Graphical abstract
Keywords: Kalanchoe pinnata, Diabetes mellitus, Enzyme inhibition, Molecular docking, Molecular dynamic and simulation, Network pharmacology
Highlights
-
•
Potent inhibitory activity of alpha-amylase and alpha-glucosidase by K. pinnata was evidenced in dose dependent manner.
-
•
Network pharmacology highlights the significant edge count for kaempferol and luteolin considering their prominence as potential therapeutic agents.
-
•
MD confirms binding affinity of ligands with AMY2A, GAA, AR and DPP4.
-
•
MDS confirmed stability and structural configuration of AMY2A with friedelin and acarbose.
Abstract
Since ancient times, bioactive phytocompounds from different parts of medicinal plants have been used to heal various disease ailments and they are now regarded as a valuable source of disease prevention globally. Kalanchoe pinnata is a member of the Crassulaceae family; it has a long history of usage in traditional ayurvedic treatment. Analysis of bioactive compounds for their potential anti-type-2 diabetes mellitus (T2DM) mechanism along with in-vitro and in-silico approaches was studied in the present research. The alpha-amylase and alpha-glucosidase inhibitory activity of methanolic extract of Kalanchoe pinnata (α-amylase: IC50 29.50 ± 0.04 μg/ml; α-glucosidase IC50 32.04 ± 0.35 μg/ml) exhibit a high degree of similarity to the standard drug acarbose (IC50 35.82 ± 0.14 μg/ml). Different biological databases were used to list phytocompounds from the plant, and ADME analysis using swissADME was carried out to screen compounds that obeyed the Lipinski rule of 5 and were employed further. STRING and KEGG pathway analysis was performed for gene enrichment analysis followed by network pharmacology to identify key target proteins involved in DM. AMY2A, NOX4, RPS6KA3, ADRA2A, CHRM5, and IL2 were identified as core targets for luteolin, kaempferol, alpha amyrin, stigmasterol compounds by modulating neuroactive ligand interaction, P13-AKT, MAPK, and PPAR signaling pathways. Molecular docking was performed to study the binding affinity among bioactive compounds of K. pinnata against aldose reductase, alpha-amylase, alpha-glucosidase, and dipeptidyl peptidase IV. Alpha-amylase-friedelin [FRI] and alpha-amylase-acarbose [STD] complexes were subjected to molecular simulation for a 200 ns duration that depicted the stability of the compounds and proteins. In the current study, employing dual approach in-silico and in-vitro enzyme assays has yielded a comprehensive and strong understanding of its potential therapeutic properties, making a significant step towards the development of novel anti-diabetic treatment.
1. Introduction
Diabetes mellitus (DM) is one of the most prevalent metabolic conditions globally, which develops predominantly as a result of a confluence of two key factors: insufficient synthesis of insulin by pancreatic cells and when insulin-sensitive tissues show resistance to insulin, these abnormalities lead to metabolic imbalance result in the pathophysiology of type 2 diabetes mellitus (T2DM) (Galicia-Garcia et al., 2020). Gestational diabetes is distinct from either type 1 or T2DM, where glucose intolerance manifests or is first identified during the second or third trimester of pregnancy (Vounzoulaki et al., 2020) these could be the main target for bioactive compounds from medicinal plants most T2DM patients are overweight or obese, and obesity inhibits the body's ability to maintain homeostatic control of systemic glucose which is a major burden on health worldwide (Banday et al., 2020). Managing diabetes mellitus entails lowering blood glucose levels by inhibiting alpha-amylase activity, which slows down the digestion and absorption of carbohydrates in the small intestine, consequently leading to an elevation in blood glucose levels (Abchir et al., 2023a). Presently, acarbose acts as an inhibitor for alpha-amylase by binding to the enzyme's active site, assisting in the regulation of blood glucose levels, this contributes to enhancing glycemic control and reducing the risk of T2DM (Abchir et al., 2023b).
Similar to other regions of the world, Southeast Asia is experiencing a higher prevalence rate; the average age of diabetes patients appears to be ten years younger in Southeast Asian individuals than in those of European descent for the productive age group (Roglic et al., 2016). According to studies, 80 % of people in underdeveloped nations depend on traditional therapies made from plants, and these are the most frequently used type of treatment for several health conditions globally for various diseases (Halayal et al., 2023). India has a long history of using ethno botanical remedies, which focus on healing with medicinal plants and their by-products, there are more than 25,000 potent plant-based medications used in conventional Indian folk medicine (Shubhashree et al., 2020). Numerous investigations have been conducted on indigenous plants used in traditional medicine to discover and innovate pharmaceuticals for treating endemic ailments, as documented in prior research (Yamari et al., 2024). The medicinal plant Kalanchoe pinnata (Bryophyllum pinnatum), also referred to as patherchat or patharkuchi, is a member of the Crassulaceae family, it has been used for a very long time in conventional ayurvedic medicine. The plant can be found in places like India, Hawaii, Australia, and tropical Africa. Stems, leaves, roots, and flowers are only a few of the various plant parts that are employed in medicine (Mehata, 2021). The existence of bioactive molecules in the plant, such as alkaloids, phenols, and flavonoids gives the plant additional properties like antiviral, antibacterial, anti-inflammatory, anticancer, antidiabetic, antilithic, and wound healing (Mehata, 2021). Several of the other chemical components found include fatty acids, triterpenoids, alkaloids, phenolic acids, saponins, tannins, glycosides, and kalanchosides that have been shown to have strong anticancer and antidiabetic potential (Kamboj and Saluja, 2009). Quercetin 3-O-α-L-arabinopyranoside-(1-2)-α-L-rhamnopyranoside, a unique quercetin glycoside a flavonoid that can be used as a chemical marker for K. pinnata (Coutinho et al., 2021). Recent research involves the ongoing identification of new therapeutic targets and medications to treat T2DM that target different proteins implicated in diabetic pathogenesis through the discovery of small molecules as either their inhibitors or activators (Chaudhury et al., 2017). The current investigation utilized the plant leaves due to their potential applications; there are several levels at which considerable anti-diabetic activity needs to be explored. We have performed a comprehensive screening of phytochemicals to determine their bioavailability, pharmacokinetics, and drug-likeness which are pivotal considerations in early-stage drug discovery and their likely interactions with targets that are involved in the onset and pathogenesis of DM (Chtita et al., 2022). In the domain of drug discovery, computational techniques such as molecular docking (MD), molecular dynamics, and simulation (MDS) (Yamari et al., 2023) play a pivotal role in accurately assessing binding affinity between ligands and target proteins, these methods not only produce reliable results but also significantly accelerate the drug development process, ultimately leading to cost and time savings (Abchir et al., 2023a). Further network pharmacology was employed to thoroughly investigate the mechanism of action of pharmaceuticals, facilitating the advancement of novel drugs through a multi-target, multi-component strategy. (Maradesha et al., 2023).
The main objective of this investigation was to identify potential ligands that inhibit alpha-glucosidase (GAA), alpha-amylase (AMY2A), dipeptidyl peptidase IV (DPP-4), and aldose reductase (AR), as well as to identify gene sets and biochemical pathways that are enriched by these ligands with in-vitro validations. However, not many reports are available on systematic in-silico approaches for anti-diabetic activity from K. pinnata.
2. Material and methods
2.1. Plant material collection and extraction
The plant specimen was gathered during its flowering season and placed in the herbarium preservation at Parapp Channappa Jabin Science College, Hubballi, the plant was chosen considering its historical usage, preclinical findings, and inclusion in polyherbal formulations currently accessible in the market. Plants reported by the informants were precisely identified and their scientific names were cross-referenced with those in the Botanical Survey's herbarium. Scientific names of plants provided additional confirmation of the scientific names of plants from NKISBS (National Knowledge and Information System for Biological Species). Fresh plant leaves were picked washed and dried in the shade before being ground. Defatting of the plant leaves for 30 min. with methanol was then followed by a Soxhlet extraction using 300 ml of methanol at 60–80 °C for a period of 4 h. The extraction was preceded by filtration with what man No. 1 filter paper. The filtered extract was then centrifuged at 7000 rpm for 30 min. followed by a rotary evaporator to concentrate the filtrates at 40 °C for 15 min. The yield of crude extract was estimated after the condensed form of the crude extract was collected. The sample was kept at 4 °C and stock solution 100 mg/ml was prepared, Dimethyl sulphoxide (DMSO) was used for further dilutions. Additionally, further different dosages were prepared in phosphate buffer saline (pH 7.4) for an in-vitro investigation.
2.2. Qualitative analysis of K. pinnata methanolic extracts
The phytochemical profiling of the methanolic leaf extract of K.pinnata involved the examination of various bioactive compounds including alkaloids, flavonoids, phenols, glycosides, steroids, terpenoids, proteins and amino acids, tannins, saponins, coumarins, quinines, anthraquinones, gum and mucilage, lipids and carbohydrates was examined using standard approaches to perform the phytochemical profiling of the methanolic leaf extract of K. pinnata (Bagewadi et al., 2019).
2.3. Spectrophotometric determination of total phytochemical compounds
The phytochemical compounds were detected using a UV–VIS spectrophotometer (LABINDIA UV 3000+). The total phenolic content of the methanolic leaf extract of K. pinnata was estimated using the Folin-Ciocalteu method. The total phenolic content was reported as gallic acid equivalents per g of extract (mg GAE/g E), with gallic acid as standard (10–100 µg/ml). The aluminum chloride colorimetric method (Balkan et al., 2018), which identified the total flavonoid concentration and reported quercetin equivalents per g of extract (mg QuE/g E), taking quercetin (5–100 µg/ml) as standard. The bromocresol green reagent (BCG) was used to calculate the total alkaloid content by the method described by (Penumala et al., 2018) where atropine (50–500 µg/ml) was used as a standard to calculate the total alkaloid content, which was expressed as atropine equivalents per g of extract (mg AE/g E).
2.4. In-vitro enzyme inhibition for antidiabetic activity
As per the methodology proposed by (Bagewadi et al., 2019), the inhibitory effect of the extract on α-amylase and α-glucosidase was investigated. Acarbose was employed as a positive control to inhibit α-amylase, enzymatic reaction includes increasing concentrations of extract and equal amounts of pancreatic α-amylase (0.5 mg/ml) and buffered starch (1.0 %) prepared in phosphate buffer pH 7.0, the mixture was first incubated for 10 min. at room temperature, further dinitro salicylic acid (DNS) was added and absorbance was recorded at 405 nm Similar to this, 1.0 ml of the extract and 0.5 ml of the enzyme were used in a reaction to block α-glucosidase, which was carried out at room temperature for 10 min. by mixing in 0.5 ml of p-nitrophenyl-d-glucopyranoside (5.0 mM) and 1 ml of 0.1 M Na2CO3, the mixture was incubated for 10 min. and the reaction was stopped and the absorbance was recorded at 405 nm. Without the extract, two enzyme controls were created and the inhibitory action against amylase and glucosidase was calculated as a percentage of inhibition.
2.5. Identification of compound using chemical databases
The research extensively explored literature sources and compound databases to acquire information on the phytochemicals present in K. pinnata, These databases included PubChem (https://pubchem.ncbi.nlm.nih.gov) (Kim et al., 2016), Indian Medicinal Plants Phytochemistry and Therapeutics (https://cb.imsc.res.in/imppat/) (Mohanraj et al., 2018), Chemical Entities of Biological Interest (https://www.ebi.ac.uk/chebi/) (Shardlow et al., 2018) and KNApSAcK (http://knapsackfamily.com/knapsack_core/top.php)(Afendi et al., 2012). Furthermore, the canonical smiles, molecular weight, and molecular formula of each phytocompound were retrieved from PubChem, providing additional structural information for further analysis and characterization.
2.6. Selection of compounds based on pharmacokinetic properties
To check whether the phytochemicals satisfied the criteria for drug-likeness, drug scans were carried out, a web-based tool called SwissADME (https://www.swissadme.ch/) (Mahanthesh et al., 2020) was used to predict ADME and pharmacokinetic fidelity of drug candidate. Lipinski's rule of five is based on lipophilicity, medicinal chemistry, water solubility, drug-likeness, physicochemical characteristics, gastrointestinal absorption, and drug permeability into the intestine were included in the projected result; hence any molecule that deviated from the threshold values was excluded from further investigation.
