Abstract
It has been proposed that changes in cell surface concentrations of coreceptors may control infections by human immunodeficiency virus type 1 (HIV-1), but the mechanisms of coreceptor function and the concentration dependencies of their activities are unknown. To study these issues and to generate stable clones of adherent cells able to efficiently titer diverse isolates of HIV-1, we generated two panels of HeLa-CD4/CCR5 cells in which individual clones express either large or small quantities of CD4 and distinct amounts of CCR5. The panels were made by transducing parental HeLa-CD4 cells with the retroviral vector SFF-CCR5. Derivative clones expressed a wide range of CCR5 quantities which were between 7.0 × 102 and 1.3 × 105 molecules/cell as measured by binding antibodies specific for CCR5 and the chemokine [125I]MIP1β. CCR5 was mobile in the membranes, as indicated by antibody-induced patching. In cells with a large amount of CD4, an unexpectedly low trace of CCR5 (between 7 × 102 and 2.0 × 103 molecules/cell) was sufficient for maximal susceptibility to all tested HIV-1, including primary patient macrophagetropic and T-cell-tropic isolates. Indeed, the titers as indicated by immunoperoxidase staining of infected foci were as high as the tissue culture infectious doses measured in human peripheral blood mononuclear cells. In contrast, cells with a small amount of CD4 required a much larger quantity of CCR5 for maximal infection by macrophagetropic HIV-1 (ca. 1.0 × 104 to 2.0 × 104 molecules/cell). Cells that expressed low and high amounts of CD4 were infected with equal efficiencies when CCR5 concentrations were above threshold levels for maximal infection. Our results suggest that the concentrations of CD4 and CCR5 required for efficient infections by macrophagetropic HIV-1 are interdependent and that the requirements for each are increased when the other component is present in a limiting amount. We conclude that CD4 and CCR5 directly or indirectly interact in a concentration-dependent manner within a pathway that is essential for infection by macrophagetropic HIV-1. In addition, our results suggest that multivalent virus-receptor bonds and diffusion in the membrane contribute to HIV-1 infections.
The membrane fusion step of infection by human immunodeficiency virus type 1 (HIV-1) requires collaboration between CD4 and coreceptors on surfaces of susceptible cells (2, 4, 15, 19, 21, 22, 26, 27). The coreceptors that have been identified normally function as G-protein-coupled receptors for proinflammatory chemokines (55). The major coreceptor for macrophagetropic (M-tropic) isolates of HIV-1 is CCR5, which responds to the chemokines RANTES, MIP1α, and MIP1β (2, 15, 19, 21, 22, 59), whereas the major coreceptor for T-cell-tropic isolates is CXCR4, which is activated by the chemokine SDF-1 (4, 6, 27). Approximately 10% of North American Caucasians carry a defective CCR5 gene (Δ32) (18, 62). The resistance of Δ32/Δ32 homozygous individuals to infection by HIV-1 strongly suggests that CCR5-dependent M-tropic viruses are critical for viral transmission (18, 45, 58, 62). In contrast, viruses that use CXCR4 accumulate late in disease progression during the demise of the immune system (17, 46, 53). HIV-1 apparently forms ternary complexes on cell surfaces with CD4 and coreceptors (30, 43, 69, 74). Several CCR5 and CXCR4 mutants defective in G-protein signaling are active in mediating HIV-1 infections (20, 24, 25, 28, 46).
Recent studies have suggested that changes in cell surface concentrations of CD4 or coreceptors may control HIV-1 infections and development of disease (33, 35, 52, 75). For example, resting T cells contain relatively little CCR5 and are resistant to infections by M-tropic HIV-1; upon activation in vitro, they synthesize CCR5 and become susceptible to infection (7, 9, 69, 75). Moreover, CCR5 is synthesized in effector/memory T cells but not in naive T cells (7, 75), in agreement with the preferential loss of memory T cells during the asymptomatic stage of HIV-1 infections (63, 71). In contrast, CXCR4 is expressed in naive T-cell populations and exhibits rapid upregulation in response to T-cell activation in vitro (7). Susceptibilities to infection by M-tropic HIV-1 of blood T-cell populations from different individuals correlate with the percentages of cells expressing CCR5 (75). Recently it has been suggested that heterozygosity for the Δ32 CCR5 mutation may delay disease progression in HIV-1-positive patients (18, 32, 51) and that lymphocytes from these individuals have reduced CCR5 expression levels and lower susceptibilities to infection by M-tropic HIV-1 (75). The fact that chemokines and their antagonists can inhibit infections by HIV-1 also has suggested that the corresponding coreceptors may be limiting for infectivity (16, 65). Similarly, we previously found that patient T-cell-tropic HIV-1 isolates can efficiently infect only cells that have very large amounts of CD4, whereas their laboratory-adapted derivatives can efficiently infect cells that have little CD4 (33, 39).
Despite the importance of these issues, little is known concerning the mechanisms of coreceptor function. Indeed, few investigations have used cells that contain known amounts of CD4 (33, 39), and none have examined the concentration dependencies of coreceptor activities. To address these issues, and to produce single-cell clones highly susceptible to infection by HIV-1 isolates of different tropisms, we made clonal panels of adherent HeLa-CD4/CCR5 cells that stably express distinct quantities of CCR5 over a broad range. HeLa cells contain CXCR4 but not CCR5 (2, 27, 41). One clonal panel was made with the HI-J clone of HeLa-CD4 cells, which uniformly expresses a large quantity of CD4 (approximately 4 × 105 molecules/cell), whereas another panel was made with the HI-R clone, which expresses much less CD4 (approximately 104 molecules/cell) (33, 38). We found that cells with a large amount of CD4 required only a trace amount of CCR5 for maximal susceptibility to infection by diverse isolates of M-tropic HIV-1, whereas cells with a low amount of CD4 required a much larger amount of CCR5 for maximal infection. Therefore, the CD4 and CCR5 concentration requirements for efficient infections by M-tropic HIV-1 are interdependent. These results have important implications for understanding receptor-coreceptor collaboration in HIV-1 infections.
MATERIALS AND METHODS
Cells and viruses.
