Summary
Structural neuroplasticity (changes in the size, strength, number, and targets of synaptic connections) can be modified by sleep and sleep disruption. However, the causal relationships between genetic perturbations, sleep loss, neuroplasticity, and behavior remain unclear. The C. elegans GABAergic DVB neuron undergoes structural plasticity in adult males in response to adolescent stress, which rewires synaptic connections, alters behavior, and is dependent on conserved autism-associated genes NRXN1/nrx-1 and NLGN3/nlg-1. We find that four methods of sleep deprivation transiently induce DVB neurite extension in day 1 adults and increase the time to spicule protraction, which is the functional and behavioral output of the DVB neuron. Loss of nrx-1 and nlg-1 prevent DVB structural plasticity and behavioral changes at day 1 caused by adolescent sleep loss. Therefore, nrx-1 and nlg-1 mediate the morphologic and behavioral consequences of sleep loss, providing insight into the relationship between sleep, neuroplasticity, behavior, and neurologic disease.
Subject areas: Behavioral neuroscience, Molecular neuroscience, Cellular neuroscience
Graphical abstract
Highlights
-
•
Adolescent sleep deprivation induces GABAergic neurite outgrowth in adult C. elegans
-
•
Sleep loss alters the synapses of the GABAergic neuron and changes behavior
-
•
Sleep loss induced morphologic/behavioral changes require neurexin and neuroligin
-
•
Adolescent sleep deprivation induced neurite outgrowth is transient in adulthood
Behavioral neuroscience; Molecular neuroscience; Cellular neuroscience
Introduction
Sleep is essential for brain function and survival across species. While the neurologic benefits of sleep are still being defined, it is known that sleep is important for memory consolidation, attention, learning, plasticity, and waste clearance/metabolic regulation for neuron preservation.1,2,3 Long-term sleep deprivation is associated with severe cognitive impairment.4,5 Additionally, sleep disruption exacerbates a number of physiological problems including neurodegenerative diseases and impedes neuronal rehabilitation after injury including stroke or traumatic brain injury.6,7,8,9,10 Further, sleep deficits are linked to neuropsychiatric and neurodevelopmental conditions11,12,13,14,15 including autism, in which sleep impairments impact core behavioral characteristics.16,17,18,19,20,21,22,23,24
Structural plasticity, or the ability of neurons to reorganize their shape and connections in response to environment and experience, appears intimately tied to sleep.11,13,25,26,27,28 Analysis in Caenorhabditis elegans, Drosophila, zebrafish, and rodents demonstrates that sleep regulates synapse strength, number, composition, and morphology, as well as learning, memory, mood, and social behaviors.29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45 However, the functional role of the synaptic changes on behaviors, and the mechanisms that bridge them remain elusive. Neuronal activity during wakefulness correlates with sleep behavior itself,46 uncovering a reciprocal relationship between sleep and plasticity. Many methods used for sleep disruption across models may also impact neuronal and circuit activity unrelated to sleep (ex. gentle handling and novel object exposure involve stress and learning, respectively).47 Additionally, disparate or contradictory findings in the number and strength of synapses after sleep and sleep disruption likely reflect differences in the brain region, organism, and sleep disruption method used.26,48 The majority of studies on sleep deprivation analyze either synaptic plasticity or behavior, but not both.30,31,32,33,34,36,37,41,42 Analyses of synapses and behavior together have mainly focused on the impact of sleep deprivation on learning-induced plasticity.38,43,44,45 The behavioral effects of specific synaptic changes in response to sleep loss beyond learning induction have been difficult to determine as the neurons and circuits that regulate distinct behaviors are still being defined. Leveraging the power of C. elegans, we can quantify how sleep deprivation at specific time windows impacts a specific neuron (DVB) and its behavioral output (spicule protraction).
One mechanism by which sleep may modify neuron and synapse structure, resulting in behavioral changes, is through synaptic adhesion molecules (SAMs). SAMs impact synapse formation and function, mediating neuron and synaptic structural changes in development and maintenance in adults. Several adhesion molecules (immunoglobulins (IG), cadherins, neurexins, neuroligins, integrins, and Eph/Ephrins) have roles in the regulation of sleep behavior.49,50,51,52,53 Neurexins are autism-associated SAM genes54,55,56 with diverse roles in synapse function and plasticity, making them particularly relevant in the context of sleep and structural plasticity.56,57 Alternatively spliced isoforms of neurexins can have discrete regulatory functions which are neuron-, synapse-, and temporal-specific.56,57,58,59,60,61,62 While there are three mammalian neurexin genes, there is only one in Drosophila and C. elegans, thereby simplifying genetic manipulations in these model systems. Deletions in Drosophila neurexin (Nrx-1) fragment sleep behavior and disrupt circadian patterns and adult specific over-expression can improve most sleep phenotypes.30 Mice lacking neuroligin (Nlgn1), the canonical binding partner of neurexins, have reduced wakefulness and increased non-rapid eye movement sleep.49 Further, sleep deprivation results in altered neuroligin and neurexin expression levels, including isoform-specific impacts.51 The role of SAMs in sleep has relied on whole brain or brain region analysis,49,53,56,63 while work on these genes beyond sleep highlights their neuron, synapse, and context specific functions. Despite the connections between sleep, SAMs, and structural neuroplasticity, how these changes are executed and the role of SAMs in sleep deprivation related behaviors are not well-characterized.