2.7. Identification of protein targets
The protein targets linked to bioactive phytocompounds derived from K. pinnata were obtained from the BindingDB database (Liu et al., 2007), which is a publicly accessible repository of measured binding affinities. This database primarily concentrates on the interactions between small drug-like compounds and proteins that are considered potential therapeutic targets, with a probability score of ≥0.7. Additional information regarding the targets, such as gene names and gene IDs, was gathered using UniProtKB https://www.uniprot.org/.
2.8. Protein-protein interactions (PPI) of potential targets
PPI analysis investigates the synchronized activity of proteins within a cell to execute biological functions; it entails an intricate configuration of interdependent proteins formed through biochemical processes. Based on the results of the aforementioned research, the interactions between the proteins represented by the differentially expressed genes were examined by String 12.0 (Szklarczyk et al., 2019).
2.9. Construction of compound-protein target pathway network
Selecting the most promising key signature molecules from a large gene network is a challenging and time-consuming task in this context, to prioritize the genes; we employed biologically-based filtering techniques, taking into account the complexity of protein–protein interactions. As a result, the interactive network was divided into a significant sub-network of DM. we utilized Cytoscape 3.10.1 software (Gustavsen et al., 2019) to construct a network between compounds and target proteins, followed by related pathways that correlate with a specific disease. Each network underwent a topology study, which helped to clarify the network's nature by providing data on the node, edge, degree, betweenness, closeness, and centrality. The shape and color of nodes in a respective network were set up to transition from lesser magnitude to bright colours.
2.10. Gene ontology and KEGG enrichment analysis
DAVID (database for annotation, visualization, and integrated discovery) is a bioinformatics tool engineered to streamline and interpret large scale biological datasets regarding gene set enrichment analysis and functional annotation clustering (https://david.ncifcrf.gov/tools.jsp), where species was set to Homo sapiens and official gene symbol was selected and genes in interesting domains have been selected out for additional functional enrichment analysis. To identify distinguishing biological characteristics, a gene ontology (GO) study was conducted on cellular components, biological processes, and molecular functions. To identify functional attributes, we performed pathway enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2023) with a significant p-value less than 0.05, in DAVID p-value adjusted to FDR values and most enriched components were analyzed graphically.
2.11. Molecular docking
Molecular docking was employed to compute the binding affinity of the protein–ligand complex and to establish initial coordinates for a molecular dynamic simulation. The structure of each ligand was obtained from PubChem in the.SDF file format, further its conversion to.PDB file format was executed employing Discovery Studio (Jejurikar and Rohane, 2021). The present investigation focuses on Aldose reductase (AR), dipeptidyl peptidase IV (DPP4), alpha-amylase (AMY2A), and alpha-glucosidase (GAA) as these four enzymes are potential therapeutic targets for DM, X-ray crystal structure of AR (PDB ID: 3RX2), GAA (PDB ID: 1UOK), DPP4 (PDB ID: 1X70) and AMY2A (PDB ID: 4W93) were all retrieved from protein data bank. The protein preparation module in Discovery Studio 2018 was employed to optimize the crystal structures for docking investigations. Co-factors, hetero-atoms, and water molecules were removed to improve the structure, and the missing atom, hydrogen bond, and charges were computed (Yaraguppi et al., 2022), (Deshpande et al., 2023). Further Autodock tool was used to perform protein–ligand interaction and binding energy was determined. The 3D grid box dimensions of binding area for AR were (20 × 20 × 20) Angstrom, with coordinates of x = 0.169, y = 5.372, z = 19.573, and the 3D grid box dimensions for AMY2A were (20 × 20 × 20) Angstrom, with coordinates of x = −7.204, y = 7.843, z = −17.482, the 3D grid box dimensions for DPP4 were (20 × 20 × 20) Angstrom, with coordinates of x = −27.446, y = 71.447, z = 38.350. The 3D grid box dimensions for GAA were (20 × 20 × 20) Angstrom, with coordinates of x = 37.282, y = 44.791, z = −2.112.
2.12. Molecular dynamics simulations (MDS)
MDS enables the prediction of temporal changes in the atomic mobility of a simulated system under the influence of a force field. This technique is commonly employed as a prodrug tool to investigate the ligand's topology (Revankar et al., 2023). To simulate and predict the dynamics of a protein molecule, the MD characteristic is used to analyze native protein–ligand complexes, the MD system inputs were generated using GROMACS 2020 and AMY2A a protein represented as APO and protein–ligand complexes i.e., amylase with friedelin (FRI) and amylase with acarbose (STD) were two complexes subjected for MDS with the CHARMM36m force field, this approach has been previously documented by (Bagewadi et al., 2023) with the aid of a Berendsen thermostat, an MD simulation of ligand in conjunction with protein was done for a total of 200 ns at 300 Kelvin and this simulation also employ 1.01325 bars of constant pressure, which mimics the body's natural temperature and pressure.
3. Results
3.1. Evaluation of phytocompounds in K. pinnata
Preliminary phytochemical screening of extract is essential to comprehend the presence of secondary metabolites i.e., bioactive molecules with medicinal properties. The methanolic leaf extract of K. pinnata contained a variety of phytochemicals, including alkaloids, saponins, flavonoids, phenols, terpenoids, carbohydrates glycosides, steroids, and proteins. The preliminary findings of the phytochemical screening have been compiled and presented in Table 1. The quantitative analysis of the extract illustrates the presence of different potential bioactive compounds, with the highest concentration being flavonoids 401.2 mg QuE/g E, subsequent to flavonoids, phenols were also detected in significant amounts 109.037 mg GAE/g E, additionally alkaloids were found in extract at a lower concentration of 9.8 mg AE/g E. the substantial flavonoids content reported in methanolic extract holds significant potential for promoting health benefits primary attributed to its antioxidant properties. Table 2 illustrates the results of the quantitative analysis.
Table 1.
Preliminary phytochemical screening of methanolic extract of K. pinnata.
| Sl.no | Phytocompound | Test | Observation |
|---|---|---|---|
| 1 | Alkaloids | Wagner | +ve |
| 2 | Flavonoids | Shinoda | +ve |
| 3 | Saponins | Frothing | −ve |
| 4 | Steroids and Triterpenoids | Salkowoski | +ve |
| 5 | Tannins | Braemers | +ve |
| 6 | Phenols | Bromine water | +ve |
| 7 | Cardiac Glycosides | Kellar-kiliani | +ve |
| 8 | Anthraquinones | Borntragens | +ve |
| 9 | Quinones | Test for quinones | +ve |
| 10 | Coumarins | Test for coumarins | +ve |
| 11 | Diterpenes | Copper acetate | +ve |
| 12 | Phobal tannins | Test for phobal tannins | −ve |
| 13 | Catechin | Matchstick | −ve |
| 14 | Anthocyanosides | Test for anthocyanosides | −ve |
| 15 | Resins | Acetone water | −ve |
| 16 | Volatile oil | Test for volatile oil | −ve |
| 17 | Acidic compounds | Test for acidic compounds | +ve |
+ve indicates presence of Phytocompounds −ve indicates absence of Phytocompounds.
Table 2.
Determination of phytocompounds from K. pinnata.
| Sl.no | Phytocompounds | Total bioactive compound |
|---|---|---|
| 1 | Alkaloid | 9.8 ± 0.01 mg AE/g E |
| 2 | Flavonoid | 401.2 ± 0.03 mg QuE/g E |
| 3 | Phenol | 109.037 ± 0.22 mg GAE/g E |
The bioactive compounds alkaloids as atropine equivalents (mg AE/g E), flavonoids as querecetin equivalents (mg QuE/g E) and phenols as gallic acid equivalents in methanolic extract of K.pinnata.
3.2. Analysis of in-vitro enzyme inhibition for anti-diabetic activity
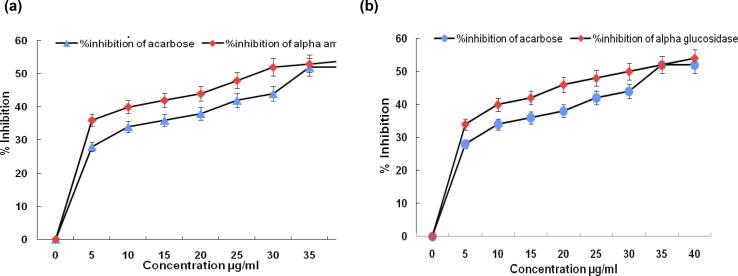
Alpha-amylase and alpha-glucosidase enzymes in the digestive system are essential for the metabolism of starch, glycogen, and disaccharides. The findings showed a notable inhibitory effect of the extract in breaking down complex sugars into simpler compounds. The synergistic tests were conducted to determine the methanolic extract's alpha-amylase and alpha-glucosidase inhibitory activities and are reported in Fig. 1. The methanolic extract's IC50 value was determined to be 29.50 ± 0.04 μg/mL for alpha-amylase activity (Fig. 1a) and 32.04 ± 0.35 μg/ml for alpha-glucosidase (Fig. 1b). The inhibitory effect of the extract increased in dose dependent manner with respect to both enzymes, the IC50 value of standard acarbose against alpha-amylase and alpha-glucosidase was found to be 35.82 ± 0.14 μg/ml and 33.21 ± 0.02 μg/ml. The IC50 value of the standard acarbose, when compared to that of the extract, suggests that the extract exhibits a higher effectiveness in inhibiting alpha-amylase and alpha-glucosidase than standard acarbose.
Fig. 1.
(a) Alpha amylase and (b) alpha glucosidase enzyme inhibiting activity of methanolic extract of Kalanchoe pinnata with standard acarbose.
3.3. Construction of bioactive phytocompounds library
Seventy-six distinct compounds were reported from K. Pinnata by referring to various databases and literature and their SDF-format 3D structures were obtained from the PubChem database. The compounds not having canonical smiles and respective disease targets were eliminated. The obtained compounds have been examined for additional ADME properties, including pharmacokinetic parameters using a combination of parameters according to Lipinski's rule of five as well the bioactive compounds were chosen primarily for their predictability for oral consumption; compounds that meet all of the aforementioned criteria were chosen for establishing networks. Totally 37 bioactive compounds from K. pinnata were listed out as shown in Table S1 in supplementary information (SI), which obey the above mentioned criteria and were further subjected to molecular docking and network construction.
3.4. Assessment of compounds and DM associated targets
We obtained 37 bioactive compounds from K. pinnata that were predicted to target 197 protein targets. The Swiss target prediction SwissADME was used to retrieve the primary target genes for the drugs that meet the pharmacokinetics requirements as shown in Table S2 in SI. Twenty of them are anticipated to target AR, 9 compounds are predicted to target AMY2A, and 12 compounds are predicted to target DPP4. In addition, gene-related data pertaining to DM was extracted from gene cards, a comprehensive database encompassing information on all known or anticipated human genes. This database includes proteomic, genomic, transcriptomic, genetic, and functional data, and was searched using the keywords “hypoglycemia,” “obesity,” “DM,” and “Homo sapiens. For additional analysis, a composite of the outcomes from the two platforms was used.
3.5. Protein-protein interaction of potential targets
Protein-protein interactions using these overlapping core target genes which include both direct and indirect relationships were thoroughly examined using the STRING database where a significant medium confidence level was set to >0.4. The PPI network had 1429 edges and 194 nodes, with a mean node degree of 14.7 and a wide local clustering coefficient on the network of 0.47, which depicts that proteins in our network have more interaction among themselves. Based on the parameters relating to degrees, betweenness, and closeness, the findings propose that the proteins are physiologically correlated as a group which is illustrated in Fig.S1 in SI. The top 5 targets in the PPI network which are considered as core targets of DM as analyzed in Cytoscape, the core targets for our network were found to be AMY2A, NOX4, RPS6KA3, ADRA2A, CHRM5, and IL2 with more number interactions.