HeLa-CD4 clones HI-J and HI-R, expressing large and small amounts of cell surface CD4 (approximately 4 × 105 and 104 molecules/cell, respectively [33, 38]), were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. We used virus produced from the retroviral expression vector SFF-CCR5 to transduce the HI-J and HI-R cells at a high multiplicity. To construct this vector, human CCR5 cDNA was isolated by PCR and used to make plasmid pKS(+)-CCR5 as described previously (41). This plasmid was cut with the restriction endonucleases BamHI and XhoI, and the coding fragment lacking the poly(A) addition signal was ligated into the pSFF vector (5) that had been cut with the same enzymes. The SFF-CCR5 plasmid was transfected into a coculture of Ψ2 ecotropic packaging cells and PA12 amphotropic packaging cells for amplification of helper-free virions, and the virions were used to infect HI-J and HI-R cells as previously described (33, 37). Clones were isolated by limiting dilution and were assayed for susceptibility to infection by the M-tropic HIV-1 isolate SF162 or Ba-L, using the immunoperoxidase focal infectivity method (12). Peripheral blood mononuclear cells (PBMCs) were isolated and maintained as described previously (13).
The M-tropic HIV-1 isolates SF162, JR-FL, ADA, and Ba-L, contributed by Jay Levy, by Irvin Chen, by Howard Gendelman, and by Suzanne Gartner, Mikulas Popovic, and Robert Gallo, respectively, were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Rockville, Md. M-tropic HIV-1 isolates were grown as described previously (39). M-tropic clone 81A and the isolate JR-CSF have been described previously (36, 68). The primary T-cell-tropic HIV-1 isolate ELI1, provided as a molecular clone, was the generous gift of Keith Peden (Laboratory of Retrovirus Research, Center of Biologics Evaluation and Research, Food and Drug Administration, Bethesda, Md.). 208K4 (K4) and the other primary isolates were isolated as described previously (13). ELI1, K4, and the laboratory-adapted T-cell-tropic isolate NL4-3 (1) were propagated as described elsewhere (39). Several of the HIV-1 primary isolates were generously supplied by Eric Daar (Cedars-Sinai Medical Center, University of California at Los Angeles).
Infectivity assays.
Infectivity assays were performed as described previously (39, 41). HIV-1 SF162, JR-FL, ADA, Ba-L, ELI1, K4, and NL4-3 viruses were titered on HeLa clones expressing large amounts of CD4 and CCR5. Doses of approximately 5,000 focus-forming units per ml were used for infections. HIV-1 infections were quantitated by a modification of the focal immunoperoxidase staining method (12, 56). Briefly, this involved plating 5 × 103 to 1.5 × 104 cells into 1-cm2 wells of a 48-well culture dish, the following day preincubating the cells for 20 min with DEAE-dextran (8 μg/ml) in serum-free medium, and then infecting them with 0.1 ml of virus that had been diluted into medium that contained 0.1% fetal bovine serum. After 2.0 h at 37°C, 0.5 ml of fresh complete medium was added, and the cultures were incubated for 48 to 72 h before fixation with 95% ethanol for 3 to 6 min and immediate rinsing with physiological saline solution containing 1 mM EDTA. Without allowing the cells to dry, immunoperoxidase staining was done as previously described, using as primary antibody, at 0.1 ml/well, 1:5 diluted supernatant fluid from the anti-HIV p24 hybridoma 183-H12-5C (14, 68), obtained from the NIH AIDS Research and Reference Reagent Program. The stained foci of infection were counted with a dissecting microscope under diffuse illumination. Fifty percent tissue culture infectious dose (TCID50) values were determined as described previously (13).
Detection of cell surface CCR5.
CCR5 expression on the surfaces of the HeLa-CD4/CCR5 cell clones was detected by immunofluorescence microscopy, using a rabbit antiserum that is specific for the amino terminus of human CCR5 (41). A 1:25 dilution of antiserum in complete culture medium was incubated for 1 h at 37°C with cells grown in four-well chamber slides (Nunc, Inc., Naperville, Ill.). After being rinsed three times with complete medium, the cells were incubated for 1 h at 37°C with fluorescein isothiocyanate-conjugated affinity-purified goat anti-rabbit immunoglobulin G serum (Biosource International, Camarillo, Calif.) diluted at 1:100 in complete medium. Cells were washed by being incubated three times for 5 min each with complete medium, rinsed with phosphate-buffered saline (PBS), fixed with cold methanol for 5 min, and rinsed again with PBS. Coverslips were mounted over drops of 50% glycerol in PBS, and slides were viewed by fluorescence microscopy.
Two methods were used to quantitatively estimate CCR5 expression. One method used a CCR5 rabbit antiserum-[125I]protein A (0.4 μCi/ml, 2 to 10 μCi/μg; NEN Life Science Products, Boston, Mass.) binding assay performed as described previously (41). Briefly, cells previously seeded at 5 × 104/well in a 24-well cluster plate 24 h prior to the assay were incubated with rabbit anti-CCR5 as described above, washed three times in complete medium, and then incubated with 0.2 ml of a saturating concentration of [125I]protein A. After a 1-h incubation at 37°C, cells were washed three times in complete medium and once in PBS, then lysed in 0.1 N NaOH, and counted in a gamma counter. Radioactive counts were normalized to sample protein content as determined by the Coomassie blue method (Bio-Rad Laboratories, Hercules, Calif.). We used a second assay to standardize the latter results to absolute levels of CCR5 expression, by binding [125I]MIP1β (2,200 Ci/mmol; NEN Life Science Products) onto cell surfaces. Briefly, 24 h prior to the assay, HeLa cells expressing large amounts of CCR5 and CD4 (clone JC.53) were seeded at 5 × 104 cells/well in a 24-well cluster plate. The assay was performed on viable cells by incubating 1 nM [125I]MIP1β and increasing concentrations of unlabeled MIP1β (Peprotech, Rocky Hill, N.J.) as a competitor at 37°C for 2 h as described previously (31, 59). The binding assay was terminated by washing cells three times in complete medium and once in PBS and then lysing cells in 0.1 N NaOH. Lysed cells were counted in a gamma counter.
RESULTS
Generation of HeLa-CD4/CCR5 clonal panels.
