Abstract
This paper provides evidence for a measles virus receptor other than CD46 on transformed marmoset and human B cells. We first showed that most tissues of marmosets are missing the SCR1 domain of CD46, which is essential for the binding of Edmonston measles virus, a laboratory strain that has been propagated in Vero monkey kidney cells. In spite of this deletion, the common marmoset was shown to be susceptible to infections by wild-type isolates of measles virus, although they did not support Edmonston measles virus production. As one would expect from these results, measles virus could not be propagated in owl monkey or marmoset kidney cell lines, but surprisingly, both a wild-type isolate (Montefiore 89) and the Edmonston laboratory strain of measles virus grew efficiently in B95-8 marmoset B cells. In addition, antibodies directed against CD46 had no effect on wild-type infections of marmoset B cells and only partially inhibited the replication of the Edmonston laboratory strain in the same cells. A direct binding assay with insect cells expressing the hemagglutinin (H) proteins of either the Edmonston or Montefiore 89 measles virus strains was used to probe the receptors on these B cells. Insect cells expressing Edmonston H but not the wild-type H bound to rodent cells with CD46 on their surface. On the other hand, both the Montefiore 89 H and Edmonston H proteins adhered to marmoset and human B cells. Most wild-type H proteins have asparagine residues at position 481 and can be converted to a CD46-binding phenotype by replacement of the residue with tyrosine. Similarly, the Edmonston H protein did not bind CD46 when its Tyr481 was converted to asparagine. However, this mutation did not affect the ability of Edmonston H to bind marmoset and human B cells. The preceding results provide evidence, through the use of a direct binding assay, that a second receptor for measles virus is present on primate B cells.
Our laboratory and another group have previously demonstrated that CD46 (also known as membrane cofactor protein) could serve as a receptor for the laboratory-adapted Edmonston strain of measles virus (13, 14, 17, 40). The Edmonston virus has been grown successfully in the laboratory for more than 30 years following adaptation of the original wild-type isolate to Vero monkey kidney cells (16). Attenuated vaccine strains of measles virus have also been generated by serial passages of the original Edmonston wild-type isolate in tissue culture with human kidney, human amnion, dog kidney, and chicken embryo cells (19, 49). However, wild-type isolates of measles virus from clinical isolates can easily be isolated in marmoset and human B-cell lines, and this process is much more efficient than adapting the virus for growth in Vero or primary monkey kidney cells (26). Measles virus is a negative-stranded RNA virus which possesses an envelope containing two glycoproteins—the hemagglutinin (H) and a membrane fusion protein (F). Attachment of the virus to a specific host cell receptor is mediated by H, while membrane fusion and penetration of the cellular plasma membrane is controlled by F (reviewed in references 19, 63, and 66).
CD46 is composed of four extracellular short consensus domains (SCR1, SCR2, SCR3, and SCR4) followed by a region rich in serine, threonine, and proline (called STP), a transmembrane region, and a short cytoplasmic domain at its carboxy terminus (34, 35). Variations in splicing of 14 exons encoding SCR domains, STP cassettes, and cytoplasmic regions yield glycoproteins which vary in size from 57 to 67 kDa (42, 43, 52). All four SCR domains are normally expressed in the higher primates, but SCR1 appears to be deleted from CD46 in the lymphocytes of South American monkeys (22). Binding of laboratory strains of measles virus to the SCR1 and SCR2 domains of CD46 has been rigorously studied in recent years (8, 9, 22, 36, 37, 57). In addition, several investigators reported that infections by the Edmonston strain of virus possessed the ability to downregulate the surface expression of CD46 on the infected cell (4, 20, 28, 41, 54, 56). However, wild-type isolates of measles virus did not produce this phenomenon (55). In addition, it has been known for many years that wild-type isolates did not have the ability to hemagglutinate African green monkey erythrocytes while laboratory strains adapted to growth in Vero cells did (16, 59, 60). Recent reports suggest that wild-type isolates do not use CD46 as a receptor but may instead interact with another receptor which is present on activated B cells (10, 30, 55). This hypothesis was based upon the inability of wild-type strains of measles virus to downregulate CD46, elicit hemagglutination of monkey erythrocytes, replicate efficiently in Vero cells, and cause fusion in infected HeLa cells.
A great deal of time and effort has been spent in sequencing genes from wild-type measles virus isolates from around the world and comparing them with those of existing vaccine strains (47–51). Based upon nucleocapsid protein and H protein sequences, year of isolation, and geographic isolation, various isolates were assigned to one of eight groups; the Edmonston wild-type and vaccine strains belong to group 1. Alignment of H proteins from 12 different vaccine strains and comparison to the same protein from more than 59 different wild-type viruses showed that several amino acids consistently differed between the two types of viruses (48–51). These corresponded to amino acids 243, 252, 276, and 481. Approximately one-third of the more recent measles virus isolates also possessed an additional N-linked glycosylation site at amino acid 416. Although sporadic changes occurred throughout the H protein, the sequences were highly conserved, with greater than 95% identity. Recently, variations of two amino acids at positions 451 and 481 of the H molecule were proposed to account for differences in hemadsorption, syncytium formation, and CD46 downregulation between vaccine and wild-type measles viruses (30).
A recent publication from our laboratory indicated that SCR1 was missing from CD46 molecules on the surface of erythrocytes and lymphocytes of New World monkeys. Based upon our findings and those of others (9, 24, 37, 57), we were aware that this region was essential for binding to the laboratory strain of Edmonston measles virus. We predicted that marmosets and tamarins from South America should be resistant to infections by the Edmonston laboratory strain of measles virus. Indeed, this appears to be the case, since marmosets inoculated with Edmonston virus developed no symptoms (2). However, other researchers have reported that the common marmoset, moustached tamarin, and squirrel monkeys are susceptible to infections by wild-type virus which causes symptoms including severe gastroenterocolitis, immunosuppression, respiratory congestion, and in some instances rash and Koplik’s spots (1, 2, 27). This basic observation, coupled with the reports that wild-type strains of measles virus were unable to hemagglutinate African green monkey erythrocytes, led us to suspect that natural isolates of the virus used a receptor other than CD46 to bind to the host cell.
In this publication, we provide evidence for the existence of a second receptor for measles virus through the use of a binding assay which was previously developed in our laboratory (22). Insect cells which expressed the wild-type H protein bound to marmoset B cells but did not adhere to Vero monkey kidney cells or rodent lines expressing CD46 on their surface. In addition, polyclonal antibodies directed against CD46 did not inhibit infections of the B cells with wild-type virus. Differences in H protein sequences between vaccine/laboratory and wild-type strains of measles virus are described, and the results are discussed in terms of receptor usage and viral pathogenesis in the infected host.
MATERIALS AND METHODS
Cell lines and virus.
HeLa, Vero, OMK, NZP-60, BJAB, 1A2, B95-8, and SML cells were purchased from American Type Culture Collection (Rockville, Md.). Sf9 insect cells were supplied by Invitrogen (San Diego, Calif.) and were grown in Grace’s medium containing 10% fetal calf serum. HeLa, Vero, and OMK cells were propagated in Dulbecco’s minimum essential medium (GIBCO/BRL, Gaithersburg, Md.) supplemented with 10% fetal calf serum. NZP-60 cells were propagated in Dulbecco’s minimum essential medium/Ham’s F12 medium (GIBCO/BRL) supplemented with 10% fetal calf serum, 10 ng of epidermal growth factor per ml, 0.005 mg of insulin per ml, 5 ng of selenium per ml, and 0.005 mg of transferrin per ml. SML, B95-8, BJAB, and 1A2 cells were propagated in RPMI 1640 medium (GIBCO/BRL) supplemented with 10% fetal calf serum. The Edmonston strain of measles virus was originally obtained from Erling Norrby (Karolinska Institute, Stockholm, Sweden) and was cultivated in Vero monkey kidney cells as previously described (18). The Montefiore 89 strain of measles virus (wild type) was obtained from Ilya Spigland and Amy Fox (Montefiore Medical Center, Bronx, N.Y.) and was amplified in B95-8 cells as previously described (26). Moraten, Zagreb, and Schwarz measles virus vaccine strains were purchased from Merck (West Point, Pa.) and SmithKline Beecham (King of Prussia, Pa.) and cultivated in Vero cells.
Antibodies.
Monoclonal antibodies directed against H (2B1-3) and polyclonal antibodies (CD46-333) directed against the entire CD46 protein were produced in our laboratory as described previously (13, 22, 44). A rabbit polyclonal antibody directed against native human CD46 was also obtained from J. P. Atkinson (Washington University, St. Louis, Mo.). Monoclonal antibodies directed against matrix (M) and H proteins of measles virus were purchased from Chemicon (Temecula, Calif.). Polyclonal antibodies directed against the SCR1 domains of moustached tamarin (Saguinus mystax) and humans were prepared in rabbits as previously reported (22). In addition, horseradish peroxidase-conjugated goat anti-mouse (IgG/IgM) antibody and fluorescein isothiocyanate-conjugated rat anti-mouse IgG heavy- plus light-chain (H+L) antibody were purchased from Jackson Laboratories (West Grove, Pa.).
SDS-polyacrylamide gel electrophoresis and immunoblot analysis.
Adherent and suspension cell lines were infected with either Edmonston strain (vaccine/laboratory strain) or Montefiore 89 strain (wild-type strain) of measles virus at a multiplicity of infection of 5 PFU per cell. The cells were harvested for 72 h postinfection, washed twice with phosphate-buffered saline (PBS) by centrifugation, and resuspended in 200 μl of sample buffer. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western immunoblot analysis were performed as previously described (22, 65). Primary antibody binding was detected with horseradish peroxidase-conjugated goat anti-mouse antibody (1:5,000 dilution) by the enhanced chemiluminescence detection method (Amersham, Arlington Heights, Ill.).