3.6. Network construction
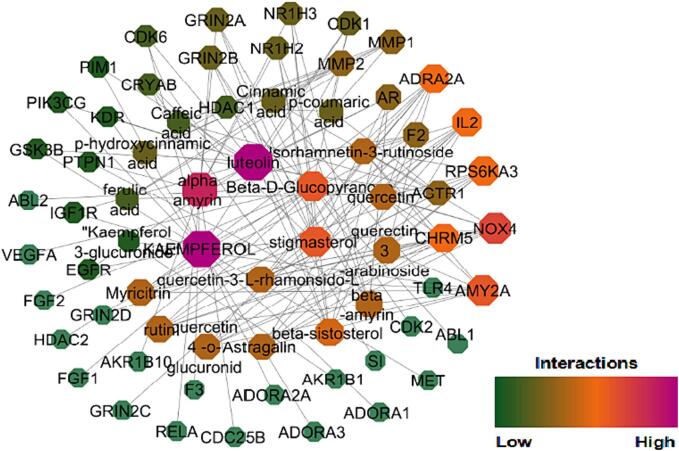
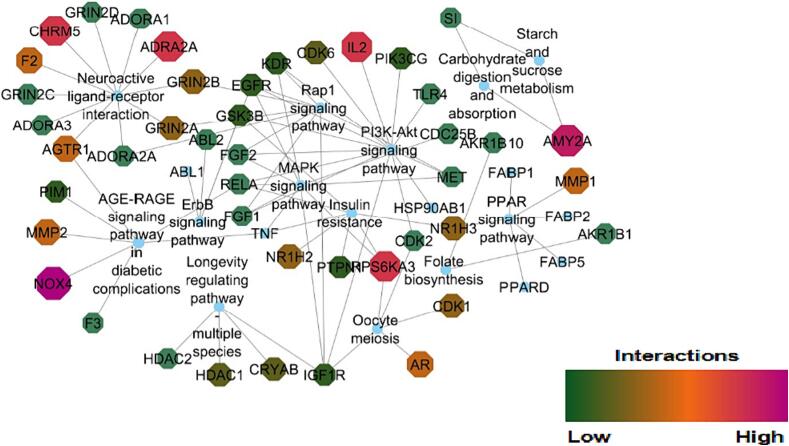
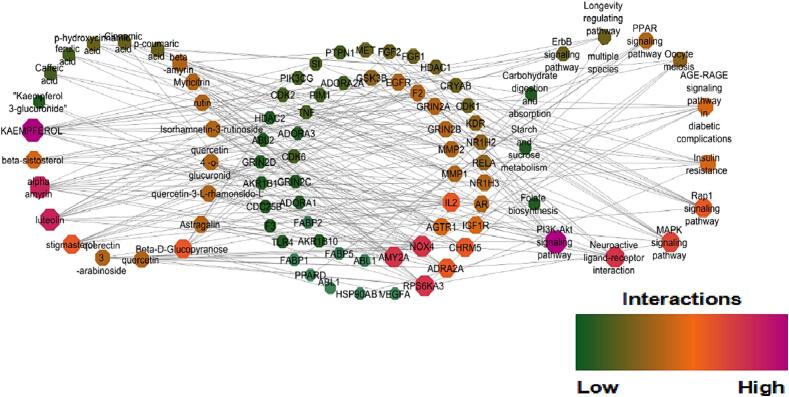
Network analysis attempts to identify the critical targets, biologically active compounds and metabolic pathways that are interconnected in the pathogenesis of DM. Kaempferol, alpha-amyrin, luteolin, beta-D-glucopyranose, stigma sterol, isorhamnetin-3-o-rutinoside, quercetin, myricetin, astragalin, quercetin-3-o-arabinoside, rutin, beta-sitosterol, and kaempferol-3-glucuronide were identified as novel bioactive compounds through examination of network that was developed between compounds and their probable protein targets as shown in Fig. 2. Protein –pathway networks are given in Fig. 3 and compound-target protein-pathway in Fig. 4 was precisely studied. The aforementioned compounds have strengthened the network by targeting significant proteins associated with T2DM namely AMY2A, NOX4, AGTR1, IL2, RPS6KA3, ADRA2A, CHRM5, IGF1R, AR, AKR1B1, NR1H3, and MMP9. The major intracellular pathways that are modulated by these proteins are P13AKT signaling pathway, the AGE-RAGE signaling pathway, the Neuroactive ligand interaction pathway, insulin resistance, the MAPK signaling pathway, and the carbohydrate digestion and absorption pathways that are involved in DM. It is significant to emphasize that, Kaempferol scored the highest edge count interaction with 15 target proteins they are ADORA1, ADORA2A, ADORA3, CDK1, CDK6, EGFR, GSK3B, MET, IGF1R, NOX4, PIK3CG, F2, PIM1, KDR, DPP 4. Similarly P13K-Akt signalling pathway modulated the highest number of molecules i.e., IL2, KDR, FGF2, CDK2, CDK6, EGFR, MET, GSK3B, HSP90AB1, TLR4, RELA, PIK3CG, FGF1, IGF1R. During the network analysis, it was found that several additional interactions were associated with the single genes. However, it is necessary to exclude these interactions during the specific network interpretation analysis, this exclusion is based on the principle of focusing on the most relevant and reliable interactions that directly contribute to the biological processes or pathways being studied.
Fig. 2.
Compound to different protein targets network interaction of K.pinnata.
Fig. 3.
Network interaction between protein targets-pathway K.pinnata.
Fig. 4.
Network interactions between compounds -protein targets-pathway of K.pinnata.
3.7. Gene ontology and KEGG enrichment analysis
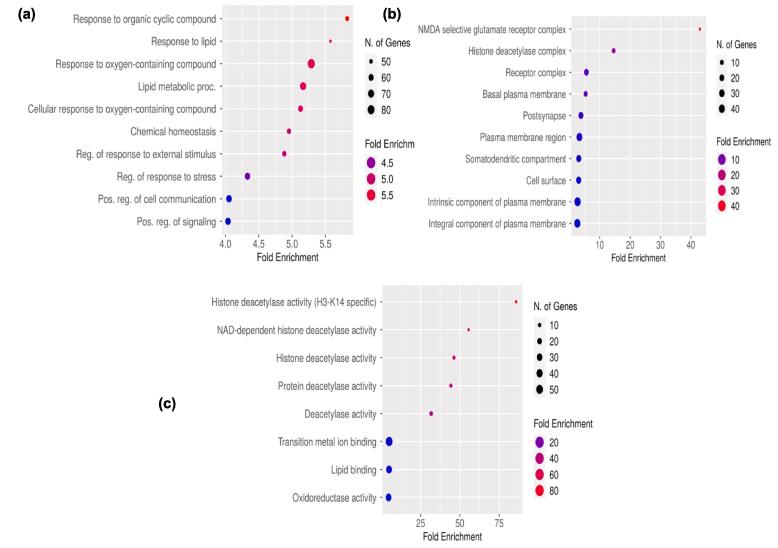
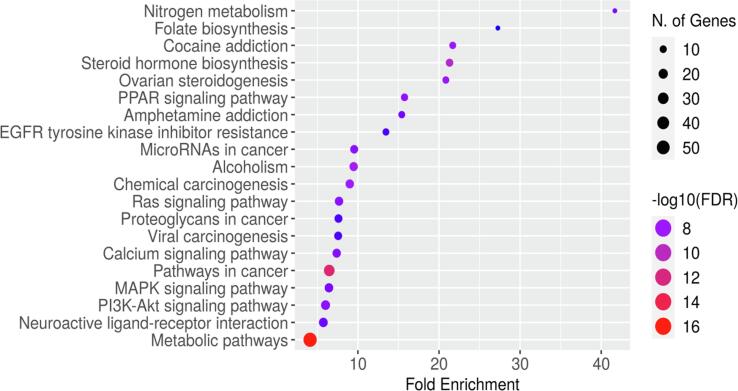
Enrichment analysis was conducted and annotated using the DAVID server, the results include the top 10 terms for each of the categories Fig. 5 represents information regarding molecular function, cellular components, and biological processes. Most of the proteins are found in the histone deacetylase receptor complex followed by slightly less abundant proteins in the plasma membrane, where 49.5 % of proteins are found in the NMDA selective glutamate complex. Although most proteins are involved in multiple biological activities inflammatory responses, followed by lipid biosynthesis and chemical homeostasis. A bubble plot was used to display the top-most enriched pathways in relation to fold enrichment of K. pinnata. KEGG enrichment analysis showed 96 pathways in Table S3 in SI which are related to protein targets identified from the bioactive of K. pinnata irrespective of diseases, further pathways responsible only for T2DM were listed, and their respective protein targets and compounds were analyzed for the study. Thirteen pathways related only to T2DM are listed out in Table 3 along with FDR value and matching proteins from the network meanwhile, pathways not linked to T2DM or lacking P-values under 0.05 were eliminated and subjected to protein to pathway network construction. Fig. 6 provides an overview of the majority of enriched metabolic pathways from targets of K.pinnata and about 50 % of the genes were derived from metabolic pathways. T2DM is a metabolic disease that results from neuroactive ligand interactions via P13- AKT, MAPK, and PPAR signaling pathways. Conclusions drawn from GO enrichment analysis, express that the phytocompounds present in K. pinnata are capable of regulating various biological processes such as protein kinase activity, cell differentiation, adhesion, and intracellular signal transduction in individuals with T2DM.
Fig. 5.
The top ten components of gene ontology enrichment analysis of Kalanchoe pinnata bioactives against DM targets (a) biological process (b) cellular component (c) molecular function.
Table 3.
KEGG pathway enrichment analysis of potential targets modulated by the phytocompounds from K.pinnata.
| KEGG ID | Pathway description | False discovery rate | Matching proteins in your network (labels) | Gene count |
|---|---|---|---|---|
| hsa03320 | PPAR signalling pathway | 2.11E-07 | FABP4,FABP2,FABP1,FABP5,PPARD,MMP1,SCD,FABP3,PPARA,NR1H3 | 10 |
| hsa04080 | Neuroactive ligand-receptor interaction | 2.98E-06 | ADORA3,GRIN2D,ADRA2A,GRIN2C,F2,GRIN2A,ADORA1,CNR1,GABBR1,CHRM5,DRD1,AGTR1,LTB4R2,TRPV1,GRIN2B,ADORA2A | 16 |
| hsa04151 | PI3K-Akt signalling pathway | 5.92E-06 | IL2,KDR,FGF2,CDK6,CDK2,EGFR,MET,GSK3B,HSP90AB1,TLR4,ITGB1,RELA,PIK3CG,VEGFC,FGF1,IGF1R | 16 |
| hsa00790 | Folate biosynthesis | 6.93E-06 | AKR1B1,CBR1,ALPI,AKR1B10,ALPL,AKR1C3 | 6 |
| hsa04015 | Rap1 signalling pathway | 1.21E-05 | KDR,FGF2,EGFR,MET,GRIN2A,CNR1,ITGB1,GRIN2B,ADORA2A,VEGFC,FGF1,IGF1R | 12 |
| hsa04933 | AGE-RAGE signalling pathway in diabetic complications | 8.40E-05 | MMP2,NOX4,F3,PIM1,RELA,TNF,AGTR1,VEGFC | 8 |
| hsa04931 | Insulin resistance | 0.00014 | GSK3B,PTPN1,RPS6KA3,RELA,PPARA,TNF,NR1H3,NR1H2 | 8 |
| hsa04010 | MAPK signalling pathway | 0.00026 | CDC25B,KDR,FGF2,EGFR,PTPN7,MET,RPS6KA3,RELA,TNF,VEGFC,FGF1,IGF1R | 12 |
| hsa04213 | Longevity regulating pathway - multiple species | 0.0146 | HDAC1,HDAC2,CRYAB,IGF1R | 4 |
| hsa00500 | Starch and sucrose metabolism | 0.018 | SI,MGAM2,AMY2A | 3 |
| hsa04114 | Oocyte meiosis | 0.0248 | CDK2,AR,RPS6KA3,CDK1,IGF1R | 5 |
| hsa04012 | ErbB signalling pathway | 0.0309 | EGFR,GSK3B,ABL1,ABL2 | 4 |
| hsa04973 | Carbohydrate digestion and absorption | 0.0354 | SI,MGAM2,AMY2A | 3 |
Fig. 6.
Bubble plot showing top 10 highly enriched pathways obtained from KEGG pathway assessment.