Structural plasticity of the GABAergic DVB neuron in adult male C. elegans
The GABAergic DVB neuron undergoes experience-dependent structural plasticity and circuit rewiring in adult male C. elegans. Increased DVB outgrowth, which occurs only in males, leads to changes in spicule protraction, a specific step of male mating behavior.64 The progressive outgrowth/branching of axon-like neurites of the DVB neuron is unlike almost any other neuron in C. elegans in its dynamic nature and variability. DVB structural plasticity is modified by experience, such as exposure to mates or manipulation of circuit activity (stimulation or inhibition with opto-/chemo-genetics).64 Interestingly, DVB outgrowth is dependent on the C. elegans ortholog of neurexins and neuroligins, nrx-1 and nlg-1. However, contrary to the canonical roles of nrx-1 and nlg-1 in many contexts, the two have antagonistic effects on DVB morphologic plasticity in adults, with NRX-1 promoting and NLG-1 restricting DVB neurite outgrowth.64
Stressors applied during an adolescent-like developmental stage (L4) lead to early DVB neurite outgrowth at day 1 of adulthood.65 While some of these stressors (starvation) result in changes in spicule protraction behavior, other stressors (UV and heat-shock) do not impact behavior despite inducing DVB morphologic plasticity.65 Additionally, while starvation induced DVB neurite extension is nrx-1 and nlg-1 dependent, neither UV nor heat-shock induced structural plasticity depend on nrx-1 and only heat-shock depends on nlg-1. Despite their antagonistic functions in adulthood, nrx-1 and nlg-1 both promote adolescent starvation stress induced DVB plasticity at day 1.65 Starvation stress coincides with the fourth larval stage and molt when C. elegans undergo a period of developmental sleep, possibly impacting sleep and leaving open the question of whether sleep may play a role in DVB plasticity.65
C. elegans sleep
Studying sleep in C. elegans is particularly advantageous given its transparency, neuronal gene conservation, and mapped 302-neuron connectome.66,67,68 Further, many cell- and gene-specific tools for sleep manipulation and technologies for measuring sleep have been developed in C. elegans.69,70,71,72,73,74,75,76,77,78 C. elegans undergo periods of behavioral quiescence during the four larval transitions, known as lethargus or developmentally timed sleep, that include all of the characteristics of sleep (cessation of movement, rapid reversibility, decreased responsiveness, stereotypical posture, reduced neuronal activity, and homeostatic regulation).79,80,81,82,83 Developmental sleep is regulated by the conserved clock Period gene82 and is promoted by the RIS neuron.84,85,86,87 Disruption of APTF-1 or LIM-6, transcription factors required to specify the RIS neuron, results in decreased sleep.88,89,90 Sleep in C. elegans has also been shown to coincide with synaptic plasticity91 and recent work has defined the neuronal and synaptic changes that link experience and sleep to memory-related behavior.29
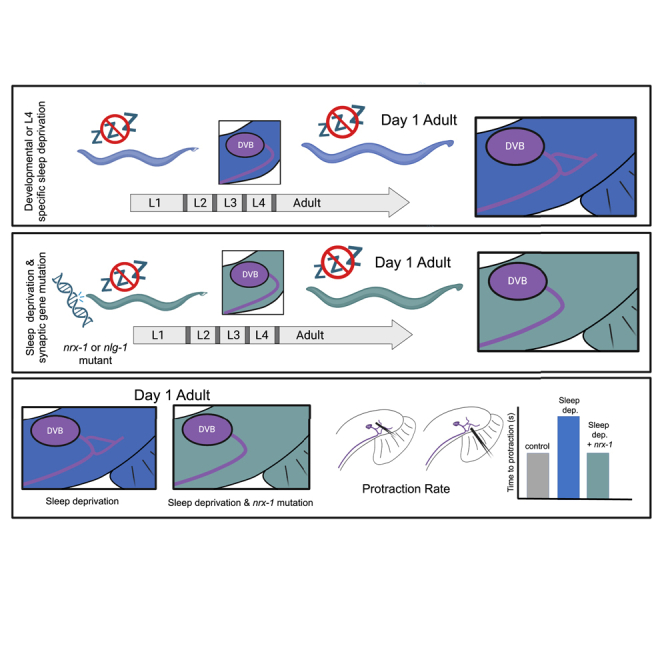
Here we use multiple methods to disrupt developmental sleep in C. elegans, including genetic, physical, and chemo-genetic manipulations. We find that sleep deprivation increases neurite outgrowth of the DVB GABAergic neuron and modifies behavior in early adulthood. We show that sleep deprivation during the adolescent-like L4 molt, but not the L2 or L3 molts, induces similar morphologic and behavioral changes to sleep deprivation throughout development, narrowing the critical timing of sleep to the last larval stage. Importantly, sleep deprivation induced morphologic and behavioral plasticity depend on the conserved SAMs, neurexin/nrx-1 and neuroligin/nlg-1, which act in the same pathway to promote DVB structural plasticity after sleep loss. These findings show that sleep is essential for proper maintenance of neuronal morphology at single neuron and behavior resolution. Additionally, our results indicate that sleep alters behavior by modifying plasticity at the level of specific synaptic and circuit connections. This work implicates neurexins and neuroligins in sleep dependent structural plasticity and identifies isoform specificity of nrx-1 alpha. Taken together, our results show that sleep disruption robustly alters DVB structural plasticity and behavior, and these changes are dependent on conserved SAMs that are associated with autism and other neurologic conditions.
Results
Developmental sleep disruption increases DVB neurite outgrowth in a nrx-1 dependent manner
To determine whether sleep deprivation alters structural plasticity, we studied neuron morphology of the sexually dimorphic GABAergic neuron, DVB, in males at day 1 of adulthood. We compared control strains, in which animals slept normally, to two strains with disrupted sleep across development (aptf-1(tm3287) and npr-1(ad609) mutants)(Figure 1A).90,92 Worms lacking APTF-1, a conserved AP2 transcription factor, are known to have profoundly decreased sleep at all periods of developmental sleep as APTF-1 is a positive regulator of FLP-11 sleep-inducing neuropeptides.88,90 The role of AP2 transcription factors in sleep regulation is conserved in Drosophila, mice, and humans.93,94,95 We found that compared to controls, aptf-1(tm3287) males had drastically decreased quiescence during the L4 to adult molt as shown by their reduced number of quiescent bins and total time quiescent (Figure S1A–S1C). Activity levels and number of quiescent bins in day 1 adult males after the L4 molt were not changed in aptf-1(tm3287) mutant C. elegans (Figures S1D and S1E).
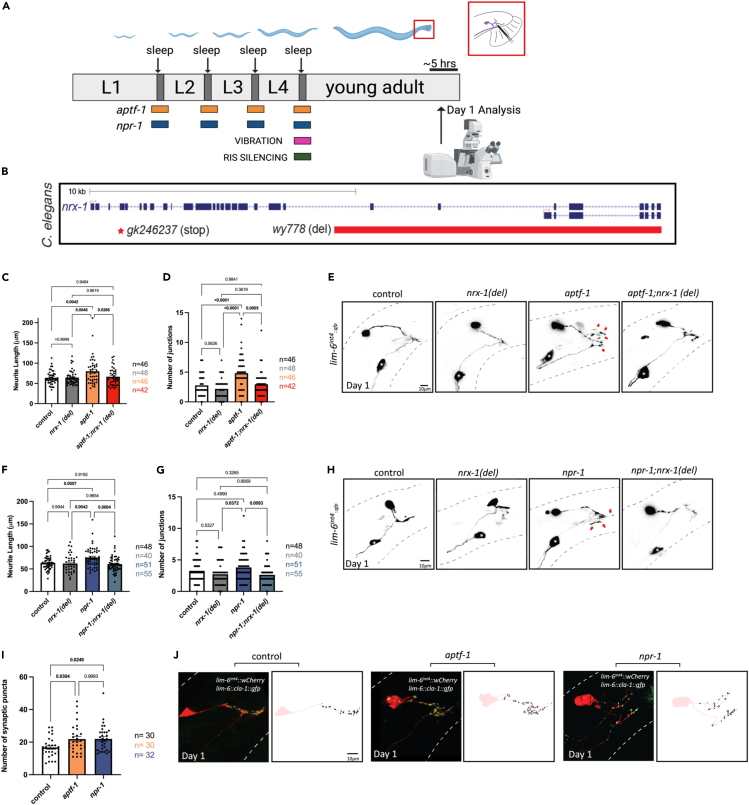
Figure 1.
