Abstract
The Y-box binding protein-1 (YBX1) gene codes for a multifunctional oncoprotein that is increasingly being linked to the regulations of many aspects of cancer cell biology. Disparities in treatment outcomes between male and female cancer patients are increasingly reported. This study aimed to examine the relationship between YBX1 expression and overall survival in male and female patients with solid tumours. Overall survival and YBX1 expression data for cohorts of male and female cancer patients obtained from freely available databases were analysed with a cox proportional hazard model with covariates of biological sex and YBX1 expression. Kaplan–Meier curves and Violin plots were constructed for segregated male and female cohorts. High YBX1 expression was significantly associated with poor survival in 2 female-only and 4 mixed-sex cancer sites. In female lung cancer patients, better survival and lower YBX1 expression were identified. The clinical importance of YBX1 expression in cancer ought to be evaluated in a sex-specific manner, especially in lung cancer.
Keywords: Y-box-binding protein 1, Survival, Cancer, Biological sex
Subject terms: Cancer, Computational biology and bioinformatics, Genetics
Introduction
YB-1, also known as DNA binding protein B (DBPB), is one of three members of the Y-box family of transcription factors1 whose impact on cancer cell biology is increasingly supported by experimental studies identifying promotion of cell proliferation and apoptosis2, regulation of DNA proliferation and repair3, stemness4 and response to treatment5. Meanwhile, the consideration of sex as a biological variable in cancer research is identifying differences in cancer cell biology mechanisms between the sexes6. YB-1 was reported to interact with the X-linked ribosomal protein S4 (RPS4X), driving cisplatin sensitivity in breast cancer cell lines7, and poor outcomes in ovarian8 and bladder cancer9. But how biological sex relate to the biological and clinical impact of major regulators of cancer cell biology such as YB-1 remains unknown.
The human YB-1 gene (YBX1) is located on chromosome 1 (1p34), contains eight exons and spans 19 kb of genomic DNA. The YB-1 gene promoter contains several E-boxes and CG-repeats that are important for YB-1 transcription into a 1.5 kb-long mRNA10 and codes for a 324 amino acid YB-1 protein normally localised in the cytoplasm where it plays a key role in the regulation of mRNA translation11,12. The detection of this protein is rapidly emerging as both a clinically useful diagnostic biomarker and a potentially viable therapeutic target in many cancer types13,14. The analysis of YBX1 mRNA levels in Head and Neck cancer patients linked high expression with poor prognosis15. But the clinical importance of the mRNA expression levels of this oncoprotein remains poorly investigated.
The Sex as a Biological Variable (SABV) policy established by the US National Institute of Health16 requires researchers to distinguish between “sex”, a term related to the presence of XX or XY chromosomes in humans, from “gender”, a term associated with the social, cultural and psychological traits of human males and females. Analysis of the Cancer Genome Atlas identified sex-biased signatures in 53% of clinically actionable genes (60/114) investigated17. Differences between the sexes are increasingly documented in the functions of both the innate and adaptive immune systems18, regulation of miRNAs and mRNA17,19, genetic polymorphism in antibody responses20, and the microbiome21. Mice studies have reported sex-specific cell death programs with males prone to PARP-1 necrosis and females to caspase-dependent apoptosis22. Others identified differences in basal redox state23, response to oxidative stress24, sensitivity to both apoptosis and autophagy25.
Taking YBX1 mRNA levels as a test case, this in silico study aimed to examine whether the segreration of cohorts of patients with solid cancer that commonly develop in both males and females according to their recorded biological sex could identify novel associations between expression and overall survival.
Materials and methods
Patient cohorts
YBX1 mRNA expression profiles and survival data of patients diagnosed with 13 cancer types that commonly develop in both males and females were accessed from various databases (Table 1). The YBX1 mRNA expression profiles and survival data of female patients with breast, ovarian and uterine endometrial cancer were included as examples of disease site where sex is a controllable biological variable. Cancer cohorts were chosen to have a minimum of 150 subjects per condition, and recorded events (deaths) ranging from 15.8% (Rectum adenocarcinoma) to 61.7% of the sample (Ovarian cancer) allowing for robust survival analysis of overall survival (OS). This data was downloaded via the KM-Plotter web-interface (http://www.kmplot.com)26, and imported in the RStudio software for analysis.
