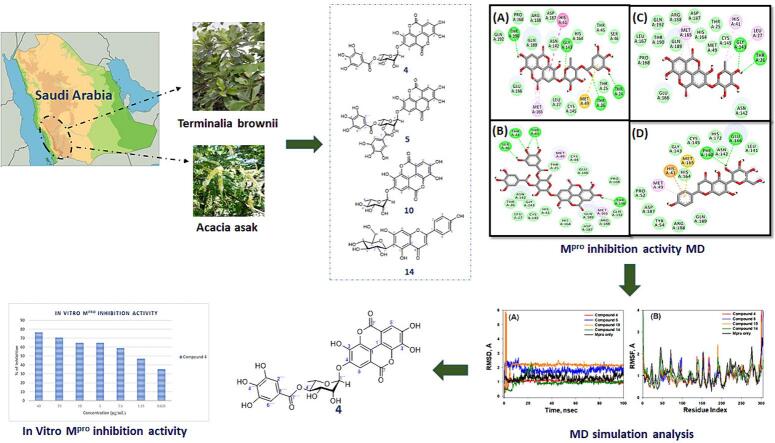
Graphical abstract
Keywords: SARS-CoV-2 Mpro, Saudi plants, Terminalia brownii, Acacia asak, Hydrolysable tannins, Flavonoid C-glycosides, Molecular docking
Abstract
The escalation of many coronavirus variants accompanied by the lack of an effective cure has motivated the hunt for effective antiviral medicines. In this regard, 18 Saudi Arabian medicinal plants were evaluated for SARS CoV-2 main protease (Mpro) inhibition activity. Among them, Terminalia brownii and Acacia asak alcoholic extracts exhibited significant Mpro inhibition, with inhibition rates of 95.3 % and 95.2 %, respectively, at a concentration of 100 µg/mL. Bioassay-guided phytochemical study for the most active n-butanol fraction of T. brownii led to identification of eleven compounds, including two phenolic acids (1, and 2), seven hydrolysable tannins (3–10), and one flavonoid (11) as well as four flavonoids from A. asak (12–15). The structures of the isolated compounds were established using various spectroscopic techniques and comparison with known compounds. To investigate the chemical interactions between the identified compounds and the target Mpro protein, molecular docking was performed using AutoDock 4.2. The findings identified compounds 4, 5, 10, and 14 as the most potential inhibitors of Mpro with binding energies of −9.3, −8.5, −8.1, and −7.8 kcal mol−1, respectively. In order to assess the stability of the protein–ligand complexes, molecular dynamics simulations were conducted for a duration of 100 ns, and various parameters such as RMSD, RMSF, Rg, and SASA were evaluated. All selected compounds 4, 5, 10, and 14 showed considerable Mpro inhibiting activity in vitro, with compound 4 being the most powerful with an IC50 value of 1.2 µg/mL. MM-GBSA free energy calculations also revealed compound 4 as the most powerful Mpro inhibitor. None of the compounds (4, 5, 10, and 14) display any significant cytotoxic activity against A549 and HUVEC cell lines.
1. . Introduction
COVID-19, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a highly contagious viral disease that has rapidly evolved into a global pandemic (Perveen et al., 2020, Berekaa, 2021). Infection with COVID-19 may entail serious health complications with a fatality rate of about 3.7 percent, according to official national statistics in China (Chakraborty et al., 2020, Landete et al., 2020, Zhu et al., 2020b). Currently, only limited approved treatment options are available for managing mild-to-moderate COVID-19 in pediatric and adult patients such as Veklury (Remdesivir), Lagevrio (molnupiravir), and Paxlovid (ritonavir tablets and nirmatrelvir tablets, co-packed for oral use), in addition to SARS-CoV-2-targeting monoclonal antibody (mAb) products, including Bebtelovimab and Evusheld (tixagevimab co-packaged with cilgavimab) (Zhu et al., 2020b). Furthermore, immune modulators are another category of drugs authorized for COVID-19 treatment, in which they help modulate immune function and suppress hyperinflammation associated with COVID-19 infection, such as Kineret (anakinra), Olumiant (baricitinib), and Actemra (tocilizumab) (U.S. Food & Drugs, 2022).
The SARS-CoV-2 virus is composed of four main structural proteins: envelope (E) protein, membrane (M) glycoproteins, spike (S) glycoprotein, and nucleocapsid (N) proteins, in terms of its structure. Of all these proteins, the S-protein holds significant importance due to its crucial role in enabling the virus to enter host cells. This is achieved through its interaction with the host protein angiotensin-converting enzyme-2 (ACE2), which serves as the receptor for SARS-CoV-2. Thus, it is considered vital to disrupt the bond between the spike protein and ACE2 in order to hinder viral entry, making it a crucial strategy in the development of medical treatments for COVID-19 infection (Satarker and Nampoothiri, 2020). Other suggested suitable tactics for medical treatment involve the utilization of direct-acting antiviral drugs that can effectively target specific enzymes responsible for viral replication. These enzymes include the main protease (Mpro) or 3C-like protease (3CLpro), papain-like protease (PLpro), non-structural protein 12 (nsp12), and RNA-dependent RNA polymerase (RdRP). By inhibiting the activity of these enzymes, the replication of the virus can be effectively hindered, providing potential avenues for the development of effective medical treatments against COVID-19.
The main protease (Mpro) plays a vital role in the virus's replication process, making it an attractive target for therapeutic intervention. Peptidomimetic inhibitors, including drugs like lopinavir and ritonavir, have been investigated as a class of inhibitors (Citarella et al., 2021). Originally developed as HIV protease inhibitors, these drugs have shown varying results in studies assessing their effectiveness against SARS-CoV-2. Although they have been considered, they are not widely recommended for the treatment of COVID-19 (Cao et al., 2020). Another approach involves small molecule inhibitors, such as remdesivir, which was initially developed for Ebola. Although not a direct Mpro inhibitor, remdesivir interferes with viral RNA synthesis and has received emergency use authorization for the treatment of COVID-19 (Garibaldi et al., 2021).
Natural products have been explored as potential Mpro inhibitors (Hossain et al., 2023). Compounds derived from medicinal plants and traditional medicines have been subjects of investigation for their antiviral properties, although their efficacy and safety profiles require comprehensive evaluation. Covalent inhibitors, exemplified by PF-07304814, represent another class (Jiang et al., 2023). Developed by Pfizer, this oral protease inhibitor is designed to bind to the active site of the Mpro enzyme, forming a stable bond and inhibiting its activity. Metal ion chelators, like disulfiram, have been studied for their potential as Mpro inhibitors (Ali et al., 2022). These compounds aim to interfere with the zinc ion, which is essential for the enzymatic activity of Mpro. Structure-based inhibitors, such as N3 and 11a, have been developed based on the crystal structure of the Mpro enzyme (Dai et al., 2020). These inhibitors are specifically tailored to fit into the enzyme's active site, disrupting its function and impeding viral replication.
The escalation of many coronavirus variants accompanied by the lack of effective cure has generated a persistent drive to seek out effective antiviral medications. Thus, the combination of discovering and designing new drug tools is one of the essential ways to follow and overcome this global crisis. In this regard, medicinal plants have been widely recognized as a valuable source of chemically diverse, structurally unique, and pharmacologically effective metabolites with a rich historical background. Therefore, the ongoing exploration of antiviral phytoconstituents, coupled with drug repositioning and designing tools, is regarded as an effective approach for the discovery of novel drug candidates (Al-Wahaibi et al., 2022).
Saudi Arabia is considered one of the wealthiest countries in terms of plant biodiversity in the Middle East. The Saudi flora contains more than 2000 plant species distributed in more than 140 families with many used in Saudi folk medicine (El-Seedi et al., 2020). Several plant species in the Saudi flora have been documented to have antiviral properties, their potential against the SARS-CoV-2 virus remains largely unexplored in scientific research.
In the current study, our objective was to investigate the in vitro inhibitory activity of 18 selected plant extracts sourced from Saudi Arabia against the SARS-CoV-2 main protease (Mpro). The selection of these plants was primarily based on their occurrence in Saudi Arabia, which signifies their availability within our ecosystem. Secondly, the well-documented and extensive range of promising antiviral activity demonstrated by these plants was also considered. We employed a bioassay-guided approach to fractionate and purify the active constituents from the most promising plant species, namely Terminalia brownii and Acacia asak.
2. Materials and methods
2.1. Apparatus and chemicals
1D and 2D NMR experiments were carried out at the College of Pharmacy, King Saud University, using an UltraShield Plus 500 and AV-700 MHz NMR spectrometers. UV spectra were acquired using a Cary 50 spectrophotometer. Mass spectroscopic analysis was performed in both negative and positive modes separately using an electrospray ionization (ESI) probe on a Waters tandem triple quadrupole mass spectrometer (TQD) (Waters, Milford, USA), with a scan range from 160 to 1500 m/z.
Thin-layer chromatography (TLC) was carried out on RP-18 F254S (Merck, Darmstadt, Germany) for reversed phase, and silica-gel 60 F254 (0.25 mm, ALUGRAM® SIL G/UV254, Macherey-Nagel, Easton, PA, USA) for normal phase. Compounds were detected by spraying with p-anisaldehyde/H2SO4 reagent, followed by heating at 110 °C for 1–2 min.
Column chromatography (CC) was performed using normal silica gel (Kieselgel 60 Å, 40–63 µm mesh size, Merck, Frankfurt, Germany) for normal phase, diaion HP-20 resin (Mitsubishi Chemical, Tokyo, Japan), and sephadex LH-20 (25–100 mm mesh size, GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
Shimadzu HPLC-LC-20 AD series was used for the reverse phase (RP)-HPLC separation equipped with a binary gradient pump, Phenomenex Jupiter Proteo column (Jupiter Proteo 90 Å, 250 × 10 mm, 4 μm) (Phenomenex, Inc., Torrance, CA, USA), and Shimadzu SPD-M20A detector (Shimadzu, Kyoto, Japan).
2.2. Plant materials
A total of 18 medicinal plants were collected from various regions of Saudi Arabia. These plants were chosen for their reported antiviral properties against DNA or RNA viruses as well as their availability in Saudi Arabia (Table 1). The authentication of plants was conducted by a taxonomic specialist at the Department of Pharmacognosy, College of Pharmacy, King Saud University, and voucher specimens have been securely deposited in the college herbarium.
Table 1.
List of the selected Saudi medicinal plants and the reported antiviral activity of their genera.
| S. No. | Plant name | Family | Voucher | Locality | Reported antiviral activity of genera | References |
|---|---|---|---|---|---|---|
| 1 | Barleria bispinosa | Acanthaceae | 15,449 | AL-Baha, KSA | HSV, HIV, RSV | (Yoosook et al., 1999) (Maregesi et al., 2010) (Chen et al., 1998) |
| 2 | Zizyphus mucronata | Rhamnaceae | 15,460 | Tabuk, KSA | IAV, HIV | (Hong et al., 2015) (Behbahani, 2014) |
| 3 | Terminalia brownii | Combretaceae | 16,031 | Abha, KSA | HCV, HIV, HSV | (Patil et al., 2022) (Ahn et al., 2002) (Kesharwani et al., 2017) |
| 4 | Acacia asak | Fabaceae | 16,387 | Madinah, KSA | HIV, HCV | (Nutan et al., 2013) (Rehman et al., 2011) |
| 5 | Hibiscus deflersii | Malvaceae | 16,257 | AL-Baha, KSA | IAV, CoxV | (Takeda, et al., 2020) (El-Shiekh et al., 2020) |
| 6 | Maerua crassifolia | Capparaceae | 15,804 | AL-Baha, KSA | Fowl pox | (Mohamed et al., 2010) |
| 7 | Ocimum forsskaolii | Lamiaceae | 16,243 | AL-Baha, KSA | H9N2, HSV, HIV | (Ghoke et al., 2018) (Kapewangolo et al., 2017) (Chiang et al., 2005) |
| 8 | Pulicaria schimperi | Asteraceae | 15,802 | AL-Baha, KSA | IAV, HBV | (Abo-Elghiet et al., 2023) (Arbab et al., 2017) |
| 9 | Bidens biternata | Asteraceae | 16,067 | Nimas, KSA | HSV | (Chiang,et al., 2003) |
| 10 | Conyza pyrrhopappa | Asteraceae | 15,302 | Al-Baha, KSA | Polio, HSV | (Ananil et al., 2000) |
| 11 | Pluchea dioscoridis | Asteraceae | 16,289 | AL-Baha, KSA | HSV | (Visintini et al., 2013) |
| 12 | Euphorbia schimperiana | Euphorbiaceae | 16,322 | Taif, KSA | HIV, HSV, RSV | (Gyuris et al., 2009) (Hohmann et al., 2000) (Huang et al., 2014) |
| 13 | Momordica balsamina | Cucurbitaceae | 16,395 | Madinah, KSA | IAV, HIV | (Pongthanapisith et al., 2013) (Fang and Ng, 2011) |
| 14 | Kalanchoe glaucescens | Crassulaceae | 16,271 | AL-Baha, KSA | CoxV, EV, HSV | (Wang et al., 2012) (Ürményi et al., 2016) |
| 15 | Rhus retinorrhoea | Anacardiaceae | 15,331 | AL-Baha, KSA | IHNV, VHSV, VACV | (Babbar et al., 1982) (Kang et al., 2012) |
| 16 | Ficus benghalensis | Moraceae | 16,080 | Riyadh, KSA | HIV | (Bunluepuech and Tewtrakul, 2009) |
| 17 | Alkanna orientalis | Boraginaceae | 16,048 | Taif, KSA | CoxV | (Elsohly et al., 1997) |
| 18 | Arnebia hispidissima | Boraginaceae | 16,066 | AL-Baha, KSA | HCV, HIV | (Li et al., 2012) (Kashiwada et al., 1995) |
Avian influenza (H9N2), Coxsackie virus (CoxV), Enterovirus (EV), Hepatitis B virus (HBV), Hepatitis C virus (HCV), Herpes simplex viruses (HSV), Human immunodeficiency virus (HIV), Infectious hematopoietic necrosis virus (IHNV), Influenza A virus (IAV), Respiratory syncytial virus (RSV), Vaccinia virus (VACV), Viral hemorrhagic septicemia virus (VHSV).
The selected plants are known for their diverse chemical composition, offering a great opportunity to study the effectiveness of different chemical classes of active constituents as antiviral agents.
2.3. Extraction, fractionation and isolation of phytoconstituents
The aerial parts of 18 medicinal plants, each weighing 200 g, were shade-dried and then coarsely powdered. They were then extracted by maceration in 80 % aqueous ethanol (1 L × 5) at room temperature. Afterward, the extracted mixture was filtered and concentrated using a rotary evaporator (Buchi Labortechnik AG, Flawil, Switzerland) under vacuum conditions.