3.8. Molecular docking
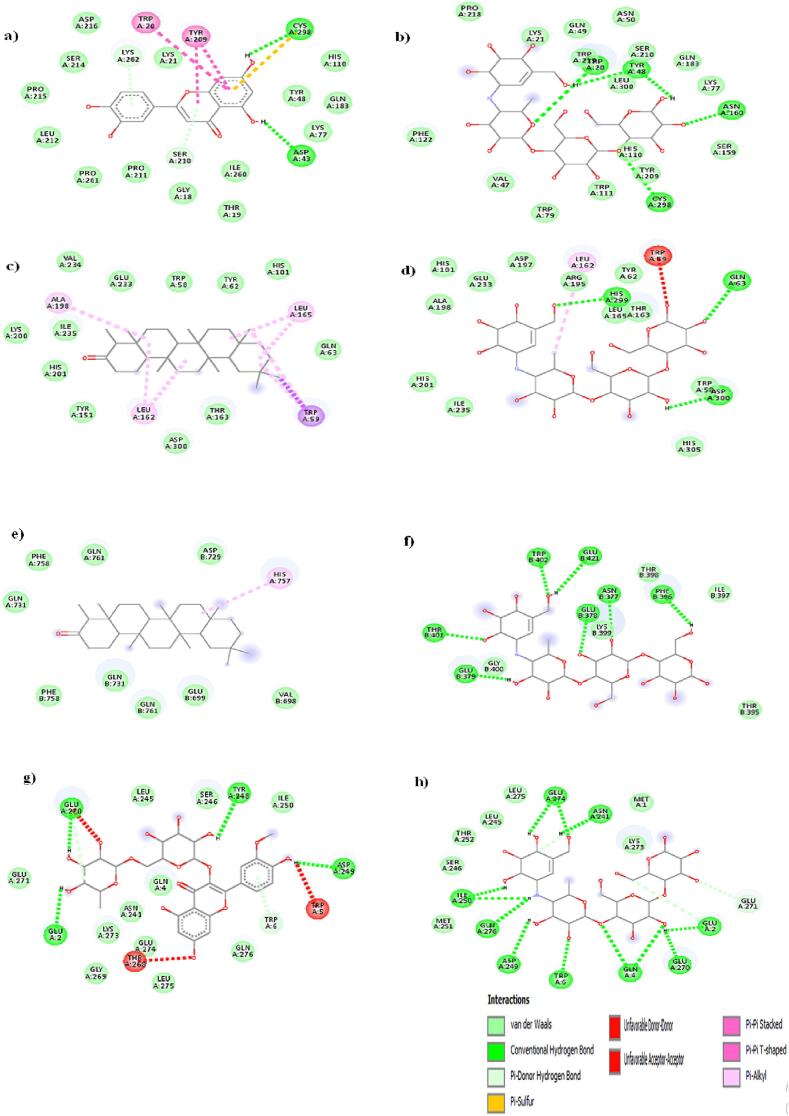
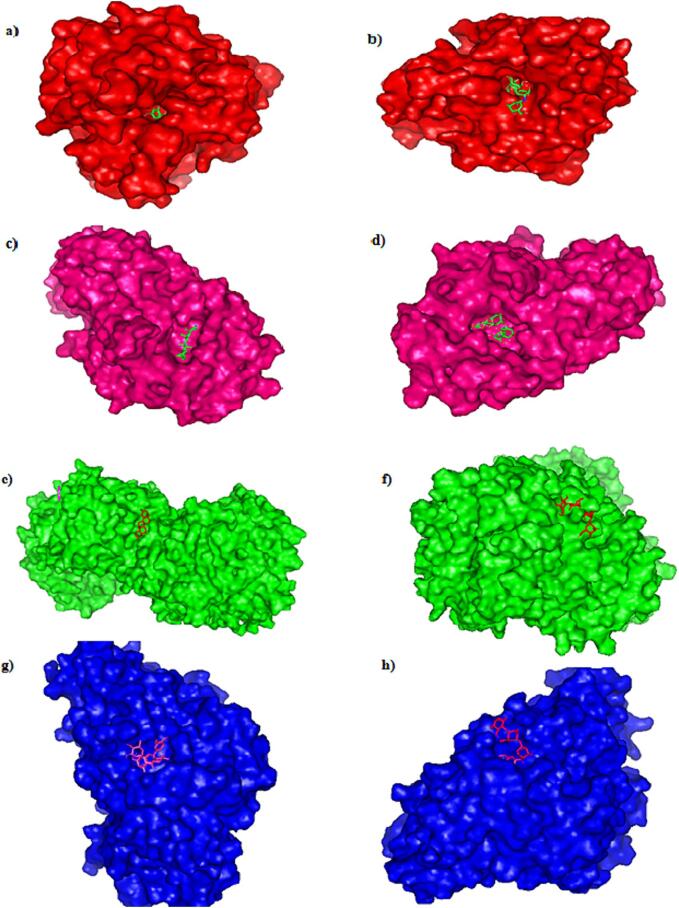
To achieve optimal conformation, ligand interactions with the target protein's active sites were investigated, along with inhibitory effects using molecular docking. Computer-aided drug design techniques were used to explore potential therapeutic compounds from K. pinnata for DM treatment. Molecular docking emerges as a viable approach to determine the positioning of molecules within the binding sites of the selected protein target. In this study, an analysis was conducted on molecular docking investigations involving 15 bioactive compounds of K. pinnata, the docking scores of these compounds in relation to acarbose are elucidated in Table S4 in SI. Only the best-docked compounds were chosen for further examination. Table 4 displays the ligands that exhibit the best binding affinity for their respective protein receptors, including hydrogen bonds, hydrophobic bonds, binding energy, salt bridges, and PI stacking information. The docking results were determined based on the docking energy (Kcal/mol) the contacts between each ligand and the functional residues of AMY2A, AR, DPP4, and GAA. The interactions between each ligand and each receptor were evaluated along with the standard acarbose, which demonstrated a lower binding affinity comparatively. AMY2A had stronger binding to friedelin (-9.4 Kcal/mol) due to hydrogen bonds with Tyr58, Ile61, Glu86, LysS87, Val88, Asn92, and Arg111, salt bridges with Lys87 and Arg111, and a PI stacking interaction with Phe209, AR had high binding to luteolin (-10 kcal/mol) through hydrogen bonds with Asp43, Tyr48, Lys77, Tyr209, Leu212, and Ser214, three hydrophobic bonds with Trp20, Tyr209, and Lys262, and a PI stacking interaction with Tyr209 in a similar way DPP4 binds with friedelin through hydrophobic interactions (His757 and Gln761) with a binding energy of −8.7 kcal/mol. GAA has a higher affinity for isorhamnetin-3-o-rutinoside with a binding energy of −8 kcal/mol. This affinity is achieved through hydrogen bonds (Tyr58, Ile59, Glu86, Lys87, Val88, Asn92, and Arg111), salt bridges (Lys307 and Arg309), and a PI stacking interaction with Phe 209. The standard acarbose binding energies are −7.5 kcal/mol for AR, −7.0 kcal/mol for AMY2A, −7.9 kcal/mol for GAA, and −5.3 kcal/mol for DPP4. This study aimed to determine how K. pinnata works as an antidiabetic agent by targeting four receptors related to DM, friedelin was found to be the most active compound targeting AMY2A, this complex is further subjected to MDS to check for stability. Fig. 7 depicts the 2D protein–ligand interaction of complexes having more binding energy along with standard acarbose. Fig. 7a and Fig. 7b illustrate the binding affinity of AR with luteolin (-10 kcal/mol) and acarbose (−7.5 kcal/mol), Fig. 7c and Fig. 7d demonstrate the binding affinity of AMY2A with friedelin (−9.4 kcal/mol) and acarbose (−7 kcal/mol). Binding of DPP4 with friedelin (−8.4 kcal/mol) and acarbose (−5.3 kcal/mol) are given in Fig. 7e and Fig. 7f. Fig. 7g. and Fig. 7h illustrate binding affinity of GAA with isorhamnatin-3-o-rutinoside (−8 kcal/mol) and GAA with acarbose (−7.9 kcal/mol). Similarly, Fig. 8 illustrates the 3D structure of an above mentioned complex.
Table 4.
Compounds and protein targets with best docking score and related molecular interactions.
| Targets | Compounds | Binding energy (Kcal/mol) |
Interactions |
|||
|---|---|---|---|---|---|---|
| Hydrophobic interactions | Hydrogen bonds | Salt bridges | Pi-stacking | |||
| Aldose reductase(AR) | Luteolin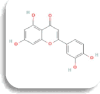
|
−10 Kcal/mol | TRP20, TYR209, LYS262 | ASP43, TYR48, LYS77, TYR209, LEU212, SER214 | TYR209 | |
| Alpha amylase (AMY2A) | Friedelin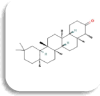
|
−9.4 Kcal/mol | TYR58, ILE61, GLU86, LYS87, VAL88, ASN92, ARG111 | LYS87, ARG111 | PHE209 | |
| Alpha glucosidase (GAA) | isorhamnetin-3-o-rutinoside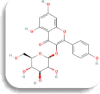
|
−8 Kcal/mol | TYR58, ILE59, GLU86, LYS87, VAL88, ASN92, ARG111, | LYS307. ARG309 | PHE209 | |
| Dipeptidyl peptidase IV(DPP4) | Friedelin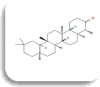
|
−8.7 Kcal/mol | HIS757, GLN761 | |||
Fig. 7.
2D interaction of target -ligand complexes (a) aldose reductase with luteolin (b) aldose reductase with acarbose (c) alpha amylase with friedelin (d) alpha amylase with acarbose (e) DPP4 with friedelin (f) DPP4 with acarbose (g) alpha glucosidase with Isorhamatin-3-o-rutinoside (h) alpha glucosidase with Isorhamatin-3-o-rutinoside.
Fig. 8.
3D interaction of target -ligand complexes (a)aldose reductase with luteolin (b)aldose reductase with acarbose (c) alpha amylase with friedelin (d) alpha amylase with acarbose (e)DPP4 with friedelin (f) DPP4 with acarbose (g)alpha glucosidase with Isorhamatin-3-o-rutinoside (h)alpha glucosidase with Isorhamatin-3-o-rutinoside.
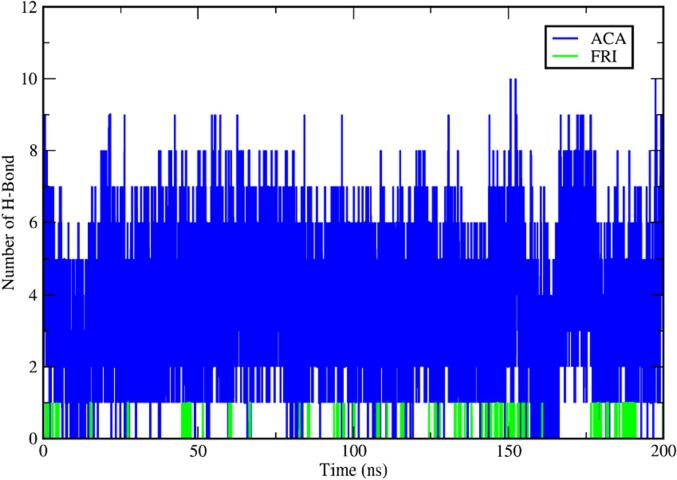
3.9. MDS
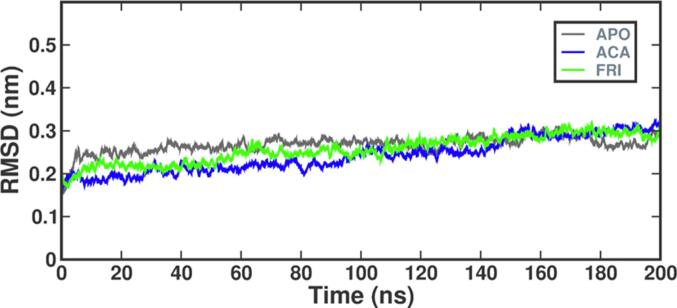
MDS is a widely used computational method for investigating the dynamic behavior and interactions of atoms and molecules through a specific period; it is particularly useful for examining the binding of small molecules to proteins, exploring different binding modes, energetic aspects, and conformational changes. Friedelin bioactive (FRI) from K. pinnata and acarbose (STD) which is standard were subjected to MDS based on the best docking score and enzyme inhibition activity that was performed and the investigation assessed the stability of the docked complex by examining a range of variables such as interactions with hydrogen bonds, solvent accessible surface area (SASA), root mean square deviation (RMSD), radius of gyration (RG) and root mean square fluctuation (RMSF). Subsequently, the study analyzed the backbone atoms in alpha-amylase (APO) and protein–ligand complexes (FRI, STD) as shown in Fig. 9 over 200 ns. Fluctuations were observed only during the initial 1–8 ns, but the complexes remained stable throughout. The RMSD value for alpha-amylase was 0.18 nm, and FRI stabilized with an acceptable RMSD range below 0.14 nm. The phytocompound had average RMSD values below the permitted range i.e. 0.22 nm (Sharma et al., 2021), and was comparable to STD with an RMSD value of 0.12 nm.