Genetic disruption of developmental sleep (via npr-1 and aptf-1) increases DVB neurite outgrowth dependent on nrx-1
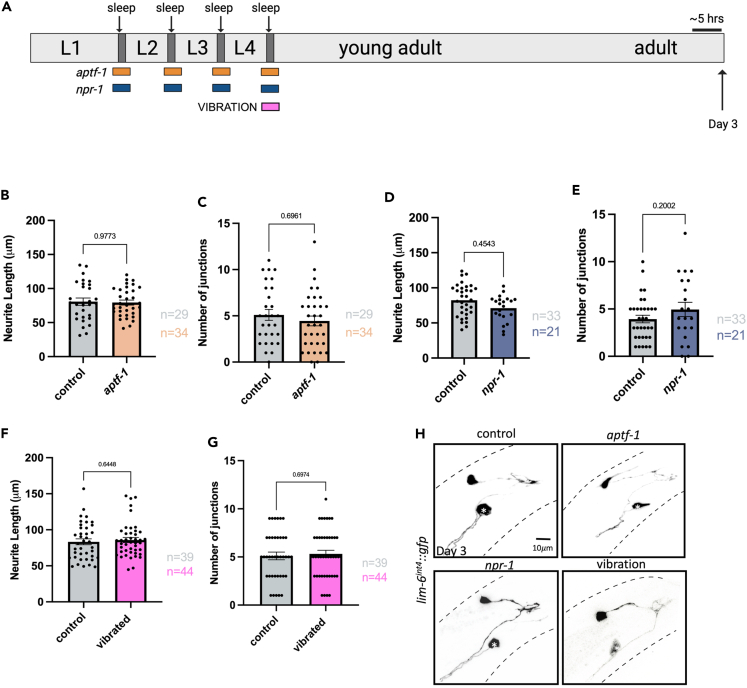
(A) Schematic of C. elegans developmental sleep stages between larval stages with timing of each sleep deprivation method indicated. aptf-1 (orange) and npr-1 (blue) animals experience impaired sleep during all larval sleep periods, while vibration (pink) and RIS silencing (green) are applied specifically during the L4 to adult transition/molt. Cartoon zoom in of the adult male tail with DVB shown.
(B) Schematic of C. elegans nrx-1 gene coding locus, red box indicates deletion in nrx-1(wy778) referred to as nrx-1(del), red star indicates premature stop in nrx-1(gk246237) referred to as nrx-1(stop). Quantification of DVB (C) total neurite length and (D) junctions in controls, nrx-1(del), aptf-1, and aptf-1; nrx-1(del) males at day 1.
(E) Representative images of DVB neuron in day 1 males using lim-6p::gfp. Red arrows indicate neurite branches. Black dashed lines represent body outline. White asterisk marks PVT neuron (not traced, see64). Quantification of DVB (F) total neurite length and (G) junctions in controls, nrx-1(del), npr-1, and npr-1; nrx-1(del) day 1 males.
(H) Representative images of DVB at day 1 in npr-1 mutant males and all controls.
(I) Quantification of number of cla-1::gfp puncta in DVB in controls and sleep deprived aptf-1 and npr-1 males at day 1.
(J) Representative images of DVB (red) and cla-1::gfp puncta (green) in controls, aptf-1, and npr-1 day 1 males. Outlines of synaptic puncta generated from FIJI particle analysis are shown (black) superimposed on a trace of the DVB neuron (light red). Number of individual animals is indicated by “n”, p values from one-way ANOVA with Tukey’s post-hoc test shown, error bars represent SEM, scale bar size as labelled.
Notably, we found an increase in DVB neurite length in aptf-1(tm3287) mutants with disrupted sleep at day 1 of adulthood (Figures 1C and 1E). Day 1 adult aptf-1(tm3287) males also had an increase in the number of DVB junctions, a proxy for neurite branching compared to controls (Figures 1D and 1E). Further, the number of quiescent bins were quantified and plotted against neurite length for individual control and aptf-1(tm3287) males. When plotting number of quiescent bins by neurite length for individual animals, we observe that aptf-1(tm3287) mutants and controls cluster separately, with aptf-1(tm3287) mutants showing both decreased quiescence and increased neurite length relative to controls. However, the number of quiescent bins of individual animals was not directly proportionate to neurite length in either control or aptf-1(tm3287) males, likely due to the variability of DVB outgrowth caused by non-sleep related experiences (Figure S1F). Mutants lacking NPR-1, a neuropeptide Y like receptor, have been shown to have decreased sleep due to increased sensory activity, mediated by the PDF-1 arousal neuropeptides.85,92,96 Though the extent of sleep deprivation was not directly assessed, we found that DVB neurite length was significantly increased in day 1 adult npr-1(ad609) mutant males compared to controls (Figures 1F–1H), similar to aptf-1(tm3287) mutants (Figures 1F–1H).
To determine whether neurexin/nrx-1 plays a role in sleep deprivation induced neurite extension, we tested a C. elegans strain carrying a nrx-1 mutation (wy778), a ∼12,000 nucleotide deletion that disrupts the long (alpha) and short (gamma) isoforms, referred to as nrx-1(del) in figures (Figure 1B). Importantly, quiescence during the L4 to adult molt in nrx-1(wy778) males, albeit slightly more variable, does not differ from that of controls (Figures S1A–S1C). We plotted time quiescent from time t = 0 to t = 300 min across genotypes and found nrx-1 males have a similar amount of sleep per 10-min bin to controls during the L4 molt but have elevated levels of quiescence between the L3 and L4 sleep periods (Figure S1C). We also observed that activity levels of day 1 adult nrx-1(wy778) males was decreased relative to controls, but did not exhibit ectopic sleep periods during day 1 of adulthood after the L4 molt (see STAR Methods)(Figures S1D and S1E). Given the reductions in activity and trend toward increased sleep in nrx-1 mutants, we directly assessed sleep in aptf-1(tm3287); nrx-1(wy778) mutants to test whether nrx-1(wy778) modifies the loss of sleep observed with aptf-1(tm3287). We found that the aptf-1(tm3287); nrx-1(wy778) mutants had the same large decrease in the number of quiescent bins and total time quiescent as aptf-1(tm3287) alone (Figures S1A and S1B). Therefore, loss of NRX-1 does not impact the ability of aptf-1(tm3287) mutation to disrupt sleep.
Notably, we found that the increase in DVB neurite outgrowth observed in aptf-1(tm3287) males at day 1 is lost in aptf-1(tm3287); nrx-1(wy778) males (Figures 1C–1E). Similarly, the increased DVB neurite outgrowth we observed in npr-1 (ad609) mutants was eliminated in npr-1(ad609); nrx-1(wy778) double mutants (Figures 1F–1H). nrx-1(wy778) alone did not impact either measure of DVB neurite outgrowth (Figures 1C, 1D, 1F, and 1G) indicating that nrx-1 is not needed for day 1 DVB morphology, but rather, is required for sleep deprivation-induced DVB neurite outgrowth, similar to previously tested stressors.64,65
We next asked whether DVB neurite outgrowth induced by sleep deprivation in male C. elegans corresponds to any changes in DVB pre-synaptic morphology. We expressed GFP tagged active zone marker, cla-1, in DVB using the lim-6 promoter.65,97 Day 1 aptf-1(tm3287) and npr-1(ad609) males have an increased number of DVB cla-1 puncta compared to controls (Figures 1I and 1J). The size of cla-1 puncta was not different in sleep deprived males compared to controls (Figure S1G). Therefore, sleep deprivation results in increased pre-synaptic puncta in the DVB neuron.