After evaluating the Mpro inhibitory activity of alcoholic extracts obtained from eighteen plants, a total of 750 g of the aerial parts of the active plants, namely T. brownii and A. asak, were completely extracted using maceration in 80 % aqueous ethanol. As a result, a combined alcoholic extract of 73 g (9.7 % w/w) for T. brownii and 66 g (8.8 % w/w) for A. asak was obtained. Subsequently, the complete alcoholic extracts underwent sequential liquid–liquid partitioning process using various organic solvents with ascending polarity: starting with n-hexane, followed by chloroform, and finally n-butanol to obtain the respective fractions. Among these fractions, the n-butanol fractions from both T. brownii and A. asak exhibited the most potent Mpro inhibition activity. Consequently, these fractions underwent bioassay-guided fractionation to isolate their active compounds.
The n-BuOH fraction obtained from T. brownii, weighing 32 g, was subjected to a series of purification steps. Firstly, it was passed through a column packed with porous-polymer Diaion HP-20 polystyrene resin. The column was eluted sequentially with water, 25 % MeOH, 50 % MeOH, and pure MeOH, each using a volume of 6 L. The fractions eluted with 25 % MeOH (6.1 g) and 50 % MeOH (7.4 g) were combined due to their similarities on thin-layer chromatography (TLC). This combined fraction was further subjected to gel filtration chromatography utilizing a Sephadex LH-20 column, with a solvent system consisting of chloroform–methanol (4:1). The elution yielded fifty-eight fractions, each collected in 100 mL portions. The fractions were then monitored on reversed-phase C18 TLC with solvent systems composed of water-acetonitrile (80:20), (70:30), and (60:40). The TLC plates were sprayed with anisaldehyde/H2SO4 in ethanol as a reagent, followed by heating.
Thirteen main groups (I-XIII) were formed by combining similar fractions. Among these groups, IV, V, VII, IX, and XII were selected for further analysis. Each selected group was subjected to high-performance liquid chromatography (HPLC) using a reversed-phase Proteo column and a gradient of 5–60 % acetonitrile (CH3CN) in water over 60 min. As a result, compound 9 (21.4 mg) was obtained from group IV, compound 8 (45.8 mg) from group V, compound 10 (19.6 mg) from group VII, compound 11 (31.5 mg) from group IX, and compound 2 (86.8 mg) from group XII.
The fraction eluted with pure methanol (MeOH) (12.4 g) underwent purification through normal silica gel column chromatography. Gradient elution was performed using a mixture of chloroform (CHCl3) and methanol (MeOH). As a result, sixty-four fractions, each with a volume of 100 mL, were collected and classified into nine main groups denoted as I-IX. Among these groups, III, VI, VII, IX, and XII were chosen for further investigation. These selected groups were subjected to high-performance liquid chromatography (HPLC) using a reversed-phase Proteo column. A gradient elution was employed with a mixture of 5–100 % acetonitrile (CH3CN) and water (H2O) over 35 min. Consequently, compound 1 weighing 53.1 mg was obtained from group III. From group V, compounds 4 (25.8 mg) and 5 (20.7 mg) were isolated. Group VIII yielded compounds 3 (16.9 mg) and 6 (27.3 mg). Lastly, compound 7 weighing 16.8 mg was obtained from group IX.
The n-BuOH fraction of A. asak (21.8 g), underwent a series of purification steps. Initially, it was passed through a column packed with porous-polymer Diaion HP-20 polystyrene resin. The column was then eluted sequentially with water, 50 % methanol (MeOH), and pure MeOH, each using a volume of 5 L. The fraction eluted with 50 % MeOH (6.1 g) was further subjected to gel filtration chromatography using a Sephadex LH-20 column. A solvent system composed of chloroform–methanol (4:1) was employed in this process.
As a result, forty-nine fractions were obtained and categorized into seven main groups. Groups IV and V were specifically chosen for additional purification. These selected groups underwent high-performance liquid chromatography (HPLC) using a reversed-phase Proteo column. A gradient elution method was employed using a mixture of 5–60 % acetonitrile (CH3CN) and water (H2O) over 40 min. This procedure yielded compound 15 weighing 36.3 mg from group IV and compound 14 weighing 23.8 mg from group V.
Finally, the pure MeOH-eluted fraction (8.6 mg) was chromatographed on a silica gel column using CHCl3–MeOH in a manner of increasing polarities to provide six main sub-fractions. Subfractions III (1.1 g) and V (0.82 g) were purified by RP Proteo HPLC using a gradient of 0–100 % CH3CN–H2O over 40 min to yield compound (13) (27.6 mg) and compound (12) (16.2 mg) respectively.
2.4. Mpro protease inhibitory assay
2.4.1. Chemical reagents and materials
To assess the Mpro inhibitory activity of plant extracts and isolated compounds, the 3CL Protease, MBP-tagged (SARS-CoV-2) Assay Kit was employed. The assay kit (catalog #79955–1) was procured from BPS Bioscience Inc. (San Diego, CA, USA) and consisted of purified MBP-tagged recombinant 3CL protease, a fluorogenic substrate, assay buffer, and 3CL inhibitor (GC376) as a positive control.
2.4.2. Fluorescent SARS-CoV-2 Mpro enzyme inhibition assay
The assay kit uses a recombinant Mpro protease enzyme tagged with MBP (maltose-binding protein) to catalyze the cleavage of a fluorogenic protease substrate, as instructed by the manufacturer. This cleavage reaction takes place within a homogeneous fluorescence resonance energy transfer (FRET)-based cleavage assay, eliminating the need for time-consuming washing steps (Li et al., 2021, Abdallah et al., 2020). when the Mpro protease enzyme cleaves the protease substrate, it emits fluorescent signals that can be detected at excitation/emission wavelengths of 360/460 nm, respectively. The activity of the Mpro protease enzyme is determined by comparing the fluorescent intensity of the test samples with that of the blank control.
In a 96-well black plate, a mixture of the test sample (10 μL) and main protease (30 μL) at different concentrations was incubated at room temperature for 30 min with slow shaking. To initiate the reaction, 10 μL of the main protease substrate was added to each well, and the plate was sealed and incubated at room temperature overnight. The fluorescence intensity of each well at an excitation wavelenght of 360 nm and an emission wavelength of 460 nm is measured using a microtiter plate reading fluorimeter (Synergy HT, BioTek Instruments; Highland Park, Winooski, Vermont, USA).
2.5. Cell viability (MTT) assay
The MTT assay was employed following the methodology described in a previous study by Nasr et al. (2020). In brief, lung cancer cell (A549; ACC 107; Braunschweig, Germany) and normal HUVEC cells (ATCC 16549; Cat# PCS-100–010, USA) were seeded in a 96-well plate at a concentration of 5 × 104 cells per well. After 24 h, the cells were exposed to tested compounds at various concentrations (1–100 µg/mL) and incubated for 48 h. Subsequently, 10 µL of freshly prepared MTT solution (5 mg/mL) was added to each well, and the plate was further incubated for 2–4 h at 37 °C. The resulting formazan product's absorbance was measured at 570 nm using an ELISA plate reader (BioTek, USA). Cell viability was then determined using the following calculation:
| % Cell Viability = Mean absorbance [(treated cells / untreated cells] × 100 |
The dose–response curve of the percentage of cell viability was used to calculate the IC50 values using OriginPro software.
2.6. Molecular docking
The molecular docking analysis was performed utilizing the PyRx software with AutoDock VINA, following the procedure outlined in Al-Shabib et al., (2018). Initially, the 2D structures of the compounds under investigation were created using ChemDraw, saved as mol2 files, and subsequently imported into PyRx. The compounds were energy minimized using the universal forcefield (UFF), and converted to pdbqt format using Open Babel. The 3D coordinates of the Mpro protein (PDB ID: 6LU7) (Jin et al., 2020) were obtained from the RCSB website (https://www.rcsb.org) and prepared by adding missing hydrogen atoms and removing non-essential foreign and water molecules. The energy of the complete system was minimized using the CHARMM36 force field. The dimensions of the grid box for Mpro were set as (−19.7 × 21.7 × 66.4) Å placed at (37.8 × 28.6 × 30.7) Å with 0.375 Å spacing. Molecular docking was performed using Lamarckian Genetic Algorithm (LGA) along with Solis and Wets local search methods (Rehman et al., 2014).
The compounds were assigned random orientations, initial positions, and torsions. Each docking run entailed a maximum of 2.5 × 106 energy calculations, where every calculation assessed the interaction energy between the ligand and the protein. This evaluation considered diverse contributions such as van der Waals forces, electrostatic interactions, and hydrogen bonding. The parameters for the docking run included a population size of 150, representing the number of individual ligand conformations generated and evaluated during the docking simulation. The translational step determined the distance by which the ligand was moved in translational space during each step of the docking algorithm. In this study, a smaller translational step size (0.2 Å) was implemented, indicating the use of a finer grid to explore translational space more precisely, facilitating accurate sampling of potential binding sites. Additionally, another docking parameter, torsion steps, was set to 5. This parameter relates to the rotation of flexible bonds in the ligand molecule. A higher number of torsion steps (in this case, 5) implies that the algorithm explores a broader range of torsional angles, allowing for increased flexibility in the ligand's conformation. Moreover, quaternions, mathematical constructs used to describe molecular rotations efficiently, were employed. In the context of molecular docking, the number of quaternions determined the granularity of rotational sampling. With a higher number of quaternions (here, 5), the algorithm considered more orientations for the ligand during the rotational search.
The obtained results were analyzed, and the final figures were created using Discovery Studio (Accelrys). The dissociation constant (Kd) of compounds for Mpro was calculated using the docking energy (ΔG) following the equation:
where R and T were the universal gas constant and temperature respectively.
2.7. Molecular dynamics (MD) simulation
To evaluate the stability of the complexes formed between the protein and ligands, 100 ns molecular dynamics (MD) simulations were performed using Desmond. The complexes underwent comprehensive preparation steps including preprocessing, optimization, and minimization procedures, utilizing OPLS3e force field for optimization (Shivakumar et al., 2012). To replicate physiological conditions, we neutralized the systems by adding 0.15 M NaCl salt and necessary counter ions. And created a suitable solvation environment using an orthorhombic box measuring 10 Å × 10 Å × 10 Å, containing TIP3P water molecules (Price and Brooks, 2004). The NPT ensemble was established at 1 atm pressure and 30 K temperature.
Before commencing the simulations, the systems underwent a relaxation process, and the Martyna-Tobias-Klein barostat, and Nose-Hoover Chain thermostat were employed to maintain NPT conditions (Martyna et al., 1994, Branka, 2000). Trajectories were recorded at 10 ps intervals with a 2 fs time step throughout the simulation for subsequent result analysis.
2.8. Free energy calculations
The Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) approach, implemented with the Prime module (Schrodinger, LLC, NY, USA), was used to calculate the free energy of protein–ligand complex formation, as described previously (Iqbal et al., 2021). The docked complexs were intially optimized using the Molecular Mechanics (MM) approach and subsequently their energies were further minimized using the OPLS3e force field in conjunction with the Generalized Born Surface Area (GBSA) continuum solvent model. The binding free energies of the protein–ligand complexes were calculated using the following relationships:
Where, ELigand, EProtein, and EComplex were the minimized energies of ligand, protein, and protein–ligand complex, respectively.
where, GSolv_GB (Ligand), GSolv_GB (Protein), and GSolv_GB (Complex) were the free energies of solvation of the ligand, protein, and protein–ligand complex, respectively.
where, GSA (Ligand), GSA (Protein), and GSA (Complex), were the surface area energies of the ligand, protein, and protein–ligand complex, respectively.
The free energy, in the Prime-MM/GBSA method, is calculated as follows:
2.9. Principal Component analysis (PCA), and PCA-based 2D free energy surface
The Principal Component Analysis (PCA) method, implemented with the Bio3D package (Grant et al., 2021), was employed to evaluate the collective motions of both proteins and their associated ligands. In this procedure, the initial step involved the elimination of translational and rotational motions of the protein. Subsequently, the covariance matrix and its corresponding eigenvectors were computed by superimposing the atomic coordinates of the protein onto a reference structure. The symmetric matrix was then diagonalized using an orthogonal transformation matrix, yielding a diagonalized matrix of eigenvalues. The covariance matrix (C) was determined according to the following equation:
where, N, xi/j and < xi/j > represent the number of Cα-atom, the Cartesian coordinate of the ith/jth Cα-atom, and the time average of all the conformations, respectively.
Further, 2D PCA-based free energy surface was determined using R package, as described previously (Loganathan et al., 2024). It helped in monitoring the crucial motions aiding protein–ligand interactions in the docked complex throughout the MD simuation time.
3. Results
3.1. Screening of plant-based extracts for inhibition of SARS-CoV-2 Mpro activity
The in vitro Mpro inhibitory activity of 18 medicinal plants was investigated using the enzymatic 3CL Protease, MBP-tagged (SARS-CoV-2) assay, with the known inhibitor GC376 serving as a positive control. The findings (Table 2) revealed that the ethanolic extracts of Terminalia brownii and Acacia asak were the most potent SARS-CoV-2 Mpro inhibitors. Their percentages of inhibition at a concentration of 100 µg/mL were 95.3 % and 95.2 % respectively. Moreover, the extract of T. brownii showed more than 50 % inhibition at a concentration of 50 µg/mL.
Table 2.
The in vitro inhibitory assays of the selected plant extracts and SARS-CoV Mpro.
| S. No. | Plant name | Family |
Eth. extract Conc. (µg /mL) |
% inhibition |
Subfractions Conc. (µg /mL) |
% inhibition |
|---|---|---|---|---|---|---|
| 1 | Barleria bispinosa | Acanthaceae |
a. 100 b. 50 |
27.7 % 13.4 % |
– | – |
| 2 | Zizyphus mucronata | Rhamnaceae |
a. 100 b. 50 |
48.8 % 31.3 % |
– | – |
| 3 | Terminalia brownii | Combretaceae |
a. 100 b. 50 |
95.3 % 51.6 % |
Hex. (50 µg/mL)Chl. (50 µg/mL) But. (50 µg/mL) |
52.9 % 47.0 % 61.8 % |
| 4 | Acacia asak | Fabaceae |
a. 100 b. 50 |
95.2 % 27.6 % |
Hex. (50 µg/mL)Chl. (50 µg/mL)But. (50 µg/mL) |
58.8 % 58.8 % 82.4 % |
| 5 | Hibiscus deflersii | Malvaceae |
a. 100 b. 50 |
31.0 % 15.8 % |
– | – |
| 6 | Maerua crassifolia | Capparaceae |
a. 100 b. 50 |
11.5 % 3.7 % |
– | – |
| 7 | Ocimum forsskaolii | Lamiaceae |
a. 100 b. 50 |
56.7 % 20.1 % |
– | – |
| 8 | Pulicaria schimperi | Asteraceae |
a. 100 b. 50 |
24.8 % 7.1 % |
– | – |
| 9 | Bidens biternata | Asteraceae |
a. 100 b. 50 |
54.3 % 26.1 % |
– | – |
| 10 | Conyza pyrrhopappa | Asteraceae |
a. 100 b. 50 |
43.0 % 22.8 % |
– | – |
| 11 | Pluchea dioscoridis | Asteraceae |
a. 100 b. 50 |
37.4 % 27.8 % |
– | – |
| 12 | Euphorbia schimperiana | Euphorbiaceae |
a. 100 b. 50 |
18.4 % 9.6 % |
– | – |
| 13 | Momordica balsamina | Cucurbitaceae |
a. 100 b. 50 |
17.8 % 10.2 % |
– | – |
| 14 | Kalanchoe glaucescens | Crassulaceae |
a. 100 b. 50 |
33.9 % 23.2 % |
– | – |
| 15 | Rhus retinorrhoea | Anacardiaceae |
a. 100 b. 50 |
43.3 % 16.8 % |
– | – |
| 16 | Ficus benghalensis | Moraceae |
a. 100 b. 50 |
65.0 % 23.6 % |
– | – |
| 17 | Alkanna orientalis | Boraginaceae |
a. 100 b. 50 |
26.6 % 21.5 % |
– | – |
| 18 | Arnebia hispidissima | Boraginaceae |
a. 100 b. 50 |
61.0 % 24.7 % |
– | – |
| GC376 | 50 µM | 100 % |
But., butanol; Chl., chloroform; Eth., Ethanol; Hex., hexane; (-), not determined.