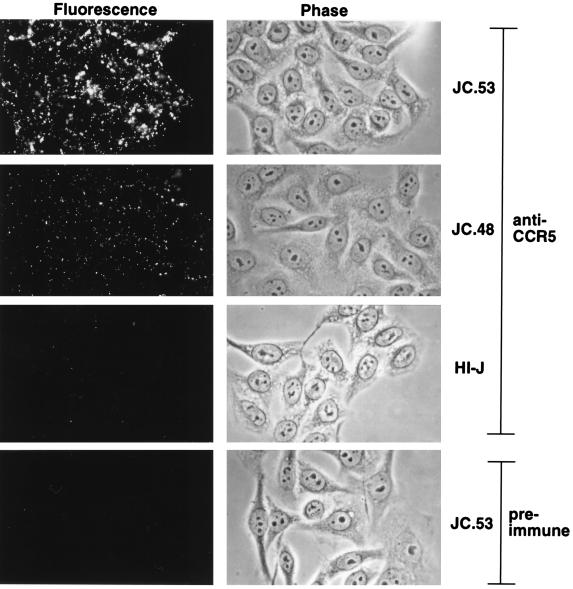
To investigate the role of CCR5 expression levels in M-tropic HIV-1 infections and to generate a cell line potentially susceptible to infection by diverse HIV-1 isolates, we transduced HeLa-CD4 cells (clone HI-J) that stably express a large amount of CD4 (approximately 4 × 105 molecules/cell [38]) with the retroviral vector SFF-CCR5. Clones were isolated by limiting dilution and were screened for CCR5 expression by infection with the SF162 M-tropic HIV-1 isolate. Of 64 clones, 34 were susceptible to infection, indicating that they expressed CCR5. Susceptible clones were then analyzed for cell surface expression of CCR5 by immunofluorescence microscopy. As shown in Fig. 1, the level of CCR5 appeared to be uniform on different cells of each clonal population. However, the clones expressed distinct quantities of CCR5, as illustrated by the difference in fluorescence intensity exhibited by clones JC.53 and JC.48. As shown in Fig. 1, the CCR5 became clustered into patches when the antibody bound to the viable cells at 37°C. In contrast, uniform staining occurred when the cells were fixed with paraformaldehyde before incubation with the antibody (results not shown). These results suggest that CCR5 is able to diffuse in the membranes and that it can be cross-linked by the antibody. Cell surface CCR5 expression levels were quantitatively measured in HeLa-CD4/CCR5 clones by the rabbit anti-CCR5-[125I]protein A binding assay. This method, which had a very low background in control HeLa-CD4 cells that lack CCR5, confirmed that the amounts of CCR5 were reproducibly widely different in different HeLa-CD4/CCR5 cell clones (Table 1). Previous evidence using other antigens indicated that this antibody-[125I]protein A method yields results that closely correlate with other quantitative measurements of relative expression levels on cell surfaces (33, 40). In addition to the HI-J panel of HeLa-CD4/CCR5 clones, a panel was created by transducing the HI-R clone of HeLa-CD4 cells that contains a low amount of CD4 (approximately 104 molecules/cell [33]) with the retroviral vector SFF-CCR5. Of 62 clones that were tested, 22 were susceptible to infection by the Ba-L M-tropic HIV-1 isolate.
FIG. 1.
Detection of CCR5 cell surface expression on viable cells by immunofluorescence microscopy. The left and right hand panels depict fluorescence and phase-contrast imaging, respectively. Representative CCR5-expressing cell clones stained with rabbit anti-CCR5 show strong fluorescence, while the parent cell line, HI-J, shows only weak reactivity. Similarly, the high-CCR5-expressing clone JC.53 shows only weak staining with preimmune serum. The two representative clones shown (JC.53 and JC.48) depict the uniformity of CCR5 expression level within a clonal population, as well as differences in CCR5 expression levels among cell clones.
TABLE 1.
CCR5-expressing clones derived from HI-J and HI-R HeLa CD4 cells
| Clonea | Mean rabbit anti-CCR5 binding (cpm/μg of protein) ± SDb | CCR5 expression (molecules/cell)c |
|---|---|---|
| HI-J control | 3 ± 1 | 0 |
| JC.20 | 4 ± 1 | <7.0 × 102 |
| JC.10 | 6 ± 1 | 2.0 × 103 |
| JC.57 | 17 ± 2 | 9.2 × 103 |
| JC.58 | 23 ± 8 | 1.3 × 104 |
| JC.37 | 26 ± 4 | 1.5 × 104 |
| JC.55 | 33 ± 3 | 2.0 × 104 |
| JC.47 | 34 ± 9 | 2.0 × 104 |
| JC.6 | 44 ± 18 | 2.7 × 104 |
| JC.54 | 49 ± 10 | 3.0 × 104 |
| JC.48 | 79 ± 9 | 5.0 × 104 |
| JC.24 | 141 ± 22 | 9.1 × 104 |
| JC.53 | 201 ± 27 | 1.3 × 105 |
| HI-R control | 3 ± 0.1 | 0 |
| RC.30 | 6 ± 2 | 2.4 × 103 |
| RC.33 | 7 ± 1 | 3.2 × 103 |
| RC.6 | 7 ± 0.4 | 3.2 × 103 |
| RC.15 | 8 ± 1 | 4.8 × 103 |
| RC.10 | 11 ± 3 | 7.1 × 103 |
| RC.56 | 11 ± 1 | 7.9 × 103 |
| RC.23 | 13 ± 3 | 8.7 × 103 |
| RC.4 | 13 ± 1 | 9.5 × 103 |
| RC.55 | 25 ± 4 | 2.1 × 104 |
| RC.28 | 39 ± 9 | 3.3 × 104 |
| RC.12 | 57 ± 10 | 4.3 × 104 |
| RC.17 | 62 ± 16 | 5.6 × 104 |
| RC.25 | 85 ± 15 | 7.8 × 104 |
| RC.49 | 92 ± 16 | 8.5 × 104 |
Cells stably expressing CCR5 were generated by retroviral transduction of HeLa-CD4 clones HI-J and HI-R. See Materials and Methods for further details.
CCR5 expression levels on cell surfaces were measured for all clones by binding rabbit antiserum to CCR5 followed by [125I]protein A as described in Materials and Methods. Protein content per sample equaled approximately 50 μg. Values represent the means of three or four assays (JC and RC clones, respectively), with each assay performed in duplicate. HI-R cells were assayed twice; the range is displayed. The RC and JC clones were analyzed at different times using different batches of [125I]protein A and different bleeds of rabbit anti-CCR5. Multiple cultures of the JC.53 clone assayed simultaneously with RC clones yielded an average value of 167 cpm/μg of protein, and the results for the RC clones were normalized relative to this internal JC.53 standard. A conversion factor of 1.2 (representing the ratio of the two anti-CCR5-specific binding values on JC.53 cells obtained while assaying JC [198 cpm/μg of protein] and RC clones [164 cpm/μg of protein]) was used to adjust RC clone cell surface CCR5 values to the JC scale. The number of CCR5 molecules per cell for the JC.53 cells did not change during this time period (see Fig. 2 and Results).