Adolescent specific sleep deprivation induces DVB neurite outgrowth
Our previous work implicated the adolescent-like stage L4, when C. elegans males sexually mature, as particularly sensitive for induction of DVB plasticity by stress.65 To ask if sleep deprivation during the L4 larval sleep period impacts DVB outgrowth in early adulthood, we applied a vibration stimulus during the L4 molt and analyzed DVB morphology in day 1 adult males (Figure 1A). Vibration was applied for 1 s every 10 min during a 6-h period overlapping with the L4 molt sleep period. L4 vibration increased DVB neurite outgrowth and number of junctions relative to controls (Figures 2A–2C). Vibration may impact DVB outgrowth unrelated to sleep through stimulation of sensory neurons or other impacts of the vibration on tissues or cells.98 However, we found that vibration only impacts DVB neurite outgrowth when applied during L4 developmental sleep; the same vibration protocol applied before or after the L4 sleep period had no impact on DVB morphology (Figures S2A–S2E). Further, we disrupted sleep using the same vibration protocol during the L2 molt and L3 molt. We find that disrupting developmental sleep during these periods did not increase DVB neurites in day 1 adult males (Figures S2A, S2F, and S2G). This result demonstrates that the L4 molt and sleep period is specifically critical for the regulation of DVB structural plasticity.
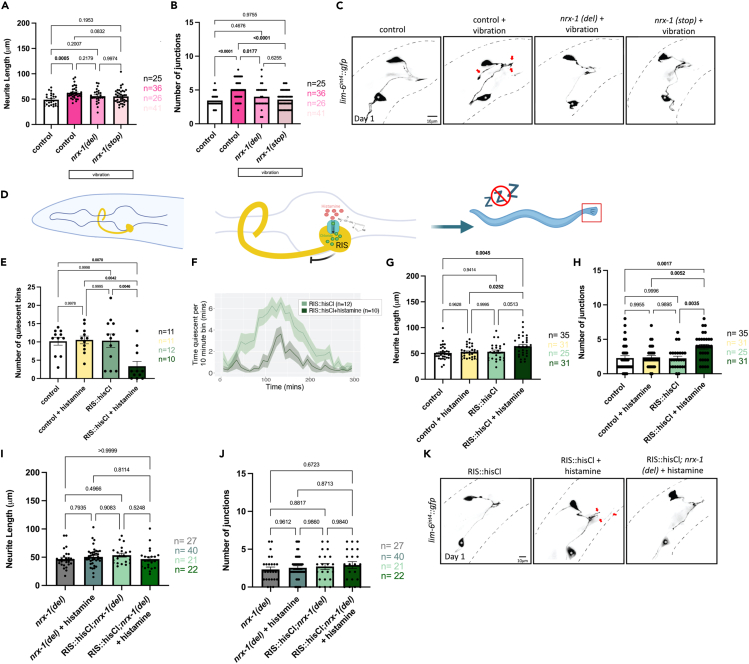
Figure 2.
Sleep deprivation specifically during adolescent-like stage (L4) induces DVB neurite outgrowth at day 1
Quantification of DVB (A) neurite length and (B) number of DVB junctions in controls, vibrated controls, and vibrated nrx-1 (del), and vibrated nrx-1 (stop) males at day 1 after vibration sleep deprivation. Vibration sleep deprivation protocol indicated with white boxes.
(C) Representative images of DVB neuron in vibration sleep deprived males and controls. White asterisk marks PVT neuron (not traced, see64).
(D) Cartoon of histamine gated chloride channel expression in RIS neuron to silence RIS neuron activity. Exogenous histamine acts to open chloride channels allowing for the influx of negative ions to hyper-polarize the neuron and prevent signaling, selectively in the RIS neuron (flp-11 promoter), referred to as RIShisCl.
(E) Graph of number of quiescent bins during L4 molt in RIShisCl transgene positive and negative males with (dark green) and without (light green) histamine application. A bin was considered quiescent if the C. elegans spent more than 2.5 min immobile per 10 min bin. Graph showing the quiescence amount in control, nrx-1, nlg-1, and aptf-1 males from t = 0 to t = 300 min, with peak quiescence aligned.
(F) Graph showing the quiescence amount in RIShisCl transgenic male C. elegans with and without (+/−) histamine application from t = 0 to t = 300 min, with peak quiescence aligned.
(G) DVB neurite length and (H) junctions in RIShisCl transgenic day 1 males +/− histamine and controls.
(I) DVB neurite length and (J) junctions in RIShisCl; nrx-1(del) +/− histamine and nrx-1(del) +/− histamine day 1 males. Experiments for G and H were conducted on separate days than I and J and therefore not plotted together or compared statistically.
(K) Representative images of DVB neuron in RIShisCl transgenic +/− histamine and RIShisCl; nrx-1(del) day 1 males. White asterisk marks PVT neuron (not traced, see64). Number of individual animals is indicated by “n”, p values from one-way ANOVA with Tukey’s post-hoc test shown, error bars represent SEM, scale bar size as labelled.
To disrupt sleep in another fashion, we next silenced the sleep promoting neuron RIS through expression of a histamine-gated chloride channel (flp-11 promoter).99 Application of histamine results in hyperpolarization of the RIS neuron, is predicted to prevent the release of GABA and the FLP-11 sleep-promoting neuropeptides, and allows for narrowing of sleep disruption to just L4 (Figure 2D). Using a WorMotel,76,100 we monitored the impact of histamine in transgenic and non-transgenic C. elegans on quiescence (Figures 2E and 2F). Overnight histamine exposure during the L4 sleep period in males expressing flp-11p::HisCl significantly reduced the number of quiescent bins compared to flp-11p::HisCl males without histamine (Figures 2E and 2F). Histamine on its own did not impact quiescence. Thus, we disrupted sleep specifically and temporally using this chemo-genetic approach.
Male C. elegans with sleep disruption via histamine at L4 displayed increased adult DVB neurite outgrowth compared to controls. We observed longer day 1 neurite length in L4 RIS-silenced animals (64.08μM) compared to untreated controls (53.24 μM)(p = 0.051, Figures 2G and 2K). RIS-silenced males had significantly increased DVB neurite length compared to control animals not carrying the HisCl transgene (N2 on histamine, N2 without histamine)(Figures 2G and 2K). RIS-silenced day 1 adult males had significantly increased DVB junctions compared to untreated HisCl transgenic animals and to controls not carrying the HisCl transgene, either untreated or treated with histamine (Figures 2H and 2K). This result confirms the importance of the L4 sleep period in regulation of DVB structural plasticity.
While our results demonstrate the importance of the L4 stage and sleep period for DVB morphology, we cannot rule out the possibility that levels of activity and movement also contributes to DVB outgrowth. To test the impact of movement, we applied vibration to paralyzed males carrying a unc-97(su110) mutation. Vibrated unc-97 males had similar DVB neurite length and junctions to non-vibrated unc-97 males (Figures S2A, S2H, and S2I). Therefore, lack of movement during sleep may contribute to DVB morphologic plasticity, although further analysis will be required to untangle this complex interaction.
nrx-1 can have isoform-specific functions101 so we asked whether DVB outgrowth after sleep disruption was dependent on any specific isoform of nrx-1. We compared the nrx-1(wy778) allele used above with nrx-1(gk246237), a premature stop allele that specifically disrupts the alpha (long) isoform, referred to as nrx-1(stop) in figures (Figure 1B). Vibration based sleep deprivation did not increase DVB neurite length in males carrying either mutant allele of nrx-1 compared to controls (Figures 2A and 2C). The number of DVB junctions was significantly increased in vibration sleep deprived males compared to nrx-1(wy778) and nrx-1 (gk246237) vibration sleep deprived males (Figures 2A–2C). This result suggests that the long isoform of nrx-1 specifically is required for DVB neurite outgrowth resulting from vibration sleep disruption.