Fig. 9.
RMSD plot for the APO and protein-ligands [FRI-STD] complex for alpha amylase protein for 200 ns. *FRI-friedelin,STD-acarbose.
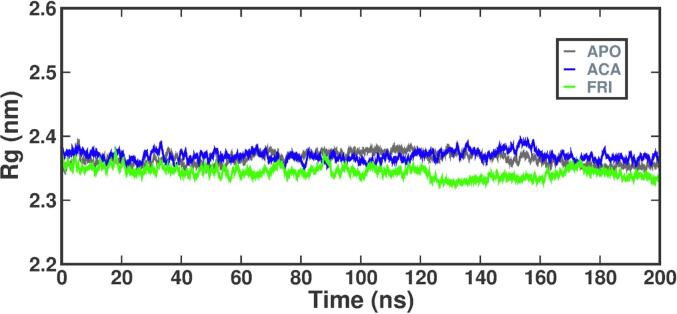
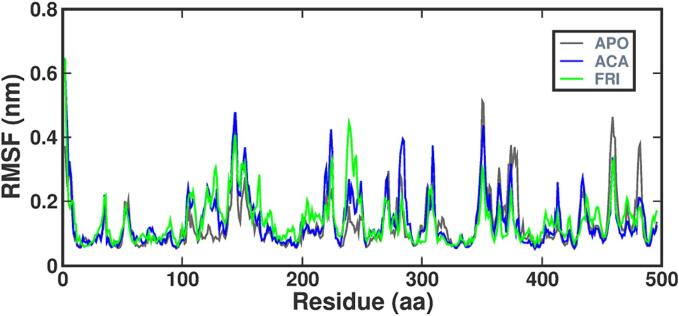
The stability of bio-molecules during MD simulation is determined by assessing the RG value. Fig. 10 shows the average RG for the APO, alpha-amylase-ligand complexes (FRI-STD) over the 200 ns trajectory. The average RG value for the alpha-amylase-FRI complex was 2.35 nm, similar to the alpha-amylase-STD i.e. RG value of 2.38 nm. The RG trajectory remained constant during the last 10 ns, indicating a consistently folded structure. A higher RMSF value indicates a more flexible region of the structure, such as turns and loops. In Fig. 11 the protein–ligand complex is depicted during 11 ns molecules showcasing the temporal equation and variations in component residues for the alpha-amylase protein. The native alpha-amylase protein, alpha-amylase-STD, and alpha-amylase-FRI exhibited average RMSF values of 0.47 nm, 0.5 nm, and 0.49 nm, respectively. The study observed variations in the residues of amino acids; the friedelin plot displayed greater RMSF in several regions that were similar to the STD and APO complexes, with minimal deviation from the APO and STD complexes.
Fig. 10.
Radius of gyration plot showing the compactness of Apo, FRI and STD complex in 200 ns duration analysis.
Fig. 11.R.
MSF plot of amino acid residues in the APO and protein–ligand [FRI-STD] complexes.
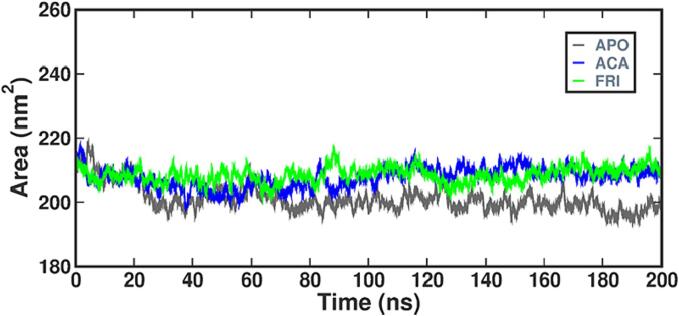
The percentage of protein surface accessible to the solvent and Protein aggregation within the system is determined using SASA, and the magnitude of conformational changes during contact are also reported. Fig. 12 shows the SASA values over time for each protein and enzyme-ligand complex. The average SASA values for APO, STD, and FRI are 219 nm2, 218 nm2, and 219 nm2, respectively, according to the analysis of hydrogen bonds, FRI and STD exhibit a stable binding to alpha-amylase. Hydrogen bonds play a crucial role in ligand binding, influencing the characteristics of pharmaceutical compounds. In Fig. 13 it is observed that there are 4 to 5 stable pairs of hydrogen bonds, friedelin demonstrates a strong affinity for binding and remains stable in the active site of alpha-amylase throughout the simulation. The results indicated that the protein complex when combined with the selected ligands enhanced the rigidity and compactness of the protein structure, thereby improving its stability.
Fig. 12.
Plot of SASA for alpha amylase-Apo, FRI and STD complex.
Fig. 13.
Number of hydrogen bonds for the trajectory of molecular dynamic simulation for alpha amylase enzyme with STD and FRI.
4. Discussion
The objective of the present investigation was to explore the biochemical process responsible for the anti-diabetic effects attributed to the K. pinnata plant commonly used in traditional Indian medicinal practices, by integration of in-vitro and in-silico methods. Diabetes is a chronic metabolic condition marked by elevated blood glucose levels, which over a prolonged period can significantly impact the cardiovascular system, blood vessels, ocular health, renal function, and neurological pathways (Shubhashree et al., 2020). Approximately 422 million individuals with diabetes predominantly reside in low and middle-income nations, and diabetes directly leads to 1.5 million mortality annually (Pradeepa and Mohan, 2021). The most recent data on the prevalence and incidence of both pre-diabetes and diabetes can be found in the National Diabetes Statistics Report, the global prevalence of diabetes in adults elevated to 8.8 % in 2017, and it is anticipated to reach 9.9 % of the worldwide population by 2045 (Standl et al., 2019). The increasing prevalence of diabetes globally imposes a substantial economic and psychological burden on both society and individual households. The medications employed, such as insulin sensitizers, insulin-secreting drugs, and insulin itself entail a range of unfavorable effects. Consequently, the pursuit of complementary and alternative therapies has gained more importance (Halayal et al., 2023).On the other hand, using a single therapy approach has not been effective in reducing all of the detrimental consequences of elevated blood sugar levels, accordingly mitigating the detrimental effects of glucose can be accomplished by employing antioxidant inhibitors, alpha-glucosidase inhibitors, aldose reductase inhibitors, and antiglycation medications, numerous studies have been demonstrated how bioactive combinations obtained from basic plant extract can help reduce blood glucose levels (Li et al., 2019). The traditional approaches of network biology and polypharmacology are widely recognized for evaluating therapeutic and mechanistic targets by integrating omics data to develop multi-target medications. As a result, network pharmacology has emerged as a recognized discipline that combines these two methodologies (Sharma and Yadav, 2022).
K. pinnata is commonly found in tropical and temperate regions of the world, it is widely acknowledged for its diverse range of pharmacological activities, particularly in the treatment of various severe illnesses that primarily affect humans. Contemporary pharmacological research has generally corroborated the traditional uses of K. pinnata and its extracts in the management of severe illnesses, inflammations, painful ulcers, fungal infections, viral diseases, microbial attacks, compromised immune system, diabetes mellitus, and insecticidal properties (Rahman et al., 2019). Natural remedies have historically played a crucial role in the management of DM, particularly in regions such as Asia, India, and Africa. In the pursuit of developing and uncovering antidiabetic medications, extensive research has been conducted on herbal remedies, specifically focusing on their hypoglycaemic effects (Li et al., 2017). In accordance with research carried out previously by (Menon et al., 2015) the use of the aqueous extract from K. pinnata has shown significant decreases in body weight, as well as notable effects in lowering blood sugar and cholesterol levels in the treatment of diabetes. Furthermore, it has been observed to effectively decrease the levels of the inflammatory cytokine (IL-6), which is often elevated in individuals with diabetes. To evaluate the supplementary influence of this plant during the initial stage, a study conducted by (Agüero-Hernández et al., 2020) validated the hypoglycaemic efficacy of K. pinnata extract when administered in conjunction with glibenclamide, demonstrating its better effectiveness comparatively. It has been reported that grape seed and peel extract could be used as a preventative measure against T2DM and it is associated with kidney and cardiac toxicity (Oueslati et al., 2016). Systematic research of pharmacological networks can provide a better understanding of the rationale, indications, and potential actions of conventional medicines. The primary principle involved in this approach is the combination of the benefits of modern research and conventional knowledge, as noted by (Chandran et al., 2015).In the current research, we employed network pharmacology to explore the material foundation and molecular mechanism of K. pinnata in treating T2DM, complex and non-linear relationships between bioactive compounds and the underlying pathophysiological mechanisms involved in T2DM were made easier.
Initially, the bioactive compounds of K. pinnata and their corresponding targets were identified and subsequently, a network was constructed to establish connections between the bioactive compounds, their targets, and the relevant KEGG pathways that are enriched by the target proteins. Through topological research, the essential pharmaceuticals for the treatment of diabetes derived from K. pinnata were determined. Furthermore, the authors have verified the existence of diverse bioactive substances in the leaf extract of this plant, particularly abundant in quercetin, a flavonoid known for its remarkable efficacy in reducing lupus arthritis in a mouse model (Indriyanti et al., 2017). As per the findings of (Ogidigo et al., 2022) ethyl acetate fractions of K. Pinnata contains significant amounts of quercetin, luteolin, isorhamnetin, and luteolin-7-glucoside, the extract exhibited a reduction in an oxidative imbalance in rats subjected to neurotoxicity induced by aluminium chloride; these results indicate a potential neuroprotective effect. According to a study (Nascimento et al., 2023), bufadienolides were identified as the key compounds accountable for the anticancer effects observed in K.pinnata extracts, bryophyllin A which is a principal compound of it has been distinguished from other compounds such as Vitamin D, stigmasterol, alpha-amyrin, and beta-amyrin compounds found in K. pinnata. These compounds were previously reported by (Rajesh and Shamsudin, 2017) to exhibit activity against pi-class Glutathione-S-transferase for lymphatic filariasis through an in-silico approach. Additionally, the natural flavonoid quercetin, present in K. pinnata, has shown therapeutic potential against T2DM by acting as an anti-inflammatory and antioxidant agent. The combination of quercetin and metformin was found to be more effective than quercetin alone (Ramon et al., 2023) in the regulation of insulin. Flavonoids play a critical role in numerous medical, pharmacological, nutraceutical, and cosmetic applications, owing to their extensive array of health-enhancing advantages they have garnered attention for their potential neuroprotective, anti-inflammatory, anti-cancer, and anti-heart disease properties (Al-Jumaili et al., 2023). Study by (Daoui et al., 2023) investigates cannabinoids and terpenes as potential EGFR-TKI candidates targeting cancer therapy. Another similar investigation by (De Araújo et al., 2018) reports kaempferol another flavonoid found in leaves of K. pinnata demonstrated antioxidant effects as well has been demonstrated to control glucose levels in cells. Alpha-glucosidase and alpha-amylase inhibition is a medicinal strategy employed in the treatment of DM by inhibiting these enzymes, the absorption of glucose is slowed down and the digestion of sugars and carbohydrates is delayed (Patil et al., 2020). In this current study, it is expected that phytoconstituents derived from K. pinnata will exhibit inhibitory effects on alpha-amylase and alpha-glucosidase enzymes, the inhibitory activity was more pronounced in the methanolic leaf extract, as indicated by its lower IC50 value compared to the standard acarbose.