Preparation of CD46 cDNAs from monkey tissues or cell lines.
Monkey tissues were completely homogenized in the presence of 5 ml of TRIzol (GIBCO/BRL) with a Polytron homogenizer (Brinkmann). B95-8 and SML cells (107 cells) were washed twice with PBS by centrifugation and resuspended in 1 ml of TRIzol. Total RNA was isolated as specified by the manufacturer. cDNA was synthesized from RNA with the First Strand Synthesis kit (Pharmacia) with the supplied random primer or a specific CD46 primer (5′-GGGACAACACAAATTACTGC-3′). Double-stranded DNA fragments were generated by nested PCR as previously described (22). Interior or nested primers corresponding to 5′-CTTCTGGCGGCCATGGTGTTG-3′ and 5′-TTTATTTTTGGAGGTGGTGTACAC-3′ were derived from Saguinus mystax or Saimiri sciureus CD46 cDNA sequences (22) and used for the final 30 rounds of PCR amplification. Double-stranded DNA fragments corresponding to CD46 cDNAs from B95-8 and SML cells were cloned into PCR Script AMP (SK+) (Stratagene, La Jolla, Calif.) and sequenced. The 5′-terminal coding regions of CD46 molecules from B95-8 and SML cells were determined with the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.) as previously described (22).
Preparation of cDNA containing the coding sequence for the HA protein from the Montefiore 89 strain of measles virus.
RNA was extracted from B95-8 cells infected with the wild-type Montefiore 89 strain of measles virus by using TRIzol, and cDNA was prepared with the First Strand Synthesis kit. Double-stranded DNA fragments of wild-type measles H were generated by 30 rounds of PCR amplification with primers derived from the H sequence (5′-GGCGGATCCACAATGTCACCACAACGAGACCGG-3′ and 5′-GAAGGATCCCTATCTGCGATTGGTTCCATCTTC-3′). H cDNA fragments from three independent PCR amplifications were cloned into the SrfI site of the PCR Script Amp (SK+) vector and sequenced.
Construction of a chimeric CD46 molecule containing SCR1 and SCR2 from Saguinus mystax fused to human SCR3 and SCR4 domains.
The SCR1 and SCR2 domains from human CD46 were replaced with SCR1 and SCR2 domains from B95-8 cells. A vaccinia virus expression vector, pTM1, containing the human CD46 coding sequence was digested with NcoI and BsrGI. SCR1 and SCR2 sequences from B95-8 cells were synthesized by PCR with the specific oligonucleotide primers described above. The amplified DNA product was digested with NcoI and BsrGI and inserted into the digested pTM1-CD46 expression vector. Recombinant vaccinia virus was prepared as previously described (22). Chimeric CD46 was expressed in mouse OST-7 cells which contained the T7 polymerase, and protein synthesis and surface expression were verified by Western immunoblotting and FACScan analysis.
Site-specific mutagenesis of measles virus H protein and expression of mutants by using baculovirus recombinants.
Specific mutations were introduced into the wild-type measles virus H molecule with the QuickChange site-directed mutagenesis kit (Stratagene) as previously described (22). Mutant plasmids were isolated, and the measles virus H inserts were completely sequenced. The mutagenized H reading frames were excised from the PCR-Script Amp (SK+) plasmid after digestion with BamHI and then inserted into the baculovirus expression vector pETL(BlueBac2), which also contains the β-galactosidase gene. Baculovirus recombinants were generated as previously described (29, 45, 64). Recombinant H protein was expressed in Sf9 insect cells, and protein synthesis and surface expression were monitored by Western immunoblotting and FACScan analysis with H-specific monoclonal antibodies.
Flow cytometry analysis of CD46 molecules and measles virus H molecules.
Mouse OST-7, B95-8, OMK, SML, and NZP-60 cells (2 × 106 cells) which expressed CD46 were suspended in 1 ml of cell dissociation buffer (Sigma, St. Louis, Mo.) and washed twice by centrifugation with fluorescence-activated cell sorter buffer (PBS containing 1% bovine serum albumin, 5 mM EDTA, and 0.1% sodium azide). Incubations were performed with a 1:100 dilution of either preimmune, polyclonal CD46(#333) or polyclonal Saguinus mystax SCR1 antibodies for 1 h on ice. The cells were washed and incubated with fluorescein isothiocyanate (FITC)-labeled anti-rabbit IgG(H+L) secondary antibodies as previously reported (22). Just before analysis, the cells were washed and suspended in 0.5 ml of fluorescence-activated cell sorter buffer, and the assays were performed on a Beckton Dickinson analyzer.
Direct binding assays between CD46 cell lines and insect cells expressing different measles virus H recombinant protein.
CD46 molecules were expressed in either mouse OST-7 cells or hamster CHO cells as previously described (13, 22). Sf9 insect cells were infected for 48 h with recombinant baculoviruses which expressed Edmonston vaccine, Montefiore 89 wild type, or mutant H proteins in addition to β-galactosidase. Assays of binding between infected insect cells and mouse or hamster cells were performed as described previously (22). Nonadherant insect cells were washed away, and binding was either visualized under the microscope in the presence of Bluogal or quantitated with the enzyme substrate o-nitrophenyl-β-d-galactopyranoside (ONPG).
Nucleotide sequence accession numbers.
The nucleotide sequences coding for the extracellular domains of B95-8 CD46 and SML CD46 molecules, which originate from moustached tamarin (Saguinus mystax) and squirrel monkey (Saimiri sciureus), respectively, were submitted to GenBank and have the following accession numbers: Saguinus mystax SCR1, AF025482; Saimiri sciureus SCR1, AF025483; Saguinus mystax SCR1-deleted ectodomain, U87918; and Saimiri sciureus SCR1-deleted ectodomain, U87919. The sequence for the cDNA coding for H protein of the Montefiore wild-type measles virus was also submitted and has accession no. AF025484.
RESULTS
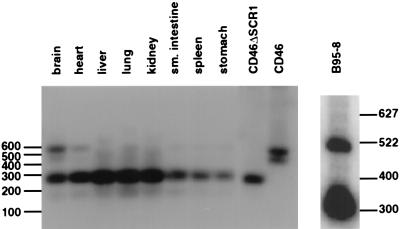
Most organs of the common marmoset contain CD46 molecules with a deletion of the SCR1 domain which blocks infections by the Edmonston laboratory strain but not wild-type strains of measles virus.
We have previously demonstrated that the CD46 molecules from lymphocytes and erythrocytes of New World monkeys contain a deletion of the SCR1 domain (22). Since this region of the receptor is critical for binding to the H protein of the Edmonston laboratory strain of measles virus (8, 24, 37), we proposed that cells and tissues from South American monkeys may be resistant to infections by measles virus. Experiments were performed to determine whether the SCR1 domain was deleted in CD46 proteins from other organs of the common marmoset (Callithrix jacchus). Brain, heart, liver, lung, kidney, small intestine, spleen, and stomach tissues were homogenized, mRNA was extracted, and cDNA was prepared. PCR was performed across the SCR1 region with oligonucleotide primers derived from the conserved signal peptide and SCR3 domains. A 300-bp product was indicative of a deleted SCR1 domain, while a 522-bp product was produced from a complete copy of the CD46 cDNA. Most organs from the marmoset contained the deleted form of CD46 (Fig. 1) but the brain and heart may contain small amounts of the undeleted species in addition to the major deleted mRNA. Marmosets inoculated with the Edmonston strain of measles virus, did not exhibit disease symptoms, and the tissues of these monkeys did not contain measles virus based on reverse transcriptase PCR RT-PCR analysis for nucleocapsid protein (Table 1). However, the animals did seroconvert, which may indicate the existence of a subclinical or local infections caused by limited uptake of the virus. These results seem to confirm previous findings (2) showing that marmosets infected intracerebrally with Edmonston virus developed encephalitis but displayed no visceral symptoms; vaccination by the intranasal and intradermal routes protected the animal, indicating seroconversion. The small amounts of undeleted forms of CD46 which we found in the marmoset brain are consistent with the ability of Edmonston measles virus to cause encephalitis. However, we were at a loss to explain why marmosets, tamarins, and squirrel monkeys, which contained deletions in CD46, were still susceptible to infections by wild-type strains of measles virus leading to severe gastroenterocolitis, immunosuppression, and respiratory distress (1, 27, 31). Productive infections with wild-type measles virus were confirmed in one of our laboratories after inoculation of the common marmoset (Callithrix jacchus) with the Pennsylvania-1 90 strain of virus (Table 1). Wild-type virus could be detected in peripheral blood cells of infected marmosets by RT-PCR, but tests for the Edmonston strain in the other group of marmosets were negative. In our experiments, the marmosets, although clearly infected with wild-type virus, did not exhibit the severe symptoms observed by Albrecht et al. (1, 2), and none of the animals died. This difference may be related to the strain of wild-type measles virus used for inoculation, the species of marmoset, or the improved nutritional state of the animals, which had been reared in captivity. However, from these studies, we began to suspect that wild-type strains of measles virus may indeed use a receptor other than CD46 during the initial stages of infection.
FIG. 1.