Table 1.
Cancer cohorts and sources.
| Cancer cohort | Database/s | Sample size (sex division) | mRNA Expression quantification technique |
|---|---|---|---|
| Bladder cancer | Pancancer27 (derived from TCGA repository) | N = 406 (m = 298, f = 108) | mRNA sequence |
| Breast cancer | GEO repository28 | N = 4929 (m = 0, f = 4929) | Genechip |
| Cervical squamous cell carcinoma | Pancancer27 (derived from TCGA repository) | N = 304 (m = 0, f = 304) | mRNA sequence |
| Gastric cancer | Gastric cancer database29 | N = 780 (m = 544, f = 236) | Genechip |
| Head-neck squamous cell carcinoma | Pancancer27 (derived from TCGA repository) | N = 499 (m = 366, f = 133) | mRNA sequence |
| Liver hepatocellular carcinoma | Pancancer27 (derived from TCGA repository) | N = 370 (m = 249, f = 141) | mRNA sequence |
| Lung cancer | caBIG/GEO/TCGA repositories30 | N = 1814 (m = 1100, f = 714) | Genechip |
| Ovarian cancer | GEO/Cancer Atlas31 | N = 1435 (m = 0, f = 1435) | Genechip |
| Pancreatic ductal adenocarcinoma | Pancancer27 (derived from TCGA repository) | N = 177 (m = 97, f = 80) | mRNA sequence |
| Renal clear cell carcinoma | Pancancer27 (derived from TCGA repository) | N = 530 (m = 344, f = 186) | mRNA sequence |
| Renal papillary cell carcinoma | Pancancer27 (derived from TCGA repository) | N = 287 (m = 211, f = 76) | mRNA sequence |
| Rectum adenocarcinoma | Pancancer27 (derived from TCGA repository) | N = 165 (m = 90, f = 75) | mRNA sequence |
| Sarcoma | Pancancer27 (derived from TCGA repository) | N = 259 (m = 118, f = 141) | mRNA sequence |
| Stomach adenocarcinoma | Pancancer27 (derived from TCGA repository) | N = 371 (m = 238, f = 133) | mRNA sequence |
| Uterine corpus endometrial carcinoma | Pancancer27 (derived from TCGA repository) | N = 542 (m = 0, f = 542) | mRNA sequence |
Analysis of overall survival with YBX1 and biological sex
The impact of both YB-1 and sex on cancer survival for the 15 cancer types was tested with a cox proportional hazard model comprising of three co-variates: YB-1 mRNA levels (a continuous measure), sex (a categorical variable), and their interaction term. This analysis was implemented in RStudio (2022.02.2 + 485 "Prairie Trillium" Release) employing the survival package, with the relevant code provided in the supplementary material (S1). In convention with best statistical practice32, all covariates were tested simultaneously. Subgroup analysis was explicitly avoided unless significant interaction between covariates was detected. A Benjamini–Hochberg procedure33 was employed to correct for multiple testing in the 15 cancer types tested and ensure a Family-wise error rate of .
Analysis of YBX1 expression according to biological sex
YBX1 expression distribution between male and female sexes was analysed with a two-sample t-test. The Šidák variation of the Bonferroni correction34 was implemented to correct for multiple comparison. The threshold significance was set by solving , where m is the number of cancer types analysed and
Kaplan Meier survival analysis
Kaplan Meier plots were constructed for cancers with significant differences after the Benjamini–Hochberg procedure was employed, to investigate sex related differences, and a significance test performed using the survival package in R implemented through RStudio (2022.02.2 + 485 "Prairie Trillium" Release, R-Version 4.2.3, https://www.r-project.org/) employing the survival package, with the relevant code provided in the supplementary material S1.
Co-expression analysis
Co-expression coefficients were generated by downloading data from the “Coexpression tab” in the cBioportal web service. The list of genes located on chromosome X was downloaded from Uniprot and validated in Human Genome Organisation (HUGO) database. The chromosome X genes that showed a Spearman’S correlation coefficient greater or lower than 0.25 and − 0.25, respectively, and q value < 0.05 (FDR < 0.5) were deemed correlated with YB-1.