The JC.53 CCR5 expression level, determined by titration with [125I]MIP1β, was approximately 1.3 × 105 molecules per cell (see Fig. 2 and Results). The cell surface CCR5 values shown were calculated by subtracting control anti-CCR5 binding values from the binding value for each clone, dividing this difference by the comparable difference obtained with clone JC.53, and then multiplying the ratio by 1.3 × 105 CCR5 molecules per cell (see Fig. 2 and results).
To quantitatively determine the number of accessible, cell surface MIP1β binding sites, we performed competitive binding assays in which [125I]MIP1β was displaced by increasing concentrations of unlabeled MIP1β, followed by Scatchard analysis of the binding data. Two independent competition assays were performed, with triplicate measurements of each data point. Figure 2 shows results of a representative analysis in which KD and MIP1β binding sites per cell of 17 nM and 1.45 × 105, respectively, were obtained. A second independent analysis gave a KD value of 22 nM and 1.2 × 105 MIP1β binding sites per cell (data not shown). We used the average number of MIP1β binding sites, 1.3 × 105 per cell, to calculate the approximate levels of CCR5 expression for the other cell clones (Table 1). The expression levels that were significantly above background spanned a broad range between approximately 2.0 × 103 and 1.3 × 105 CCR5 molecules per cell for the JC clones (Table 1) and 2.4 × 103 and 8.5 × 104 molecules/cell for the RC clones. One clone, JC.20, expressed virtually undetectable levels of CCR5, using the immunological means described above (implying fewer than 7 × 102 molecules per cell). However, reverse transcriptase PCR analysis demonstrated that these cells contain CCR5 mRNA (data not shown), suggesting that they had been transduced with SFF-CCR5 and that they expressed the CCR5 protein at a very low level.
FIG. 2.
Scatchard and competitive binding analysis of [125I]MIP1β onto HeLa-CD4/CCR5 clone JC.53. At the time of assay, the cells had grown to approximately 7 × 104/well. Specific [125I]MIP1β binding in counts/minute was calculated by subtracting the nonspecific binding (approximately 300 cpm) of [125I]MIP1β measured in the presence of 1 μM unlabeled MIP1β. Background binding of [125I]MIP1β, measured on HI-J cells lacking CCR5, gave binding values of approximately 300 cpm at all concentrations of unlabeled MIP1β. A Scatchard analysis of the binding data (inset) was performed to determine the number of CCR5 molecules/cell. The x intercept yields the number of accessible MIP1β binding sites/cell and equals approximately 1.45 × 105 CCR5 molecules/cell. The competition curve yields an 50% inhibitory concentration value of 15 nM, in close agreement with a KD of 17 nM which was obtained when the same binding data were used to generate a Scatchard plot with the x axis presented in molar concentration. Data points represent the means of triplicate assays.
CCR5 expression level requirements for infection by M-tropic HIV-1 in cells expressing large amounts of CD4.
Figure 3 shows representative examples of the foci of immunoperoxidase-stained p24 antigen occurring in the HeLa-CD4/CCR5 cell cultures after infection with M-tropic HIV-1 clone 81A (68) (Fig. 3A), JR-CSF (Fig. 3B), and an isolate derived from an acutely infected patient (Fig. 3C). As seen previously in infected macrophages (14, 68) and consistent with recent independent evidence (72), HeLa-CD4/CCR5 cells infected with M-tropic HIV-1 formed large numbers of syncytia. The sizes of syncytia depended on the virus isolates.
FIG. 3.

Foci of infection of HeLa-CD4/CCR5 cells by M-tropic HIV-1 clones or isolates. HIV-1 p24 antigen in infected cells was detected by indirect immunoperoxidase staining. p24-positive foci consisted of single or grouped multinucleated giant cells, with occasional adjacent mononuclear cells. (A) M-tropic HIV-1 clone 81A, containing the V1, V2, and V3 regions of the env gene of clone Ba-L in the background of clone NL4-3 (68). One small syncytium with four nuclei and three large syncytia containing greater than 20 nuclei are shown. (B) Field showing 10 typical foci of cells infected by M-tropic clone JR-CSF containing 1 to 10 nuclei each. (C) Numerous small foci of cells infected by an HIV-1 isolate obtained from a patient on day 7 of illness due to primary HIV-1 infection.
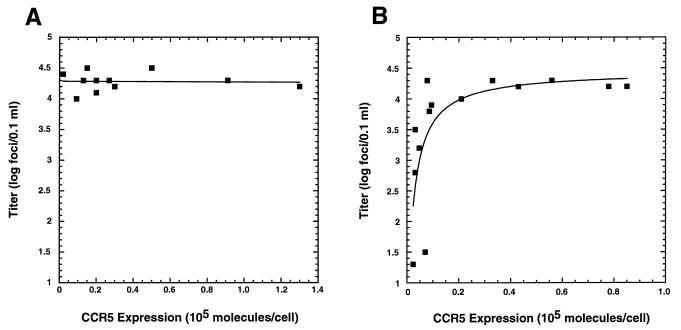
HIV-1 susceptibilities of selected JC clones expressing distinct amounts of CCR5 are depicted in Fig. 4. The T-cell-tropic HIV-1 strain NL4-3 infected HeLa-CD4 cells with or without CCR5 expression with equal efficiencies (Fig. 4). Thus, the untransduced HI-J cells were infected as efficiently as the CCR5-expressing clones. In contrast, M-tropic viruses did not infect HeLa-CD4 cells lacking CCR5; however, they were maximally infectious for HeLa-CD4 clones that had as few as 2.0 × 103 CCR5 molecules per cell (Fig. 4). Increases in CCR5 expression above this level did not significantly enhance infection by any of the tested M-tropic isolates of HIV-1. Additionally, HIV-1 titers on JC cells were equal within experimental error to the TCID50 values measured on PBMCs, illustrating the exquisite sensitivity of the JC clones to HIV-1 infection. As indicated previously, clone JC.20 had only a background level of CCR5 expression, consistent with fewer than 7.0 × 102 CCR5 molecules per cell. With these cells T-cell-tropic HIV-1 was detected at normal levels, but M-tropic HIV-1 gave detectable foci at 300-fold-lower levels than on the other CCR5-positive clones (Fig. 4). Thus, CCR5 expression in JC.20 cells was below the threshold level required for maximum sensitivity to infection by M-tropic HIV-1 strains.
FIG. 4.