A compound to target protein then to pathway network was constructed to investigate the involvement of multiple target genes involved in T2DM; the findings indicate the significant efficacy of K. pinnata medicinal potential against T2DM, as it is associated with 13 signaling pathways, 51 targets, and 23 bioactive compounds related to DM. The significant interactions observed among quercetin, kaempferol, and luteolin within the network indicate their potential as crucial multifunctional bioactive compounds of K. pinnata in combating T2DM. We have identified Akt1 as the most significant protein associated with the onset and advancement of DM within our network. Furthermore, the PI3K/AKT pathway, which mediates the pleiotropic effects of insulin on metabolic processes, also supports cellular growth and function. The integrity of the PI3K/AKT pathway as a whole is crucial for sustaining the equilibrium of energy therefore, it is crucial to fully comprehend its pathophysiological involvement in the development of obesity (Savova et al., 2023). According to prior research, luteolin enhances the PI3K-Akt signaling pathway, which prevents insulin resistance, a state brought on by disorders (diabetes and metabolic syndrome) linked to the beginning of menopause (Huang and Zhang, 2021).IL2 is a fundamental target protein within our network that is enriched in the P13-Akt signaling pathway, specifically due to the presence of the compounds quercetin and quercetin-3-arabinoside this finding is substantiated with research conducted by (He et al., 2019). It is worth noting that this cytokine serves as a crucial early inflammatory biomarker for assessing the risks and consequences associated with T2DM, as patients with T2DM have elevated degrees of interleukin-10 (IL-10), IL-1B, and IL-16, when exposed to elevated glucose, leading to increased oxidative stress and impaired pancreatic function. This stress also increases the activity of matrix metalloproteinases MMP1, MMP2, and MMP9 resulting in decreased insulin production, reduced glucose uptake, insulin resistance, and abnormal glucose synthesis (Yin et al., 2020), this finding is consistent with our research which indicates that kaempferol-3-glucuronide, caffeic acid, ferulic acid, and cinnamic acid share a common target namely MMP1 which is enriched by the PPAR signaling pathway.
Heat-shock proteins (HSPs) protect cells from oxidative damage, inflammation, and apoptosis, a study by (XU et al., 2020) found a relationship between changes in the HSP90AB1 gene expression and cellular damage caused by high blood sugar these changes were observed in diabetic complications. This protein is correlated with toxifolin and dihydroquercetin, which are bioactive compounds of K.pinnata, PPAR ligands possess the potential to treat T2DM and the metabolic syndrome, encompassing obesity and insulin resistance. Additionally, the development of an antagonist for the AGE (RAGE) receptor could prove to be an efficacious treatment for T2DM, as the activation of this receptor is a distinct clinical consequence of the disease (Oh et al., 2020). It should be noted that one compound has the ability to influence numerous proteins and signaling pathways, and its method of action may differ from that of a collection of related compounds, for example in the present study PPAR signaling pathway, PI3K-Akt signaling pathway, neuroactive ligand interaction pathway, insulin resistance pathway were primarily modulated by phytocompounds kaempferol, friedelin and luteolin. This implies that rather than a single successful molecule, the choice of appropriate compounds to target the specific protein should be followed by the regulation of several proteins implicated in the pathophysiology of interest (Khanal and Patil, 2020).
In the present study, we used the “lock and key” design principle to synthesize ligand molecules that selectively target specific receptors. Docking studies were conducted to elucidate the ligand affinity and optimal match of bioactive compounds with their respective target molecules. A total of 15 phytocompounds were selected to target four potential therapeutic targets, namely AMY2A, GAA, AR, and DPP4. The binding potency of these compounds was compared to that of the standard drug acarbose. Among the compounds tested, luteolin exhibited the strongest ligand affinity with AR, which is known to interact with multiple protein targets and pathways. Luteolin is a natural flavonoid found in many edible plants and has been shown to possess significant anti-oxidative and anti-inflammatory properties (Agarwal & Shanmugam, 2019). Moreover, it has been demonstrated to improve insulin resistance in diet-induced obese mice (Zang et al., 2016). Excellent enzyme activity inhibition was demonstrated by in-silico molecular analyses of luteolin interactions with the active sites of the enzyme glucosidase, demonstrating that luteolin is one of the potential candidates for use as an anti-diabetic compound (Kahksha et al., 2023). The potential binding of AMY2A and DPP4 with friedelin, similar to the study conducted by (Smruthi et al., 2016) is evident, despite the low binding energy it has been observed that the phytoconstituent friedelin from the jamun tree exhibits a more effective inhibition of alpha-amylase compared to acarbose. The GAA exhibits a notable affinity for Isorhamnetin-3-o-rutinoside in comparison to fifteen other phytocompounds. According to (Sabiu et al., 2021) report, the phenolic compound isorhamnetin-3-o-rutinoside exhibited a robust docking interaction with alpha-glucosidase in comparison to acarbose. (Abchir et al., 2023a) conducted a similar investigation, examining computationally the stability of cannabis extract as a potential alpha-amylase inhibitor with acarbose as a reference drug where, bioactive compounds have shown better docking score compared to acarbose. Nevertheless, it is important to note that docking is only an initial stage in evaluating the appropriateness of a ligand for a receptor's binding pocket, therefore further assessments of binding energy calculations and molecular dynamics simulations were conducted on the binding orientations of the investigated phenolics and flavonoids.
Docking results show differences in enzyme binding sites between the phytocompound and acarbose. Luteolin binds to alpha glucosidase through TRP20, TYR209, CYS298, and ASP43, while acarbose binds through TRP20, TYR48, ASN160, and CYS298. Acarbose is predicted to be a competitive inhibitor, consistent with (Yan et al., 2014) research. Friedelin occupies alpha amylase's active site through ALA198, LEU168, and TRP59, while acarbose binds through TRP59, HIS299, ARG195, and ASP300. Green colour lines indicate hydrogen bonds in all complexes shown in Fig. 7 it confirms the role of H-bonds in the binding of ligands to protein.
The complexes of alpha-amylase with friedelin and alpha-amylase with acarbose were subjected to MDS analysis, along with the apoenzyme for 200 ns simulation. The absence of significant fluctuations indicates that the protein and ligand have a strong binding affinity and do not significantly impact the protein's conformation within the 200 ns timeframe. Furthermore, the results obtained from the actual enzymatic assays were supported by this In-silico data. Although the traditional medical system provides numerous prospects, they have not yet been fully explored for the treatment of many diseases. Potential findings can be obtained if one uses contemporary computational chemistry tools to investigate the possibilities of the traditional pharmaceutical system (Kaushik et al., 2014).
5. Conclusion
As a concluding remark, the identified compounds from Kalanchoe pinnata have been demonstrated as notable potential inhibitors of GAA, AMY2A, AR, and DPPH. While the results of the in-vitro research provided some insight into K. pinnata's promising potential antidiabetic properties inhibiting alpha-amylase and alpha-glucosidase enzymes responsible for blood sugar homeostasis which is accountable for the development of T2DM. These findings suggest that luteolin, alpha amyrin, friedelin, and isorhamnetin-3-o-rutinoside may have the potential to be formulated as curative agents for T2DM, these compounds exert their effects through the modulation of multiple pathways, and by interacting with a variety of relevant targets associated with T2DM. Further research and clinical investigation are necessary to confirm their efficacy and safety in the treatment of T2DM. To comprehend the complicated, non-linear relationships between bioactive compounds and underlying pathophysiological processes, network pharmacology is a helpful technique. Additional molecular docking and molecular dynamic simulations were conducted to validate the binding affinity and stability of the phytocompound friedelin with the alpha-amylase enzyme.
Author contributions
RH and ZB were involved in the development of conceptual research design. RH has carried out the experiments related to biological activities and in-silico analysis using network pharmacology, molecular docking, molecular dynamics and simulations. RH and ZB have executed the data analysis and interpretation and have written the manuscript. NA, IS and AK has reviewed the manuscript and all authors agreed on the contents of the manuscript. ZB has supervised the entire research work.
CRediT authorship contribution statement
Rekha Y. Halayal: Conceptualization, Methodology, Investigation, Validation, Writing – original draft, Writing – review & editing. Zabin K. Bagewadi: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Supervision, Resources, Project administration. Nayef Abdulaziz Aldabaan: Formal analysis. Ibrahim Ahmed Shaikh: Funding acquisition, Formal analysis, Writing – review & editing. Aejaz Abdullatif Khan: Formal analysis, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The first author and the corresponding author thank KLE Technological University, Hubballi, for providing support under the Ph.D Fellowship Program. The authors are thankful to the Deanship of Scientific Research at Najran University for funding this work under the Research Group Funding program grant code NU/RG/MRC/12/14.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2024.102026.
Contributor Information
Rekha Y. Halayal, Email: 01fe21rbt002@kletech.ac.in.
Zabin K. Bagewadi, Email: zabin@kletech.ac.in.
Nayef Abdulaziz Aldabaan, Email: naaldabaan@nu.edu.sa.
Ibrahim Ahmed Shaikh, Email: iashikh@nu.edu.sa.