Accordingly, the alcoholic extracts of T. brownii and A. asak were subjected to biological assay guided fractionation and isolation for bioactive entities. Subsequent fractionations of the alcoholic extracts were achieved to n-hexane, chloroform and then n-butanol. The strongest activity was observed with the n-butanol fractions of both plants with percentage of inhibition of 61.8 % for T. brownii and 82.4 % for A. asak at a concentration of 50 µg/mL.
Furthermore, the results indicated that Zizyphus mucronata, Ocimum forsskaolii, Bidens biternata, Ficus benghalensis, and Arnebia hispidissima exhibited moderate inhibitory activities, with inhibition percentages of 48.8 %, 56.7 %, 54.3 %, 65 %, and 61 % respectively, at a concentration of 100 µg/mL. On the other hand, the remaining medicinal plants, including Barleria bispinosa, Hibiscus deflersii, Maerua crassifolia, Pulicaria schimperi, Conyza pyrrhopappa, Pluchea dioscoridis, Euphorbia schimperiana, Momordica balsamina, Kalanchoe glaucescens, Rhus retinorrhoea, and Alkanna orientalis displayed low inhibitory activity (<45 %) at a concentration of 100 µg/mL (Table 2).
3.2. Phytochemical studies of the n-butanol fractions of the active plants
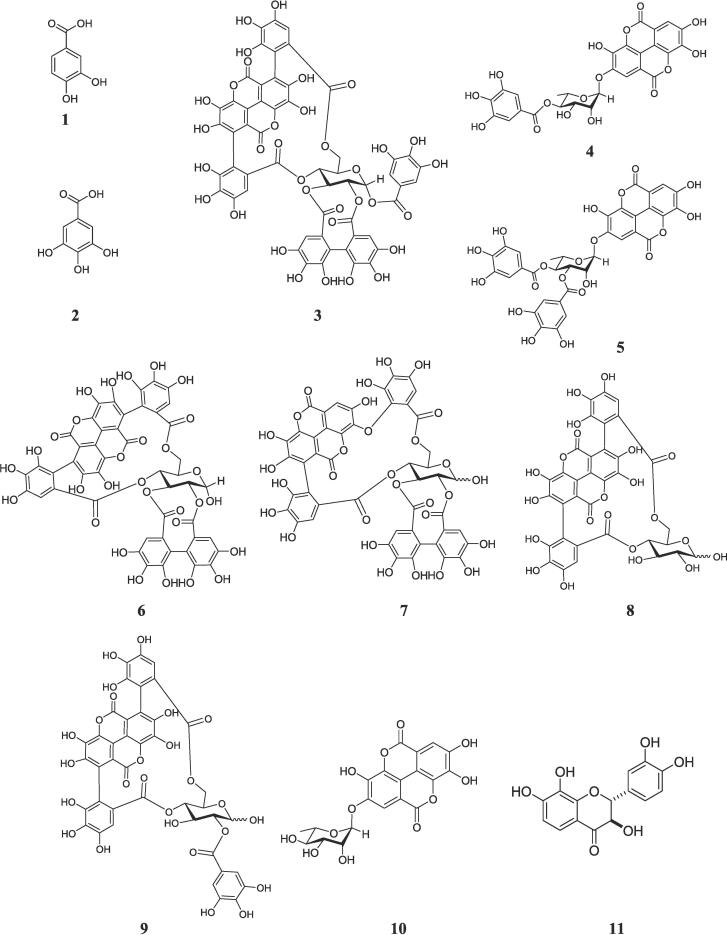
Through bioassay-guided isolation procedures, the n-butanol fractions of T. brownii were subjected to phytochemical investigations, resulting in the identification of eleven compounds (1–11). These compounds were classified as hydrolysable tannins, flavonoids, and phenolic acids. Additionally, four compounds (12–15) were isolated from A. asak, which belonged to the categories of free flavonoids and C-glycoside derivatives. The structures of the isolated compounds (Fig. 1, Fig. 2) were established by different NMR techniques, mass spectrometry, and comparison with the literature data ((Tables S1-S14) Figures S3-S95).
Fig. 1.
Chemical structures of the isolated compounds (1–11) from Terminalia brownii.
Fig. 2.
Chemical structures of the isolated compounds (12–15) from Acacia asak.
3.2.1. Identification of T. Brownii active compounds
Eleven compounds were identified from the n-butanol fraction of T. brownii including two aromatic acids, 3,4-Dihydroxy benzoic acid (1) (Abdullah et al., 2016), and gallic acid (2) (López-Martínez et al., 2015); eight hydrolysable tannins, combreglutinin (3) (Jossang et al., 1994), 4-O-(4′'-O-galloyl-α-L-rhamnopyranosyl)-ellagic acid (4) (Pfundstein et al., 2010), 4-O-(3′',4′'-di-O-galloyl-α-L-rhamnopyranosyl)-ellagic acid (5) (Pfundstein et al., 2010), α-punicalagin (6) (Doig et al., 1990, Kraszni et al., 2013), α/β-isoterchebulin (7) (Conrad et al., 2001), α-/β-punicalin (8) (Kraszni et al., 2013, El-Toumy and Rauwald, 2002), 2-O-galloyl α-/β-punicalin (9) (Tanaka et al., 1986), ellagic acid-2-O-α-L-rhamnopyranoside (eschweilenol C) [10] (Yang et al., 1998); and one flavonoid, (2R,3R)-7,8,3′,4′ -tetrahydroxydihydro-flavonol (11) (Lai, 1987) (Fig. 1).
3.2.2. Identification of A. Asak active compounds
Four flavonoids derivatives were identified from the n-butanol fraction of A. Asak using bioassay guided fractionation as 7,8,3′,4′-Tetrahydroxy-flavonol (melanoxetin) (12) (Banerjee et al., 2013), luteolin (13) (Huang et al., 2013), apigenin-6-C-β-D-glucoside (isovitexin) (14) (El-Hela et al., 2023), and luteolin-6-C-β-D-glucoside (isoorientin) (15) (El-Hela et al., 2023). All isolated compounds are firstly reported from A. asak (Fig. 2).
3.3. Analysis of molecular docking of T. Brownii and A. Asak active compounds
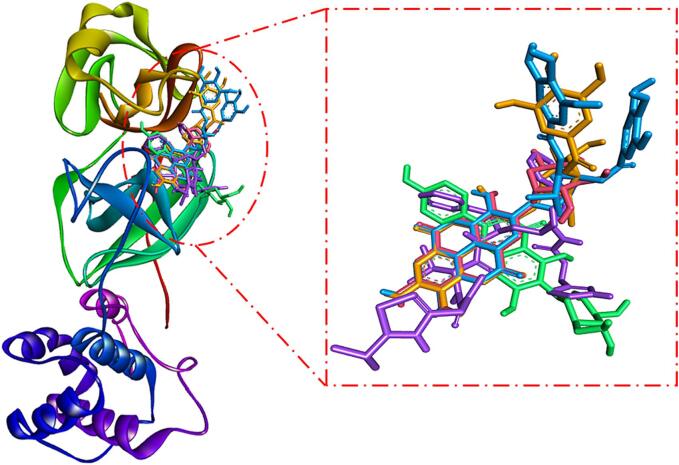
Using the molecular docking approach, the potential inhibitory effects of the isolated compounds (1–15) against Mpro of SARS-CoV-2 were evaluated. The findings revealed that all compounds were actively able to bind to the Mpro substrate binding site, except compounds 3, 6, 7, 8, and 9 (Fig. 3). The binding energies of all isolated compounds against Mpro were calculated and presented in Table 3. According to Table 3, the binding energies ranged from −5.3 to −9.3 kcal mol−1, with the most potent compounds (≤-7.5 kcal mol-1) being 4, 5, 10 and 14 with docking energies of −9.3, −8.5, −8.1, and −7.8 kcal mol−1, respectively. Previously, the docking energy of the positive control ritonavir has been reported as −8.2 kcal mol−1 (Al-Wahaibi et al., 2021). Also, to acknowledge the binding of compounds 4, 5, 10 and 14 to the substrate binding site or active site of Mpro, an overlay of compounds 4, 5, 10 and 14 along with control (ritonavir) was performed (Fig. 4). The overlay of compounds confirmed that they occupied a similar binding site on Mpro as employed by ritonavir (control). In order to further confirm our results, we performed molecular docking of the most potent shortlisted compounds (4, 5, 10, and 14) using Glide software (Schrodinger, LLC, NY, USA) under standard-precision (SP) and extra-precision (XP) modes, and the results are given in Table 3. We found out that the SP and XP docking energies of the shortlisted compounds (4, 5, 10, and 14) were comparable to that obtained from the AutoDock Vina-enabled PyRx software. The Glide SP docking scores of compounds 4, 5, 10, and 14 were −9.6, −7.7, −8.2, and −8.3 kcal mol−1 respectively, while the Glide XP docking scores of compounds 4, 5, 10, and 14 were −10.1, −8.8, −8.9, and −8.8 kcal mol−1 respectively (Table 3).
Fig. 3.
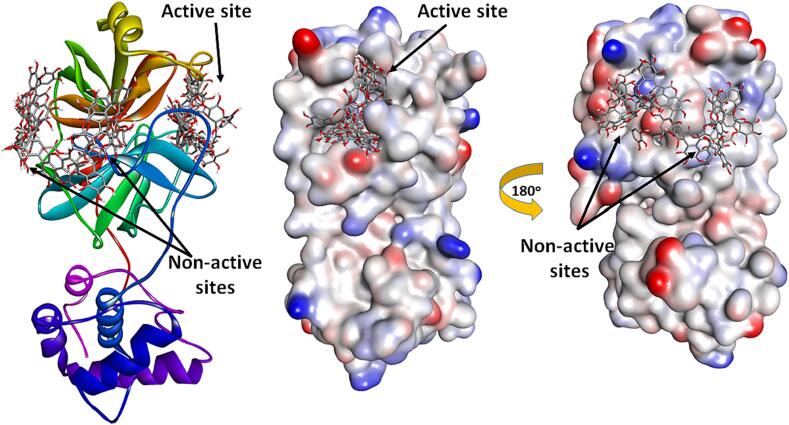
Binding of ligands to different parts of Mpro. Some ligands (e.g. 4, 5, 10, 14) occupied the substrate binding site (or active site) of Mpro, while others (e.g. 3, 6, 7, 8, and 9) were located outside.
Table 3.
Molecular docking parameters of the interaction between Mpro and T. brownii and A. asak compounds (1–15).
| Compound |
ΔG, kcal/mol |
Receptor amino acids |
Unfavorable amino acids Interaction |
ΔG (SP), kcal/mol |
ΔG (XP), kcal/mol |
MM-GBSA, kcal/mol |
|---|---|---|---|---|---|---|
| 1 | −5.3 | Gln189, His172, Glu166, Met165, His163, Ser144, Gly143, Asn142, Leu141, Phe140 | Cys145 | −5.1 | −4.4 | −10.465 |
| 2 | −5.5 | Gln189, Glu166, Met165, His163, Cys145, Ser144, Gly143, Asn142, Leu141, Phe140 | – | −4.0 | −3.5 | −23.298 |
| 3 | −7.8 | Glu178, Val104, Phe103, Lys102, Tyr101, Lys90, Lys88, Asn84, Gln83, Ser81, Tyr37, Val35, Asp34, Asp33 | Asn180 | −4.1 | −4.6 | −27.357 |
| 4 | −9.3 | Gln192, Thr190, Gln189, Arg188, Asp187, Pro168, Glu166, Met165, His164, Cys145, Gly143, Asn142, Met49, Ser46, Thr45, His41, Leu27, Thr26, Thr25, Thr24 | – | −6.9 | −8.8 | −49.034 |
| 5 | −8.5 | Gln192, Thr190, Gln189, Arg188, Asp187, Pro168, Glu166, Met165, His164, Cys145, Gly143, Asn142, Met49, Ser46, Thr45, Cys44, His41, Leu27, Thr26, Thr25, Thr24 | – | −7.6 | −8.0 | −47.075 |
| 6 | −7.2 | Val186, Pro184, Gly183, Phe181, Asn180, Gly179, Phe134, Arg105, Cys85, Asn84, Arg40 | – | −3.2 | −3.9 | −22.253 |
| 7 | −7.1 | Gln189, Pro168, Glu166, His164, His163, Cys145, Ser144, Gly143, Asn142, Leu141, Phe140, Tyr118, Met49, Ser46, Leu27, Thr26, Thr25 | His41 | −4.3 | −4.7 | −26.741 |
| 8 | −7.7 | Arg188, Val186, Phe185, Pro184, Phe181, Asn180, Arg105, Cys85, Asn84, Tyr54, Arg40 | Gly183 | −0.3 | −1.3 | −27.919 |
| 9 | −7.7 | Glu178, Phe103, Lys102, Tyr101, Lys100, Lys90, Leu89, Lys88, Gln83, His80, Tyr37, Val36, Val35, Asp33 | Ser81 | −2.2 | −3.0 | −23.299 |
| 10 | −8.1 | Gln192, Thr190, Gln189, Arg188, Asp187, Pro168, Leu167, Glu166, Met165, His164, Cys145, Gly143, Asn142, Met49, His41, Leu27, Thr26, Thr25 | – | −5.5 | −7.0 | −38.182 |
| 11 | −7.3 | Gln192, Thr190, Gln189, Arg188, His172, Glu166, Met165, His164, Cys145, Ser144, Gly143, Asn142, Leu141, Phe140, Met49, His41 | His163 | −2.0 | −6.2 | −10.414 |
| 12 | −7.4 | Gln189, Arg188, Asp187, His172, Glu166, Met165, His164, His163, Cys145, Ser144, Leu141, Phe140, Tyr54, Met49, His41 | – | −5.0 | −5.6 | −20.124 |
| 13 | −7.4 | Gln192, Thr190, Gln189, Arg188, Glu166, Met165, His163, Cys145, Ser144, Gly143, Asn142, Leu141, Phe140 | – | −5.0 | −3.9 | −15.923 |
| 14 | −7.8 | Gln189, Arg188, Asp187, His172, Glu166, Met165, His164, Cys145, Gly143, Asn142, Leu141, Phe140, Tyr54, Pro52, Met49, His41 | – | −5.3 | −6.7 | −30.814 |
| 15 | −7.4 | Gln189, His172, Glu166, Met165, His163, Cys145, Ser144, Gly143, Asn142, Leu141, Phe140, Met49, Ser46, Thr45, Cys44, Thr25, Thr24 | Thr26 | −4.8 | −6.2 | −21.713 |
| Ritonavir (Control) |
−8.2 | Gln192, Ala191, Thr190, Gln189, Arg188, Asp187, His172, Pro168, Leu167, Glu166, Met165, His164, His163, Ser144, Gly143, Asn142, Leu141, Phe140, Tyr54, Met49, His41, Leu27, Thr26, Thr25 | – | – | – | – |
Arg: Arginine; Asn: Asparagine; Asp: Aspartic acid; Cys: Cysteine; Glu: Glutamic acid; Gln: Glutamine; Gly: Glycine; His: Histidine; Leu: Leucine; Lys: Lysine; Met: Methionine; Phe: Phenylalanine; Pro: Proline; Ser: Serine; Thr: Threonine; Tyr: Tyrosine; Val: Valine. ΔG (SP), and ΔG (XP): docking scores obtained under standard-precision, and extra-precision docking modes using Glide software (Schrodinger, LLC, NY, USA).