Ethics declaration
All data generated or analysed during this study was downloaded from the freely accessible databases outlined in Table 1. This data was irrevocably anonymous and is deposited on these open access platforms. All methods were carried out in accordance with relevant guidelines and regulations.
Results
High YBX1 expression is associated with reduced overall survival
We first analysed available survival data in all 15 identified cancer cohorts using a Cox proportional hazard model. Application of the Benjamini–Hochberg procedure to keep the false discovery rate at for all cohorts yielded a threshold significance value of . A significant relationship between YBX1 expression and survival was detected in 6 cancer types: breast, liver, lung, renal papilloma, uterine cancer, and sarcoma (Table 2). In all these sites, the hazard ratio for YBX1 was > 1, indicating that higher expression levels were associated with poorer survival (Table 2 and Fig. 1). As expected, biological sex did not affect survival in female only cancers (Breast, uterine cancer). In cancer sites affecting both males and females, biological sex did not interact with YBX1 expression and did not affect survival in this analysis (Hazard ratio = 1) (data not shown).
Table 2.
YBX1 expression and biological sex survival analysis in 15 cancer cohorts.
| Cancer cohort | Sample size (sex division) | YBX1 expression profile (mean, min–max values) | YBX1 expression significance level | YBX1 hazard ratio* (95% confidence) |
|---|---|---|---|---|
| Significant after Benjamini–Hochberg procedure for multiple comparisons | ||||
| Breast cancer | N = 4929 (m = 0, f = 4929) | GeneChip (9370, 96–33945) | 1.049 (1.039–1.059) | |
| Liver cancer | N = 370 (m = 249, f = 141) | mRNA sequence (6649, 1591–26649) | 1.145 (1.110–1.191) | |
| Lung cancer | N = 1814 (m = 1100, f = 714) | GeneChip (10975, 173–33991) | 1.038 (1.022–1.054) | |
| Renal papilloma | N = 287 (m = 211, f = 76) | mRNA sequence (7743, 1501–23551) | 1.287 (1.151–1.439) | |
| Uterine cancer | N = 542 (m = 0, f = 542) | mRNA sequence (13243, 436–44756) | 1.051 (1.018–1.084) | |
| Sarcoma | N = 259 (m = 118, f = 141) | mRNA sequence (14586, 3929–46500) | 1.089 (1.028–1.154) | |
| Non-significant after Benjamini–Hochberg procedure for multiple comparisons | ||||
| Stomach cancer | N = 371 (m = 238, f = 133) | mRNA sequence (12902, 3259–29802) | 0.994 (0.898–0.993) | |
| Ovarian cancer | N = 1435 (m = 0, f = 1435) | GeneChip (15042, 170–43850) | 1.009 (0.999–1.020) | |
| Cervical cancer | N = 304 (m = 0, f = 304) | mRNA sequence (13757, 2707–40652) | 1.041 (0.990–1.094) | |
| Bladder cancer | N = 406 (m = 298, f = 108) | mRNA sequence (13457, 1223–192954) | 1.007 (0.998–1.016) | |
| Renal clear cell | N = 530 (m = 344, f = 186) | mRNA sequence (7975, 861–19403) | 1.063 (0.977–1.157) | |
| Rectal cancer | N = 165 (m = 90, f = 75) | mRNA sequence (14727, 4704–31516) | 0.923 (0.816–1.044) | |
| Pancreatic cancer | N = 177 (m = 97, f = 80) | mRNA sequence (8027, 2463–38335) | 1.019 (0.967–1.074) | |
| Gastric cancer | N = 780 (m = 544, f = 236) | GeneChip (12443, 3167–25195) | 1.003 (0.968–1.039) | |
| Head and neck cancer | N = 499 (m = 366, f = 133) | mRNA sequence (13105, 3534–56844) | 1.000 (0.971–1.031) | |
*Hazard ratios are given per 1000 units of gene expression Quoted p-values and hazard ratios refer to YB-1 expression. Direct sex effects did not reach significance threshold and are not included here.
Figure 1.
Log of the Hazard ratio against YB-1 expression levels for cancers with significant expression effects in Table 2. The shaded region depicts the 95% confidence interval. Note the varying axes limits for both log hazard ratio and expression level.