Aejaz Abdullatif Khan, Email: aejaz@ibnsina.edu.sa.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abchir O., Daoui O., Nour H., Yamari I., Elkhattabi S., Errougui A., Chtita S. Cannabis constituents as potential candidates against diabetes mellitus disease using molecular docking, dynamics simulations and ADMET investigations. Scientific African. 2023 https://sciencedirect.com/science/article/pii/S2468227623002016 [Google Scholar]
- Abchir O., Yamari I., Nour H., Daoui O., ElKhattabi S., Errougui A., Chtita S. Structure-Based Virtual Screening, ADMET analysis, and Molecular Dynamics Simulation of Moroccan Natural Compounds as Candidates α-Amylase Inhibitors. ChemistrySelect. 2023;8(26):e202301092. doi: 10.1080/14786419.2023.2281002. https://chemistry-europe.onlinelibrary.wiley.com/doi/abs/10.1002/slct.202301092 [DOI] [PubMed] [Google Scholar]
- Afendi F.M., Okada T., Yamazaki M., Hirai-Morita A., Nakamura Y., Nakamura K., Kanaya S. KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53(2):e1. doi: 10.1093/pcp/pcr165. https://academic.oup.com/pcp/article/53/2/e1/1868095 [DOI] [PubMed] [Google Scholar]
- Agarwal H., Shanmugam V.K. Anti-inflammatory activity screening of Kalanchoe pinnata methanol extract and its validation using a computational simulation approach. Inf. Med. Unlocked. 2019;14:6–14. https://www.sciencedirect.com/science/article/pii/S2352914818301989 [Google Scholar]
- Agüero-Hernández A.L., Rosales-López C., Herrera C., Vargas-Picado A., Muñoz R., Abdelnour-Esquivel A. Hypoglycemic Effect of Kalanchoe pinnata (Lam) Pers. Leaf Extract. Pharmacognosy J. 2020;12(3) https://phcogj.com/article/1145 [Google Scholar]
- Al-Jumaili M.H.A., Siddique F., Abul Qais F., Hashem H.E., Chtita S., Rani A., Almzaien K.A. Analysis and prediction pathways of natural products and their cytotoxicity against HeLa cell line protein using docking, molecular dynamics and ADMET. J. Biomol. Struct. Dyn. 2023;41(3):765–777. doi: 10.1080/07391102.2021.2011785. [DOI] [PubMed] [Google Scholar]
- Bagewadi Z.K., Muddapur U.M., Madiwal S.S., Mulla S.I., Khan A. Biochemical and enzyme inhibitory attributes of methanolic leaf extract of Datura inoxia Mill. Environ. Sustain. 2019;2:75–87. https://link.springer.com/article/10.1007/s42398-019-00052-6 [Google Scholar]
- Bagewadi Z.K., Khan T.Y., Gangadharappa B., Kamalapurkar A., Shamsudeen S.M., Yaraguppi D.A. Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach. Saudi J. Biol. Sci. 2023;30(9) doi: 10.1016/j.sjbs.2023.103753. https://www.sciencedirect.com/science/article/pii/S1319562X23001985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkan I.A., Doğan H.T., Zengin G., Colak N., Ayaz F.A., Gören A.C., Yeşilada E. Enzyme inhibitory and antioxidant activities of Nerium oleander L. flower extracts and activity guided isolation of the active components. Ind. Crop. Prod. 2018;112:24–31. https://www.sciencedirect.com/science/article/abs/pii/S0926669017307422 [Google Scholar]
- Banday M.Z., Sameer A.S., Nissar S. Pathophysiology of diabetes: an overview. Avicenna J. Med. 2020;10(04):174–188. doi: 10.4103/ajm.ajm_53_20. https://www.thieme-connect.com/products/ejournals/html/10.4103/ajm.ajm_53_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran U., Mehendale N., Tillu G., Patwardhan B. Network pharmacology of ayurveda formulation Triphala with special reference to anti-cancer property. Comb. Chem. High Throughput Screen. 2015;18(9):846–854. doi: 10.2174/1386207318666151019093606. https://www.ingentaconnect.com/content/ben/cchts/2015/00000018/00000009/art00004 [DOI] [PubMed] [Google Scholar]
- Chaudhury A., Duvoor C., Reddy Dendi V.S., Kraleti S., Chada A., Ravilla R., Mirza W. Clinical review of antidiabetic drugs: implications for type 2 diabetes mellitus management. Front. Endocrinol. 2017;8:6. doi: 10.3389/fendo.2017.00006. https://www.frontiersin.org/articles/10.3389/fendo.2017.00006/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtita S., Fouedjou R.T., Belaidi S., Djoumbissie L.A., Ouassaf M., Qais F.A., Lakhlifi T. In silico investigation of phytoconstituents from Cameroonian medicinal plants towards COVID-19 treatment. Struct. Chem. 2022;33(5):1799–1813. doi: 10.1007/s11224-022-01939-7. https://link.springer.com/article/10.1007/s11224-022-01939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho M.A.S., Casanova L.M., Nascimento L.B.D.S., Leal D., Palmero C., Toma H.K., Costa S.S. Wound healing cream formulated with Kalanchoe pinnata major flavonoid is as effective as the aqueous leaf extract cream in a rat model of excisional wound. Nat. Prod. Res. 2021;35(24):6034–6039. doi: 10.1080/14786419.2020.1817012. https://www.tandfonline.com/doi/abs/10.1080/14786419.2020.1817012 [DOI] [PubMed] [Google Scholar]
- Daoui O., Mali S.N., Elkhattabi K., Elkhattabi S., Chtita S. Repositioning Cannabinoids and Terpenes as Novel EGFR-TKIs Candidates for Targeted Therapy Against Cancer: a virtual screening model using CADD and biophysical simulations. Heliyon. 2023;9(4) doi: 10.1016/j.heliyon.2023.e15545. https://www.cell.com/heliyon/pdf/S2405-8440(23)02752-4.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Araújo E.R.D., Guerra G.C.B., Araújo D.F.D.S., De Araújo A.A., Fernandes J.M., de Araújo Júnior R.F., Zucolotto S.M. Gastroprotective and antioxidant activity of Kalanchoe brasiliensis and Kalanchoe pinnata leaf juices against indomethacin an2hanol-induced gastric lesions in rats. Int. J. Mol. Sci. 2018;19(5):1265. doi: 10.3390/ijms19051265. https://www.mdpi.com/1422-0067/19/5/1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande S.H., Muhsinah A.B., Bagewadi Z.K., Ankad G.M., Mahnashi M.H., Yaraguppi D.A., Roy S. In Silico Study on the Interactions, Molecular Docking, Dynamics and Simulation of Potential Compounds from Withania somnifera (L.) Dunal Root against Cancer by Targeting KAT6A. Molecules. 2023;28(3):1117. doi: 10.3390/molecules28031117. https://www.mdpi.com/1420-3049/28/3/1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., Martín C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020;21(17):6275. doi: 10.3390/ijms21176275. https://www.mdpi.com/1422-0067/21/17/6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsen J.A., Pai S., Isserlin R., Demchak B., Pico A.R. RCy3: Network Biology Using Cytoscape from within R. F1000Research. 2019:8. doi: 10.12688/f1000research.20887.1. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6880260/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halayal R.Y., Bagewadi Z.K., Maliger R.B., Al Jadidi S., Deshpande S.H. Network pharmacology based anti-diabetic attributes of bioactive compounds from Ocimum gratissimum L. through computational approach. Saudi J. Biol. Sci. 2023;30(9) doi: 10.1016/j.sjbs.2023.103766. https://www.sciencedirect.com/science/article/pii/S1319562X23002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Huang J.H., Zhang Z.Y., Du Q., Peng W.J., Yu R., Qin Y.H. A network pharmacology-based strategy for predicting active ingredients and potential targets of LiuWei DiHuang pill in treating type 2 diabetes mellitus. Drug Des. Devel. Ther. 2019:3989–4005. doi: 10.2147/DDDT.S216644. https://www.tandfonline.com/doi/full/10.2147/DDDT.S216644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhang X. Luteolin alleviates polycystic ovary syndrome in rats by resolving insulin resistance and oxidative stress. Am. J. Physiol. Endocrinol. Metab. 2021;320(6):E1085–E1092. doi: 10.1152/ajpendo.00034.2021. https://journals.physiology.org/doi/full/10.1152/ajpendo.00034.2021 [DOI] [PubMed] [Google Scholar]
- Indriyanti N., Soeroso J., Khotib J. The benefits of active compounds in Kalanchoe Pinnata (LMK) pers ethyl acetate fraction on lupus arthritis mice. Asian J. Pharm. Clin. Res. 2017 https://www.researchgate.net/profile/Niken-Indriyanti/publication/320804819_The_benefits_of_active_compounds_in_Kalanchoe_Pinnata_LMK_pers_ethyl_acetate_fraction_on_lupus_arthritis_mice/links/59fd1947aca272347a23cb07/The-benefits-of-active-compounds-in-Kalanchoe-Pinnata-LMK-pers-ethyl-acetate-fraction-on-lupus-arthritis-mice.pdf [Google Scholar]
- Jejurikar, B.L., Rohane, S.H., 2021. Drug designing in discovery studio. https://www.indianjournals.com/ijor.aspx?target=ijor:ajrc&volume=14&issue=2&article=008.
- Kahksha, Alam O., Al-Keridis L.A., Khan J., Naaz S., Alam A., et al. Evaluation of Antidiabetic Effect of Luteolin in STZ induced diabetic rats: molecular docking, molecular dynamics, in vitro and in vivo studies. J. Funct. Biomater. 2023;14(3):126. doi: 10.3390/jfb14030126. https://www.mdpi.com/2079-4983/14/3/126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj A., Saluja A. Bryophyllum pinnatum (Lam.) Kurz.: Phytochemical and pharmacological profile: a review. Pharmacogn. Rev. 2009;3(6):364. https://d1wqtxts1xzle7.cloudfront.net/99745360/PhcogRev-3-6-364-libre.pdf?1678634372=&response-content-disposition=inline%3B+filename%3DBryophyllum_pinnatum_Lam_Kurz_Phytochemi.pdf&Expires=1704106838&Signature=VUJhQAx8jpirVhqnY5gauZ∼Z4Eh7MvlShmBBjOYkLwcmSz8KFmMCX-FZaTMx2PY6XEFBYTwoyM3∼VXhW9li8C4k45SiYj5Iuhw∼niHCA9F5EcdbkPVHGwvisjE-XXBRd3M∼CZ2-enkTKrC0jJHAr6zZIXOTciC3ljnLkHkulpSKhSdzhsbFIT∼rGdxqm∼7lVC679t8tWIBIVXq8aTo0L2NcHPQqlWnaj62AD1Ri1nJ6WJnKvzBS∼cywonBrUSPYcq5Uv2KiixiPN9H7T5j8nLFOjCb0snkVI-FPPV9tADHfXjqQQXDxbqCe9ZS5-AA9wIq6kzbp6KdVyR3J2s0XBpA__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA [Google Scholar]
- Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. doi: 10.1093/nar/gkac963. https://academic.oup.com/nar/article/51/D1/D587/6775388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, P., Lal Khokra, S., Rana, A.C., Kaushik, D., 2014. Pharmacophore modeling and molecular docking studies on Pinus roxburghii as a target for diabetes mellitus. Adv. Bioinform., 2014. https://downloads.hindawi.com/archive/2014/903246.pdf. [DOI] [PMC free article] [PubMed]
- Khanal P., Patil B.M. Gene ontology enrichment analysis of α-amylase inhibitors from Duranta repens in diabetes mellitus. J. Diabetes Metab. Disord. 2020;19:735–747. doi: 10.1007/s40200-020-00554-9. https://link.springer.com/article/10.1007/s40200-020-00554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Bryant S.H. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. https://academic.oup.com/nar/article/44/D1/D1202/2503131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Huang Y., Zhao S., Guo Q., Zhou J., Han W., Xu Y. Based on network pharmacology to explore the molecular mechanisms of astragalus membranaceus for treating T2 diabetes mellitus. Ann. Transl. Med. 2019;7(22) doi: 10.21037/atm.2019.10.118. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6944577/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yuan G., Pan Y., Wang C., Chen H. Network pharmacology studies on the bioactive compounds and action mechanisms of natural products for the treatment of diabetes mellitus: a review. Front. Pharmacol. 2017;8:74. doi: 10.3389/fphar.2017.00074. https://www.frontiersin.org/articles/10.3389/fphar.2017.00074/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Lin Y., Wen X., Jorissen R.N., Gilson M.K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35(suppl_1):D198–D201. doi: 10.1093/nar/gkl999. https://academic.oup.com/nar/article/35/suppl_1/D198/1119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanthesh M.T., Ranjith D., Yaligar R., Jyothi R., Narappa G., Ravi M.V. Swiss ADME prediction of phytochemicals present in Butea monosperma (Lam.), Taub. J. Pharmacognosy Phytochem. 2020;9(3):1799–1809. https://www.phytojournal.com/archives/2020.v9.i3.11579/swiss-adme-prediction-of-phytochemicals-present-in-butea-monosperma-lam-taub [Google Scholar]