Fig. 4.
Overlay of compounds 4, 5, 10, and 14 over the binding site of ritonavir (control). The compounds are represented as sticks, and color-coded for compounds 4, 5, 10, 14, and ritonavir (control) as Golden, Blue, Pink, Green, and Magenta respectively.
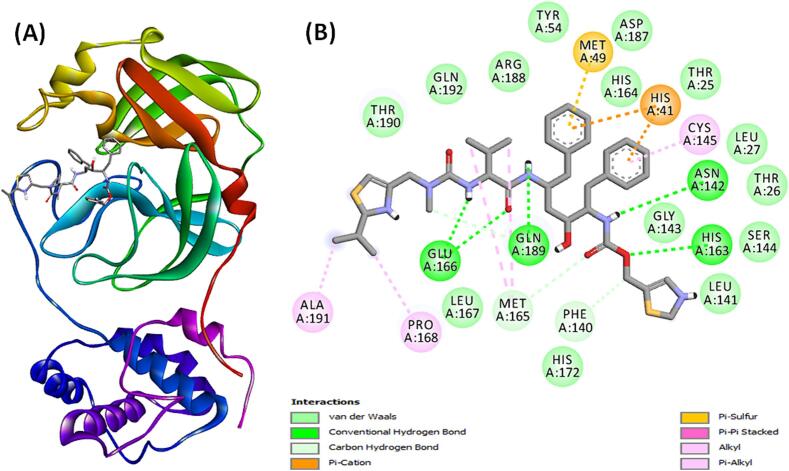
The Mpro-ritonavir complex exhibited stability through three carbon hydrogen bonds with Gln189, Met165, and Phe140 along with five conventional hydrogen bonds with Gln189, Glu166, His163, and Asn142 (Fig. 5A,B and Table 3). Additionally, the complex formed a Pi-Sulfur interaction with Met49, and two Pi-Cation electrostatic interactions with the active site residue His41. Also, there were four alkyl hydrophobic interactions with Ala191, Pro168, and Met165 (two interactions) along with one Pi-Pi Stacked hydrophobic interaction with the active site residue His41, and one Pi-alkyl hydrophobic interaction with the active site residue Cys145. Furthermore, van der Waals interaction occurred between ritonavir and Mpro amino acid residues such as Gln192, Thr190, Arg188, Asp187, His172, Leu167, His164, Ser144, Gly143, Leu141, Tyr54, Leu27, Thr26, and Thr25. The docking energy of ritonavir towards Mpro was −8.2 kcal mol−1, and the dissociation was 1.03 × 106 M−1 (Table 3).
Fig. 5.
(A) Binding of ritonavir (control) to the substrate binding site of Mpro, and (B) 2D diagram depicting the molecular interaction between Mpro and ritonavir (control).
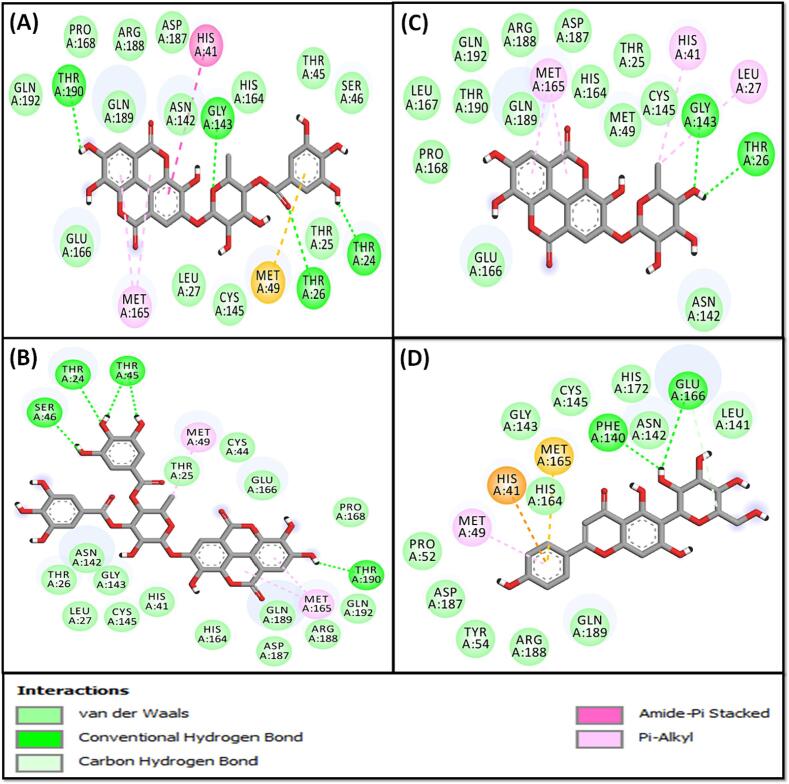
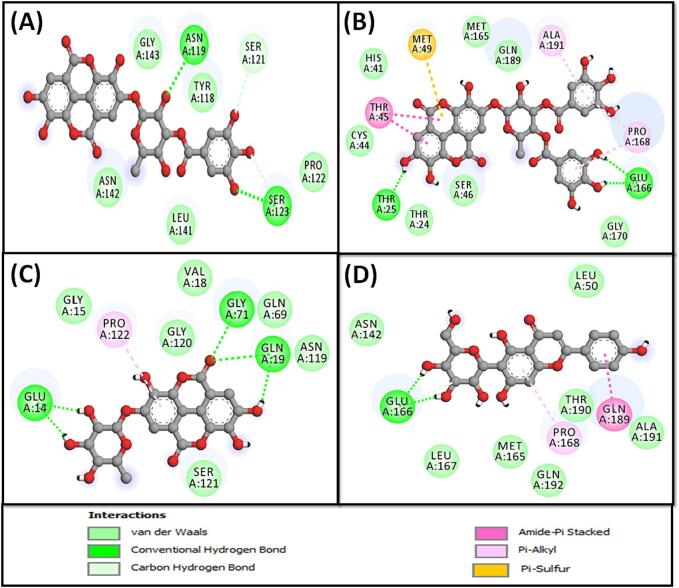
The Mpro-compound 4 complex exhibited stability through three hydrophobic interactions with the active site residue His41, and Met165 (two interactions), as well as four conventional hydrogen bonds with Thr190, Gly143, Thr26, and Thr24 (Fig. 6A and Table 3). Additionally, it participated in van der Waals interactions with specific amino acid residues such as Gln192, Gln189, Arg188, Asp187, Pro168, Glu166, His164, Cys145, Asn142, Ser46, Thr45, Leu27, and Thr25. The estimated docking energy and binding affinity of compound 4 towards Mpro were estimated to be −9.3 kcal mol−1 and 6.62 × 106 M−1, respectively (Table 3).
Fig. 6.
Molecular interaction between Mpro and compounds 4 (A), 5 (B), 10 (C) and 14 (D).
The stability of the Mpro-compound 5 complex was maintained through five conventional hydrogen bonds with Thr24, Ser46, Thr190, and Thr45 (two interactions). Additionally, compound 5 interacted with Mpro via two Pi-Alkyl hydrophobic interactions with Met165 and one alkyl hydrophobic interaction with Met49 (Fig. 6B and Table 3). Furthermore, compound 5 participated in van der Waals interactions with specific amino acid residues such as Gln192, Gln189, Arg188, Asp187, Pro168, Glu166, His164, Cys145, Gly143, Asn142, Cys44, His41, Leu27, Thr26, and Thr25. The estimated docking energy and binding affinity of compound 5 toward Mpro were −8.5 kcal mol−1 and 1.72 × 106 M−1, respectively (Table 3).
The estimation of the Mpro-compound 10 interaction revealed the complex’s stabilization through one alkyl hydrophobic interaction with Leu27, three Pi-Alkyl hydrophobic interactions with Met165 (two interactions) and active site residue His41, and two conventional hydrogen bonds with Gly143 and Thr26 (Fig. 6C and Table 3). Additionally, compound 10 formed a network of van der Waals interactions with Gln192, Thr190, Gln189, Arg188, Asp187, Pro168, Leu167, Glu166, Cys145, His164, Asn142, Met49, and Thr25. The estimated docking energy and dissociation constant of compound 10 toward Mpro were estimated to be −8.1 kcal mol−1 and 8.73 × 105 M−1, respectively (Table 3).
The complex between Mpro and compound 14 was stabilized by one carbon hydrogen bond with Glu166, two conventional hydrogen bonds Glu166 and Phe140, along with the formation of one hydrophobic interaction with Met49, and a Pi-Sulfur interaction with Met165. Additionally, the Mpro-compound 14 complex was stabilized by one electrostatic interaction with one of the active site residue His41 (Fig. 6D and Table 3). Importantly, van der Waals interactions occured between compound 14 and Gln189, Arg188, Asp187, His172, His164, Cys145, Gly143, Asn142, Leu141, Tyr54, and Pro52. The estimated docking energy and binding affinity of compound 14 toward Mpro were estimated to be −7.8 kcal mol−1and 5.26 × 105 M−1, respectively (Table 3).
3.4. Analysis of moelcular dynamics simulation (MDS)
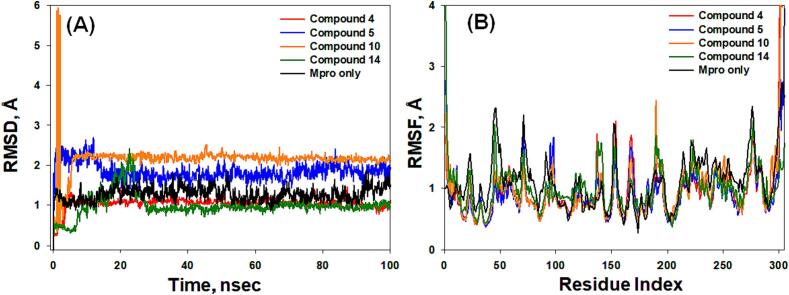
In this study, MDS was conducted over a duration of 100 ns to gain insights into the binding mechanism, structural behavior and flexibility of Mpro in the presence of selected compounds (4, 5, 10, and 14). Key parameters, including RMSD, RMSF, RG, and SASA were computed and based on the 100 ns dynamics trajectory (Fig. 7, Fig. 8).
Fig. 7.
The MD trajectory analysis, (A) Root mean square deviation (RMSD) and (B) Root mean square fluctuation (RMSF), of the Mpro alone and in the presence of compounds 4, 5, 10, and 14.
Fig. 8.
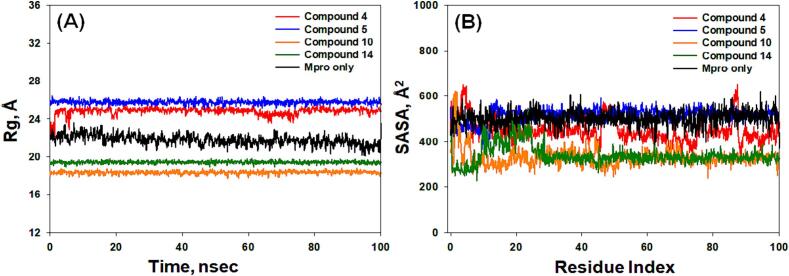
The MD trajectory analysis, (A) Radius of gyration (Rg) and (B) Solvent accessible surface area (SASA), of the Mpro alone and in the presence of compounds 4, 5, 10, and 14.
3.4.1. Root mean square deviation (RMSD)
RSMD measures the deviation in a protein's structure with or without a ligand, relative to its initial structure, during of a simulation. This metric provides insights into the stability of the system (Jabir et al., 2021, Bowers et al., 2006). In this investigation, RMSDs were calculated for Cα-atoms of Mpro in the presence of selected compounds (4, 5, 10, and 14) over 100 ns (Fig. 7A). The RMSD for Cα-atoms of Mpro without any ligand exhibited fluctuations within the range of 1.02–1.73 Å, with an average RMSD of 1.31 ± 0.08 Å. Additionally, the RMSDs for Cα-atoms of Mpro in the presence of compounds 4, 5, 10, and 14 initially showed fluctuations and then stabilized within the respective ranges of 1.03–1.35 Å, 1.53–2.84 Å, 1.22–2.31 Å, and 0.48–2.23 Å. The average RMSDs (during 30–100 ns) for Mpro-4, Mpro-5, Mpro-10, and Mpro-14 complexes were determined as 1.19 ± 0.04 Å, 1.87 ± 0.08 Å, 2.22 ± 0.05 Å, and 0.98 ± 0.04 Å, respectively (Fig. 7A). The observed RMSD values were within the acceptable limits, signifying the formation of stable Mpro-ligand complexes (Rehman et al., 2021).
3.4.2. Root mean square fluctuation (RMSF)
In MD simulation, the impact of ligand binding on protein side-chain dynamics is evaluated by examining RMSF (AlAjmi et al., 2021). In this study, RMSF values for Mpro, both in isolation and in conjunction with compounds 4, 5, 10, and 14, were tracked over the course of the simulation (Fig. 7B). The RMSF profiles for the Mpro-4, Mpro-5, Mpro-10, and Mpro-14 complexes closely mirrored that of Mpro in isolation, indicating a lack of substantial alterations in Mpro's conformation attributed to its interactions with the compounds (Fig. 7B). Any minor fluctuations observed in the RMSF plots could be attributed to ligand–protein binding events.