YBX1 expression and biological sex
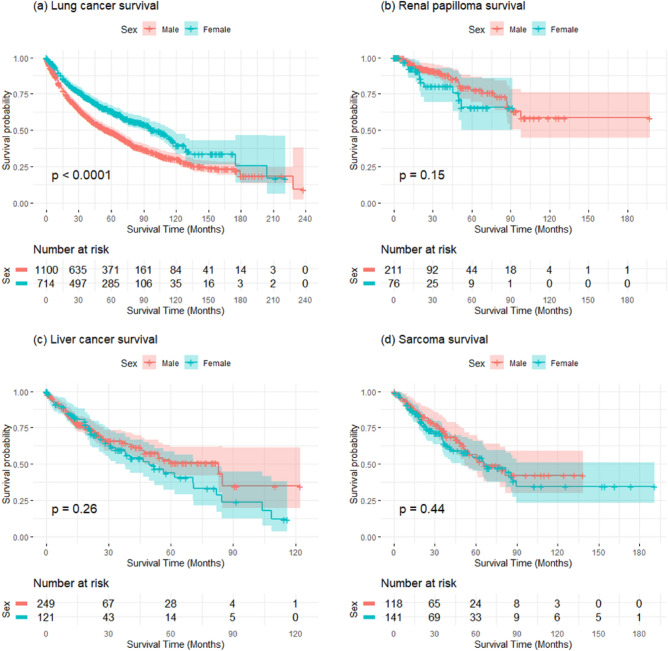
We next focused on the 4 cancer sites that affect both males and females, where YBX1 expression was identified to significantly affect survival: liver, lung, renal papilloma, and sarcoma. First, we constructed Kaplan–Meier curves to compare the survival of segregated male and female patient cohorts (Fig. 2). Lung was the only cancer type displaying a highly significant difference in survival when the data was analysed according to sex.
Figure 2.
Kaplan–Meier survival curves for male and female patients cohorts in lung, renal papilloma, liver and sarcoma. The p-values for the differences between male and female cohort survival is given in the figure for each cancer type.
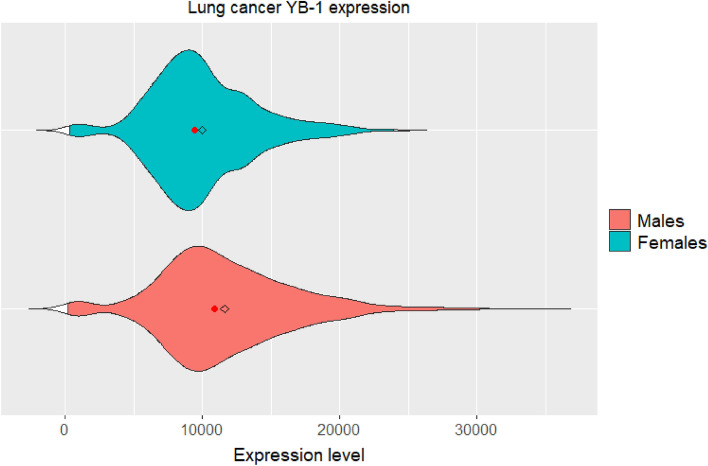
Second, we examined the distributions of YBX1 expression levels in both male and female cohorts and confirmed that these were approximately normal, with two-tailed t-tests. Finally, we compared YBX1 expression between sexes (Table 3). A Šidák variation of the Bonferroni correction yielded a threshold of . At this threshold, sex differences in YBX1 expression for the Lung cancer cohort were highly significant with a Cohen’s D of 0.363, indicating a medium to large effect size. A violin plot of the YBX1 expression distribution indicates higher expression in males, compared to females in lung cancer (Fig. 3).
Table 3.
Sex differences in YB-1 expression.
| Cancer type | Sex difference in expression significance | Cohen’s D |
|---|---|---|
| Significant after Šidák correction for multiple comparisons | ||
| Lung cancer | 0.363 | |
| Non-significant after Šidák correction for multiple comparisons | ||
| Renal papilloma | 0.245 | |
| Liver cancer | 0.267 | |
| Sarcoma | 0.162 | |
Figure 3.
Sex differences in YBX1 expression distribution for Lung cancer. The red dot indicates distribution medians, and the diamond indicates distribution means.