Southern blot of PCR amplification spanning the SCR1 region of the common marmoset (Callithrix jacchus). The brain, heart, liver, lung, kidney, small intestine, spleen, and stomach of a common marmoset were isolated and homogenized in TRIzol, mRNA was extracted, and cDNA was prepared with RT. PCR was performed across the SCR1 region with oligonucleotide primers derived from the conserved signal peptide and SCR3 domains. A 300-bp product was indicative of a deleted SCR1 domain, while a 522-bp product was produced from a complete copy of the CD46 cDNA. Most organs from the marmoset contained the deleted form of CD46, but the brain and heart may contain small amounts of the undeleted species in addition to the major deleted mRNA. B95-8 marmoset B cells were homogenized, and mRNA was extracted and treated in a similar manner to that from the marmoset organs. Undeleted and deleted forms of CD46 were present in the B95-8 cells. PCR analysis was also performed on cDNA clones which had been prepared from mRNA isolated from B95-8 cells and inserted into the pCR Script AMP (SK+) vector. The PCR products from the deleted clone (CD46ΔSCR1) and the nondeleted clone (CD46) templates were also analyzed. PCR products were resolved by agarose gel electrophoresis, transferred to nitrocellulose, probed with 32P-labelled fragments derived from the SCR2 and SCR3 regions of CD46, and subjected to autoradiography with Royal X-OMAT film for 24 h.
TABLE 1.
Infection of marmosets with wild-type and vaccine strains of measles virus
| Group (no. of subjects) | No. of mice infected/total no.a
|
|||||
|---|---|---|---|---|---|---|
| MV-IgGpreb | MV-IgMpreb | MV-IgGpostc | MV-IgMpostc | MV-PCRd | Virus rescuee | |
| MV vaccine (3) | 0/3 | 0/3 | 3/3 | 3/3 | 0/3 | ND |
| MV wild type (3) | 0/3 | 0/3 | 3/3 | 3/3 | 2/3 | 2/3 |
| Naive/exposed (3) | 0/3 | 0/3 | 3/3 | 3/3 | 3/3 | 3/3 |
Animals were anesthetized and inoculated intranasally with 104 50% tissue culture infective doses of virus. For the last group, naive animals were housed in cages adjoining the group inoculated with WT virus beginning on the day of inoculation. The Attenuvax (Moraten) vaccine strain, a derivative of Edmonston virus, was adapted to grow in Vero cells. The wild-type strain was Pennsylvania-1 90 and was cultivated in phytohemagglutinin-stimulated marmoset peripheral blood mononuclear cells to avoid contamination by Epstein-Barr virus, which is present in B95-8 cells.
An enzyme-linked immunosorbent assay specific for measles virus (MV) IgG or IgM was performed as previously described (23). Preinoculation serum samples were obtained 24 h before inoculation or exposure.
Postinoculation serum samples were positive for measles virus IgG and IgM by 14 to 21 days after inoculation or exposure.
RT-PCR to detect measles virus RNA was performed on RNA extracted from peripheral blood mononuclear cells and collected at weekly intervals from days 7 to 60 after inoculation or exposure by using oligonucleotides specific for the nucleocapsid protein.
Peripheral blood mononuclear cells collected at weekly intervals after inoculation were inoculated into B95-8 cultures.
Marmoset B cells (B95-8 cells) and squirrel monkey lung (SML) cells can be infected with the Edmonston strain of measles virus, while owl monkey kidney (OMK) and marmoset kidney (NZP-60) cells are resistant to infection.
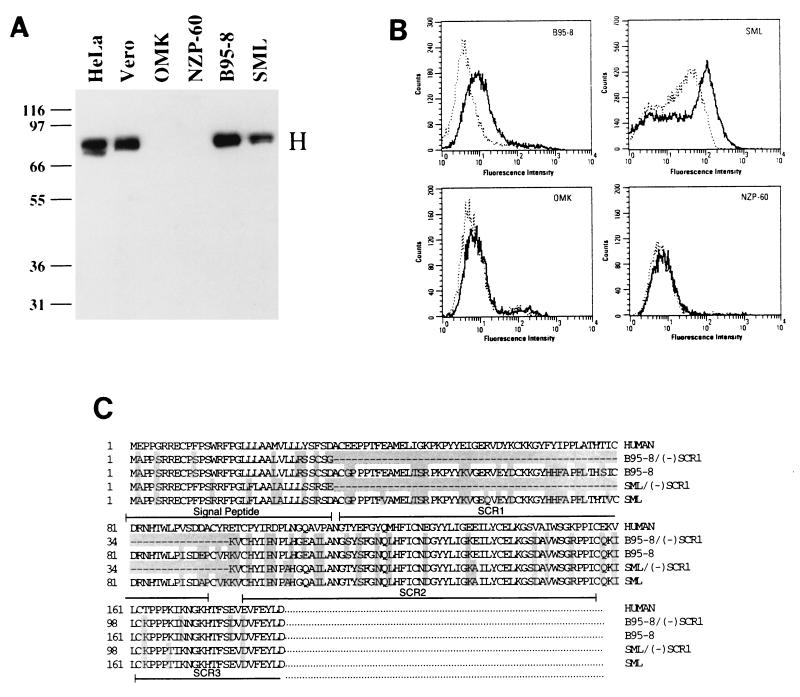
A B-cell line (B95-8) transformed with Epstein-Barr virus was originally derived from the moustached tamarin (Saguinus mystax) (39). B95-8 cells were previously shown to support the growth of both wild-type measles virus and virus which had been adapted to growth in Vero cells (26). Through RT-PCR, we determined that B95-8 cells contain both the deleted and undeleted forms of CD46 mRNA (Fig. 1). The SCR1 deletion corresponded to a missing exon 2 in mRNA derived from the CD46 gene, and the exon was previously shown to be present in marmoset chromosomal DNA (22, 42, 43, 52). It is possible that under certain circumstances, such as viral transformation, exon 2 is correctly spliced to yield full-length CD46 mRNA. We subsequently tested two Old World and four New World primate cell lines for their ability to support Edmonston measles virus infection. Viral protein synthesis was demonstrated by the presence of the measles virus H protein in immunoblot analysis with a monoclonal antibody directed against H. As expected, the Old World primate cell lines, HeLa (human cervical carcinoma) and Vero (African green monkey kidney), supported Edmonston measles virus infection, as indicated by the presence of 79-kDa bands corresponding to the molecular mass of the H protein (Fig. 2A). The New World monkey cell lines, OMK (owl monkey kidney) and NZP-60 (Callithrix argentata kidney), did not support Edmonston measles virus infection. On the other hand, the other two New World monkey cell lines, B95-8 and SML (squirrel monkey lung transformed with a simian retrovirus), did support Edmonston measles virus infection. Again, mRNA from these cell lines was isolated, cDNA was prepared, and RT-PCR was performed across the SCR1 region with primers derived from the signal peptide of CD46 and SCR3. Southern blot hybridization of the PCR products revealed that the OMK and NZP-60 cell lines yielded only one band corresponding to the SCR1-deleted form of CD46 mRNA while the B95-8 and SML cell lines produced two products corresponding to deleted and nondeleted forms of CD46 mRNA (46). Vero and HeLa cells yielded one PCR product which corresponded to the nondeleted form of CD46, as expected.
FIG. 2.
Growth of the Edmonston strain of measles virus in New World monkey cell lines is impaired when SCR1 is deleted. (A) Human cervical carcinoma (HeLa), African green monkey kidney (Vero), owl monkey kidney (OMK), marmoset kidney (NZP-60), marmoset B (B95-8), and squirrel monkey lung (SML) cells were infected with the Edmonston strain of measles virus which had previously been adapted for growth in Vero cells. The cells were inoculated with 5 PFU of virus per cell, and infections were allowed to proceed for 72 h, after which the infected cells were subjected to immunoblot analysis with monoclonal antibodies to measles virus H protein. Viral protein synthesis was not observed in the OMK and NZP-60 cell lines, but measles virus H protein was detected in B95-8 and SML cells. (B) FACScan analysis was performed on B95-8, OMK, SML, and NZP-60 cells with an antibody to the SCR1 domain of the moustached tamarin (Saguinus mystax) and detected with goat anti-rabbit antibodies which had been conjugated to fluorescein isothiocyanate (solid line). The cells were also tested with rabbit preimmune antisera (dotted line). Shifts in fluorescence were observed in B95-8 and SML cells but not in OMK and NZP-60 cells. (C) mRNA was extracted from B95-8 and SML cells, cDNA was prepared, and PCR products spanning the signal peptide, SCR1, SCR2, and SCR3 domains were prepared and sequenced. The predicted amino acid sequence is shown and was derived from three independent amplification reactions for each sequence. Both deleted and nondeleted forms of mRNA were present in the two cell lines.
FACScan analysis was also performed on B95-8, OMK, SML, and NZP-60 cells with an antibody directed against the SCR1 domain of Saguinus mystax. Cells were also tested with rabbit preimmune antisera and a polyclonal antibody directed against the entire human CD46 molecule (46). Each of the cell lines exhibited a shift in fluorescence due to the presence of CD46 on their surfaces, but only the B95-8 and SML cells possessed a fluorescent signal specific for the SCR1 domain (Fig. 2B). Both the SCR1 and SCR2 domains have previously been implicated in binding to measles virus which had been grown in Vero cells (8, 9, 22, 37), and the fluorescence cytometry shown in Fig. 2B supports the results of the viral infections in Fig. 2A.