3.4.3. Radius of gyration (Rg)
The evaluation of protein–ligand complex compactness and its associated stability commonly relies on monitoring the Radius of Gyration (Rg) during a simulation (Muteeb et al., 2020). In our study, we analyzed the Rg of Mpro in isolated and complexed forms with compounds 4, 5, 10, and 14 over a 100 ns simulation period (Fig. 8A). The Rg values for Mpro alone fluctuated between 21.3 and 24.0 Å, with an average of 22.7 ± 0.06 Å. Similarly, the Rg values for the Mpro-4, Mpro-5, Mpro-10, and Mpro-14 complexes fluctuated within the ranges of 23.4–25.4 Å, 25.8–26.4 Å, 18.5–18.9 Å, and 1.90–1.96 Å, respectively, with average values of 24.8 ± 0.05 Å, 26.2 ± 0.04 Å, 18.7 ± 0.03 Å, and 19.3 ± 0.03 Å (Fig. 8A). These findings unequivocally show that the compounds are firmly located within the binding pocket of their respective proteins, contributing to the formation of robust and stable protein–ligand complexes.
3.4.4. Solvent accessible surface area (SASA)
SASA, an indicator of the exposure of a protein–ligand complex to the surrounding solvent molecules, serves as a measure of complex stability (AlAjmi et al., 2018). In our investigation, we evaluated SASA for Mpro in both its bound, its unbound state, and in the presence of compounds 4, 5, 10, and 14 (Fig. 8B). The SASA values for isolated Mpro exhibited fluctuations from 406 to 604 Å2, with an average value of 510 ± 14 Å2 (Fig. 8B). For the Mpro-4, Mpro-5, Mpro-10, and Mpro-14 complexes, SASA values showed variations within the ranges of 326–627 Å2, 383–594 Å2, 267–612 Å2, and 227–577 Å2, respectively, with average values of 589 ± 21 Å2, 534 ± 9 Å2, 327 ± 11 Å2, and 346 ± 12 Å2, respectively (Fig. 8B). These results strongly support the assertion that the compounds are securely located within the binding pockets of their respective proteins, contributing to the establishment of stable protein–ligand complexes.
3.4.5. Analysis of first and last frames of MD simulation
Examining both the initial and final frames of molecular dynamics (MD) simulations provides crucial insights into the dynamics and stability of biomolecular systems. The first frame captures the system's starting configuration before simulation dynamics are applied, acting as a baseline reference for understanding its evolution. Conversely, the last frame reflects the system's final conformation after running for a specific duration, incorporating the cumulative effects of molecular motions and interactions. Analyzing the last frame enables assessment of overall stability, structural changes, and binding interactions. In this study, we compared the first and last frames to gain a comprehensive understanding of the system's behavior throughout the simulation period (Supplementary Figures S111-S114). In the initial frame of the MD simulation, compound 4 established conventional hydrogen bonds with Thr25 (two interactions), Glu162 (two interactions), Gln185, and Asn138. Additionally, it formed a carbon hydrogen bond with Asn138 and a Pi-Sulfur bond with Met46. However, in the final frame, while retaining hydrogen bonds with Thr25, Glu162, and Gln185, the Pi-Sulfur bond with Met46 was lost, and two new carbon hydrogen bonds emerged with Thr42 and Ser43. For compound 5, the first frame displayed conventional hydrogen bonds with Thr25, Glu162 (two interactions), Pro164, Gln185, and Thr186, along with a Pi-Amide stacked interaction with GLN185. In contrast, the last frame showcased significant changes, with the emergence of new Pi-Alkyl interactions between compound 5 and Met46 (four interactions). Additionally, new Pi-Alkyl interactions with Ala187 and Pro164, alongside hydrogen bonds with Thr42 and Ser43, were observed. Nevertheless, the hydrogen bonds with Thr25 and Glu162 persisted throughout the MD simulation. Similarly, in the first frame of MD simulation, compound 10 formed two conventional hydrogen bonds with Thr25 and Gln185, along with a Pi-Sulfur bond with Met46. However, in the last frame, it established two conventional hydrogen bonds with Thr25 and a Pi-Sigma interaction with Ser43. Regarding compound 14, the first frame revealed two conventional hydrogen bonds and one carbon hydrogen bond with Glu166, along with Pi-Alkyl and Amide-Pi stacked interactions with Pro168 and Gln189, respectively. In the last frame of the MD simulation, while the Pi-Alkyl interaction with Pro164 persisted, the Amide-Pi stacked interaction shifted to Gln185. Additionally, Glu162 formed two conventional hydrogen bonds and one carbon hydrogen bond instead of Glu166.
3.5. Analysis of free energy calculations (MM-GBSA)
To evaluate the binding strength between Mpro and compounds 4, 5, 10, and 14, the free energy, which reflects the protein–ligand interaction within the solvent environment, was assessed using the MM-GBSA method. Table 4 presents the results, indicating that the Mpro-compound 4 complex exhibited the lowest free energy (−35.07 kcal mol−1), followed by the Mpro-compound 5 (−34.59 kcal mol−1), Mpro-compound 10 (−31.44 kcal mol−1), and Mpro-compound 14 (−15.38 kcal mol−1). In every scenario, the primary factors contributing to the establishment of stable Mpro-ligand complexes were the Coulombic energy (ΔGCoulomb) and van der Waals energy (ΔGvdW). Conversely, the primary opposing force to Mpro-ligand interactions was the polar solvation energy (ΔGSolv or ΔGSolGB). These outcomes align with the molecular docking and in vitro findings, emphasizing that compound 4 exhibited notable inhibitory potential against SARS-CoV-2 Mpro (Table 4).
Table 4.
Free energy (MM-GBSA) analysis of SARS-CoV-2 Mpro interaction with compounds 4,5,10 and 14.
| Compound |
ΔG or ΔGBind |
ΔGCoulomb | ΔGCovalent | ΔGH-bond |
ΔGSA or ΔGSol_Lipo |
ΔGSolv or ΔGSolGB |
ΔGPacking | ΔGvdW |
|---|---|---|---|---|---|---|---|---|
| 4 | −35.07 | −33.32 | 2.08 | −4.18 | −4.54 | 24.86 | −1.27 | −18.70 |
| 5 | −34.59 | −23.70 | 2.28 | −3.97 | −7.85 | 30.94 | −2.78 | −29.51 |
| 10 | −31.44 | −21.82 | 1.68 | −2.14 | −4.07 | 12.02 | −1.47 | −15.64 |
| 14 | −15.38 | −15.53 | 6.80 | −3.29 | −4.59 | 22.23 | −1.45 | −19.55 |
ΔGCovalent, ΔGvdW, ΔGSolv or ΔGSolGB, ΔGCoulomb, ΔGH-bond, ΔGPacking, ΔGSA or ΔGSol-Lipo, and ΔG or ΔGBind stands for minimized molecular mechanics energies namely covalent binding energy, van der Waals’ energy, solvation energy, coulomb energy, energy due to H-bonds, packing energy, lipophilic energy, and binding energy, respectively. All the energies are in kcal/mol.
To further delve into the analysis, thermal MM-GBSA calculations were conducted on each frame, totaling 1000 frames for each compound, to assess and scrutinize the frame with the lowest energy. Thermal MM-GBSA computations were executed using the Python script of Prime (Schrodinger, LLC, NY, USA) across 1000 frames. MM-GBSA was computed utilizing the MD simulation trajectories for 6lu7-4, 6lu7-5, 6lu7-10, and 6lu7-14. The minimum ΔG_bind energies for each complex were determined to unveil the most stable structure and assess the stability of the protein–ligand complexes post the 100 ns simulation. Each frame was sorted based on the lowest ΔG_bind energy. For the 6lu7-4 complex, energies in the initial frames (ranging from 1 to 40) spanned from −15.56 to −48.96 kcal mol−1. However, with the passage of time and frames, it stabilized until the conclusion of the 100 ns simulation. Energies from frame 40 to 1000 ranged from −15.76 to −69.28 kcal mol−1. The minimum structure of 6lu7-4, along with its lowest ΔG_bind and 2D interaction, is depicted in Fig. 9A. Mpro formed conventional hydrogen bonds with Asn119 and Ser123 along with a carbon hydrogen bond with Ser121. In addition, compound 4 formed van der Waals’ interactions with Gly143, Thr118, Pro122, Asn142, and Leu141. The minimum frame energy was observed at the 615th frame out of the total 1000 frames, and it did not decrease below −40.05 kcal mol−1 until the end of the simulation, indicating full stabilization of the complex. Similarly, for the 6lu7-5 complex, energies were calculated for 1000 frames. The range of ΔG_bind energies varied from −40.99 to −77.42 kcal mol−1. The lowest ΔG_bind energy was −77.42 kcal mol−1 at the 229th frame, but from then onwards, until frame 1000, it ranged from −42.91 to −76.87 kcal mol−1. The minimum structure of 6lu7-5, along with its lowest ΔG_bind and 2D interaction, is illustrated in Fig. 9B. Compound 5 interacted with Mpro through Amide-Pi stacked hydrophobic interaction with Thr45 (two interactions), and Pi-Alkyl hydrophobic interactions with Ala191 (one interaction), and Pro168 (two interactions). In addition, compound 5 formed one Pi-Sulfur bond with Met49, and conventional hydrogen bonds with Glu166 (two interactions), and Thr25. Several van der Waals’ interactions also existed between compound 5 and Mpro residues such as active site residue His41, Cys44, Thr24, Ser46, Met165, Gly170, and Gln189 (Fig. 9B). Likewise, for the 6lu7-10 complex, the binding free energies ranged from −2.97 to −60.03 kcal mol−1. The frame with the lowest energy was observed at the 494th frame (Fig. 9C), after which it remained within the range of −28.82 to −56.89 kcal mol−1. Compound 10 formed conventional hydrogen bonds with Glu14 (two interactions), Gln19 (two interactions), and Gly71. It also formed a Pi-Alkyl hydrophobic interaction with Pro122, and van der Waals’ interactions with Gly15, Val18, Gln69, Asn119, Gly120, and Ser121 (Fig. 9C). For the 6lu7-14 complex, energies in the initial frames (1–100) ranged from −40.20 to −75.31 kcal mol−1. Subsequently, energies for frames 101–500 remained constant, ranging from −40.87 to −68.71 kcal mol−1. From frames 501–1000, energies ranged from −40.74 to −64.89 kcal mol−1. The lowest binding energy was observed at the 105th frame, representing the minimum structure in the entire simulation trajectory. This structure and its 2D diagram are illustrated in Fig. 9D. Compound 14 interacted with Mpro through two conventional hydrogen bonds with Glu166, one Amide-Pi stacked hydrophobic interaction with Gln189, and a Pi-Alkyl hydrophobic interaction with Pro168. In addition, compound 14 interacted with several residued of Mpro such as Leu50, Asn142, Met165, Leu167, Thr190, Ala191, and Gln192 (Fig. 9D).
Fig. 9.
Molecular interaction between Mpro and compounds 4 (A), 5 (B), 10 (C) and 14 (D), showing minimum energy state during MD simulation.
3.6. Principal Component analysis (PCA), and PCA-based 2D free energy surface
PCA is a commonly employed method for tracking the global motion of a protein in the absence or presence of a ligand. In our study, we investigated the conformational sampling of Cα-atoms along PC1 and PC2 of Mpro with and without compounds 4, 5, 10, and 14, as depicted in Supplementary Figure S110. Each red and black dot signifies a unique conformational state of the protein, and the clusters of red and black dots indicate distinct regions of energetically favorable conformations. When Mpro was analyzed alone, it explored a conformational subspace ranging from −15 to + 15 along PC1 (accounting for 22.01 % of the variance) and −15 to + 15 along PC2 (accounting for 14.87 % of the variance) (Supplementary Figure S110A). In the presence of compounds 4, and 5, Mpro's conformational space extended from −20 to + 30 along PC1 (16.78 %) and −30 to + 20 along PC2 (14.57 %), and −20 to + 30 along PC1 (30.71 %), and −15 to + 15 along PC2 (9.67 %) respectively (Supplementary Figures S110B,C). Similarly, the conformational space of Mpro extended from −20 to + 30 along PC1 (31.15 %) and −20 to + 20 along PC2 (12.07 %) for compound 10, and −30 to + 40 along PC1 (29.36 %), and −20 to + 20 along PC2 (21.65 %) for compound 14 (Supplementary Figures S110D,E). Notably, the combined first three eigenvalues of Mpro alone and in the presence of compounds 4, 5, 10, and 14 accounted for 46.5 %, 42.5 %, 48.2 %, 50.1 %, and 58.5 % of the conformational variances, respectively (Supplementary Figures S110F-J). These findings suggest a marginal increase in the flexibility of Mpro upon the introduction of compounds 4, 5, and 10, whereas the presence of compound 14 led to a substantial increase in Mpro's flexibility. These results provide further evidence of the favorable interaction between compounds 4, 5, and 10 with Mpro.
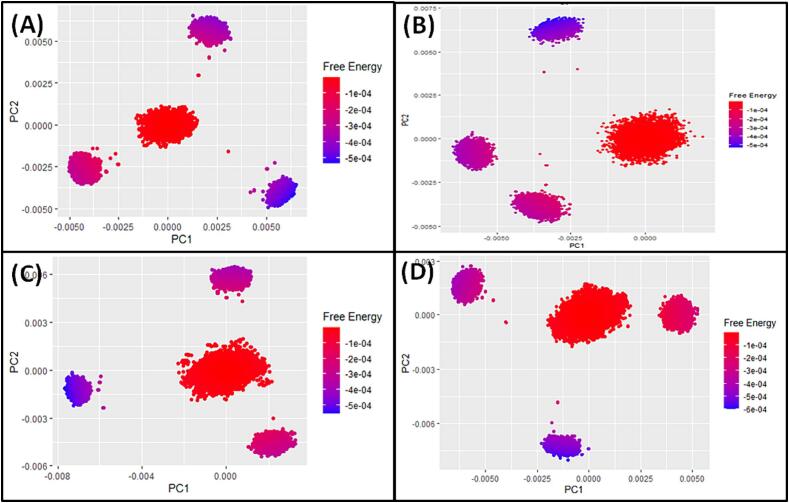
Furthermore, PCA-based 2D free energy surface was generated to understand the thermodynamically stable conformations of Mpro in the presence of compounds 4, 5, 10, and 14, as shown in Fig. 10. Gibb’s free energy surface inspects the direction of the fluctuation in the two systems (PC1 and PC2) for all Cα atom of protein/ligand complexes. Among the various Mpro and compounds complexes, lowest Gibb’s free energy cluster (purple color) was found in Mpro-compounds 4 complex, followed by Mpro-compounds 5, 10, and 14 compelxes, hence these complexes are found to be thermodynamically stable. It indicates that these complexes followed energetically stable transition from one conformation to another, and that the binding of the compounds 4, 5, 10, and 14 stabilized the protein/ligand complexes throughout the simulation.