YBX1 expression and the X chromosome
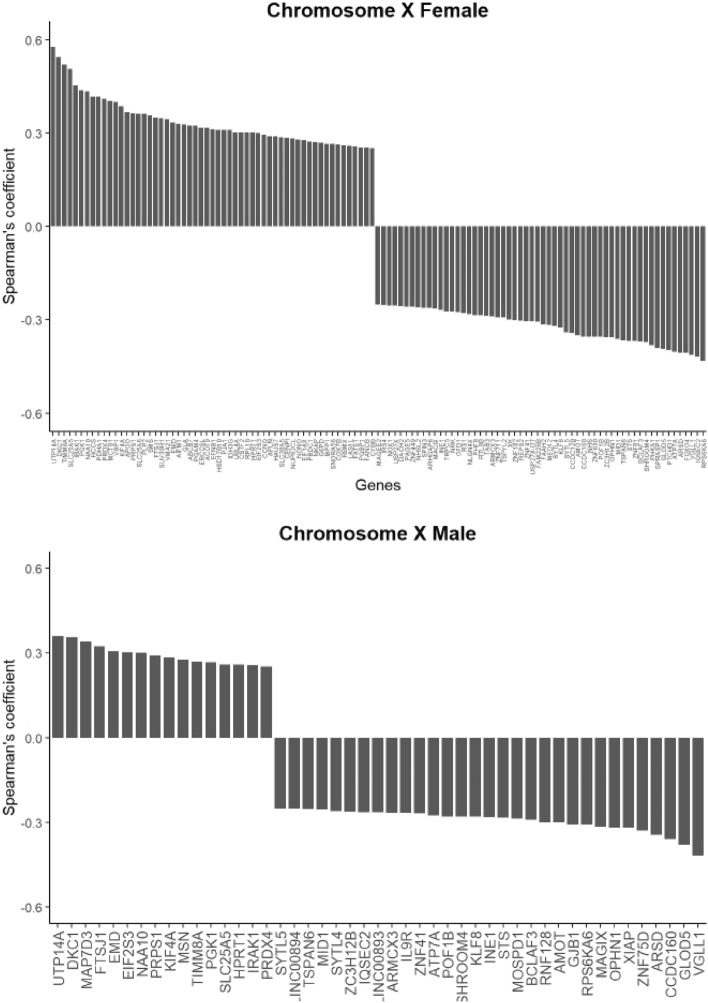
We next focused on one cancer site that is known to affect both males and females differently35–38, where YBX1 expression was not identified to significantly affect survival in our analysis: bladder cancer. We generated correlation coefficients for the expression of YBX1 and individual X-linked genes in both male and female patients. In total 47 (male) and 115 (female) chromosome X genes were identified to co-express positively or negatively with YBX1 (Fig. 4). N = 37 were common to both sexes (Supplementary material S2). Of those, DKC1 held the highest positive correlation coefficient (0.36) and VGLL1 the lowest (− 0.42). Kaplan–Meier analysis identified an association between expression and overall survival in both male and female cohort for VGLL1 but not DKC1 (data not shown). Of the 78 genes uniquely identified in the female cohort (Supplementary material S2), VBP1 held the highest positive correlation coefficient (0.4) and FOXO4 the lowest (− 0.40). In these female patients, low VBP1 was associated with poorer overall survival (HR = 1.87 (1.05–3.01), p = 0.03). No association was detected for FOXO4. In the male cohort (Supplementary material S2), all but MOSPD1 appeared associated with overall survival on Kaplan–Meier analysis (data not shown).
Figure 4.
Waterfall plots of the X-linked genes identified as associated with YB-1 on male and female bladder cancer patients.
Discussion
Sex is a fundamental biological variable increasingly studied as a factor influencing cancer treatment response39. Cancer affects men and women40; but we treat patients. This sex-neutral approach results from the belief that circulating sex hormones dominate sexual differentiation biology41 and the practice of sex data pooling39. Our efforts, however, yield unequal success between the sexes42. The predicted rise in the 19.3 million annual new cancer cases40 will worsen the clinical and societal impact of treatment resistance and innovation in cancer management is a clinical priority.