We next tested nrx-1(wy778) mutants with the flp-11p::HisCl transgene in the presence or absence of histamine application. In alignment with our other findings, RIS-silenced nrx-1(wy778) mutants did not have increased neurite length or junctions relative to non-transgenic nrx-1(wy778) mutants on histamine (Figures 2I–2K). This result confirms and extends our results from genetic and vibration sleep deprivation that nrx-1 regulates the morphological changes caused by L4 sleep deprivation.
Sleep deprivation induced DVB structural plasticity alters behavior
Increased DVB neurite outgrowth in adulthood leads to changes in spicule protraction behavior, which is the readout of the balance between excitation from cholinergic SPC neurons and inhibition from the GABAergic DVB neuron (Figure 3A).102 We induced spicule protraction by application of aldicarb, an acetylcholinesterase inhibitor that promotes spicule protraction.64 The aldicarb spicule protraction assay is a proxy for the natural behavior of spicule protraction during mating or upon stimulation of the spicule protraction circuit. As a crucial step of mating, spicule protraction timing correlates with mating efficiency as defined by successful sperm transfer.64 We monitored spicule protraction at day 1 on aldicarb (Figures 3A and 3B)64 and found that aptf-1(tm3287) males took longer to protract their spicules compared to controls (Figure 3C). Day 1 npr-1(ad609) males also showed an increase in time to spicule protraction relative to controls (Figure 3D). As was the case with aptf-1 and npr-1 sleep-deficient mutants, we found that sleep disruption at the L4 molt via vibration significantly increased time to spicule protraction at day 1 (Figure 3E). Previously, we showed that nrx-1(wy778) day 1 males do not alter time to spicule protraction.65
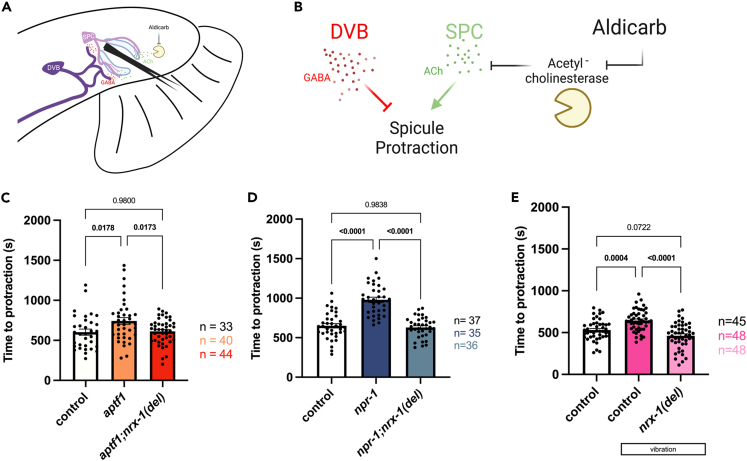
Figure 3.
Sleep deprivation alters DVB-dependent spicule protraction behavior
(A) Cartoon of neurons and muscles controlling spicule protraction in male tail. DVB is shown in dark purple and SPC is shown in light purple.
(B) Cartoon depicting mechanism for aldicarb induced spicule protraction with opposing functions of DVB GABAergic inhibition (red) and SPC cholinergic excitation (green).
(C) Time to spicule protraction following aldicarb exposure in control, aptf-1, and aptf-1; nrx-1(del) day 1 males.
(D) Time to spicule protraction following aldicarb exposure in control, npr-1, and npr-1; nrx-1(del) day 1 males.
(E) Time to spicule protraction following aldicarb exposure in control day 1 males and control and nrx-1(del) day 1 males that underwent L4 vibration sleep deprivation. Number of individual animals is indicated by “n”, p values from one-way ANOVA with Tukey’s post-hoc test shown, error bars represent SEM.
To determine any role for nrx-1 in behavioral changes following sleep deprivation we tested aptf-1(tm3286); nrx-1(wy778), npr-1(ad609); nrx-1(wy778) double mutants, and nrx-1(wy778) mutants that underwent L4 vibration. We found that the increased time to spicule protraction in the sleep mutants and after L4 vibration is suppressed by loss of nrx-1 (Figures 3C–3E). These results show that sleep deprivation impacts DVB-dependent behavior and indicate that neurite outgrowth induced by sleep loss has functional consequences. Further, we show that neurexin/nrx-1, which is required for sleep deprivation induced morphologic plasticity, is also required for behavioral plasticity.
DVB neurite outgrowth and behavior after sleep deprivation is dependent on nlg-1
To determine whether neurexin’s canonical binding partner, neuroligin/nlg-1, also contributes to sleep deprivation induced DVB plasticity and behavior, we tested the impact of a large deletion allele, nlg-1(ok259) (Figure 4A). nlg-1(ok259) males did not differ from controls in any measure of quiescence during the L4 molt or in activity/quiescence after the L4 molt (Figures S1A–S1E), therefore we did not assess the impact of nlg-1(ok259) on sleep deprivation. Total time quiescent was significantly lower in nlg-1(ok259) males compared to nrx-1(wy778) males (Figure S1B), potentially due to the reduction in activity of nrx-1(wy778) mutants. We found that increased DVB neurite length in npr-1 mutant males was suppressed by nlg-1(ok259) (Figures 4B–4D). npr-1(ad609); nlg-1(ok259) males and nlg-1(ok259) mutants alone were not significantly different from controls (Figures 4B–4D). nlg-1(ok259) mutation also suppressed the increased latency to spicule protraction observed in npr-1 single mutants (Figure 4E). These results demonstrate that nlg-1, like nrx-1, regulates both DVB structure and function in response to sleep deprivation without altering sleep.
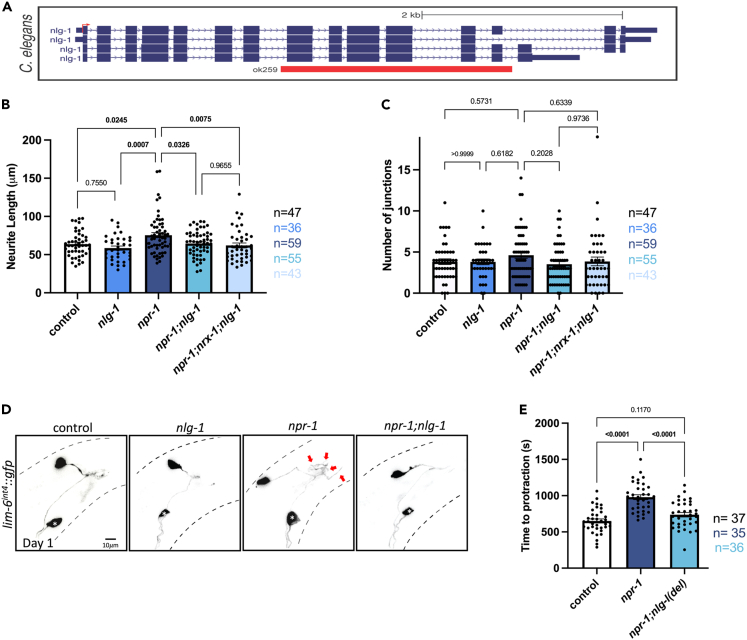
Figure 4.
nlg-1 is required for sleep deprivation induced DVB neurite outgrowth and spicule protraction behavioral changes
(A) Schematic showing nlg-1 C. elegans gene locus with nlg-1(ok259) deletion indicated with red box.