Fig. 10.
Free energy of Mpro in the presence of (A) compound 4, compound 5 (B), compound 10 (C), and compound 14 (D).
3.7. In vitro Mpro inhibition assay for compounds (4, 5, 10 and 14)
The strongest MD proposed Mpro inhibitors (4, 5, 10, and 14) were subjected to the in vitro Mpro inhibition flouorescence-based assay using the broad-spectrum antiviral compound GC376 as a positive control. The findings (Table 5) revealed that all compounds demonstrated significant Mpro inhibitory activity. Among them, compound 4 emerged as the most potent compound, with an IC50 value of 1.2 µg/mL. These results align well with the finding of the in silico data that compound 4 had the strongest Mpro inhibition activity.
Table 5.
Mpro inhibition activity of compounds 4, 5, 10 and 14.
| Compounds | IC50 (µg/mL) |
|---|---|
| 4 | 1.2 |
| 5 | 20.1 |
| 10 | 10.0 |
| 14 | 9.4 |
| GC376 | 0.42 µM |
3.8. Cell viability evaulation for compounds (4, 5, 10 and 14)
Cytotoxicity of candidate compounds in respect of a target cell is a vital criterion for its appropriateness as antiviral compounds. Generally, the candidate compounds should be effective against the virus without causing any cytotoxic or cellular metabolism effects of target cells (Simões et al., 1999). To address this concern, cell viability was evaluated for the extracts/fractions and compound 4, 5, 10, and 14 at effective inhibitory concentrations to assess their effect on the viability of normal cells (HUVEC) and lung cancer cells (A549) (Table 6). Our results showed that none of the compounds 4, 5, 10, and 14 exhibited cytotoxic activity against normal and lung cancer cells, at concentrations corresponding to their viral inhibition activity reflecting their safe use as antiviral agents.
Table 6.
Cytotoxic activity of active compounds (4, 5, 10 and 14) by MTT assay.
| Compound |
IC50 (µg/mL) |
|
|---|---|---|
| A549 | HUVEC | |
| 4 | >100 | >100 |
| 5 | >100 | >100 |
| 10 | >100 | >100 |
| 14 | >100 | >100 |
| Doxorubicin | 1.45 ± 0.05 | 2.5 ± 0.1 |
Values are represented as the average ± SD (n = 3).
4. Discussion
A total of eighteen medicinal plants native to Saudi Arabia were carefully chosen based on their availability within the country and the previously reported antiviral activities of their species or genera. These selected plants were further assessed for their potential to inhibit the activity of the SARS-CoV-2 main protease (Mpro) using a fluorescent Mpro FRET inhibition assay. The findings indicated that the ethanolic extracts of T. brownii and A. asak displayed noteworthy inhibitory activity against Mpro among all the tested extracts.
According to literatures review, Terminalia brownii (Combretaceae) is a popular deciduous tree that is native to sub-tropical and tropical regions of East and Central Africa. The previous phytochemical studies of T. brownii extracts exhibited various compounds of significant pharmaceutical value, such as tannins, flavonoids, chromones, pentacyclic triterpenoids, which possess notable properties such as antifungal, anti-malarial, antibacterial, antioxidant, and antidiabetic properties, as well as immunoregulatory effects. Numerous interventions are currently accessible for the treatment of bacterial and fungal infections (Negishi et al., 2011, Salih et al., 2017, Salih et al., 2018, Biruk et al., 2020, Salih et al., 2022).
The hydrolysable tannins of Tarminalia species displayed well documented antiviral activities against different DNA and RNA such as Herpes Simplex Virus (HSV) (Kesharwani et al., 2017), Human Immune Difficiency virus (HIV) (Maregesi et al., 2010), Hepatitis C virus (HCV) (Ajala et al., 2014), Measles and Mumps viruses (Sumithira et al., 2012), and Zika virus (Priya et al., 2018). The reported antiviral activity of tannins includes preventing binding, penetration, and cell-to-cell spread, as well as secondary infection by blocking interactions between cell surface glycosaminoglycans and viral glycoproteins, as in HSV and HIV (Lin et al., 2011). Furthermore, tannins can suppress viral replication, as reported with HIV, by inhibiting HIV-reverse transcriptase, -protease and -integrase enzymes (Martino et al., 2002, Notka et al., 2004).
The assessment of in vitro Mpro inhibition activity of T. brownii sub-fractions demonstrated that the n-butanol extract exhibited the most potent activity. Subsequently, the phytochemical analysis of the n-butanol fraction unveiled a high abundance of phenolic compounds, primarily hydrolysable tannins. The investigation of potential Mpro inhibitory activity through molecular docking demonstrated that all identified compounds were capable of actively binding to the substrate binding site of Mpro, except for compounds 3, 6, 7, 8, and 9. These compounds were unable to fit into the Mpro substrate active site properly due to their high molecular weight, causing them to be positioned outside. Notably, compounds 4, 5, and 10 exhibited the strongest binding stability among the other compounds.
Based on the information in Table 3, the structural activity relationship (SAR) was concluded that tannins exhibited higher binding affinity than flavonoids and free phenolic acids. Among the tannin, it was evident that the presence of a sugar moiety had a significant impact on the activity. Moreover, glycosylation with ellagic acid at OH-1, as demonstrated in compound 10, resulted in improved binding stability compared to glycosylation with gallagic acid of a higher molecular weight, as observed in compound 8. Compound 4, with a single acylation of one galloyl moiety at OH-4, showed the highest level of activity compared to compound 5, with double acylation involving another galloyl moiety at OH-3, which exhibited reduced binding stability and inhibition activity. This suggests that increased acylation, by galloyl moiety or any higher molecular weight substructure as hexahydroxydiphenoyl (HHDP), as demonstrated in compounds 3, 6 and 7 resulted in diminished activity due to steric hindrance and unfavorable interactions. These factors could impede the proper fitting and interaction of these compounds with the active site of Mpro.
Acacia Asak is a tree belonging to family Fabaceae. It is native to regions spanning from Tropical Africa to the Arabian Peninsula. Historically, it has been employed for its medicinal benefits in addressing gastric ulcers, skin diseases, and as an antiseptic (Al-Fatimi et al., 2007). As far as our knowledge extends, this study marks the inaugural documentation of the constituents within the herbal extract of A. asak and their associated biological activities.
The phytochemical investigations of the n-butanol fraction using bioassay guided isolations procedures resulted in the isolation of four compounds, including both free flavonoids and C-glycoside derivatives. Notably, this is the first documented instance of these compounds being isolated from the A. asak extract.
The investigation into the potential Mpro inhibitory activity through molecular docking demonstrated that all compounds effectively bound to the Mpro substrate binding site, with compound 14 identified as the most potent. While it was evident that C-glycosylation at C-6 could enhance the activity among the identified flavonoids, drawing definitive conclusions about the structural-activity relationship was challenging due to the limited number of isolated compounds.
Collectively, four compounds (4, 5, 10, and 14) were identified as the most potent Mpro inhibitors from the alcoholic extract of T. brownii and A. asak with binding energies (≤−7.5 kcal mol−1). The order of binding stability strength was identified as follows: 4 > 5 > 10 > 14.
An insight into the binding modes of bioactive compounds to SARS-CoV-2 Mpro was gained by performing molecular docking and molecular dynamics simulation. It's worth noting that the active site of Mpro can be subdivided into distinct sub-sites, namely S1‘, S1, and S2. The S1‘ subsite is defined by Thr25 and Thr22, while the S1 subsite comprises Glu166, His163, Asn142, Leu141, and Phe140. Similarly, the S2 subsite is shaped by Asp187, Met165, Tyr54, Met49, and His41. Furthermore, a hydrophobic patch is formed by Gln192, Gln189, Phe185, Leu167, and Met165. Together, these sub-sites and the hydrophobic patch contribute to substrate accommodation and facilitate the hydrolytic process.
Based on the data in Table 3, it was evident that tannin compounds (4, 5, and 10) displayed favorable interactions with the crucial residues Thr24 and Thr25, integral components of the S1‘ subsite of Mpro. In contrast, the flavonoid (14) did not interact with these residues, potentially explaining its lower binding stability compared to tannin compounds in inhibiting Mpro. Also, compounds (4, 5, 10, and 14) interacted with crucial residues of Mpro such as Asn142, and Glu166 (S1 subsite residues), and active site residue His41, Met165 and Asp187 (S2 subsite residues). Notably, the results of the molecular dynamics simulation, as indicated by parameters such as RMSD, RMSF, Rg, and SASA, aligned with and provided further support for the molecular docking findings. Further in-depth analysis of interaction between Mpro and compounds 4, 5, 10, and 14 through PCA and thermal MM-GBSA suggested that compound 4 formed van der Waals’ interactions with S1 subsite residue of Mpro such as Leu141 and Asn142, while compound 5 interacted with S1‘, S1, and S2 subsite residues Thr25, Glu166, and Met49 respectively. In addition, compound 5 formed van der Waals’ interactions with S1‘subsite residue Thr24, and S2 subsite residues His41 (active site residue), and Met165. Similarly, compound 14 interacted with S1 subsite residues Asn142, Glu166, and Met165, which constitute S2 subsite of Mpro active site. However, compound 5 did not interact with any of the active site residues of Mpro in the lowest energy state. All these results complement the in vitro finding that compound 4 is the most potent agents against Mpro of SARS-CoV-2.
Previously, research findings have demonstrated the potential inhibitory effect of tannic acid on SARS-CoV-2 Mpro, as indicated by Wang et al. (2020) and Haddad et al., 2022 (Wang et al., 2020, Haddad et al., 2022). The inhibitory concentration (IC50) of tannic acid has been reported to range from 1 to 13.4 µM. Additionally, tannic acid has been identified as an inhibitor of TMPRSS2, a crucial cell surface protease involved in processing the viral spike protein (S protein) of SARS-CoV-2. This processing enhances the binding of the spike protein to the cell surface receptor ACE2, facilitating cell entry through membrane fusion (Zhu et al., 2020a). The dual inhibitory activity of tannic acid, targeting both the viral main protease and the cellular TMPRSS2 protease, suggests the potential to impede the cellular entry of SARS-CoV-2. Notably, among plant-derived polyphenols, tannic acid exhibits a high potential for inhibiting SARS-CoV-2. Similar inhibitory activities against Mpro have been observed in dimeric proanthocyanidins, punicalagin, and mixtures of tannic acid with other polyphenols in various in vitro studies (Zhu and Xie, 2020, Tito et al., 2021, Nguyen et al., 2021). Green tea-derived tannins have also demonstrated the inhibition of SARS-CoV-2 replication in vitro, persisting in the pharyngeal mucosa even one hour after administration by throat spray. Hydrolysable tannins, such as pedunculagin, tercatain, and castalin, have shown potential as inhibitors of SARS-CoV-2. These natural compounds are predicted to be able to bind to the catalytic dyad residues of Mpro/3CLpro and inhibit the enzyme activity (Khalifa et al., 2020). Of significance is the anti-COVID-19 activity demonstrated through oral administration of highly-purified isomers of tannic acid, showcasing efficacy against both the omicron and delta variants.
Finally, the in vitro study outcomes validated the in silico data, confirming substantial Mpro inhibitory activity for the chosen compounds (4, 5, 10, and 14), with the most potent compound being 4, with an IC50 value of 1.2 µg/mL. Additionally, the cytotoxicity analysis revealed that none of these compounds exhibited any cytotoxic effects on normal and lung human cells at concentrations corresponding to their antiviral activity. These findings affirm the potential of these compounds as safe antiviral agents.
5. Conclusions
In conclusion, 18 Saudi Arabian medicinal plants were evaluated in vitro for SARS-CoV-2 Mpro inhibition activity using an enzymatic fluorescence Mpro inhibition FRET assay. Among the screened plants, T. brownii and A. asak displayed significant Mpro inhibition activity. To delve deeper into these plants, we employed a bioassay-guided fractionation and isolation procedure, resulting in the identification of fifteen compounds from the most active n-butanol fractions of both plants.
Moreover, in silico molecular docking studies of the isolated compounds revealed notable docking scores for compounds 4, 5, 10, and 14, indicating the strongest affinity among the tested compounds. Molecular dynamics simulations confirmed the formation of stable protein complexes by assessing RMSD, RMSF, Rg, SASA, and MM-GBSA parameters. The in vitro Mpro inhibition activity aligned with the in silico findings, demonstrating significant inhibitory activity for all tested compounds. Remarkably, compound 4 emerged as the most potent Mpro inhibitor. Importantly, none of the selected active compounds (4, 5, 10, and 14) exhibited significant cytotoxic activity, suggesting their safety for use as antiviral agents.
Fianlly, our results pinpoint plant extracts and compounds, employing bioassay-guided procedures, that hold promise in the quest for COVID-19 treatments. This serves as a foundation for subsequent in vivo and clinical trials, paving the way for continued exploration and the potential development of effective treatments for COVID-19. Molecular docking and simulation techniques, while instrumental in understanding molecular interactions at the atomic level, possess inherent limitations that must be acknowledged when attempting to predict in vivo efficacy. Challenges include inaccuracies in scoring functions and force fields, oversimplification of molecular flexibility, neglect of solvent effects, and difficulties in parameterizing complex ligands. Computational constraints limit the simulation of biologically relevant time scales, potentially missing critical events. Additionally, the predictive stability of protein–ligand complexes may not align with biological stability, and cellular and tissue contexts are often overlooked. While these methods offer insights, experimental validation remains crucial for a comprehensive understanding, highlighting the importance of integrating computational and experimental approaches in drug discovery studies.
CRediT authorship contribution statement
Yousef T.M. Alharbi: Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Wael M. Abdel-Mageed: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Supervision, Project administration. Omer A. Basudan: Conceptualization, Methodology Validation, Investigation, Data curation, Writing – original draft, Writing – review & editing, Supervision. Ramzi A. Mothana: Conceptualization, Validation, Formal analysis, Investigation, Writing – review & editing, Supervision, Project administration, Funding acquisition. Md Tabish Rehman: Methodology, Software, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Ali A. ElGamal: Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Ali S. Alqahtani: Methodology, Formal analysis, Resources, Writing – review & editing, Supervision. Omer I. Fantoukh: Formal analysis, Resources, Writing – original draft, Writing – review & editing. Mohamed F. AlAjmi: Formal analysis, Resources, Writing – original draft, Writing – review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Researchers Supporting Program through Project number (RSP2024R119), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Program for funding this research work through the Project number (RSP2024R119), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2024.102023.
Contributor Information
Wael M. Abdel-Mageed, Email: wabdelmageed@ksu.edu.sa.