PCR products from the previous experiments with B95-8 and SML cells were cloned and sequenced by three independent amplification reactions. The deduced polypeptide sequences containing the signal peptide and short consensus regions (SCR1 and SCR2) of CD46 were aligned with the Clustal program from Lasergene (Fig. 2C). The larger cDNAs of B95-8 and SML CD46 molecules contained the SCR1 domain, while the smaller isoforms did not. The SCR1 regions from B95-8 cells (Saguinus mystax) and SML cells (Saimiri sciureus) were very similar and had 90% identity to the human sequence. Since the SCR1 coding region was present in the mRNA from both the B95-8 and SML cell lines, one might expect that these two New World monkey cell lines should support infections by the laboratory strain of Edmonston measles virus while cell lines lacking SCR1 (OMK and NZP-60) would not. These sequences corroborate our previous results from RT-PCR assays and FACS analysis and provide one explanation why B95-8 and SML cells can support infection by the Edmonston laboratory strain of measles virus.
A wild-type strain of measles virus (Montefiore 89) will infect B95-8 cells but cannot grow in other cell lines.
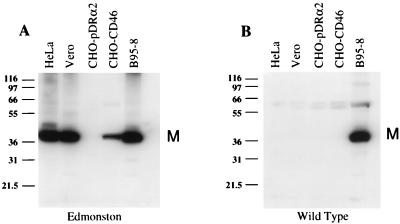
Several different CD46-positive cell lines were inoculated with either Edmonston measles virus (adapted to growth in Vero cells) or the wild-type Montefiore 89 strain of measles virus to test their susceptibility to these viruses. Cells were incubated for 72 h with either strain of virus, and infection was monitored by the synthesis of the measles virus M protein by immunoblot analysis. As expected, HeLa, Vero, CHO-CD46, and B95-8 cells supported infection by the Edmonston strain of virus and 40-kDa bands corresponding to the M protein were present on immunoblots prepared from infected-cell lysates (Fig. 3A). CHO cells, which do not express CD46 on their surface, were transfected with the expression vector alone and did not support infection. HeLa, Vero, and CHO-CD46 cells which were inoculated with Montefiore 89 wild-type measles virus, did not show any evidence of viral infection. However, B95-8 cells clearly supported infection by either the Montefiore 89 or Edmonston strain, as indicated by the presence of viral M protein (Fig. 3B). These results confirm the findings of others (26) and in addition suggest that wild-type strains of measles virus may use a receptor other than CD46, which appears to be present on transformed marmoset B cells.
FIG. 3.
Infection of cell lines with Edmonston laboratory and Montefiore 89 wild-type strains of measles virus. HeLa cells, Vero cells, Chinese hamster ovary cells transfected with an empty expression vector (CHO-pDRα2), Chinese hamster ovary cells expressing human CD46 (CHO-CD46), and a marmoset B-cell line (B95-8) were inoculated with either Edmonston or wild-type Montefiore 89 strains of measles virus. The cells were incubated for 72 h with either strain of virus, and infection was monitored by immunoblot analysis with a monoclonal antibody to the measles virus M protein (40 kDa). (A) HeLa, Vero, CHO-CD46, and B95-8 cells supported infection by the Edmonston strain, while CHO cells transfected with the expression vector alone (CHO-pDRα2) were not infected. (B) HeLa, Vero, CHO-pDRα2, and B95-8 cells were inoculated with the wild-type Montefiore 89 strain, and infections were allowed to proceed for 72 h. Only the B95-8 cells supported infection. Protein standards (in kilodaltons) are shown at the left of each panel.
CD46 polyclonal antibody blocks infections by the Edmonston strain in Vero cells but does not inhibit infections by the Montefiore 89 strain of measles virus in B95-8 cells.
We previously demonstrated that a polyclonal antibody directed against the entire human CD46 molecule could both recognize CD46 proteins from different monkeys and inhibit infections by the Edmonston strain of measles virus in HeLa cells (13, 22). In addition, the polyclonal antibody at dilutions as low as 1:400 effectively neutralized infections by the Edmonston strain in Vero monkey kidney cells (Fig. 4E). Polyclonal antibodies directed against CD46 and the marmoset SCR1 were also tested for their ability to inhibit infections by either the Edmonston laboratory strain or the wild-type Montefiore strain of measles virus in marmoset B95-8 cells. The B95-8 cells were preincubated with dilutions of polyclonal anti-CD46/SCR1 serum or preimmune antiserum ranging from 1:10 to 1:400 for 1 h, the Edmonston strain or Montefiore strain of measles virus was added and allowed to adsorb for another hour, and virus was subsequently removed and replaced with fresh medium containing CD46 antibodies. Cells were inoculated with measles virus at a multiplicity of infection of 1 PFU/cell, the infection was allowed to proceed for 36 h in the presence of antibody, and the cytopathic effects found in infected cells were examined under the microscope. Virus-dependent syncytium formation was clearly observed in B95-8 cells which were infected with either the Edmonston or Montefiore 89 strain of measles virus in the presence of preimmune antibodies (Fig. 4B and D). On the other hand, treatment of B95-8 cells with anti-CD46/SCR1 polyclonal antiserum at dilutions as low as 1:10 appeared to reduce infections by Edmonston measles virus but failed to fully protect the cells from infection (Fig. 4C). Infections of B95-8 cells by the Montefiore 89 strain were not inhibited by anti-CD46/SCR1 (Fig. 4A). These observations were confirmed by immunoblot analysis with monoclonal antibodies directed against H or M (46). No viral protein synthesis could be detected in Vero cells infected with the Edmonston strain of measles virus when polyclonal antibodies directed against CD46 were present, but viral proteins could be detected in B95-8 cells infected with the Montefiore strain when even low dilutions (1:10) of anti-CD46 were present. In addition, preincubation of B95-8 cells with 1:10 dilutions of CD46 polyclonal antibodies reduced but could not abolish viral protein synthesis in cells inoculated with the Edmonston virus. The preceding data provide further evidence that CD46 may not function as a receptor for the wild-type strain of measles virus, since CD46-specific antibodies had no effect upon infections by the Montefiore 89 strain. Wild-type virus presumably binds to an as yet unidentified receptor which is present on marmoset B cells. In addition, the Edmonston strain was only partially inhibited by CD46 antibodies during infections of B95-8 cells, and it, too, may use this new hypothetical receptor under certain circumstances.
FIG. 4.
Polyclonal antibody to CD46 does not inhibit infections by the Montefiore 89 wild-type strain of measles virus. Antibodies to CD46 and the marmoset SCR1 were combined and tested for their ability to inhibit infections by Montefiore 89 and Edmonston measles virus in B95-8 and Vero cells. Cells were treated with CD46 and SCR1 immune antibodies (A, C, and E) or preimmune serum (B, D, and F). CD46 antibodies at dilutions of 1:10 had no effect upon infections of B95-8 cells by the wild-type Montefiore 89 virus (A) but partially inhibited infections of the same type of cells by the Edmonston strain of virus (C). The same antibodies at dilutions as low as 1:400 completely inhibited the infection of Vero cells by the Edmonston virus (E). Infections were assessed by the formation of syncytia or multinucleated cells. Bar, 3 μm.
The H protein from the Montefiore 89 wild-type strain of measles virus does not interact with CD46 in a direct binding assay.