- Maradesha T., Martiz R.M., Patil S.M., Prasad A., Babakr A.T., Silina E., Ramu R. Integrated network pharmacology and molecular modeling approach for the discovery of novel potential MAPK3 inhibitors from whole green jackfruit flour targeting obesity-linked diabetes mellitus. PLoS One. 2023;18(1):e0280847. doi: 10.1371/journal.pone.0280847. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0280847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehata M.S. Green synthesis of silver nanoparticles using Kalanchoe pinnata leaves (life plant) and their antibacterial and photocatalytic activities. Chem. Phys. Lett. 2021;778 https://www.sciencedirect.com/science/article/abs/pii/S0009261421004437 [Google Scholar]
- Menon N., Sparks J., Omoruyi F. Hypoglycemic and hypocholesterolemic activities of the aqueous preparation of Kalanchoe pinnata leaves in streptozotocin-induced diabetic rats. Asian Pac. J. Trop. Biomed. 2015;5(1):3–9. https://www.sciencedirect.com/science/article/pii/S2221169115301623 [Google Scholar]
- Mohanraj, K., Karthikeyan, B.S., Vivek-Ananth, R.P., Chand, R.B., Aparna, S.R., Mangalapandi, P., Samal, A., 2018. IMPPAT: A curated database of I ndian M edicinal P lants, P hytochemistry A nd T herapeutics. Scientific reports, 8(1), 4329. j, K., Karthikeyan, B. S., Vivek-Ananth, R. P., Chand, R. B., Aparna, S. R., Mangalapandi, P., Samal, A. (2018). IMPPAT: A curated database of I ndian M edicinal P lants, P hytochemistry A nd T herapeutics. Sci. Rep., 8(1), 4329. https://www.nature.com/articles/s41598-018-22631-z. [DOI] [PMC free article] [PubMed]
- Nascimento L.B.D.S., Casanova L.M., Costa S.S. Bioactive compounds from Kalanchoe genus potentially useful for the development of new drugs. Life. 2023;13(3):646. doi: 10.3390/life13030646. https://www.mdpi.com/2075-1729/13/3/646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogidigo J.O., Anosike C.A., Joshua P.E., Ibeji C.U., Nwanguma B.C., Nwodo O.F.C. Neuroprotective effect of Bryophyllum pinnatum flavonoids against aluminum chloride-induced neurotoxicity in rats. Toxicol. Mech. Methods. 2022;32(4):243–258. doi: 10.1080/15376516.2021.1995557. https://www.tandfonline.com/doi/abs/10.1080/15376516.2021.1995557 [DOI] [PubMed] [Google Scholar]
- Oh K.K., Adnan M., Cho D.H. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0240873. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0240873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueslati N., Charradi K., Bedhiafi T., Limam F., Aouani E. Protective effect of grape seed and skin extract against diabetes-induced oxidative stress and renal dysfunction in virgin and pregnant rat. Biomed. Pharmacother. 2016;83 doi: 10.1016/j.biopha.2016.07.024. https://www.sciencedirect.com/science/article/abs/pii/S0753332216305054 [DOI] [PubMed] [Google Scholar]
- Patil V.S., Deshpande S.H., Harish D.R., Patil A.S., Virge R., Nandy S., Roy S. Gene set enrichment analysis, network pharmacology and in silico docking approach to understand the molecular mechanism of traditional medicines for the treatment of diabetes mellitus. J. Proteins Proteomics. 2020;11:297–310. https://link.springer.com/article/10.1007/s42485-020-00049-4 [Google Scholar]
- Penumala M., Zinka R.B., Shaik J.B., Mallepalli S.K.R., Vadde R., Amooru D.G. Phytochemical profiling and in vitro screening for anticholinesterase, antioxidant, antiglucosidase and neuroprotective effect of three traditional medicinal plants for Alzheimer’s Disease and Diabetes Mellitus dual therapy. BMC Complement. Altern. Med. 2018;18(1):113. doi: 10.1186/s12906-018-2140-x. https://bmccomplementmedtherapies.biomedcentral.com/articles/10.1186/s12906-018-2140-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepa R., Mohan V. Epidemiology of type 2 diabetes in India. Indian J. Ophthalmol. 2021;69(11):2932. doi: 10.4103/ijo.IJO_1627_21. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8725109/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R., Al-Sabahi J.N., Ghaffar A., Nadeem F., Umar A. Phytochemical, morphological, botanical, and pharmacological aspects of a medicinal plant: Kalanchoe pinnata–A review article. Int. J. Chem. Biochem. Sci. 2019;16:5–10. https://www.iscientific.org/wp-content/uploads/2020/11/2-IJCBS-19-16-2.pdf [Google Scholar]
- Rajesh A., Shamsudin M. In silico molecular docking studies on phytocompounds from the plant Kalanchoe pinnata targeting the pi-class glutathione-s-transferase of Wuchereria bancrofti. Int J Zool Appl Biol. 2017;2(5):258–265. https://d1wqtxts1xzle7.cloudfront.net/88399099/8._IJZAB_ID_No._262-libre.pdf?1657374509=&response-content-disposition=inline%3B+filename%3DIN_SILICO_MOLECULAR_DOCKING_STUDIES_ON_P.pdf&Expires=1704107799&Signature=BkEitnfUyhYbmW7xu-svYjgfIO8xh1rH06sRPDvMk2-mtBwaQAli39kKSQP9lV5lbiVyfpJLlWgYyhywvWwx07zFmUg6EpIGIGJkhFISaEz3e1C-kAUQn64I5h0OSZKvgaAN2Prkih-8Nm2UR6r0kEisFf06BlYPwmNMGriKR7N-EIaaovoQYa3l4W4rA2NNuP53y0GkFZMcMzpSrW8F8lb4BHZRr4ma2PLWJDE0WU2-8nqU-5Dzii2KbuDzD5-Wtfn4rk0oRftfaJc2oPu8eHfU1fQl0ZPuGrgF0HulO6hWCt0HvRZQVCtxWdT∼RU12KqPK2bz0ndA92Si6EDLrTw__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA [Google Scholar]
- Ramon P., Bergmann D., Abdulla H., Sparks J., Omoruyi F. Bioactive Ingredients in K. pinnata Extract and Synergistic Effects of Combined K. pinnata and Metformin Preparations on Antioxidant Activities in Diabetic and Non-Diabetic Skeletal Muscle Cells. Int. J. Mol. Sci. 2023;24(7):6211. doi: 10.3390/ijms24076211. https://www.mdpi.com/1422-0067/24/7/6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revankar A.G., Bagewadi Z.K., Shaikh I.A., Mannasaheb B.A., Ghoneim M.M., Khan A.A., Asdaq S.M.B. In-vitro and computational analysis of Urolithin-A for anti-inflammatory activity on Cyclooxygenase 2 (COX-2) Saudi J. Biol. Sci. 2023;30(11) doi: 10.1016/j.sjbs.2023.103804. https://www.sciencedirect.com/science/article/pii/S1319562X23002498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roglic G., Varghese C., Thamarangsi T. Diabetes in South-East Asia: burden, gaps, challenges and ways forward. WHO South-East Asia J. Public Health. 2016;5(1):1–4. doi: 10.4103/2224-3151.206546. https://iris.who.int/bitstream/handle/10665/329626/seajphv5n1p1.pdf [DOI] [PubMed] [Google Scholar]
- Sabiu S., Balogun F.O., Amoo S.O. Phenolics profiling of Carpobrotus edulis(L.) NE Br. and insights into molecular dynamics of their significance in type 2 diabetes therapy and its retinopathy complication. Molecules. 2021;26(16):4867. doi: 10.3390/molecules26164867. https://www.mdpi.com/1420-3049/26/16/4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savova M.S., Mihaylova L.V., Tews D., Wabitsch M., Georgiev M.I. Targeting PI3K/AKT signaling pathway in obesity. Biomed. Pharmacother. 2023;159 doi: 10.1016/j.biopha.2023.114244. https://www.sciencedirect.com/science/article/pii/S075333222300032X [DOI] [PubMed] [Google Scholar]
- Shardlow, M., Nguyen, N., Owen, G., O’Donovan, C., Leach, A., McNaught, J., et al., 2018, May. A new corpus to support text mining for the curation of metabolites in the ChEBI database. In Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018). https://aclanthology.org/L18-1042.pdf.
- Sharma P., Joshi T., Joshi T., Chandra S., Tamta S. Molecular dynamics simulation for screening phytochemicals as α-amylase inhibitors from medicinal plants. J. Biomol. Struct. Dyn. 2021;39(17):6524–6538. doi: 10.1080/07391102.2020.1801507. https://www.tandfonline.com/doi/abs/10.1080/07391102.2020.1801507 [DOI] [PubMed] [Google Scholar]
- Sharma B., Yadav D.K. Metabolomics and network pharmacology in the exploration of the multi-targeted therapeutic approach of traditional medicinal plants. Plants. 2022;11(23):3243. doi: 10.3390/plants11233243. https://www.mdpi.com/2223-7747/11/23/3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubhashree M.N., Shanta T.R., Thakur R., Rao V.R. Significance of identification of ayurvedic drugs with its different sources. Res. J. Pharmacol. Pharmacodynamics. 2020;12(3):117–129. https://www.indianjournals.com/ijor.aspx?target=ijor:rjppd&volume=12&issue=3&article=003 [Google Scholar]
- Smruthi G., Mahadevan V., Vadivel V., Brindha P. Docking studies on antidiabetic molecular targets of phytochemical compounds of Syzygium cumini (L.) skeels. Asian J. Pharm. Clin. Res. 2016;9(3):287–293. https://d1wqtxts1xzle7.cloudfront.net/69706575/admin_2C_Journal_manager_2C_60_AJPCR_14920_RA.pdf_filename_UTF-8admin_2C_Journal_manager_2C_60_AJPCR_14920_RA-libre.pdf?1631721018=&response-content-disposition=inline%3B+filename%3DDocking_Studies_on_Antidiabetic_Molecula.pdf&Expires=1704106962&Signature=EitO7o1l0vm8TN20WvsKuYZhW1pLqKBSr1wx5NAP0IsJP2d5RTXXOmGwcKDdEOWJCNqSMRb2qr7B0D8ETosY7pmvWGeWMMfXSC4∼hJpVIAbLi∼Pkzh1iuBMG6ymrH6IyJxv5rN∼7OcDkQKDClXR4fmWTo∼73yBYhsILeFUYYGzUq98YQ∼e0hKXlagOF9xpFZFP6C55c0eACBjRazOA48∼hy9∼8bfzhOVcFPvVPrzD2GI8qScNhS43fUtaOMjzzlvfj0QVp1N5qMLEN8DQk5∼7Q2JhkMZxEOJZuJj984vmckpnp∼-xqY4MW0qq9rjHVWRFucTgWVXIYvCtgXkk4iw__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA [Google Scholar]
- Standl E., Khunti K., Hansen T.B., Schnell O. The global epidemics of diabetes in the 21st century: current situation and perspectives. Eur. J. Prev. Cardiol. 2019;26(2_suppl):7–14. doi: 10.1177/2047487319881021. https://academic.oup.com/eurjpc/article/26/2_suppl/7/5925429 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Mering C.V. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. https://academic.oup.com/nar/article/47/D1/D607/5198476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vounzoulaki, E., Khunti, K., Abner, S.C., Tan, B.K., Davies, M.J., Gillies, C.L., 2020. Progression to type 2 diabetes in women with a known history of gestational diabetes: systematic review and meta-analysis. Bmj, 369. https://www.bmj.com/content/369/bmj.m1361.full. [DOI] [PMC free article] [PubMed]
- Xu, M., Li, Z., Yang, L., Zhai, W., Wei, N., Zhang, Q., et al., 2020. Elucidation of the mechanisms and molecular targets of sanhuang xiexin decoction for type 2 diabetes mellitus based on network pharmacology. BioMed Res. Int., 2020. https://www.hindawi.com/journals/bmri/2020/5848497/. [DOI] [PMC free article] [PubMed]
- Yamari I., Mouhib A., Es-Sounni B., Nejjari R., Mazoir N., Bakhouch M., Chtita S. Oxidative functionalization of triterpenes isolated from Euphorbia resinifera latex: semisynthesis, ADME-Tox, molecular docking, and molecular dynamics simulations. Chem. Phys. Impact. 2023;7 https://www.sciencedirect.com/science/article/pii/S2667022423002116 [Google Scholar]
- Yamari I., Abchir O., Siddique F., Zaki H., Errougui A., Talbi M., Chtita S. The anticoagulant potential of Lippia Alba extract in inhibiting SARS-CoV-2 Mpro: Density functional calculation, molecular docking analysis, and molecular dynamics simulations. Scientific African. 2024;23:e01986. https://www.sciencedirect.com/science/article/pii/S2468227623004404 [Google Scholar]
- Yan J., Zhang G., Pan J., Wang Y. α-Glucosidase inhibition by luteolin: Kinetics, interaction and molecular docking. Int. J. Biol. Macromol. 2014;64:213–223. doi: 10.1016/j.ijbiomac.2013.12.007. https://www.sciencedirect.com/science/article/abs/pii/S0141813013006570 [DOI] [PubMed] [Google Scholar]
- Yaraguppi D.A., Bagewadi Z.K., Deshpande S.H., Chandramohan V. In silico study on the inhibition of UDP-N-acetylglucosamine 1-carboxy vinyl transferase from Salmonella typhimurium by the lipopeptide produced from Bacillus aryabhattai. Int. J. Pept. Res. Ther. 2022;28(3):80. https://link.springer.com/article/10.1007/s10989-022-10388-z [Google Scholar]
- Yin, B., Bi, Y.M., Fan, G.J., Xia, Y.Q., 2020. Molecular mechanism of the effect of Huanglian Jiedu decoction on type 2 diabetes mellitus based on network pharmacology and molecular docking. J. Diabetes Res., 2020. https://www.hindawi.com/journals/jdr/2020/5273914/. [DOI] [PMC free article] [PubMed]
- Zang Y., Igarashi K., Li Y. Anti-diabetic effects of luteolin and luteolin-7-O- glucoside on KK-A y mice. Biosci. Biotech. Bioch. 2016;80(8):1580–1586. doi: 10.1080/09168451.2015.1116928. https://academic.oup.com/bbb/article/80/8/1580/5938170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.