Infectivities of three HIV-1 strains in JC clones and PBMCs. M-tropic HIV-1 strain JR-CSF and Ba-L and T-tropic HIV-1 NL4-3 were used to infect nine HeLa-CD4/CCR5 JC clones with differing levels of CCR5 expression, as well as the CCR5-negative CD4-positive parental clone HI-J and PBMCs. JC clones with CCR5 expression ranging from 2 × 103 to 1.3 × 105 molecules/cell gave similar titers for the two M-tropic strains. In contrast, HI-J was negative and clone JC.20, with background CCR5 expression (<7 × 102 molecules/cell), had markedly reduced titers. Although M-tropic HIV-1 did not infect HI-J cells, titers are indicated as <5 to account for the virus dilutions used in the assays. Titers in PBMCs expressed as TCID50/0.05 ml were numerically similar to titers (foci/0.05 ml) seen on the highly sensitive JC clones.
Effect of low CD4 expression on CCR5 concentration requirements for infection by M-tropic HIV-1.
The results presented above indicate that HeLa-CD4 cells with a large amount of CD4 and trace amounts of CCR5 are maximally sensitive to infection by M-tropic HIV-1. We next wished to assess the CCR5 expression level requirements for HIV-1 infection in cells that expressed a low amount of CD4. Such cells may be more representative of HIV-1 target cells in vivo. For that purpose, we used the RC panel of HeLa-CD4/CCR5 cells that contained approximately 104 CD4 molecules/cell (33). In contrast to results obtained with the JC panel (Fig. 4 and 5A), titers of M-tropic HIV-1 on the RC panel were reproducibly dependent on CCR5 expression levels below a threshold of approximately 1.0 × 104 to 2.0 × 104 molecules per cell (Fig. 5B). RC clones expressing amounts of CCR5 above this threshold level were maximally susceptible to infection by M-tropic HIV-1 (Fig. 5B). Similar results were obtained with the M-tropic HIV-1 isolates SF162, JR-FL, and ADA (data not shown). Thus, CD4 expression levels influence the amount of cell surface CCR5 that is required for efficient infections by M-tropic HIV-1. Additionally, the maximal viral titers were identical in the JC and RC panels (i.e., 1.0 × 104 to 3.0 × 104 foci/0.1 ml in Fig. 5), suggesting that the differences in CD4 quantities did not influence efficiencies of infection by M-tropic HIV-1 when the CCR5 quantities were substantial. In contrast, when levels of CCR5 were below 1.0 × 104 to 2.0 × 104/cell, the infectivity titers were dependent on the quantities of CD4 expressed (compare Fig. 5A and B).
FIG. 5.
M-tropic HIV-1 infectivity on JC (high CD4) and RC (low CD4) clonal panels. Infectivity assays were performed as described in Materials and Methods on the 11 JC clones (A) and the 14 RC clones (B) whose CCR5 expression was above background (Table 1). Approximately 500 focus-forming units of diluted HIV-1 Ba-L was added per well (1 cm2), in 0.1 ml. Titers were calculated by multiplying foci/well by the dilution factor at which the virus was used. Points represent the mean of duplicate assays.
Previously, we found that infections of primary T-cell-tropic HIV-1 are directly dependent on CD4 cell surface concentrations, whereas laboratory-adapted T-cell-tropic viruses can efficiently infect cells independently of CD4 levels (33, 39). The results in Fig. 5 suggested that M-tropic titers may be independent of CD4 expression levels when the CCR5 concentrations are high. We tested this by directly comparing M-tropic, primary T-cell-tropic, and laboratory-adapted T-cell-tropic HIV-1 infectivities on JC.24 and RC.49 clones. CCR5 levels in these cell lines allowed maximal M-tropic HIV-1 infection, as demonstrated in Fig. 5. As shown in Fig. 6, the M-tropic isolates SF162, JR-FL, Ba-L, and ADA all infected JC.24 and RC.49 clones with equal efficiencies, whereas these viruses were unable to infect HI-J and HI-R clones of HeLa-CD4 cells that lack CCR5. In contrast, the primary T-cell-tropic isolates ELI1 and K4 showed a marked decrease in titer on RC.49 cells relative to JC.24 cells (approximately 8- and 13-fold decreases for ELI1 and K4, respectively). As expected, these T-cell-tropic primary virus isolates could also infect the HI-J and HI-R cells that lacked CCR5, and the titers of ELI1 and K4 were 12- and 24-fold higher, respectively, on HI-J compared to HI-R cells. Consistent with our previous results (33, 39), the laboratory-adapted T-cell-tropic virus NL4-3 infected the cells in Fig. 6 regardless of CCR5 expression and independently of the differences in CD4 expression.
FIG. 6.
Infectivities of M-tropic and T-cell-tropic HIV-1 isolates on HeLa-CD4/CCR5 cells. Infectivity assays were performed as described in Materials and Methods. M-tropic (SF162, JR-FL, Ba-L, and ADA), primary T-cell-tropic (ELI 1 and K4), and laboratory-adapted T-cell-tropic (NL4-3) titers were determined on clones RC.49 and JC.24, expressing low CD4/high CCR5 and high CD4/high CCR5 cell surface amounts, respectively, and on parental HI-R and HI-J clones, expressing low and high amounts of CD4, respectively. Titers are expressed as percent titer obtained on JC.24 cells. Approximately 500 focus-forming units of diluted HIV-1 isolates were added per well, in 0.1 ml. Assays were performed in duplicate except for SF162 and JR-FL infections of JC.24, which are presented as single assays. Error bars represent the range of values obtained.
Infectivity of HIV-1 patient isolates on JC.37 cells.
The results presented above suggest that cell surface concentrations of CCR5 and CD4 can determine the efficiencies of HIV-1 infections. Limitations in HIV-1 patient isolate infection can occur due to insufficient CD4 expression or to inadequate CCR5 expression in a low CD4 context. To learn whether our HeLa-CD4/CCR5 cell clones that have optimal amounts of CD4 and CCR5 might serve as a sensitive assay system for patient HIV-1 isolates, we tested JC.37 cells for their susceptibilities to infection by numerous primary patient clade B HIV-1 isolates. With all viruses, abundant foci of infection were detected. Representative results are shown in Table 2. Several of the isolates from patients late in disease could also infect HeLa-CD4 cells that lack CCR5, as expected for T-cell-tropic or dualtropic viruses. For all HIV-1 viruses that we examined, the titers on the JC.37 cells were as high as the TCID50 values measured in PBMCs (results not shown). Our JC clones have been grown for many months without apparent changes in their properties.