YB-1 is a multifunctional protein involved in both the transcriptional and translational regulation of gene expression43. The detection of this oncoprotein in tumour specimens is increasingly linked to poor patient outcomes. But the importance of YBX1 gene expression remains poorly documented. In the Prognoscan database44, YBX1 expression is associated with an increased hazard ratio for overall survival with Breast cancer, Lung cancer and prostate cancer (data not shown). We used available data for 15 cancer types to examine the link between YBX1 expression and survival outcomes. Our analysis identifies that high YBX1 expression is associated with poor survival in 6 cancer types.
YB-1 controls almost all DNA and mRNA dependent processes in the cell such as cellular differentiation, proliferation and stress response43. The regulation of these critical processes is increasingly linked to biological sex. This fundamental biological variable is defined by the presence of genetic information provided by the X and Y chromosomes, whose regulation and loss are proving relevant to cancer biology and treatment outcomes7–9,36,38,45–47. Lack of sex analysis in preclinical and interventional studies was proposed to increase the risk for an effect being lost or claimed where it only applies to one sex16. In biomedical research analysis of the literature identified the underrepresentation of female animals and a lack of sex-specific reporting48. Our analysis expands earlier report that a correlation between the expression of YB-1 and X-linked genes exists7–9. In both male and female patients with bladder cancer, we identified 37 interactions common to both sexes, 10 limited to male patients and 78 to female patients. The relevance of biological sex in this disease is increasingly reported and could affect disease classification, and immune responses36,38,47. Further characterisation of the relevance of X-linked genes to the behaviour of malignant diseases is warranted.
This study aimed to determine whether the relationship between YBX1 expression and overall survival is affected by the biological sex categorisation of the patient cohorts49. Cox proportional hazard analysis of available data failed to identify biological sex as a co-variate significantly affecting survival in all 15 cancer sites examined. Similarly, meta-analysis of YB-1 protein expression, survival and clinicopathological features indicated that overexpression correlates with worse overall survival, but no association was identified with sex on multi-variate analysis13. Yet, Kaplan Meier curves were significantly different between male and female lung cancer patients. In lung cancer, the analysis of gene expression signatures according to the sex of the patients included revealed distinct cluster groups50. Our analysis of YBX1 expression identified a significant difference between the expression distributions of the male and the female cohorts in the case of lung cancer, which might be related to the stark differences in mortality between sexes. While we were unable to find a suitable data set for male-specific disease like prostate cancer, the prognoscan database44 suggests that YBX1 expression increases hazard ratio in prostate cancer survival, and future work is needed to elucidate why this might be the case.
This work serves to highlight that the generation of sex‐based analysis could refine the relevance of candidate genetic markers and emerging therapeutic targets. Further evaluation of the biological and clinical implications of our findings is needed. Future studies aimed as assessing he biological functions and clinical importance of YBX1, and its protein product in cancer ought to consider the biological sex of their models and patients, especially in lung cancer. This could be of particular relevance to the development of senolytic drugs, such as the YB-1 inhibitor fisetin, for the treatment of cancer51. In lung cancer, several reports already indicate the capacity of this drug to affect lung cancer cell growth, migration and apoptosis52–54, but unfortunately this effect was only tested in male lung cancer models.
Supplementary Information
Author contributions
All authors contributed to the preparation of this manuscript.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-57771-y.