Ramzi A. Mothana, Email: rmothana@ksu.edu.sa.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abdallah H.M., El-Halawany A.M., Sirwi A., El-Araby A.M., Mohamed G.A., Ibrahim S.R.M., et al. Repurposing of some natural product isolates as SARS-COV-2 main protease inhibitors via in vitro cell free and cell-based antiviral assessments and molecular modeling approaches. Pharmaceuticals. 2020;2021(14):213. doi: 10.3390/ph14030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah N.H., Salim F., Ahmad R. Chemical constituents of Malaysian U. Cordata var. Ferruginea and their in vitro α-Glucosidase inhibitory activities. Molecules. 2016;21:525. doi: 10.3390/molecules21050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo-Elghiet F., Rushdi A., Ibrahim M.H., Mahmoud S.H., Rabeh M.A., Alshehri S.A., El Menofy N.G. Chemical profile, antibacterial, antibiofilm, and antiviral activities of Pulicaria crispa most potent fraction: an in vitro and in silico study. Molecules. 2023;28:4184. doi: 10.3390/molecules28104184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M.J., Kim C.Y., Lee J.S., Kim J., Kim S.H., Lee C.K., et al. Inhibition of HIV-1 integrase by galloyl glucose from Terminalia chebula and flavonol glycoside gallates from Euphorbia pekinensis. Planta Med. 2002;68:457–459. doi: 10.1055/s-2002-32070. [DOI] [PubMed] [Google Scholar]
- Ajala O.S., Jukov A., Ma C.M. Hepatitis C virus inhibitory hydrolysable tannins from the fruits of Terminalia chebula. Fitoterapia. 2014;99:117–123. doi: 10.1016/j.fitote.2014.09.014. [DOI] [PubMed] [Google Scholar]
- AlAjmi M.F., Rehman M.T., Hussain A., Rather G.M. Pharmacoinformatics approach for the identification of polo-like kinase-1 inhibitors from natural sources as anti-cancer agents. Int. J. Biol. Macromol. 2018;116:173–181. doi: 10.1016/j.ijbiomac.2018.05.023. [DOI] [PubMed] [Google Scholar]
- AlAjmi M.F., Azhar A., Owais M., Rashid S., Hasan S., Hussain A., Rehman M.T. Antiviral potential of some novel structural analogs of standard drugs repurposed for the treatment of COVID-19. J. Biomol. Struct. Dyn. 2021;39:6676–6688. doi: 10.1080/07391102.2020.1799865. [DOI] [PubMed] [Google Scholar]
- Al-Fatimi M., Wurster M., Schröder G., Lindequist U. Antioxidant, antimicrobial and cytotoxic activities of selected medicinal plants from Yemen. J. Ethnopharmacol. 2007;111:657–666. doi: 10.1016/j.jep.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Ali H.M., Soliman A.G., Elfiky H.G.A.G. SAR and QSAR of COVID-19 Main protease-inhibitor interactions of recently X-ray crystalized complexes. Proc. Natl. Acad. Sci. India Sect. B. 2022;92:281–291. doi: 10.1007/s40011-021-01338-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shabib N.A., Khan J.M., Malik A., Alsenaidy M.A., Rehman M.T., AlAjmi M.F., et al. Molecular insight into binding behavior of polyphenol (rutin) with beta lactoglobulin: spectroscopic, molecular docking and MD simulation studies. J. Mol. Liq. 2018;269:511–520. doi: 10.1016/j.molliq.2018.07.122. [DOI] [Google Scholar]
- Al-Wahaibi L.H., Rehman M.T., Al-Saleem M.S.M., Basudan O.A., El-Gamal A.A., AlAjmi M.F., et al. Phenolics from the heartwood of Tecoma mollis as potential inhibitors of COVID-19 virus main protease and spike proteins: an in silico study. Pharmacogn. Mag. 2021;17:s278–s286. doi: 10.4103/pm.pm_35_21. [DOI] [Google Scholar]
- Al-Wahaibi L.H., Rehman M.T., Al-Saleem M.S.M., Basudan O.A., El-Gamal A.A., Abdelkader M.S.A., et al. Virtual screening and molecular dynamics simulation study of abyssomicins as potential inhibitors of COVID-19 virus main protease and spike protein. J. Biomol. Struct. Dyn. 2022;2022:1–17. doi: 10.1080/07391102.2022.2139295. [DOI] [PubMed] [Google Scholar]
- Ananil K., Hudson J.B., de Souzal C., Akpaganal K., Tower G.H.N., Arnason J.T., Gbeassor M. Investigation of medicinal plants of Togo for antiviral and antimicrobial activities. Pharm. Biol. 2000;38:40–45. doi: 10.1076/1388-0209(200001)3811-BFT040. [DOI] [PubMed] [Google Scholar]
- Arbab A.H., Parvez M.K., Al-Dosari M.S., Al-Rehaily A.J. In vitro evaluation of novel antiviral activities of 60 medicinal plants extracts against hepatitis B virus. Exp. Ther. Med. 2017;14:626–634. doi: 10.3892/etm.2017.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbar O.P., Joshi M.N., Madan A.R. Evaluation of plants for antiviral activity. Indian J. Med. Res. 1982;76:54–65. [PubMed] [Google Scholar]
- Banerjee, D., Hazra, A.K., Seal, T., Sur, T., Bhattacharya, D., Ray, J., et al., 2013. Antioxidant and anti-inflammatory activities of different solvent extracts and isolated compounds of Ipomoea pescaprae (L) sweet of Sunderban mangrove eco-complex. Asian J. Chem. 25, 4997–5000. 10.14233/ajchem.2013.14226.
- Behbahani M. Anti-HIV-1 activity of eight monofloral Iranian honey types. PLoS One. 2014;9:e108195. doi: 10.1371/journal.pone.0108195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berekaa M.M. Insights into the COVID-19 pandemic: origin, pathogenesis, diagnosis, and therapeutic interventions. Front. Biosci. (elite Ed) 2021;13:117–139. doi: 10.2741/874. [DOI] [PubMed] [Google Scholar]
- Biruk, H., Sentayehu, B., Alebachew, Y., Tamiru, W., Ejigu, A., Assefa, S., 2020. In vivo antimalarial activity of 80% methanol and aqueous bark extracts of Terminalia brownii Fresen. (Combretaceae) against Plasmodium berghei in Mice. Biochem. Res. Int. 2020, 9749410. 10.1155/2020/9749410. [DOI] [PMC free article] [PubMed]
- Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., et al. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters, SC’06; p. 43. [Google Scholar]
- Branka A.C. Nose-Hoover chain method for nonequilibrium molecular dynamics simulation. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 2000;61:4769–4773. doi: 10.1103/physreve.61.4769. [DOI] [PubMed] [Google Scholar]
- Bunluepuech K., Tewtrakul S. Anti-HIV integrase activity of Thai medicinal plants. Songklanakarin J. Sci. Technol. 2009;31:289–292. [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan J., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Eng. J. Med. 2020;382:787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Sharma G., Bhattacharya M., Lee S.S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Blanc P., Stoddart C.A., Bogan M., Rozhon E.J., Parkinson N., et al. New iridoids from the medicinal plant Barleria prionitis with potent activity against respiratory syncitial virus. J. Nat. Prod. 1998;61:1295–1297. doi: 10.1021/np980086y. [DOI] [PubMed] [Google Scholar]
- Chiang C.-C., Chang J.-S., Chen C.-C., Ng L.-T., Lin C.-C. Anti-herpes simplex virus activity of Bidens pilosa and hottuynia cordata. Am. J. Chin. Med. 2003;31:355–362. doi: 10.1142/S0192415X03001090. [DOI] [PubMed] [Google Scholar]
- Chiang L.C., Ng L.T., Cheng P.W., Chiang W., Lin C.C. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin. Exp. Pharmacol. Physiol. 2005;32:811–816. doi: 10.1111/j.1440-1681.2005.04270.x. [DOI] [PubMed] [Google Scholar]
- Citarella A., Scala A., Piperno A., Micale N. SARS-CoV-2 mpro: a potential target for peptidomimetics and small-molecule inhibitors. Biomolecules. 2021;2021(11):607. doi: 10.3390/biom11040607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad J., Vogler B., Reeb S., Klaiber I., Papajewski S., Roos G., et al. Isoterchebulin and 4,6-O-isoterchebuloyl-D-glucose, novel hydrolyzable tannins from Terminalia macroptera. J. Nat. Prod. 2001;2001(64):294–299. doi: 10.1021/np000506v. [DOI] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig A.J., Williams D.H., Oelrichs P.B., Baczynskj L. Isolation and structure elucidation of punicalagin, a toxic hydrolysable tannin, from Terminalia oblongata. J. Chem. Soc. Perkin Trans. I. 1990;1990:2317–2321. doi: 10.1039/P19900002317. [DOI] [Google Scholar]
- El-Hela A.A., Bakr M.S.A., Hegazy M.M., Dahab M.A., Elmaaty A.A., Ibrahim A.E., et al. Phytochemical characterization of pterocephalus frutescens with in-silico evaluation as chemotherapeutic medicine and oral pharmaco-kinetics prediction study. Sci. Pharm. 2023;91:7. doi: 10.3390/scipharm91010007. [DOI] [Google Scholar]
- El-Seedi H.R., Kotb S.M., Musharraf S.G., Shehata A.A., Guo Z., Alsharif S.M., et al. Saudi Arabian Plants: a powerful weapon against a plethora of diseases. Plants. 2020;11:3436. doi: 10.3390/plants11243436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shiekh R.A., Abdelmohsen U.R., Ashour H.M., Ashour R.M. Novel antiviral and antibacterial activities of Hibiscus schizopetalus. Antibiotics. 2020;9:756. doi: 10.3390/antibiotics9110756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsohly H.N., El-Feraly F.S., Joshi A.S., Walker L.A. Antiviral flavonoids from Alkanna orientalis. Planta Med. 1997;1997(63):384. doi: 10.1055/s-2006-957713. [DOI] [PubMed] [Google Scholar]
- El-Toumy S.A., Rauwald H.W. Two ellagitannins from Punica granatum heartwood. Phytochemistry. 2002;61:971–974. doi: 10.1016/s0031-9422(02)00435-1. [DOI] [PubMed] [Google Scholar]
- Fang E.F., Ng T.B. Bitter gourd (Momordica charantia) is a cornucopia of health: a review of its credited antidiabetic, anti-HIV, and antitumor properties. Curr. Mol. Med. 2011;11:417–436. doi: 10.2174/156652411795976583. [DOI] [PubMed] [Google Scholar]
- Garibaldi B.T., Wang K., Robinson M.L., Zeger S.L., Bandeen-Roche K., Wang M.-C., et al. Comparison of time to clinical improvement with vs without remdesivir treatment in hospitalized patients with COVID-19. JAMA Netw. Open. 2021;4:e213071. doi: 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoke S.S., Sood R., Kumar N., Pateriya A.K., Bhatia S., Mishra A., et al. Evaluation of antiviral activity of Ocimum sanctum and Acacia arabica leaves extracts against H9N2 virus using embryonated chicken egg model. BMC Complement. Altern. Med. 2018;18:174. doi: 10.1186/s12906-018-2238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.J., Skjaerven L., Yao X.Q. The Bio3D packages for structural bioinformatics. Protein Sci. 2021;30:20–30. doi: 10.1002/pro.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris A., Szlávik L., Minárovits J., Vasas A., Molnár J., Hohmann J. Antiviral activities of extracts of Euphorbia hirta L. against HIV-1, HIV-2 and SIVmac251. In Vivo. 2009;23:429–432. [PubMed] [Google Scholar]
- Haddad M., Gaudreault R., Sasseville G., Nguyen P.T., Wiebe H., van de Ven T., et al. Molecular interactions of tannic acid with proteins associated with SARS-CoV-2 infectivity. Int. J. Mol. Sci. 2022;23:2643. doi: 10.3390/ijms23052643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann J., Redei D., Mathe I., Molnar J., Musi I., Evanics F., Dombi G. Diterpene polyesters with antiviral activity from Euphorbia peplus and Euphorbia serrulata. Phytomedicine. 2000;7:85. doi: 10.1016/S0040-4020(00)00278-7. [DOI] [Google Scholar]
- Hong E.H., Song J.H., Kang K.B., Sung S.H., Ko H.J., Yang H. Anti-influenza activity of betulinic acid from Zizyphus jujuba on influenza A/PR/8 virus. Biomol. Ther. 2015;23:345–349. doi: 10.4062/biomolther.2015.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain A., Rahman M.E., Rahman M.S., Nasirujjaman K., Matin M.N., Faruqe M.O., Rabbee M.F. Identification of medicinal plant-based phytochemicals as a potential inhibitor for SARS-CoV-2 main protease (Mpro) using molecular docking and deep learning methods. Comput. Biol. Med. 2023;157 doi: 10.1016/j.compbiomed.2023.106785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.S., Luo S.H., Li Y.L., Li C.H., Hua J., Liu Y., et al. antifeedant and antiviral diterpenoids from the fresh roots of Euphorbia jolkinii. Nat. Prod. Bioprospect. 2014;4:91–100. doi: 10.1007/s13659-014-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.Y., Zhong L.J., Xie J.M., Wang F., Zhang Y.H. A new taraxastane-type triterpene from Vitex trifolia var. simplicifolia. Helv. Chim. Acta. 2013;96:2040–2045. doi: 10.1002/hlca.201200614. [DOI] [Google Scholar]
- Iqbal D., Rehman M.T., Bin Dukhyil A., Rizvi S., Al Ajmi M.F., Alshehri B.M., et al. High-throughput screening and molecular dynamics simulation of natural product-like compounds against alzheimer's disease through multitarget approach. Pharmaceuticals. 2021;14:937–955. doi: 10.3390/ph14090937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir N.R., Rehman M.T., Tabrez S., Alserihi R.F., AlAjmi M.F., Khan M.S., et al. Identification of butyrylcholinesterase and monoamine oxidase B targeted ligands and their putative application in alzheimer's treatment: a computational strategy. Curr. Pharm. Des. 2021;27:2425–2434. doi: 10.2174/1381612827666210226123240. [DOI] [PubMed] [Google Scholar]
- Jiang H., Zou X., Zeng P., Zeng X., Zhou X., Wang J., et al. Crystal structures of main protease (Mpro) mutants of SARS-CoV-2 variants bound to PF-07304814. Mol. Biomed. 2023;4:23. doi: 10.1186/s43556-023-00134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jossang A., Pousset J.-L., Bodo B. Combreglutinin, a hydrolyzable tannin from Combretum glutinosum. J. Nat. Prod. 1994;57:732–737. doi: 10.1021/np50108a008. [DOI] [Google Scholar]
- Kang S.Y., Kang J.Y., Oh M.J. Antiviral activities of flavonoids isolated from the bark of Rhus verniciflua stokes against fish pathogenic viruses in vitro. J. Microbiol. 2012;50:293–300. doi: 10.1007/s12275-012-2068-7. [DOI] [PubMed] [Google Scholar]
- Kapewangolo P., Kandawa-Schulz M., Meyer D. Anti-HIV activity of Ocimum labiatum extract and isolated pheophytin-a. Molecules. 2017;22:1763. doi: 10.3390/molecules22111763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada Y., Nishizawa M., Yamagishi T., Tanaka T., Nonaka G., Cosentino L.M., et al. Anti-AIDS agents, 18. sodium and potassium salts of caffeic acid tetramers from Arnebia euchroma as anti-HIV agents. J. Nat. Prod. 1995;58:392–400. doi: 10.1021/np50117a007. [DOI] [PubMed] [Google Scholar]