TABLE 2.
Susceptibility of HeLa-CD4/CCR5 cells (clone JC.37) to patient isolates of HIV-1
| Patient groupa | Patient no. | HIV titer (foci/0.1 ml of virus stock)b
|
|
|---|---|---|---|
| JC.37 cells (CCR5+) | HI-J cells (CCR5−) | ||
| Plasma isolates from AIDS patients | |||
| with advanced disease | 1 | 3.4 × 103 | <40 |
| 2 | 6.0 × 103 | <40 | |
| 3 | 4.6 × 103 | <40 | |
| 4 | 3.0 × 103 | <40 | |
| 5 | 7.2 × 102 | 1.2 × 102 | |
| 7 | 1.4 × 103 | 2.4 × 102 | |
| 8 | 3.6 × 103 | <40 | |
| 9 | 4.0 × 103 | <40 | |
| Blood isolates from AIDS patients | |||
| with advanced disease | 11 | 2.0 × 103 | <40 |
| 12 | 8.0 × 103 | <40 | |
| 13 | 2.0 × 103 | <40 | |
| 14 | 7.4 × 103 | 1.3 × 103 | |
| 15 | 1.0 × 103 | <40 | |
| 16 | 7.6 × 102 | <40 | |
| 17 | 6.2 × 103 | 1.4 × 103 | |
| 18 | 1.0 × 104 | <40 | |
| 19 | 2.8 × 104 | <40 | |
| Blood isolates from acute HIV | |||
| infection (days 7–47 of illness) | 21 | 2.0 × 103 | <40 |
| 22 | 6.2 × 103 | <40 | |
| 23 | 5.6 × 103 | <40 | |
| 24 | 6.6 × 103 | <40 | |
| 25 | 8.0 × 103 | <40 | |
| 26 | 1.7 × 103 | <40 | |
| 28 | 7.6 × 103 | <40 | |
| 29 | 7.2 × 102 | <40 | |
Isolates were passaged once on PBMCs after original isolation and stocks produced were titered on CD4-positive HeLa cells expressing or lacking the CCR5 coreceptor.
Most of the primary patient isolates cause no foci on the CCR5-negative HI-J cells. The titers in these cases are given as <40 to account for the virus sample dilutions used in these assays.
DISCUSSION
HeLa-CD4/CCR5 clonal panels with diverse quantities of CD4 and CCR5.
These HeLa-CD4/CCR5 cell clones appear to provide a sensitive and stable system for quantitatively analyzing infectivities by diverse M-tropic and T-cell-tropic isolates of HIV-1, including primary patient isolates. The ability to quantitatively compare the titers of viruses of distinct tropisms in a common cellular background is a major advantage and may be especially useful for drug testing and for neutralization testing using patient sera. Viruses grown and assayed in different cells are difficult to compare because HIV-1 promoters and accessory genes often function in a cell-specific manner (42, 73) and because cellular factors can influence the compositions and properties of the virions that are produced (47, 61, 70). Furthermore, changes in the cells or their plating efficiencies can perturb the comparisons. Focal infectivity assays are also advantageous compared with infectivity assays that depend on multiple cycles of replication and spread within a culture. HeLa-CD4/CCR5 cell clones grow as adherent monolayers, and their foci of infection are clear and easy to quantitate simply by staining for viral proteins (Fig. 3). Because many of our cell clones contain substantial amounts of both CD4 and CCR5, they are highly susceptible to all primary patient HIV-1 isolates that we have tested (Table 2 and Fig. 6). Consistent with other recent evidence (72), M-tropic HIV-1 isolates form abundant syncytia in our HeLa-CD4/CCR5 cell clones (Fig. 3), although they are nonsyncytium inducing in CD4-positive T cells. Therefore, the ability of M-tropic HIV-1 and CCR5 to participate in syncytium formation must depend on additional factors that vary in different target cells (8).
Other cell lines made using HeLa-MAGI and human osteosarcoma (HOS) cells that express CD4 and coreceptors appear to also be useful for titering diverse HIV-1 isolates, including HIV-1 isolates of different clades (10, 11, 19, 34, 72). Chackerian et al. found that their HeLa-MAGI-CCR5 indicator cell lines, similar to the HeLa-CD4/CCR5 clones described here, were as sensitive to M-tropic HIV-1 infection as PBMCs (10). In contrast to our investigation, the HeLa-MAGI cells used in one of the above-cited investigations and HOS-CD4 coreceptor expressing cells were pooled populations of drug-resistant colonies rather than pure clones (19, 72). Nonclonality may have contributed to the instability of CCR5 expression observed by Vodicka et al. in their HeLa-MAGI/CCR5 cells (72). For the HeLa-MAGI and HOS cell populations, the absolute numbers of CD4 and coreceptors expressed on the cell surfaces are not known, in contrast to the HeLa-CD4/CCR5 clonal panels described here.
Interaction of CD4 and coreceptors in HIV-1 infections.
Our results suggest that the quantity of CCR5 required for maximal infection of HeLa-CD4/CCR5 cells by M-tropic isolates of HIV-1 is a function of the CD4 expression level on the cells (Fig. 4 and 5). In the HI-J clonal panel that contains a substantial quantity of CD4 (approximately 4.5 × 105 CD4/cell), a surprisingly low trace amount of CCR5 estimated to be between 700 and 2 × 103 CCR5/cell suffices for maximal infectivity. In contrast, the HI-R clonal panel contains a much smaller quantity of CD4 (ca. 104 CD4/cell), and in this case a correspondingly increased amount of 1.0 × 104 to 2.0 × 104 CCR5/cell is necessary for maximal infectivity by M-tropic HIV-1. In both panels, however, the maximum levels of infectivity were identical to each other and also to the TCID50 values measured using the same virus preparations in human peripheral blood mononuclear cultures (Fig. 4 and 5). Thus, a high concentration of either CD4 or CCR5 can compensate for a low concentration of the other component, resulting in efficient infection of the cell. These results strongly suggest that CD4 and CCR5 must directly or indirectly interact in a concentration-dependent manner within a pathway that is essential for infection by M-tropic HIV-1. Because the efficiency of this pathway depends on CD4 and CCR5 concentrations but not on the precise amounts of either component, we infer that it behaves as expected for a mass action process. Accordingly, we propose that it may depend on diffusion of these components in the membrane and on the frequencies of their spontaneous interactions to form virus-CD4-CCR5 ternary complexes.