References
- 1.Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Q, et al. YB-1 promotes cell proliferation and metastasis by targeting cell-intrinsic PD-1/PD-L1 pathway in breast cancer. Int. J. Biochem. Cell Biol. 2022;153:106314. doi: 10.1016/j.biocel.2022.106314. [DOI] [PubMed] [Google Scholar]
- 3.Sangermano F, Delicato A, Calabro V. Y box binding protein 1 (YB-1) oncoprotein at the hub of DNA proliferation, damage and cancer progression. Biochimie. 2020;179:205–216. doi: 10.1016/j.biochi.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Alkrekshi A, Wang W, Rana PS, Markovic V, Sossey-Alaoui K. A comprehensive review of the functions of YB-1 in cancer stemness, metastasis and drug resistance. Cell Signal. 2021;85:110073. doi: 10.1016/j.cellsig.2021.110073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lettau K, et al. Targeting the Y-box binding protein-1 axis to overcome radiochemotherapy resistance in solid tumors. Int. J. Radiat. Oncol. Biol. Phys. 2021;111:1072–1087. doi: 10.1016/j.ijrobp.2021.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Wilson MA, Buetow KH. Novel mechanisms of cancer emerge when accounting for sex as a biological variable. Cancer Res. 2020;80:27–29. doi: 10.1158/0008-5472.CAN-19-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garand C, et al. An integrative approach to identify YB-1-interacting proteins required for cisplatin resistance in MCF7 and MDA-MB-231 breast cancer cells. Cancer Sci. 2011;102:1410–1417. doi: 10.1111/j.1349-7006.2011.01948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsofack SP, et al. Low expression of the X-linked ribosomal protein S4 in human serous epithelial ovarian cancer is associated with a poor prognosis. BMC Cancer. 2013;13:303. doi: 10.1186/1471-2407-13-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paquet ER, et al. Low level of the X-linked ribosomal protein S4 in human urothelial carcinomas is associated with a poor prognosis. Biomark. Med. 2015;9:187–197. doi: 10.2217/bmm.14.115. [DOI] [PubMed] [Google Scholar]
- 10.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: Functions and regulation. Wiley Interdiscip. Rev. RNA. 2014;5:95–110. doi: 10.1002/wrna.1200. [DOI] [PubMed] [Google Scholar]
- 11.Minich W, Korneyeva N, Berezin Y, Ovchinnikov L. A special repressor/activator system controls distribution of mRNA between translationally active and inactive mRNPs in rabbit reticulocytes. FEBS Lett. 1989;258:227–229. doi: 10.1016/0014-5793(89)81659-X. [DOI] [PubMed] [Google Scholar]
- 12.Minich WB, Maidebura P, Ovchinnikov LP. Purification and characterization of the major 50-kDa repressor protein from cytoplasmic mRNP of rabbit reticulocytes. Eur. J. Biochem. 1993;638:633–638. doi: 10.1111/j.1432-1033.1993.tb17701.x. [DOI] [PubMed] [Google Scholar]
- 13.Jiang L, et al. Positive expression of Y-box binding protein 1 and prognosis in non-small cell lung cancer: A meta-analysis. Oncotarget. 2017;8:55613–55621. doi: 10.18632/oncotarget.14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurya PK, et al. Role of Y Box Protein-1 in cancer: As potential biomarker and novel therapeutic target. J. Cancer. 2017;8:1900–1907. doi: 10.7150/jca.17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan Y, et al. YB1 associates with oncogenetic roles and poor prognosis in nasopharyngeal carcinoma. Sci. Rep. 2022;12:3699. doi: 10.1038/s41598-022-07636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller LR, et al. Considering sex as a biological variable in preclinical research. FASEB J. 2017;31:29–34. doi: 10.1096/fj.201600781R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan Y, et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell. 2016;29:711–722. doi: 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein SL, Flanagan KL. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Zhang Q, Ma X, Wang J, Liang T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep. 2017;7:39812. doi: 10.1038/srep39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scepanovic P, et al. Human genetic variants and age are the strongest predictors of humoral immune responses to common pathogens and vaccines. Genome Med. 2018;10:59. doi: 10.1186/s13073-018-0568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzetto L, Fava F, Tuohy KM, Selmi C. Connecting the immune system, systemic chronic inflammation and the gut microbiome: The role of sex. J. Autoimmun. 2018;92:12–34. doi: 10.1016/j.jaut.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Jog NR, Caricchio R. Differential regulation of cell death programs in males and females by Poly (ADP-Ribose) Polymerase-1 and 17beta estradiol. Cell Death Dis. 2013;4:e758. doi: 10.1038/cddis.2013.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malorni W, Campesi I, Straface E, Vella S, Franconi F. Redox features of the cell: A gender perspective. Antioxid. Redox Signal. 2007;9:1779–1801. doi: 10.1089/ars.2007.1596. [DOI] [PubMed] [Google Scholar]