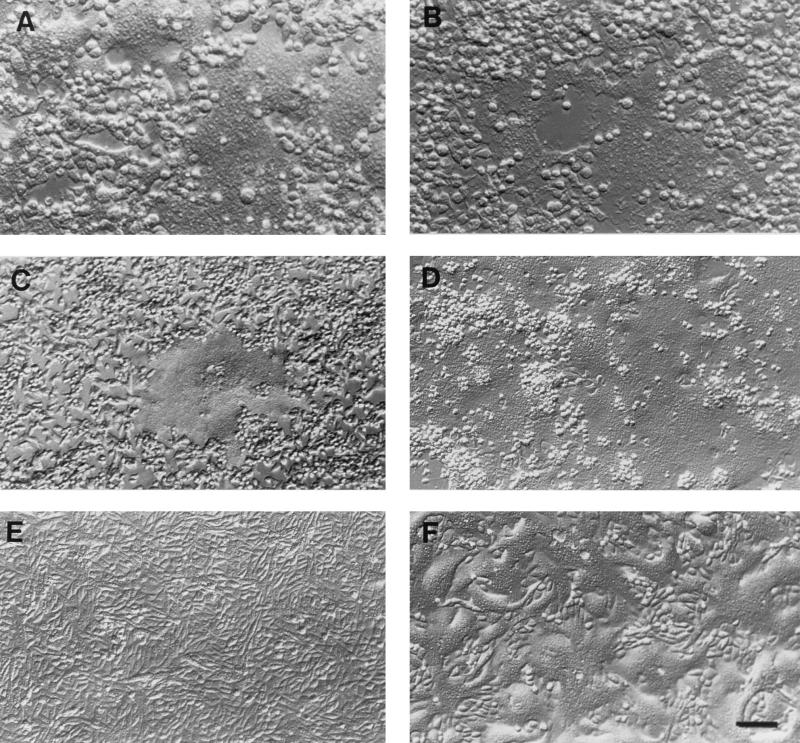
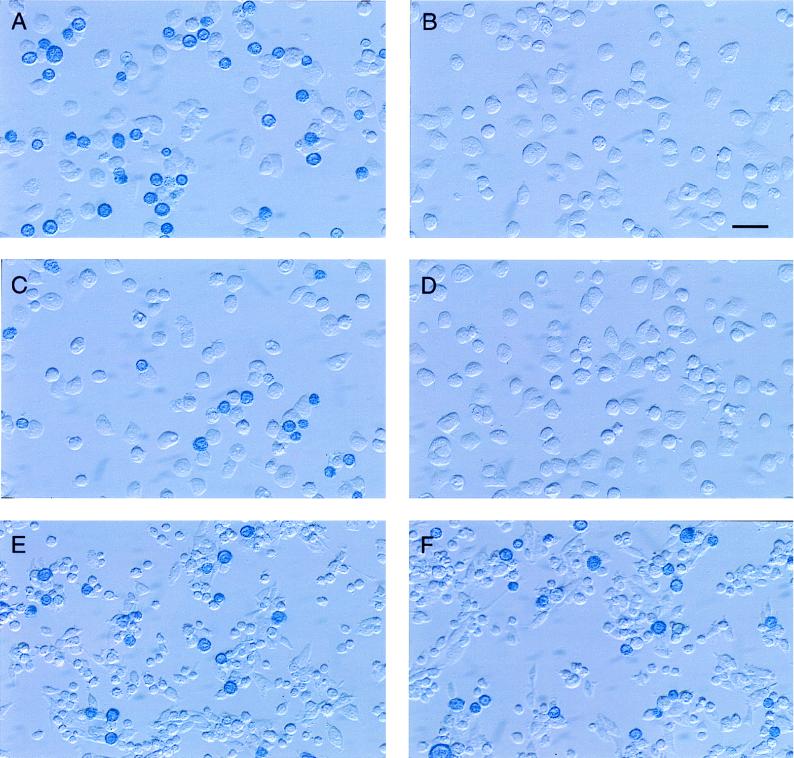
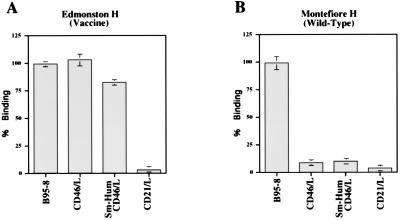
The ability of Montefiore 89 wild-type and Edmonston H proteins to interact directly with CD46 or a receptor on immortalized B cells was measured in a binding assay that we have described previously (22). The cDNA for the H protein of the Montefiore 89 strain was cloned and expressed on the surface of Sf9 insect cells by using the recombinant baculovirus system. In addition, a marmoset/human CD46 chimeric molecule was constructed by using the SCR1 and SCR2 domains from B95-8 cells and the SCR3, SCR4, STP, transmembrane, and cytoplasmic domains of human of CD46. The chimera was expressed in the mouse OST-7 cells by using the recombinant vaccinia virus system in the same way that human CD46 was previously expressed in this mouse L-cell line (22). Expression of chimeric CD46 on the surface of mouse OST-7 cells was confirmed by FACScan analysis as previously described (22). The purpose of constructing the chimeric molecule was to determine whether subtle differences between human and marmoset CD46 molecules could account for the altered tropism of wild-type virus for human and B95-8 cell lines. Sf9 insect cells expressing either Edmonston H or Montefiore 89 H in addition to β-galactosidase were stained blue by the addition of the enzyme substrate Bluogal. The blue insect cells were incubated with OST-7 cells expressing human CD46, marmoset/human chimeric CD46, or human CD21 molecules. Insect cells expressing H were also incubated with the marmoset B95-8 cell line. Sf9 cells which did not adhere to the target cells were washed away, and binding of insect cells was first evaluated under the microscope (Fig. 5). Insect cells which expressed the Edmonston H protein remained attached to mouse cells expressing human or chimeric CD46, as well as to the marmoset B95-8 cell line (Fig. 5A, C, and E). As we previously demonstrated (13, 22), the Edmonston H protein did not bind to OST-7 cells, CHO cells, or a mouse B-cell line (46). On the other hand, the wild-type Montefiore H protein did not bind to mouse cells expressing human CD46 or chimeric CD46 but did adhere to the marmoset B-cell line (Fig. 5B, D, and F). Similar results were found with human lymphoma B-cell lines grown in our laboratory, where insect cells expressing wild-type H or Edmonston H also bound to human BJAB and human 1A2 cells (46). Binding of insect cells expressing wild-type H to B95-8 cells could be inhibited by preincubating the target cells with Montefiore 89 virus (46), indicative of the saturation binding properties which characterize a receptor. The results of the preceding experiments were quantitated (and are summarized in Fig. 6) by measuring the hydrolysis of the β-galactosidase substrate ONPG. We concluded that the H protein of the Edmonston vaccine strain of virus could bind to human CD46 or chimeric CD46 which was expressed on the OST-7 mouse L cells. The Edmonston H protein could also bind to receptors on the B95-8 cells (Fig. 6A). On the other hand, the H protein of the Montefiore wild-type virus did not bind to human CD46, chimeric CD46, or CD21 but did attach to some receptor which was present on B95-8 cells (Fig. 6B). This receptor was not CD21, a molecule with similar properties to CD46, which is known to bind Epstein-Barr virus. These direct binding assays clearly establish that wild-type H protein interacts with some receptor, other than CD46, which is present on the surface of marmoset B95-8 cells.
FIG. 5.
Assays of binding of H proteins from Montefiore 89 and Edmonston measles virus to mouse OST-7 cells expressing CD46 and marmoset B95-8 cells. H proteins from the Montefiore 89 and Edmonston strains of measles virus were cloned and expressed on the surface of Sf9 insect cells by using the recombinant baculovirus system. Sf9 cells also expressed β-galactosidase and were stained blue by the addition of Bluogal substrate. The blue insect cells were incubated with mouse cells expressing human CD46 (A and B), mouse cells expressing marmoset/human chimeric CD46 (C and D), or B95-8 cells (E and F), and loosely adsorbed Sf9 cells were washed away. Insect cells which expressed the Edmonston H protein remained attached to mouse cells expressing human or chimeric CD46 as well as to the marmoset B95-8 cell line (A, C, and E). The wild-type Montefiore H protein did not bind to mouse cells expressing human CD46 or chimeric CD46, but it did adhere to the marmoset B cell line (B, D, and F). Bar, 2.5 μm.
FIG. 6.
Quantitation of Edmonston H and Montefiore H binding to mouse cells expressing human CD46, marmoset/human chimeric CD46, or CD21 or to B95-8 marmoset cells. Sf9 insect cells expressing Edmonston (A) or Montefiore 89 (B) H proteins were incubated with B95-8 marmoset cells or mouse L cells expressing CD46, chimeric CD46, or CD21 as described in the legend to Fig. 5. Loosely attached cells were washed away, and binding was measured colorimetrically with the ONPG substrate for β-galactosidase. Edmonston H bound to marmoset B95-8 cells and mouse cells expressing human CD46 and chimeric CD46. It did not bind to mouse cells expressing CD21 Montefiore H bound to marmoset B95-8 cells but did not adhere to mouse cells expressing human CD46, chimeric CD46, or CD21. Binding is expressed as a percentage relative to either Edmonston H binding or Montefiore H binding to B95-8 cells.
Amino acid residue 481 of the H molecule determines the ability of the viral protein to bind to CD46 and reveals another receptor which is present on B95-8 cells.
Measles virus isolates have been classified into eight groups based on nucleic acid sequence, year of isolation, and country of isolation (46, 50, 51). The measles virus Montefiore 89 strain was isolated at the Montefiore Medical Center/Albert Einstein Medical College and appears to belong to group 2 based upon its similarity (99.5 to 100% identity) to Chicago 1, San Diego 89, Illinois 89, Pennsylvania 90, Texas 89, and California 90 strains of measles viruses, which were isolated between 1989 and 1990. The Montefiore 89 H sequence was 100% identical to that of California 90 measles virus. We aligned different H protein sequences from 59 wild-type and 12 vaccine/laboratory strains of measles virus based upon data which had previously been entered in GenBank. H proteins were aligned with the MegAlign Clustal program marketed by Lasergene (46). The wild-type strains had been propagated in marmoset B95-8 cells, whereas the vaccine/laboratory strains were amplified in Vero monkey kidney cells. When the two different types of isolates were compared, differences were consistently observed at positions 211, 243, 276, and 481. Amino acids 451 and 481 had previously been implicated in determining hemadsorption, cell fusion, and CD46 downregulation caused by vaccine strains of measles virus (30, 59). Our alignments demonstrated that 97% of both wild-type and vaccine strains contained a valine residue at residue 451 and only five wild-type viruses contained glutamic acid at this position. We concluded that this residue appeared to be irrelevant in determining the wild-type virus phenotype. About one-third of the wild-type strains possessed an additional potential glycosylation site at position 416. The results of our alignments were related to the ability of individual isolates to infect either Vero or B95-8 cells. The results of these findings are summarized in Table 2. Wild-type virus H proteins possessed an Asn residue at position 481 instead of the Tyr present in vaccine/laboratory virus H proteins. In all cases, viruses which contained a tyrosine at residue 481 produced cytopathic effects in Vero cells while those with an asparagine at this position yielded syncytia and caused damage in B95-8 cells but not the monkey kidney cell line. This amino acid change has previously been observed during successive passages of wild-type measles virus isolates in Vero cells and may parallel the ability of virus to agglutinate monkey erythrocytes (30, 59). The gradual mutation of N481 to Y during adaptation of a Montefiore 89 wild-type isolate to Vero cells over five passages was also observed by one of our laboratories (61). On the other hand, amino acid differences at positions 416 and 451 did not seem to affect the growth of wild-type virus in Vero cells (Table 2) and appeared to correlate with H group-specific changes. The N481Y mutation, which occurs between the H proteins of wild-type and vaccine/laboratory strains of measles virus, can be attributed to a single nucleotide change (AAC→TAC). It is not yet known whether this change also occurs in vivo during natural infections.
TABLE 2.