Previously, we reported that M-tropic isolates of HIV-1 can efficiently infect cells that have either low or high concentrations of CD4 on their surfaces (39). However, our new results indicate that this is correct only for cells that have sufficient CCR5 (Fig. 5 and 6). Specifically, when the CCR5 cell surface concentrations are low, the infectivities of M-tropic HIV-1 isolates are highly dependent on the CD4 concentrations (compare Fig. 5A and B). Thus, both the CD4 and CCR5 concentration requirements for M-tropic HIV-1 infections are interdependent.
It is intriguing that in HeLa-CD4/CCR5 cells with large amounts of CD4, the infectivities of M-tropic isolates of HIV-1 are independent of the levels of CCR5 expression above a low trace quantity. As reported earlier, this limitation in the maximum titer is not caused by depletion of infectious virus from the medium (33, 57). Previous studies have suggested that adsorption of HIV-1 onto cell surfaces generally requires CD4 but does not require the presence of a functional coreceptor. For example, HIV-1 binds onto mouse fibroblasts that express human CD4 (48). Moreover, gp120 binding to coreceptors is dramatically enhanced by CD4, indicating that CD4 induces a conformational change in gp120 that exposes the coreceptor binding site (30, 69, 74). Accordingly, our results imply that M-tropic HIV-1 that initially binds onto cell surface CD4 can efficiently infect cells that contain a large excess of CD4 and only relatively few CCR5 molecules. Specifically, the clones of our HI-J panel contain approximately 4 × 105 CD4/cell (38). This number is several hundred times in excess of the CCR5 molecules present on surfaces of JC.10 cells (2 × 103 CCR5/cell) that are efficiently infected by M-tropic isolates of HIV-1. Based on this stoichiometry, it is evident that the initial virus-CD4 complexes would not necessarily be in proximity to a CCR5 molecule, implying that CCR5 must enter these complexes secondarily. This strongly suggests that membrane fluidity is important for CCR5 coreceptor function, in agreement with our proposal that HIV-1 infections depend on a mass action process that involves diffusion of CD4 and/or CCR5 in the membranes.
We believe that our results would be compatible with two models for CD4 and CCR5 function. One model is that the efficiency of HIV-1 adsorption onto cell surfaces may depend on both receptor and coreceptor concentrations and that a low concentration of either component can be compensated for by an increase in the other. According to physical chemical evidence, a virus that diffuses into contact with a cell will efficiently contact receptors even when the receptor concentration is very low (3, 64). However, this will result in irreversible adsorption only if the virus-CD4 bond is strong or if secondary receptors or coreceptors diffuse into the site to augment the adhesion. Therefore, if the initial virus-CD4 bond is weak, a high concentration of either CD4 or CCR5 could enhance the efficiency of HIV-1 adsorption. According to the second model, the initial monovalent virus-CD4 bond may be strong enough to ensure efficient adhesion, but the postbinding steps may be inefficient when both CD4 and CCR5 concentrations are low. Presumably, a cell with a low concentration of CD4 would contain predominately monovalent virus-CD4 complexes, whereas a cell with abundant CD4 would develop a cluster of CD4 that interacts with the virus. Because CCR5 can bind effectively only to gp120 molecules that are in contact with CD4 (30, 69, 74), CCR5 would bind to a multivalent virus-CD4 complex much more strongly than to a monovalent virus-CD4 complex. Consequently, a low concentration of CCR5 would suffice for efficient infection only if the CD4 concentration were substantial. Conversely, a high concentration of CCR5 would be necessary when the CD4 concentration was low. In agreement with these models, previous evidence also suggests that multivalent virus-CD4 complexes can contribute to HIV-1 infections of specific cells (23, 44, 60).
Relevance to natural HIV-1 infections.
As discussed above, there is substantial indirect evidence that concentrations of CD4 or coreceptors can limit the efficiencies of HIV-1 infections in specific cells and that such limitations may be important in the pathogenesis of AIDS (33, 35, 52, 75). Our results may be particularly relevant to previous studies of the natural cellular targets of HIV-1 which document variations in CD4, CXCR4, and CCR5 expression in T lymphocytes during differentiation and activation (7, 67, 69, 75) and in amounts of CD4 on macrophages in response to lipopolysaccharides, tumor necrosis factor alpha, and interleukin-1β (29). Unfortunately, precise data concerning the concentrations of CD4 and coreceptors on these cells in their normal environments are not available. However, recent evidence has implied that activated CD4-positive human T cells contain an average of approximately 2 × 104 CCR5/cell, high levels of CD4 (ca. 105 to 106/cell), and quantities of CXCR4 that are several times higher than the amounts in HeLa cells (49). This could possibly explain the sensitivity of these cells to infections by primary patient and laboratory-adapted T-cell-tropic and M-tropic isolates of HIV-1. In contrast, macrophage susceptibility to HIV-1 infections may be especially sensitive to changes in coreceptor expression because of the low CD4 expression in these cells (29, 54). Although macrophages contain detectable CXCR4 as well as CCR5, they are generally considered to be relatively resistant to T-cell-tropic isolates of HIV-1 (50). However, this resistance appears to depend on the differentiation state of the cells and on the culture conditions (66). Additional studies will be required to more thoroughly evaluate these issues.
ACKNOWLEDGMENTS
This research was supported by NIH grant CA67358 (to D.K.). E.J.P. was supported by NRSA postdoctoral fellowship IF32AID9735 from the NIH.