- 24.Penaloza C, et al. Sex of the cell dictates its response: Differential gene expression and sensitivity to cell death inducing stress in male and female cells. FASEB J. 2009;23:1869–1879. doi: 10.1096/fj.08-119388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lista P, Straface E, Brunelleschi S, Franconi F, Malorni W. On the role of autophagy in human diseases: A gender perspective. J. Cell Mol. Med. 2011;15:1443–1457. doi: 10.1111/j.1582-4934.2011.01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanczky A, Gyorffy B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021;11:6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021;19:4101–4109. doi: 10.1016/j.csbj.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szasz AM, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, Ware JH. Detecting moderator effects using subgroup analyses. Prev. Sci. 2013;14:111–120. doi: 10.1007/s11121-011-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 34.Šidák Z. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 1967;62:626–633. doi: 10.1080/01621459.1967.10482935. [DOI] [Google Scholar]
- 35.Doshi B, Athans SR, Woloszynska A. Biological differences underlying sex and gender disparities in bladder cancer: Current synopsis and future directions. Oncogenesis. 2023;12:44. doi: 10.1038/s41389-023-00489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jong JJ, et al. Distribution of molecular subtypes in muscle-invasive bladder cancer is driven by sex-specific differences. Eur. Urol. Oncol. 2020;3:420–423. doi: 10.1016/j.euo.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Toren P, et al. The sex gap in bladder cancer survival—a missing link in bladder cancer care? Nat. Rev. Urol. 2023 doi: 10.1038/s41585-023-00806-2. [DOI] [PubMed] [Google Scholar]
- 38.Abdel-Hafiz HA, et al. Single-cell profiling of murine bladder cancer identifies sex-specific transcriptional signatures with prognostic relevance. iScience. 2023;26:107703. doi: 10.1016/j.isci.2023.107703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton JA. Applying the new SABV (sex as a biological variable) policy to research and clinical care. Physiol. Behav. 2018;187:2–5. doi: 10.1016/j.physbeh.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020 doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 41.Rubin JB, et al. Sex differences in cancer mechanisms. Biol. Sex. Differ. 2020;11:17. doi: 10.1186/s13293-020-00291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff I, Brookman-May S, May M. Sex difference in presentation and outcomes of bladder cancer: Biological reality or statistical fluke? Curr. Opin. Urol. 2015;25:418–426. doi: 10.1097/MOU.0000000000000198. [DOI] [PubMed] [Google Scholar]
- 43.Kosnopfel C, Sinnberg T, Schittek B. YB1, a prognostic marker and target in tumour therapy. Eur. J. Cell Biol. 2013;93:61–70. doi: 10.1016/j.ejcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kido T, Lau YF. Roles of the Y chromosome genes in human cancers. Asian J. Androl. 2015;17:373–380. doi: 10.4103/1008-682X.150842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng S, Stoner SA, Zhang DE. Sex chromosome loss and the pseudoautosomal region genes in hematological malignancies. Oncotarget. 2016;7:72356–72372. doi: 10.18632/oncotarget.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdel-Hafiz HA, et al. Y chromosome loss in cancer drives growth by evasion of adaptive immunity. Nature. 2023;619:624–631. doi: 10.1038/s41586-023-06234-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 2011;35:565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505:612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mostertz W, et al. Age- and sex-specific genomic profiles in non-small cell lung cancer. JAMA. 2010;303:535–543. doi: 10.1001/jama.2010.80. [DOI] [PubMed] [Google Scholar]
- 51.Chandrakar L, Ambatwar R, Khatik GL. Cellular senescence and senolytic agents: Recent updates on their role and applications. Curr. Top. Med. Chem. 2024;24:157–178. doi: 10.2174/0115680266273698231107110956. [DOI] [PubMed] [Google Scholar]
- 52.Renault-Mahieux M, et al. Co-encapsulation of fisetin and cisplatin into liposomes: Stability considerations and in vivo efficacy on lung cancer animal model. Int. J. Pharm. 2024;651:123744. doi: 10.1016/j.ijpharm.2023.123744. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Huang S. Fisetin inhibits the growth and migration in the A549 human lung cancer cell line via the ERK1/2 pathway. Exp. Ther. Med. 2018;15:2667–2673. doi: 10.3892/etm.2017.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabarwal A, et al. A novel 4′-brominated derivative of fisetin induces cell cycle arrest and apoptosis and inhibits EGFR/ERK1/2/STAT3 pathways in non-small-cell lung cancer without any adverse effects in mice. FASEB J. 2022;36:e22654. doi: 10.1096/fj.202200669RR. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.