(B) Graph displaying quantification of DVB neurite length and (C) DVB junctions in controls, nlg-1, npr-1, npr-1; nlg-1, and npr-1; nrx-1; nlg-1 day 1 males.
(D) Representative images of DVB neuron in controls, nlg-1, npr-1, and npr-1; nlg-1 day 1 males. White asterisk marks PVT neuron (not traced, see64).
(E) Time to spicule protraction following aldicarb application of control, npr-1, and npr-1; nlg-1 day 1 males. Number of individual animals is indicated by “n”, p values from one-way ANOVA with Tukey’s post-hoc test shown, error bars represent SEM, scale bar size as labelled.
nrx-1 and nlg-1 may act in concert to control sleep deprivation induced plasticity of DVB in which case there would be no further suppression of the sleep deprivation induced phenotypes in males lacking both nrx-1 and nlg-1. DVB neuronal morphology in npr-1(ad609); nrx-1(wy778); nlg-1(ok259) triple mutants did not differ from npr-1(ad609) mutants with single mutations in either nrx-1 or nlg-1 (Figures 4B–4D). This result suggests that nrx-1 and nlg-1 act in the same molecular pathway to regulate sleep deprivation induced DVB morphologic plasticity. Another possibility is that npr-1; nrx-1; nlg-1 triple mutants do not show a further decease in neurite length due to a floor effect; however, we observe that DVB branching and length in these day 1 adults is higher than both L4 animals and hermaphrodites.64
DVB neurite outgrowth after developmental sleep deprivation is transient
DVB neurite outgrowth increases from day 1 to day 5 of adulthood and adolescent stress leads to lasting changes in DVB morphology at least to day 3 of adulthood.64,65 We asked if DVB neurite outgrowth at day 1 after sleep deprivation persists into later days of adulthood. Disruption of developmental sleep via aptf-1(tm3287) or npr-1(ad609) mutations, which increases neurite outgrowth at day 1, had no difference in DVB neurite outgrowth when analyzed at day 3 (Figures 5A–5E, 5H, and S3A–S3D). Additionally, neurite outgrowth induced by vibration, while different from controls at day 1, was indistinguishable from controls at day 3 of adulthood (Figures 5A, 5F–5H, S3E, and S3F). Thus, the DVB morphologic changes following developmental sleep deprivation observed at day 1 are transient and not long lasting, distinguishing sleep deprivation from other adolescent stressors, which have long-term changes in DVB neurite morphology.65
Figure 5.
DVB neurite outgrowth induced using genetic developmental sleep disruption is transient
(A) Schematic of experimental set-up for day 3 imaging of DVB neuron in sleep deprivation mutants, aptf-1 (orange) and npr-1(blue), and L4 vibration sleep deprived males (pink).
(B) Graph showing DVB neurite length and (C) junctions in day 3 males in controls and aptf-1 males.
(D) Day 3 DVB neurite length and (E) junctions in controls and npr-1 males.
(F) Graph of neurite length and (G) number of junctions in day 3 control males and males after application of vibration during L4 to adult molt.
(H) Representative images of day 3 DVB neurons in control, aptf-1, npr-1, and L4 vibrated males. The effect of nrx-1 and nlg-1 mutations on day 3 DVB neurite outgrowth was not tested as there was no difference between sleep deprived and control C. elegans. White asterisk marks PVT neuron (not traced, see64).Number of individual animals is indicated by “n”, p values from two-tailed unpaired t-test shown, error bars represent SEM, scale bar size as labelled.
Discussion
We previously described structural plasticity of the GABAergic neuron DVB in male C. elegans during the first 5 days of adulthood64 and altered plasticity following adolescent stress.65 Changes in DVB morphology and behavior are dependent on the single neurexin (nrx-1) and neuroligin (nlg-1) in C. elegans, two highly conserved synaptic adhesion molecules (SAMs) with links to autism.54,55,56 How sleep, structural neuroplasticity, autism genes, and behavior influence one another requires a system in which to simultaneously manipulate and analyze each interaction. Here, we disrupted developmental and adolescent sleep in C. elegans and measured the impact on the morphologic and behavioral plasticity of the DVB neuron.64,65 We found that all four methods of sleep disruption tested (encompassing genetic, physical, and chemo-genetic manipulations) altered plasticity of the DVB neuron, resulting in changes in morphology and behavior at day 1 of adulthood. Sleep disruption specifically during larval stage 4 induced neurite outgrowth and behavioral changes. Disruption of sleep before or after the L4 molt or in the L2 or L3 molt did not induce changes in DVB morphologic plasticity. Changes in plasticity have been previously shown to occur during the L4 period, including at GABAergic neuromuscular junctions in C. elegans, thus highlighting this as a particularly dynamic period of remodeling.91,103 Additionally, we found that the neuronal changes induced by sleep loss depend on nrx-1 and nlg-1, acting in the same molecular pathway. Neither nrx-1 nor nlg-1 impact the average time males spend quiescent during the L4 to adult molt, though sleep depth was not directly assessed.
We previously found that UV, heat-shock, and starvation during larval stage 4 affect DVB outgrowth at day 1, and that starvation stress depends on nrx-1 and nlg-1. We show that sleep deprivation impacts DVB morphologic and behavioral plasticity and that sleep deprivation induced plasticity changes rely on both nrx-1 and nlg-1, distinct from UV or heat-shock.65 The similarity in the mechanistic findings between sleep deprivation and starvation stress65 without further characterization beyond their dependence on nrx-1 and nlg-1 is a limitation of this work. Still, in striking contrast to other adolescent stressors that impact DVB morphology at day 3,65 sleep deprivation induced DVB morphologic changes are transient. Our results indicate that sleep (developmentally and during L4 lethargus) is an important factor in regulating structural plasticity and behavior and that autism-associated gene mutations alter the neuronal response to sleep loss at day 1 specifically.
Sleep in many animals can be identified and measured in part by a lack of activity and purposeful movement. Here we quantify sleep and the disruption of sleep using sequential lack of movement of animals. While we have attempted to specifically target and measure sleep with our methods, they are intimately tied together and influence sleep as well as movement of animals. Vibration during L4 sleep in unc-97 males did not impact DVB neurite length, which suggests that the movement induced during sleep disruption may be crucial for the impact on DVB. However, unc-97 mutants may have altered sleep on their own, which we cannot analyze with our methods, or may be frail in a way that changes the impact of vibration. The unc-97 males are sickly and may be slightly delayed, which could impact the ability to target the L4 period to induce sleep dependent neurite outgrowth in day 1 animals. We have shown comparable changes in DVB plasticity using four distinct and increasingly specific methods of sleep deprivation; however, we must consider that increased developmental or L4 specific activity during otherwise quiescent periods is involved.