The following isolates of HIV-1 were provided by the NIH AIDS Research and Reference Reagent Program: macrophagetropic isolates JR-FL, SF162, Ba-L, and ADA, contributed by Irvin Chen, by Jay Levy, by Suzanne Gartner, Mikulas Popovic, and Robert Gallo, and by Howard Gendelman, respectively. We are grateful to our coworkers and colleagues Susan Kozak, Navid Madani, and Chetankumar Tailor for encouragement and advice and to Jay Nelson for donating several primary patient HIV-1 isolates. We thank Ali Nouri for performing the reverse transcriptase PCR analysis. Special thanks are due to Seth Pincus (University of Montana), who independently corroborated several of our results with these HeLa-CD4/CCR5 cell clones and informed us of his results.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Berg H C, Purcell E M. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bestwick R K, Kozak S L, Kabat D. Overcoming interference to retroviral superinfection results in amplified expression and transmission of cloned genes. Proc Natl Acad Sci USA. 1988;85:5404–5408. doi: 10.1073/pnas.85.15.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 7.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broder C C, Berger E A. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll R G, Riley J L, Levine B L, Feng Y, Kaushal S, Ritchey D W, Bernstein W, Weislow O S, Brown C R, Berger E A, June C H, St. Louis D C. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 10.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesebro B, Wehrly K, Metcalf J, Griffin D E. Use of a new CD4-positive HeLa cell clone for direct quantitation of infectious human immunodeficiency virus from blood cells of AIDS patients. J Infect Dis. 1991;163:64–70. doi: 10.1093/infdis/163.1.64. [DOI] [PubMed] [Google Scholar]
- 14.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 16.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 19.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P D, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 20.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Earl P L, Doms R W, Moss B. Multimeric CD4 binding exhibited by human and simian immunodeficiency virus envelope protein dimers. J Virol. 1992;66:5610–5614. doi: 10.1128/jvi.66.9.5610-5614.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1beta-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 26.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 27.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 28.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C L, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herbein G, Doyle A G, Montaner L J, Gordon S. Lipopolysaccharide (LPS) down-regulates CD4 expression in primary human macrophages through induction of endogenous tumour necrosis factor (TNF) and IL-1 beta. Clin Exp Immunol. 1995;102:430–437. doi: 10.1111/j.1365-2249.1995.tb03801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horuk R, Huang J J, Covington M, Newton R C. A biochemical and kinetic analysis of the interleukin-1 receptor. Evidence for differences in molecular properties of IL-1 receptors. J Biol Chem. 1987;262:16275–16278. [PubMed] [Google Scholar]
- 32.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 33.Kabat D, Kozak S L, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitchen S G, Zack J A. CXCR4 expression during lymphopoiesis: implications for human immunodeficiency virus type 1 infection of the thymus. J Virol. 1997;71:6928–6934. doi: 10.1128/jvi.71.9.6928-6934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 37.Kozak S L, Kabat D. Ping-pong amplification of a retroviral vector achieves high-level gene expression: human growth hormone production. J Virol. 1990;64:3500–3508. doi: 10.1128/jvi.64.7.3500-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak, S. L., and D. Kabat. Unpublished data.
- 39.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak S L, Siess D C, Kavanaugh M P, Miller A D, Kabat D. The envelope glycoprotein of an amphotropic murine retrovirus binds specifically to the cellular receptor/phosphate transporter of susceptible species. J Virol. 1995;69:3433–3440. doi: 10.1128/jvi.69.6.3433-3440.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacoste J, D’Addario M, Roulston A, Wainberg M A, Hiscott J. Cell-specific differences in activation of NF-κ B regulatory elements of human immunodeficiency virus and beta interferon promoters by tumor necrosis factor. J Virol. 1990;64:4726–4734. doi: 10.1128/jvi.64.10.4726-4734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 44.Layne S P, Merges M J, Dembo M, Spouge J L, Nara P L. HIV requires multiple gp120 molecules for CD4-mediated infection. Nature. 1990;346:277–279. doi: 10.1038/346277a0. [DOI] [PubMed] [Google Scholar]
- 45.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 46.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luban J. Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell. 1996;87:279–282. doi: 10.1016/s0092-8674(00)81811-5. [DOI] [PubMed] [Google Scholar]
- 48.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 49.McKeating, J. Personal communication.
- 50.McKnight A, Wilkinson D, Simmons G, Talbot S, Picard L, Ahuja M, Marsh M, Hoxie J A, Clapham P R. Inhibition of human immunodeficiency virus fusion by a monoclonal antibody to a coreceptor (CXCR4) is both cell type and virus strain dependent. J Virol. 1997;71:1692–1696. doi: 10.1128/jvi.71.2.1692-1696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 52.Moore J P. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276:51–52. doi: 10.1126/science.276.5309.51. [DOI] [PubMed] [Google Scholar]
- 53.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 54.Moscicki R A, Amento E P, Krane S M, Kurnick J T, Colvin R B. Modulation of surface antigens of a human monocyte cell line, U937, during incubation with T lymphocyte-conditioned medium: detection of T4 antigen and its presence on normal blood monocytes. J Immunol. 1983;131:743–748. [PubMed] [Google Scholar]
- 55.Murphy P M. The molecular biology of leukocyte chemokine receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 56.Pincus S H, Wehrly K, Chesebro B. Use of a focal infectivity assay for testing susceptibility of HIV to antiviral agents. BioTechniques. 1991;10:336–342. [PubMed] [Google Scholar]
- 57.Platt E J, Madani N, Kozak S L, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H H, Du J G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 60.Rieber E P, Federle C, Reiter C, Krauss S, Gurtler L, Eberle J, Deinhardt F, Riethmuller G. The monoclonal CD4 antibody M-T413 inhibits cellular infection with human immunodeficiency virus after viral attachment to the cell membrane: an approach to postexposure prophylaxis. Proc Natl Acad Sci USA. 1992;89:10792–10796. doi: 10.1073/pnas.89.22.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakai H, Shibata R, Sakuragi J, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 63.Schnittman S M, Lane H C, Greenhouse J, Justement J S, Baseler M, Fauci A S. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci USA. 1990;87:6058–6062. doi: 10.1073/pnas.87.16.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz M. The adsorption of coliphage lambda to its host: effect of variations in the surface density of receptor and in phage-receptor affinity. J Mol Biol. 1976;103:521–536. doi: 10.1016/0022-2836(76)90215-1. [DOI] [PubMed] [Google Scholar]
- 65.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 66.Stent G, Joo G B, Kierulf P, Åsjö B. Macrophage tropism: fact or fiction? J Leukocyte Biol. 1997;62:4–11. doi: 10.1002/jlb.62.1.4. [DOI] [PubMed] [Google Scholar]
- 67.Terstappen L W, Huang S, Picker L J. Flow cytometric assessment of human T-cell differentiation in thymus and bone marrow. Blood. 1992;79:666–677. [PubMed] [Google Scholar]
- 68.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 69.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 70.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 71.Vanham G, Kestens L, De Meester I, Vingerhoets J, Penne G, Vanhoof G, Scharpe S, Heyligen H, Bosmans E, Ceuppens J L, et al. Decreased expression of the memory marker CD26 on both CD4+ and CD8+ T lymphocytes of HIV-infected subjects. J Acquired Immune Defic Syndr. 1993;6:749–757. [PubMed] [Google Scholar]
- 72.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 73.Winslow B J, Trono D. The blocks to human immunodeficiency virus type 1 Tat and Rev functions in mouse cell lines are independent. J Virol. 1993;67:2349–2354. doi: 10.1128/jvi.67.4.2349-2354.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 75.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]