Correlations of sequence variations in the H proteins from vaccine and wild-type strains of measles virus and their ability to grow in Vero monkey kidney or B95-8 marmoset B-cell lines
| Virus | Amino acid at positionc:
|
Cytopathic effectd in:
|
|||
|---|---|---|---|---|---|
| 451 | 481 | 416 | B95-8 | Vero | |
| Vaccine strainsa | V | Y | D | + | + |
| Edmonston (Vero cell adapted) | V | Y | D | + | + |
| Group 1 wild typeb (China 93-5) | V | N | D | + | − |
| Group 2 wild typeb (Montefiore 89, Chicago-1) | V | N | N | + | − |
| Group 3 wild typeb (Colorado 1994) | V | N | N | + | − |
| Group 4 wild typeb (New Jersey 94) | V | N | D | + | − |
| Group 5 wild typeb (Tennessee) | K | N | D | + | − |
| Group 8 wild typeb (China 93-1) | V | N | D | + | − |
Moraten, Rubeovax, Zagreb, Schwarz, and AIK-C vaccine strains were tested.
All wild-type viruses were initially isolated with B95-8 cells.
Predicted amino acids in the H protein including the potential glycosylation site at residue 416.
B95-8 and Vero cells were infected and examined for the presence (+) or absence (−) of cytopathic effect.
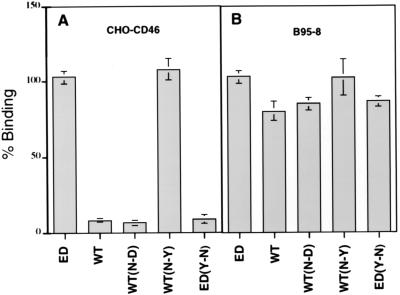
To determine whether the N481Y change controls the ability of the measles virus H protein to bind either to CD46 or to a new unidentified receptor which is present on B95-8 cells, mutations were introduced into the H protein at this position by site-specific mutagenesis. Mutated H proteins from wild-type and Edmonston vaccine strains of measles virus were expressed in Sf9 insect cells and incubated with Vero, HeLa, CD46-CHO, or marmoset B95-8 cells. The binding of insect cells expressing the mutated H proteins, Montefiore 89 wild-type H, or Edmonston H was assayed with CD46-CHO cells and the marmoset B-cell line and was subsequently quantitated by the ONPG assay for β-galactosidase (Fig. 7). The Edmonston H protein bound to both the CD46-CHO and B95-8 cells, while the wild-type H protein attached only to the B95-8 cells. Mutations N416D, which abolished the potential glycosylation site, had little effect on binding of the wild-type H to either CD46-CHO or B95-8 cells. However, the N481Y mutation, which was introduced into wild-type H protein, now permitted this glycoprotein to bind to CD46-CHO cells (Fig. 7A). Similarly, when the Y481 was changed to N in the Edmonston H protein, attachment to CD46-CHO cells was abolished (Fig. 7A). However, the Y481N change had no effect upon the binding of Edmonston H protein to B95-8 cells, and it would appear that this mutation did not perturb the interaction with the putative receptor which is present on marmoset B95-8 cells (Fig. 7B). Similar results were reproduced in mouse OST-7 cells infected with CD46 recombinant vaccinia virus, Vero cells, or HeLa cells following incubation with insect cells expressing mutant H proteins (46). We concluded that the tyrosine residue at position 481 in the measles virus H molecule was a key determinant of H protein binding to the CD46 receptor. However, mutations at amino acid 481 did not impair binding to the marmoset B-cell receptor, and these results suggest that another region of the measles virus H protein may interact with this as yet unidentified cell surface protein.
FIG. 7.
The Tyr481Asn mutation inhibits the binding of Edmonston and Montefiore 89 H proteins to CD46. Edmonston H (ED), Montefiore H (WT), and mutated forms of these proteins were expressed in Sf9 insect cells and incubated with CHO cells containing human CD46 (A) or marmoset B95-8 (B) cells. As expected, insect cells expressing Edmonston H (ED) bound to both CHO-CD46 and B95-8 cells, while wild-type Montefiore 89 H (WT) protein bound only to B95-8 cells. The N416D mutation introduced into Montefiore 89 H [WT(N-D)] had no effect on binding to either cell line. However, an N481Y mutation in the wild-type H [WT(N-Y)] converted the protein to a CD46-binding phenotype. In addition, when the Y481N mutation was placed in Edmonston H [ED(Y-N)], binding to CHO-CD46 cells was abolished. None of the mutations affected the binding of either Edmonston or wild-type H proteins to marmoset B95-8 cells. Binding was measured by quantitating β-galactosidase activity and was expressed as a percentage relative to the binding observed for Edmonston H protein.
DISCUSSION
This article provides convincing evidence for the existence of another measles virus receptor, in addition to CD46, which is present on marmoset B cells. Through sequence analysis and binding assays with H molecules from wild-type and Vero cell-adapted isolates of measles virus, it was apparent that Tyr481 of the H protein was critical in determining whether this protein bound to CD46. H proteins from attenuated vaccine strains and laboratory isolates of measles virus which had been propagated in Vero cells all contained this Tyr at position 481, while wild-type isolates which had been propagated in marmoset B95-8 cells produced a protein with Asn481. Binding assays indicated that wild-type H molecules did not interact with human or marmoset CD46 molecules but, rather, bound to another receptor which was present on B95-8 cells. Although polyclonal antibodies directed against CD46 and SCR1 reduced infections by the Edmonston laboratory strain, they could not totally inhibit viral infections in B95-8 cells, even at very high concentrations. A decrease in infection by the Edmonston virus in these cells, when antibody is present, may reflect a smaller number of functional receptors and reduced binding affinity to the second receptor for the attenuated virus. Further studies will eventually clarify this situation. The same antibodies had no effect on wild-type viral infections in the B95-8 cell line. The fact that CD46 antibodies were very effective in blocking infections by the Edmonston virus in Vero monkey kidney cells, coupled with the ability of the H protein containing the Y481N mutation to retain its binding properties for B95-8 cells, appeared to indicate that the Edmonston H protein may bind to either CD46 or the wild-type virus receptor. There may be two separate sites on the Edmonston H protein which could bind to either CD46 or the unidentified receptor on marmoset B cells, but this remains to be confirmed.
Amino acids 211, 243, and 276 of the measles virus H protein consistently vary between attenuated laboratory and wild-type strains of virus. The role of these amino acids in binding to the host cell is still not apparent. One publication has suggested that amino acids 211 to 214 may contribute to the CD46 binding site (53) while amino acids 451 to 617 appear to constitute the primary receptor binding site which is involved in hemagglutination (53, 59). Antibodies directed against regions spanning amino acids 185 to 195 have also been reported to inhibit hemagglutination (66) but may not be involved directly in binding to CD46. However, our evidence indicates that amino acids 211, 243, and 276, which differ between attenuated and wild-type strains, may not be extremely important for binding to CD46, since wild-type H containing the mutation N481Y binds just as efficiently as Edmonston H to cells expressing CD46 on their surface. The role of these amino acids in binding to the wild-type viral receptor remains to be determined. Other investigators have suggested that the binding site for CD46 lies between residues 451 and 617 (12, 17, 53, 59), and this seems to agree with our findings.
Results indicating the existence of a second receptor for measles virus which are presented in this paper partially explain why marmosets are susceptible to infections by measles virus in spite of the deleted SCR1 domain in their CD46 molecules. Our data also explains why wild-type measles virus grown in B95-8 cells does not hemagglutinate African green monkey erythrocytes even though these cells have CD46 on their surface. This second receptor obviously plays a critical role during infections initiated by wild-type isolates of measles virus. The virus is normally spread as an aerosol to the nasopharynx. However, the primary cellular target for the virus is not known for certain. During the acute phase of measles virus infection, the virus undergoes primary replication in the respiratory tract and disseminates throughout the body via the reticuloendothelial system. The virus has been reported to undergo a round of secondary replication in lymphoid tissues, and it efficiently infects monocytes (15). Isolates of wild-type virus were normally obtained as throat swabs containing mucosal epithelial cells and lymphocytes, which were subsequently propagated in B95-8 cells. Tracheal and bronchial epithelial cells are thought to be the primary target cells of measles virus, but lymphocytes and monocytes in the local lymph nodes are also infected by the virus at very early stages. Virus spreads to the thymus, spleen, skin, conjunctivae, kidneys, lungs, gastrointestinal tract, respiratory mucosa, small blood vessels, and liver. Endothelial cells, epithelial cells, monocytes, macrophages, and lymphocytes are all target cells which support virus growth in vivo (19, 38). The characteristic rash is caused by infiltration of macrophages to areas of skin endothelium that have been infected by the virus. The role that two different receptors play in natural measles virus infections remains to be determined. Since CD46 normally prevents complement lysis of the host cell (34, 35) and has been shown to be downregulated on the surface of the infected cell (4, 20, 28, 41, 54, 55), it might not be advantageous for the virus to use this receptor under certain circumstances.