Our findings lead us to a model in which overall DVB neurite outgrowth is determined by the early balance of DVB neurite extension and retraction, as supported by our previous work which shows these dynamic changes in DVB neurites (Figure 6A).64 Selective pruning and strengthening of synapses by neurons allows for tuning of neural circuits and can impact learning.45,104 During adulthood, projections to nonessential targets may form and get pruned back while projections to other targets, like SPC and spicule muscles, are stabilized or increased.64 In turn, males become more efficient at mating, likely reflecting improved coordination of spicule protraction. In support of this, day 1 males show aberrant spicule protractions during defecation that is ameliorated by day 3 due to increased projections and inhibition by DVB.64
Figure 6.
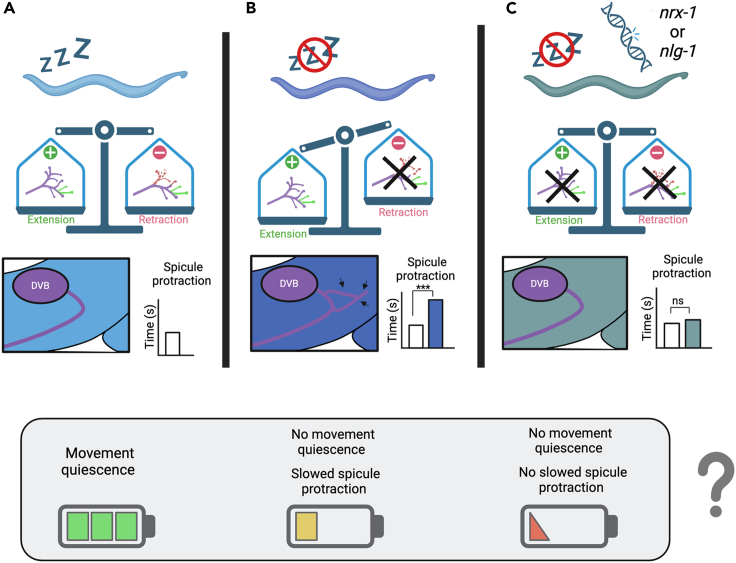
Model for sleep regulation of DVB neurite outgrowth and behavior
(A) We propose that DVB morphology in day 1 male C. elegans is determined by the balance of neurite extension and retraction (or pruning) after the L4 transition to adult. DVB neurite outgrowth leads to inhibition from DVB, which determines the time to spicule protraction. C. elegans with undisturbed sleep have a balance in extension and retraction of neurites. The movement quiescence during lethargus may result in conservation of energy.
(B) We suggest that DVB undergoes neurite extension without retraction in male C. elegans that experience sleep loss, with pruning being dependent on sleep, thus resulting in an imbalance between extension and retraction and an overall increase in DVB neurite outgrowth. The increase in DVB neurites results in an increase in DVB inhibition and a longer time to spicule protraction. While these males do not experience quiescence, therefore increasing energy expenditure, the increase in inhibition on spicule neurons and muscles, observed as slower spicule protraction, may conserve energy.
(C) In day 1 animals lacking sleep with mutations in nrx-1 or nlg-1, we hypothesize that sleep deprivation induced DVB neurite extension is not stabilized, leading to rebalance in extension and retraction, as both processes are affected (retraction impacted by lack of sleep). Therefore, DVB morphology is similar to controls and the time to spicule protraction is not increased. Lack of energy conservation due to impaired sleep and potential energy expenditure in spicule protraction neurons and muscles may lead to further loss of energy, and altered response to sleep deprivation may be detrimental.
The question of why animals sleep has yet to be answered; however, there are many supported hypotheses, including energy regulation105,106,107,108,109 and neural plasticity.25,26,27,28,38,39,40,110,111,112 Sleep deprived C. elegans undergo less movement quiescence89,90,92 and likely have increased energy expenditures during a typical period of energy conservation.99 One possible explanation for DVB structural changes in response to sleep deprivation may be to prevent circuit processes or behaviors that have higher energetic demands, such as spicule protraction. This may explain why paralyzed unc-97 vibration sleep deprived males, which lack muscle contraction and likely have less energetic demand, do not show differences in DVB morphology compared to control unc-97 males. Another proposed role for sleep is in synaptic downscaling, circuit consolidation, and other plasticity mechanisms.27,45,110,111,112 This role for sleep fits with our hypothesis that DVB morphology is determined by the balance of neurite extension and retraction, where animals with less sleep, and therefore decreased neurite retraction, have overall increased neurite outgrowth and synapses (Figure 6B).
We propose that DVB neurite length in animals lacking nrx-1 or nlg-1 at day 1 do not differ from controls, as processes which are not properly stabilized without these essential synaptic adhesion molecules are likely retracted in C. elegans during sleep. Our model predicts that in sleep deprived nrx-1 or nlg-1 mutant animals, neurites that would normally grow out in response to sleep loss do not do so as they may not be able to be stabilized (Figure 6C). This lack of DVB extension therefore prevents slowing of spicule protraction, potentially leading to excess energy drain or other consequences in nrx-1 or nlg-1 animals compared to controls. (Figure 6C). Therefore, the altered neuronal and behavioral changes after sleep loss in animals without nrx-1 or nlg-1 may have detrimental impacts.
Neurexin/NRXN1 and neuroligin/NLGN3 are both associated with autism and a number of other neurodevelopmental and neuropsychiatric disorders and conditions.55 Between 50 and 85% of individuals with autism report sleep problems compared to 12–40% in neurotypical individuals,21,23 underscoring the need for further research into the effect of sleep loss in this condition. Additionally, our identification of sexual maturation as critical for sleep deprivation induced neuroplasticity is particularly interesting given the many connections between neurological conditions and adolescence. In fact, individuals with autism have heightened responses to stress during adolescence and increased variability in cortisol levels, including increases at night.113 Increased stress reactivity in adolescence may increase the likelihood of comorbid anxiety and sleep disorders.113 Other neuropsychiatric conditions including schizophrenia (NRXN1 associated) have an average age of onset in mid to late adolescence or shortly after, suggesting selective vulnerability during this period. Further, the specific involvement of nrx-1 alpha in regulating structural response to sleep deprivation is particularly relevant as most human NRXN1 mutations affect the long (alpha) isoform.114
There is clear evidence for a relationship between sleep and autism; however, the directionality between these two is uncertain. Sleep loss may modify characteristic behaviors of autism (social impairment, repetitive and fixated behavior) and, concurrently, sleep problems may be intensified by alterations in anxiety, sensory sensitivity or integration,.16,17,18,19,20,21,22,23,24 Our results identify autism-associated SAM genes that contribute to the response of neuronal circuits and behavior to sleep deprivation through modification of plasticity mechanisms. Therefore, we may expect altered neuronal response to sleep loss in individuals with variants in SAM genes, leading to altered or additional behavioral changes. This is supported by evidence that sleep interventions can modify behavior in autism.23,24 Our findings present a model system in which to continue studying the directional relationships between sleep, autism genes, and relevant behaviors.
Here, we provide insights into the structural and functional effects of sleep deprivation on neurons that depend on the autism-associated genes neurexin and neuroligin. This work overcomes numerous methodological caveats in the study of sleep dependent plasticity in more complex systems by using four specific and complementary methods to disrupt sleep in C. elegans. Additionally, our results integrate analysis of sleep dependent plasticity at every level, including neurons, synapses, and behaviors. These findings highlight the importance of conserved SAMs in the regulation of sleep dependent structural neuroplasticity and the behavioral output of a single neuron. This work provides examination, at an unprecedented resolution, into the interplay of sleep, structural plasticity, neuronal function, and behavior, with potential translational relevance for the impact of sleep disruption in neurodevelopmental conditions.