- Kesharwani A., Polachira S.K., Nair R., Agarwal A., Mishra N.N., Gupta S.K. Anti-HSV-2 activity of Terminalia chebula Retz extract and its constituents, chebulagic and chebulinic acids. BMC Complement. Altern. Med. 2017;17:110. doi: 10.1186/s12906-017-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa I., Zhu W., Mohammed H.H.H., Dutta K., Li C. Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CL(pro): an in silico approach with 19 structural different hydrolysable tannins. J. Food Biochem. 2020;e13432 doi: 10.1111/jfbc.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraszni M., Marosi A., Larive C.K. NMR assignments and the acid-base characterization of the pomegranate ellagitannin punicalagin in the acidic pH-range. Anal. Bioanal. Chem. 2013;405:5807–5816. doi: 10.1007/s00216-013-6987-x. [DOI] [PubMed] [Google Scholar]
- Lai Y.F. Configuration and conformation of dihydroflavonols from Acacia melanoxylon. Phytochemistry. 1987;26:813–817. doi: 10.1016/S0031-9422(00)84793-7. [DOI] [Google Scholar]
- Landete P., Quezada Loaiza C.A., Aldave-Orzaiz B., Muñiz S.H., Maldonado A., Zamora E., et al. Clinical features and radiological manifestations of COVID-19 disease. World J. Radiol. 2020;12:247–260. doi: 10.4329/wjr.v12.i11.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.M., Tang Y.L., Zhang Z.H., Liu C.J., Li H.Z., Li R.T., Xia X.S. Compounds from Arnebia euchroma and their related anti-HCV and antibacterial activities. Planta Med. 2012;78:39–45. doi: 10.1055/s-0031-1280266. [DOI] [PubMed] [Google Scholar]
- Li H., Xu F., Liu C., Cai A., Dain J.A., Li D., et al. Inhibitory effects and surface plasmon resonance-based binding affinities of dietary hydrolyzable tannins and their gut microbial metabolites on SARS-CoV-2 main protease. J. Agric. Food Chem. 2021;69:12197–12208. doi: 10.1021/acs.jafc.1c03521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.T., Chen T.Y., Chung C.Y., Noyce R.S., Grindley T.B., McCormick C., et al. Hydrolyzable tannins (chebulagic acid and punicalagin) target viral glycoprotein-glycosaminoglycan interactions to inhibit herpes simplex virus 1 entry and cell-to-cell spread. J. Virol. 2011;85:4386–4398. doi: 10.1128/JVI.01492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loganathan C., Ameen F., Sakayanathan P., Islam M.A., Thayumanavan P. Exploring the interaction of phytochemicals from Hibiscus rosa-sinensis flowers with glucosidase and acetylcholinesterase: an integrated in vitro and in silico approach. Comput. Biol. Chem. 2024;108 doi: 10.1016/j.compbiolchem.2023.107996. [DOI] [PubMed] [Google Scholar]
- López-Martínez L.M., Santacruz-Ortega H., Navarro R.E., Sotelo-Mundo R.R., González-Aguilar G.A. A 1H NMR investigation of the interaction between phenolic acids found in mango (manguifera indica cv ataulfo) and papaya (Carica papaya cv maradol) and 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radicals. PLoS One. 2015;10:e0140242. doi: 10.1371/journal.pone.0140242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maregesi S., Van Miert S., Pannecouque C., Feiz Haddad M.H., Hermans N., Wright C.W., et al. Screening of Tanzanian medicinal plants against Plasmodium falciparum and human immunodeficiency virus. Planta Med. 2010;76:195–201. doi: 10.1055/s-0029-1186024. [DOI] [PubMed] [Google Scholar]
- Martino V.S., López P., Martinez Irujo J.J., Sanromán M., Cuevas M.T., Santiago E., et al. Inhibitory effect against polymerase and ribonuclease activities of HIV-reverse transcriptase of the aqueous leaf extract of Terminalia triflora. Phytother. Res. 2002;16:778–780. doi: 10.1002/ptr.1065. [DOI] [PubMed] [Google Scholar]
- Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- Mohamed I.E.T., El Nur E.B.E.S., Abdelrahman M.E.N. The antibacterial, antiviral activities and photochemical screening of some Sudanese medicinal plants. Eurasian. J. Biosci. 2010;4:8–16. doi: 10.5053/ejobios.2010.4.0.2. [DOI] [Google Scholar]
- Muteeb G., Alshoaibi A., Aatif M., Rehman M.T., Qayyum M.Z. Screening marine algae metabolites as high-affinity inhibitors of SARS-CoV-2 main protease (3CLpro): an in silico analysis to identify novel drug candidates to combat COVID-19 pandemic. Appl. Biol. Chem. 2020;63:79. doi: 10.1186/s13765-020-00564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasr F.A., Shahat A.A., Alqahtani A.S., Ahmed M.Z., Qamar W., Al-Mishari A.A., Almoqbil A.N. Centaurea bruguierana inhibits cell proliferation, causes cell cycle arrest, and induces apoptosis in human MCF-7 breast carcinoma cells. Mol. Biol. Rep. 2020;47:6043–6051. doi: 10.1007/s11033-020-05679-x. [DOI] [PubMed] [Google Scholar]
- Negishi H., Maoka T., Njelekela M., Yasui N., Juman S., Mtabaji J., et al. New chromone derivative terminalianone from African plant Terminalia brownii Fresen (Combretaceae) in Tanzania. J. Asian Nat. Prod. Res. 2011;13:281–283. doi: 10.1080/10286020.2011.552431. [DOI] [PubMed] [Google Scholar]
- Nguyen T.T.H., Jung J.H., Kim M.K., Lim S., Choi J.M., Chung B., et al. The inhibitory effects of plant derivate polyphenols on the main orotease of SARS coronavirus 2 and their structure-activity relationship. Molecules. 2021;26:1924. doi: 10.3390/molecules26071924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notka F., Meier G., Wagner R. Concerted inhibitory activities of Phyllanthus amarus on HIV replication in vitro and ex vivo. Antiviral. Res. 2004;64:93–102. doi: 10.1016/j.antiviral.2004.06.010. PMID: 15498604. [DOI] [PubMed] [Google Scholar]
- Nutan, Modi, M., Dezzutti, C.S., Kulshreshtha, S., Rawat, A.K., Srivastava, S.K., et al., 2013. Extracts from Acacia catechu suppress HIV-1 replication by inhibiting the activities of the viral protease and Tat. Virol. J. 10, 309. 10.1186/1743-422X-10-309. [DOI] [PMC free article] [PubMed]
- Patil V.S., Harish D.R., Vetrivel U., Roy S., Deshpande S.H., Hegde H.V. Hepatitis C virus NS3/4A inhibition and host immunomodulation by tannins from Terminalia chebula: a structural perspective. Molecules. 2022;27:1076. doi: 10.3390/molecules27031076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perveen S., Orfali R., Azam M.S.U., Aati H.Y., Bukhari K., Bukhari S.I., Al-Taweel A. Coronavirus nCOVID-19: A pandemic disease and the Saudi precautions. Saudi Pharm. J. 2020;28:888–897. doi: 10.1016/j.jsps.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfundstein B., El Desouky S.K., Hull W.E., Haubner R., Erben G., Owen R.W. Polyphenolic compounds in the fruits of Egyptian medicinal plants (Terminalia bellerica, Terminalia chebula and Terminalia horrida): characterization, quantitation and determination of antioxidant capacities. Phytochemistry. 2010;71:1132–1148. doi: 10.1016/j.phytochem.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Pongthanapisith, V., Ikuta, K., Puthavathana, P., Leelamanit, W., 2013. Antiviral Protein of Momordica charantia L. Inhibits Different Subtypes of Influenza A. Evid. Based Complement. Alternat. Med. 2013, 729081. 10.1155/2013/729081. [DOI] [PMC free article] [PubMed]
- Price D.J., Brooks C.L., 3rd. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- Priya S., Kumar N.S., Hemalatha S. Antiviral phytocompounds target envelop protein to control Zika virus. Comput. Biol. Chem. 2018;77:402–412. doi: 10.1016/j.compbiolchem.2018.08.008. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., Shamsi H., Khan A.U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharm. 2014;11:1785–1797. doi: 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., AlAjmi M.F., Hussain A. Natural compounds as inhibitors of SARS-CoV-2 Main protease (3CLpro): a molecular docking and simulation approach to combat COVID-19. Curr. Pharm. Des. 2021;27:3577–3589. doi: 10.2174/1381612826999201116195851. [DOI] [PubMed] [Google Scholar]
- Rehman S., Ashfaq U.A., Riaz S., Javed T., Riazuddin S. Antiviral activity of Acacia nilotica against Hepatitis C Virus in liver infected cells. Virol. J. 2011;8:220. doi: 10.1186/1743-422X-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih E.Y.A., Fyhrquist P., Abdalla A.M.A., Abdelgadir A.Y., Kanninen M., Sipi M., et al. LC-MS/MS tandem mass spectrometry for analysis of phenolic compounds and pentacyclic triterpenes in antifungal extracts of Terminalia brownii (Fresen) Antibiotics. 2017;6:37. doi: 10.3390/antibiotics6040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih E.Y.A., Julkunen-Tiitto R., Lampi A.M., Kanninen M., Luukkanen O., Sipi M., et al. Terminalia laxiflora and Terminalia brownii contain a broad spectrum of antimycobacterial compounds including ellagitannins, ellagic acid derivatives, triterpenes, fatty acids and fatty alcohols. J. Ethnopharmacol. 2018;227:82–96. doi: 10.1016/j.jep.2018.04.030. [DOI] [PubMed] [Google Scholar]
- Salih E.Y.A., Julkunen-Tiitto R., Luukkanen O., Fyhrqvist P. Anti-Candida activity of extracts containing ellagitannins, triterpenes and flavonoids of Terminalia brownii, a medicinal plant growing in semi-arid and Savannah woodland in Sudan. Pharmaceutics. 2022;14:2469. doi: 10.3390/pharmaceutics14112469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome Coronavirus-2. Arch. Med. Res. 2020;2020(51):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar D., Harder E., Damm W., Friesner R.A., Sherman W. Improving the prediction of absolute solvation free energies using the next generation OPLS force field. J. Chem. Theory Comput. 2012;8:2553–2558. doi: 10.1021/ct300203w. [DOI] [PubMed] [Google Scholar]
- Simões C.M., Amoros M., Girre L. Mechanism of antiviral activity of triterpenoid saponins. Phytother. Res. 1999;13:323–328. doi: 10.1002/(SICI)1099-1573(199906)13:4<323::AID-PTR448>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Sumithira P., Dhivya Mangala S., Sophie A.M., Padma Latha C. Antiviral and antioxidant activities of two medicinal plants. Int. J. Curr. Sci. 2012:256–261. [Google Scholar]
- Takeda Y., Okuyama Y., Nakano H., Yaoita Y., Machida K., Ogawa H., Imai K. Antiviral activities of Hibiscus sabdariffa L. tea extract against human influenza a virus rely largely on acidic pH but partially on a low-pH-independent mechanism. Food Environ. Virol. 2020;12:9–19. doi: 10.1007/s12560-019-09408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Nonaka G.-I., Nishioka I. Tannins and related compounds. XL. Revision of the structures of punicalin and punicalagin, and isolation and characterization of 2-O-galloyl punicalin from the bark of Punica granatum L. Chem. Pharmaceut. Bull. 1986;34:650–655. doi: 10.1248/cpb.34.650. [DOI] [Google Scholar]
- Tito A., Colantuono A., Pirone L., Pedone E., Intartaglia D., Giamundo G., et al. Pomegranate peel extract as an inhibitor of SARS-CoV-2 spike binding to human ACE2 receptor (in vitro): a promising source of novel antiviral drugs. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.638187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food & Drugs: Coronavirus (COVID-19) Drugs. [Accessed December 12, 2022]. Available from URL: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs.
- Ürményi F.G., Saraiva G.D., Casanova L.M., Matos A.D., de Magalhães Camargo L.M., Romanos M.T., Costa S.S. Anti-HSV-1 and HSV-2 flavonoids and a new kaempferol triglycoside from the medicinal plant Kalanchoe daigremontiana. Chem. Biodivers. 2016;13:1707–1714. doi: 10.1002/cbdv.201600127. [DOI] [PubMed] [Google Scholar]
- Visintini Jaime M.F., Redko F., Muschietti L.V., Campos R.H., Martino V.S., Cavallaro L.V. In vitro antiviral activity of plant extracts from asteraceae medicinal plants. Virol. J. 2013;10:245. doi: 10.1186/1743-422X-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.C., Chen Y., Wang Y.C., Wang W.J., Yang C.S., Tsai C.L., et al. Tannic acid suppresses SARS-CoV-2 as a dual inhibitor of the viral main protease and the cellular TMPRSS2 protease. Am. J. Cancer Res. 2020;10:4538–4546. [PMC free article] [PubMed] [Google Scholar]
- Wang C.Y., Huang S.C., Zhang Y., Lai Z.R., Kung S.H., Chang Y.S., Lin C.W. Antiviral ability of Kalanchoe gracilis leaf extract against enterovirus 71 and coxsackievirus A16. Evid. Based Complement. Alternat. Med. 2012;2012 doi: 10.1155/2012/503165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Zhou B.N., Wisse J.H., Evans R., van der Werff H., Miller J.S., Kingston D.G. Three new ellagic acid derivatives from the bark of Eschweilera coriacea from the Suriname rainforest. J. Nat. Prod. 1998;61:901–906. doi: 10.1021/np980046u. [DOI] [PubMed] [Google Scholar]
- Yoosook C., Panpisutchai Y., Chaichana S., Santisuk T., Reutrakul V. Evaluation of anti-HSV-2 activities of Barleria lupulina and Clinacanthus nutans. J. Ethnopharmacol. 1999;67:179–187. doi: 10.1016/s0378-8741(99)00008-2. [DOI] [PubMed] [Google Scholar]
- Zhu H., Wei L., Niu P. The novel coronavirus outbreak in Wuhan, China. Glob. Health Res. Policy. 2020;5:6. doi: 10.1186/s41256-020-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Xie D.Y. Docking characterization and in vitro inhibitory activity of flavan-3-ols and dimeric proanthocyanidins against the main protease activity of SARS-Cov-2. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Zhu C., Zhu Y., Sun F. Minireview of progress in the structural study of SARS-CoV-2 proteins. Curr. Res. Microb. Sci. 2020;1:53–61. doi: 10.1016/j.crmicr.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.