The shift in tropism which accompanies the N481Y mutation in wild-type H molecules as they adapt to growth in culture requires a minor one nucleotide change (AAC→TAC). It is unclear whether this single base mutation also occurs in vivo during measles virus infections. Changes in the H protein could determine whether infections of lymphocytes, epithelial cells, or endothelial cells were favored. A precedent for a change in cellular tropism during an ongoing infection exists, since it was recently shown that many human immunodeficiency virus type 1 infections are initially macrophage tropic but, through minor changes in the V3 loop of the envelope protein, shift to become T-cell tropic and subsequently use the CXCR4 coreceptor for entry (reviewed in references 7, 11, and 32). Minor changes in the viral attachment proteins of other viruses have also been associated with changes in receptor usage. Coxsackie B viruses can use either CD55 (5) or CAR (6) as receptors depending on whether they have been adapted to growth in a rhadomyosarcoma cell line or HeLa cells, respectively. Ross River virus is a member of the togavirus family and varies its tropism among small mammals, chicken fibroblasts, mosquitoes, and human with single amino acid changes in the E1 and E2 viral membrane proteins (25, 62). Finally, two amino acid changes in the S protein of transmissible gastroenteritis coronavirus of pigs were recently found to abolish enteric tropism to favor respiratory infections (3). Thus, small changes in viral glycoproteins routinely dictate which receptor can be used and influence the type of cell which can be infected. It is quite possible that wild-type isolates of measles virus contain a mixture of virions which can use either CD46 or a new receptor found on B cells. In addition, small changes in the H protein also associated with neurovirulent strains of measles virus in rats (33), which do not have a CD46 analog, and this may reflect altered receptor usage in the brain. Since all wild-type measles virus strains are currently being isolated with B95-8 marmoset B cells or activated human B-cell lines, virions which characteristically bind to the new unidentified B-cell receptor are now being selected in the laboratory.
The involvement of more than one cellular molecule in virus attachment and penetration in measles virus infections seems more than likely. The envelope glycoprotein of human immunodeficiency virus type 1 attaches to CD4 and a variety of chemokine receptors (reviewed in reference 7). This may also be the case with coxsackie A viruses, which were recently shown to tightly bind to ICAM-1 but also interact with the low-affinity receptor CD55. Coimmunoprecipitation and chemical cross-linking studies seemed to indicate that CD55 and ICAM-1 are closely associated on the cell surface (58). Thus, one might expect other proteins to interact with CD46, which could act to pull the measles virus closer to the cell membrane in order to facilitate fusion with the cell membrane.
The new unidentified receptor for measles virus appears to be on marmoset B cells which have been immortalized and transformed by Epstein-Barr virus. We tested immortalized human B cells, and they also bound the H protein from Montefiore 89 wild-type virus, whereas mouse B cells were less efficient in this process (46). Previous investigators have claimed that measles virus can infect mouse B cells, which do not express CD46, and this suggests that the newly identified receptor may also be present on mouse lymphocytes (21). The identity of this new receptor for measles virus is unknown, but we are searching for and attempting to further characterize it in our laboratory. It also remains to be determined whether other coreceptors participate in the membrane fusion and internalization process of measles virus.
ACKNOWLEDGMENTS
We thank Laarni Antonio and Jenny Krum of the Amgen DNA sequencing facility at Thousand Oaks, Calif., for sequencing the different marmoset and tamarin CD46 clones and measles H protein mutants. The help of Marees Harris-Brandts in purifying SCR1 polypeptides for generation of polyclonal antisera is also acknowledged. A polyclonal antibody directed against human CD46 was kindly supplied by John P. Atkinson, Washington University, St. Louis, Mo. The assistance of Robert Lerch, Pramila Walpita, and Haiping Wang in the original isolation and gene sequencing of the Montefiore 89 virus strain is also acknowledged.
This work was supported by an operating grant (MA10638) from the Medical Research Council of Canada and a University of Toronto Graduate Student Open Scholarship awarded to E.C.H.
REFERENCES
- 1.Albrecht P, Lorenz D, Klutch M J, Vickers J H, Ennis F A. Fatal measles infection in marmosets pathogenesis and prophylaxis. Infect Immun. 1980;27:969–978. doi: 10.1128/iai.27.3.969-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht P, Lorenz D, Klutch M J. Encephalitogenicity of measles virus in marmosets. Infect Immun. 1981;34:581–587. doi: 10.1128/iai.34.2.581-587.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballesteros M L, Sanchez C M, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz R, Brinckmann U, Dunster L M, Rima B, ter Meulen V, Schneider-Schaulies J. Mapping amino acids of the measles virus hemagglutinin responsible for receptor (CD46) down-regulation. Virology. 1996;224:334–337. doi: 10.1006/viro.1996.0538. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson J M, Mohanty J G, Crowell R L, St. John N F, Lublin D M, Finberg R W. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55) J Virol. 1995;69:1903–1906. doi: 10.1128/jvi.69.3.1903-1906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergelson J M, Cunningham J A, Droguett G, Kurtjones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 7.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16. [PubMed]
- 8.Buchholz C J, Schneider U, Devaux P, Gerlier D, Cattaneo R. Cell entry by measles virus: long hybrid receptors uncouple binding from membrane fusion. J Virol. 1996;70:3716–3723. doi: 10.1128/jvi.70.6.3716-3723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz C J, Koller D, Devaux P, Mumenthaler C, Schneider-Schaulies J, Braun W, Gerlier D, Cattaneo R. Mapping of the primary binding site of measles virus to its receptor CD46. J Biol Chem. 1997;272:22072–22079. doi: 10.1074/jbc.272.35.22072. [DOI] [PubMed] [Google Scholar]
- 10.Buckland R, Wild T F. Is CD46 the cellular receptor for measles virus? Virus Res. 1997;48:1–9. doi: 10.1016/s0168-1702(96)01421-9. [DOI] [PubMed] [Google Scholar]
- 11.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaux P, Loveland B, Christiansen D, Milland J, Gerlier D. Interactions between the ectodomains of haemagglutinin and CD46 as a primary step in measles virus entry. J Gen Virol. 1996;77:1477–1481. doi: 10.1099/0022-1317-77-7-1477. [DOI] [PubMed] [Google Scholar]
- 13.Dörig R E, Marcil A, Chopra A, Richardson C D. The human CD46 is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 14.Dörig R E, Marcil A, Richardson C D. CD46, a primate-specific receptor for measles virus. Trends Microbiol. 1994;2:312–318. doi: 10.1016/0966-842x(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 15.Esolen L M, Ward B J, Moench T R, Griffin D E. Infection of monocytes during measles. J Infect Dis. 1993;168:47–52. doi: 10.1093/infdis/168.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Fraser K B, Martin S J. Measles virus and its biology. London: Academic Press Inc. (London) Ltd.; 1978. [Google Scholar]
- 17.Gerlier D, Varior-Krishnan G, Devaux P. CD46-mediated virus entry: a first key to host-range specificity. Trends Microbiol. 1995;3:338–345. doi: 10.1016/s0966-842x(00)88972-6. [DOI] [PubMed] [Google Scholar]
- 18.Graves M, Silver S, Choppin P W. Measles virus polypeptide synthesis in infected cells. Virology. 1978;86:254–263. doi: 10.1016/0042-6822(78)90025-9. [DOI] [PubMed] [Google Scholar]
- 19.Griffin D E, Bellini W J. Measles virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven Publishers; 1996. pp. 1267–1312. [Google Scholar]
- 20.Hirano A Y, Iwata K, Kortesarfaty J, Seya T, Nagasawa S, Wong T C. Human cell-receptor CD46 is down-regulated through recognition of a membrane-proximal region of the cytoplasmic domain in persistent measles virus infection. J Virol. 1996;70:6929–6936. doi: 10.1128/jvi.70.10.6929-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvat B, Rivailler P, Varior-Krishnan G, Cardoso A, Gerlier D, Rabourdin-Combe C. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infections. J Virol. 1996;70:6673–6681. doi: 10.1128/jvi.70.10.6673-6681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu E C, Dörig R E, Sarangi F, Marcil F, Iorio C, Richardson C D. Artificial mutations and natural variations in the CD46 molecules from human and monkey cells define regions important for measles virus binding. J Virol. 1997;71:6144–6154. doi: 10.1128/jvi.71.8.6144-6154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel K B, Erdman D D, Heath J, Bellini W J. Baculovirus expression of the nucleoprotein gene of measles virus and utility of the recombinant protein in diagnostic enzyme immunoassays. J Clin Microbiol. 1992;30:2874–2880. doi: 10.1128/jcm.30.11.2874-2880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata K, Seya T, Yanagi Y, Pesando J M, Johnson P M, Okabe M, Ueda S, Ariga H, Nagasawa S. Diversity of sites for measles virus binding and for inactivation of complement C3b and C4b on membrane cofactor protein CD46. J Biol Chem. 1995;270:15148–15152. doi: 10.1074/jbc.270.25.15148. [DOI] [PubMed] [Google Scholar]
