable prognosis, Mod Pathol, vol. 9, no. 2, pp. 95 – 8, Feb 1996. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/8657726 [PubMed]
- 11.Kawate S, Fukusato T, Ohwada S, Watanuki A, Morishita Y (1999) Amplification of c-myc in Hepatocellular Carcinoma: Correlation with Clinicopathologic Features, Proliferative Activity and p53 Overexpression, Oncology, vol. 57, no. 2, pp. 157–163, 10.1159/000012024. Karger Publishers [DOI] [PubMed]
- 12.Fang Y, Huang B, Liang Q, Li H, Huang C (2001) [Clinical significance of c-myc oncogene amplification in primary hepatocellular carcinoma by interphase fluorescence in situ hybridization], Zhonghua Bing Li Xue Za Zhi, vol. 30, no. 3, pp. 180-2, Jun [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/11866973 [PubMed]
- 13.Peng SY, Lai PL, Hsu HC Amplification of the c-myc gene in human hepatocellular carcinoma: biologic significance, J Formos Med Assoc, vol. 92, no. 10, pp. 866 – 70, Oct 1993. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/7511953 [PubMed]
- 14.Xu Z et al (2019) The mTORC2-Akt1 Cascade Is Crucial for c-Myc to Promote Hepatocarcinogenesis in Mice and Humans, Hepatology, vol. 70, no. 5, pp. 1600–1613, Nov 10.1002/hep.30697 [DOI] [PMC free article] [PubMed]
- 15.Edge S, Byrd D, Compton C, Fritz A, Greene F, Trotti A (eds) (2010) AJCC Cancer Staging Manual. Springer, New York, NY [Google Scholar]
- 16.Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. The French METAVIR Cooperative Study Group, Hepatology, vol. 20, no. 1, pp. 15–20, Jul 1994. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/8020885 [PubMed]
- 17.Llovet JM, Bru C, Bruix J (1999) Prognosis of hepatocellular carcinoma: the BCLC staging classification, Semin Liver Dis, vol. 19, no. 3, pp. 329 – 38, 10.1055/s-2007-1007122 [DOI] [PubMed]
- 18.Nault JC et al (2013) A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection, Gastroenterology, vol. 145, no. 1, pp. 176–187, Jul 10.1053/j.gastro.2013.03.051 [DOI] [PubMed]
- 19.Erstad DJ, Tanabe KK (May 2019) Prognostic and therapeutic implications of Microvascular Invasion in Hepatocellular Carcinoma. Ann Surg Oncol 26(5):1474–1493. 10.1245/s10434-019-07227-9 [DOI] [PubMed]
- 20.Silva MA, Hegab B, Hyde C, Guo B, Buckels JA, Mirza DF (2008) Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: a systematic review and meta-analysis, Gut, vol. 57, no. 11, pp. 1592-6, Nov 10.1136/gut.2008.149062 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Danish Data Protection Agency does not allow open access to the data included in this study. However, reasonable requests for additional analyses on the dataset can be made to the corresponding author.