- 25.Kerr P J, Weir R C, Dalgarno L. Ross River virus variants selected during passage in chick embryo fibroblasts: serological, genetic and biological changes. Virology. 1993;193:446–449. doi: 10.1006/viro.1993.1143. [DOI] [PubMed] [Google Scholar]
- 26.Kobune F, Sakata H, Sugiura A. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J Virol. 1990;64:700–705. doi: 10.1128/jvi.64.2.700-705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobune F, Takahashi H, Terao K, Ohkawa T, Ami Y, Suzaki Y, Nagata N, Sakata H, Yamanouchi K, Chieko K. Nonhuman primate models of measles. Lab Anim Sci. 1996;46:315–320. [PubMed] [Google Scholar]
- 28.Krantic S, Gimenez C, Rabourdin-Combe C. Cell-to-cell contact via measles virus haemagglutinin-CD46 interaction triggers CD46 downregulation. J Gen Virol. 1995;76:2793–2800. doi: 10.1099/0022-1317-76-11-2793. [DOI] [PubMed] [Google Scholar]
- 29.Lalumiére M, Richardson C D. Production of recombinant baculoviruses using rapid screening vectors that contain the gene for β-galactosidase. In: Richardson C D, editor. Baculovirus expression protocols. Totowa, N.J: Humana Press; 1995. pp. 161–177. [DOI] [PubMed] [Google Scholar]
- 30.Lecouturier V, Fayolle J, Caballero M, Carabaña J, Celma M L, Fernandez-Muñoz R, Wild T F, Buckland R. Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J Virol. 1996;70:4200–4204. doi: 10.1128/jvi.70.7.4200-4204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy B M, Merkovic R R. An epizootic of measles in a marmoset colony. Lab Animal Sci. 1971;21:33–39. [PubMed] [Google Scholar]
- 32.Levy J A. HIV and the pathogenesis of AIDS. Washington, D.C: ASM Press; 1994. pp. 35–36. [Google Scholar]
- 33.Liebert U G, Flanagan S G, Loffler S, Baczko K, ter Meulen V, Rima B K. Antigenic determinants of measles virus hemagglutinin associated with neurovirulence. J Virol. 1994;68:1486–1493. doi: 10.1128/jvi.68.3.1486-1493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liszewski M K, Post T W, Atkinson J P. Membrane cofactor protein (MCP of CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 35.Liszewski M K, Atkinson J P. Membrane cofactor protein. Curr Top Microbiol Immunol. 1992;178:45–60. doi: 10.1007/978-3-642-77014-2_4. [DOI] [PubMed] [Google Scholar]
- 36.Maisner A, Liszewski M K, Atkinson J P, Herrler G. Oligosaccharide in SCR-2 is required for measles virus binding and infection. J Virol. 1996;70:4973–4977. doi: 10.1128/jvi.70.8.4973-4977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manchester M, Valsamakis A, Kaufman R, Liszewski M K, Alvarez J, Atkinson J P, Lublin D M, Oldstone M B A. Measles virus and C3 binding sites are distinct on membrane cofactor protein (CD46) Proc Natl Acad Sci USA. 1995;92:2303–2307. doi: 10.1073/pnas.92.6.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McChesney M B, Miller C J, Rota P A, Zhu Y D, Antipa L, Lerche N W, Ahmed R, Bellini W J. Experimental measles. I. Pathogenesis in the normal and the immunized host. Virology. 1997;233:74–84. doi: 10.1006/viro.1997.8576. [DOI] [PubMed] [Google Scholar]
- 39.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naniche D, Varior-Krishnan G, Cervoni F, Wild T F, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naniche D, Wild T F, Rabourdin-Combe C, Gerlier D. Measles virus haemagglutinin induces down-regulation of gp57/67, a molecule involved in virus binding. J Gen Virol. 1993;74:1073–1079. doi: 10.1099/0022-1317-74-6-1073. [DOI] [PubMed] [Google Scholar]
- 42.Post T W, Liszewski M K, Adams E M, Tedja I, Miller E A, Atkinson J P. Membrane cofactor protein of the complement system: alternative splicing of serine/threonine/proline-rich exons and cytoplasmic tails produce multiple isoforms that correlate with protein phenotype. J Exp Med. 1991;174:93–102. doi: 10.1084/jem.174.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell D F J, Russell S M, Deacon N J, Brown M A, Hooker D J, McKenzie I F C. Alternatively spliced RNAs encode several isoforms of CD46 (MCP), a regulator of complement activation. Immunogenetics. 1991;33:335–344. doi: 10.1007/BF00216692. [DOI] [PubMed] [Google Scholar]
- 44.Richardson C D, Berkovich A, Rozenblatt S, Bellini W J. Use of antibodies directed against synthetic peptides for identifying cDNA clones, establishing reading frames, and deducing the gene order of measles virus. J Virol. 1985;54:186–193. doi: 10.1128/jvi.54.1.186-193.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson C D, Banville M, Lalumiére M, Vialard J, Meighen E A. Bacterial luciferase produced with rapid screening baculovirus vectors in a sensitive reporter for infection of insect cells and larvae. Intervirology. 1992;34:213–227. doi: 10.1159/000150285. [DOI] [PubMed] [Google Scholar]
- 46.Richardson, C. D., and E. C. Hsu. Unpublished data.
- 47.Rima B K, Earle J A P, Baczko K, Rota P A, Bellini W J. Measles virus strain variations. In: ter Meulen V, Billeter M A, editors. Measles virus. Heidelberg, Germany: Springer-Verlag KG; 1995. pp. 65–83. [DOI] [PubMed] [Google Scholar]
- 48.Rota J S, Hummel K B, Rota P A, Bellini W J. Genetic variability of the glycoprotein genes of current wild-type measles isolates. Virology. 1992;188:135–142. doi: 10.1016/0042-6822(92)90742-8. [DOI] [PubMed] [Google Scholar]
- 49.Rota J S, De-Wang Z, Rota P A, Bellini W J. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 1994;31:317–330. doi: 10.1016/0168-1702(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 50.Rota J S, Heath J L, Rota P A, King G E, Celma M L, Carabana J, Fernandez-Munoz R, Brown D, Jin L, Bellini W J. Molecular epidemiology of measles virus: identification of pathways of transmission and implications for measles elimination. J Infect Dis. 1996;173:32–37. doi: 10.1093/infdis/173.1.32. [DOI] [PubMed] [Google Scholar]
- 51.Rota P A, Rota J S, Bellini W J. Molecular epidemiology of measles virus. Semin Virol. 1995;5:379–386. [Google Scholar]
- 52.Russell S M, Sparrow R L, McKenzie I F, Purcell D F. Tissue-specific and allelic expression of the complement regulator CD46 is controlled by alternative splicing. Eur J Immunol. 1992;22:1513–1518. doi: 10.1002/eji.1830220625. [DOI] [PubMed] [Google Scholar]
- 53.Saito H, Sato H, Abe M, Harata S, Amano K, Suto T, Morita M. Cloning and characterization of the cDNA encoding the HA protein of a hemagglutination-defective measles virus strain. Virus Genes. 1994;8:107–113. doi: 10.1007/BF01703609. [DOI] [PubMed] [Google Scholar]
- 54.Schneider-Schaulies J, Dunster L M, Kobune F, Rima B, ter Meulen V. Differential downregulation of CD46 by measles virus strains. J Virol. 1995;69:7257–7259. doi: 10.1128/jvi.69.11.7257-7259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider-Schaulies J, Schnorr J-J, Brinnckmann U, Dunster L M, Baczko K, Liebert U G, Schneider-Schaulies S, ter Meulen V. Receptor usage and differential downregulation of CD46 by measles virus wild-type and vaccine strains. Proc Natl Acad Sci USA. 1995;92:3943–3947. doi: 10.1073/pnas.92.9.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider-Schaulies J, Schnorr J-J, Schlender J, Dunster L M, Schneider-Schaulies S, ter Meulen V. Receptor (CD46) modulation and complement mediated lysis of uninfected cells after contact with measles virus-infected cells. J Virol. 1996;70:255–263. doi: 10.1128/jvi.70.1.255-263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seya T, Kurita M, Hara T, Iwata K, Semba T, Hatanaka M, Masumoto M, Yanagi Y, Ueda S, Nagasawa S. Blocking measles virus infection with a recombinant soluble form of, or monoclonal antibodies against, membrane cofactor protein of complement (CD46) Immunology. 1995;84:619–625. [PMC free article] [PubMed] [Google Scholar]
- 58.Shafren D R, Dorahy D J, Ingham R A, Burns G F, Barry R D. Coxsackie virus A21 binds decay-accelerating factor but requires intracellular adhesion molecule 1 for cell entry. J Virol. 1997;71:4736–4743. doi: 10.1128/jvi.71.6.4736-4743.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibahara K, Hotta H, Katayama Y, Homma M. Increased binding activity of measles virus to monkey red blood cells after long-term passage in Vero cell cultures. J Gen Virol. 1994;75:3511–3516. doi: 10.1099/0022-1317-75-12-3511. [DOI] [PubMed] [Google Scholar]
- 60.Shirodaria P V, Dermott E, Gould E A. Some characteristics of salt-dependent haemagglutinating measles viruses. J Gen Virol. 1976;33:107–115. doi: 10.1099/0022-1317-33-1-107. [DOI] [PubMed] [Google Scholar]
- 61.Sidhu, M., and S. A. Udem. Unpublished data.
- 62.Strauss J H, Rumenapf T, Weir R C, Kuhn R J, Wang K S, Strauss E G. Cellular receptors for alphaviruses. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 141–163. [Google Scholar]
- 63.ter Meulen V, Billeter M A. Measles virus. Curr Top Microbiol Immunol. 1995;191:1–193. doi: 10.1007/978-3-642-78621-1_7. [DOI] [PubMed] [Google Scholar]
- 64.Vialard J, Lalumiére M, Vernet T, Briedis D, Alkhatib G, Henning D, Levin D, Richardson C D. Synthesis of the membrane fusion and hemagglutinin proteins of measles virus, using a novel baculovirus vector containing the β-galactosidase gene. J Virol. 1990;64:37–50. doi: 10.1128/jvi.64.1.37-50.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vialard J E, Richardson C D. The 1629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J Virol. 1993;67:5859–5866. doi: 10.1128/jvi.67.10.5859-5866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wild T F, Buckland R. Functional aspects of envelope-associated measles virus proteins. In: ter Meulen V, Billeter M A, editors. Measles virus. Heidelberg, Germany: Springer-Verlag KG; 1995. pp. 51–64. [DOI] [PubMed] [Google Scholar]