Limitations of the study
We find that early adulthood DVB neuronal and behavioral plasticity is impacted by levels of adolescent sleep, and in part these changes are mediated by neurexin and neuroligin. We recognize that this is highly similar to our previous findings with other adolescent stressors, and we do not define where or when these genes are acting for sleep disruption induced changes. We have yet to define how sleep levels are signaled to the DVB neuron, which is important to truly understand how sleep links to plasticity and the function of these genes. Future work is needed to fully disentangle whether or how much the changes in DVB neuron plasticity we observe are due to disruption of sleep or the increase in activity levels that results from all of our sleep disruption methods.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| ALDICARB PESTANAL, ANALYTICAL | EMSCO/FISHER | Cat#111019528 |
| HISTAMINE DIHYDROCHLORIDE | SIGMA | Cat#H7250 |
| Deposited data | ||
| Data included in 'Structural neuroplasticity after sleep loss modifies behavior and requires neurexin and neuroligin' | Digital Commons Data | https://doi.org/10.17632/d74xzc9kz7.1 |
| Experimental models: Organisms/strains | ||
| C. elegans: him-8(e1489) IV; otIs525[lim-6int4::gfp] | Hart & Hobert 2018 | OH15098 |
| C. elegans: aptf-1(tm3287) II; him-8(e1489) IV; otIs525[lim-6int4::gfp] | this study | MPH35 |
| C. elegans: aptf-1(tm3287)II; unc-119(ed3)III; nrx-1(wy778[unc-119(+)]III)V; him-8(e1489)IV; otIs525[lim-6int4::gfp] | this study | MPH36 |
| C. elegans: unc-119(ed3) III; nrx-1(wy778[unc-119(+)] III) V; him-8(e1489) IV; otIs525[lim-6int4::gfp] | Hart & Hobert 2018 | OH15111 |
| C. elegans: him-8(e1489) IV; otIs525[lim-6int4::gfp]; hmpEx1[lim-6int4::cla-1::gfp, myo-2p::mCherry] | Hart 2019 | MPH1 |
| C. elegans: aptf-1(tm3287) II; him-8(e1489) IV; otIs541[lim-6int4::wCherry]; hmpEx1[lim-6int4::cla-1::gfp,myo-2p::mCherry] | this study | MPH37 |
| C. elegans: npr-1(ad609) II; him-8(e1489) IV; otIs541[lim-6int4::wCherry]; hmpEx1[lim-6int4::cla-1::gfp,myo-2p::mCherry] | this study | MPH38 |
| C. elegans: npr-1(ad609) X;him-8(e1489) IV; otIs525[lim-6int4::gfp] | this study | MPH39 |
| C. elegans: npr-1(ad609) X; unc-119(ed3); nrx-1(wy778[unc-119(+)III]) V; him-8(e1489) IV; otIs525[lim-6int4::gfp] | this study | MPH40 |
| C. elegans: nrx-1(gk246237) V; him-8(e1489) IV; otIs525[lim-6int4::gfp] | Hart & Hobert 2018 | OH15116 |
| C. elegans: him-8(e1489) IV; otIs525[lim-6int4::gfp]; qnEx643[flp-11p::HisCl::SL2::mCherry, myo-2p::mCherry] | this study | MPH41 |
| C. elegans: unc-119(ed3) III; nrx-1((wy778[unc-119(+)III] V); him-8(e1489) IV; otIs525[lim-6int4::gfp]; qnEx643[flp-11p::HisCl::SL2::mCherry, myo-2p::mCherry] | this study | MPH42 |
| C. elegans: nlg-1(ok259) X; him-8(e1489) IV; otIs525[lim-6int4::gfp] | Hart 2019 | MPH3 |
| C. elegans: npr-1(ad609)X;nlg-1(ok259)X;him-8(e1489) IV; otIs525[lim-6int4::gfp] | this study | MPH43 |
| C. elegans: npr-1(ad609) X; unc-119(ed3)III; nrx-1(wy778[unc-119(+)III])V; nlg-1(ok259) X; him-8(e1489) IV; otIs525[lim-6int4::gfp] | this study | MPH44 |
| C. elegans: unc-97(su110); him-5(e1490); otIs541[lim-6int4::wCherry] | Hart & Hobert 2018 | OH15107 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael P. Hart (hartmic@pennmedicine.upenn.edu).
Materials availability
All strains can be requested by contacting Michael P. Hart (hartmic@pennmedicine.upenn.edu). List of strains is located in the key resources table.
Data and code availability
-
•
All data have been deposited in Mendeley and are publicly available as of the date of publication. Mendeley Data: https://doi.org/10.17632/d74xzc9kz7.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
C. elegans strain maintenance
Male C. elegans were used for this work. All strains tested had a him mutation (him-8 or him-5) to increase the proportion of male C. elegans. All strains were group-housed and maintained at room temperature on NGM-agar filled plates and seeded with OP50 bacteria as a food source. Experiments were performed only on males since DVB adult structural plasticity occurs only in males.64 Hermaphroditic (self-reproducing female) C. elegans were utilized for crosses and strain propagation/maintenance. Transgenic worms used in this study were previously published and either existed in the lab or were provided by others.65,99 Strains, mutant alleles, and transgenics included are listed in key resources table by figure order. Developmental stage of animals is listed for each experiment. Briefly, sleep deprivation occurred throughout development, during Larval Stage 2, 3 or 4 and imaging was performed when males were day 1 or day 3 adults as indicated.
Method details
Confocal microscopy
For day 1 experiments, C. elegans were picked onto new plates at larval stage 4 based on morphology and size, then subjected to confocal imaging the following day (day 1 adult) or three days later (day 3 adults). Males were imaged on a Leica SP8 point scanning confocal microscope using a 63x objective with glycerol immersion fluid (type G). Worms were mounted on an agar pad made of 5% agarose and paralyzed in a 100mM sodium azide solution in M9. The DVB neuron was imaged at 2x zoom. Z-stacks were set such that imaging spanned the entire neuron at a slice size of 0.6μM. Z-stacks were approximately 30-75 slices. Representative image figures were made using FIJI image J and Adobe Illustrator.
Quiescence and activity analysis
Quiescence and activity were monitored using the WormMotel set-up developed by Tau Scientific. The center 24 wells of a 48-well grid WorMotel were filled with NGM and 1.5uL of concentrated OP50 bacteria was pipetted on top of each well. Once the food was dry, individual L4 males were placed on each well. WorMotels were run overnight for a recording period of sixteen hours with a picture taken every ten seconds. A MATLAB GUI76 was used to apply pixel subtraction between images to determine activity and quiescence of worms.
For quiescence, the peak quiescent period (highest value) was identified and the two-hours before and two-hours after were analyzed in ten-minute bins. An individual ten-minute bin was called as quiescent if it contained a value higher than 2.5. Number of quiescent bins was calculated and plotted in GraphPad Prism 9. We additionally plotted total time quiescent in minutes for each genotype by summing the values for each quiescent bin.
For graphs showing the quiescent time per bin, the peak quiescence was aligned and the average time quiescent and standard error of the mean per ten-minute bin were plotted in R studio for a time period spanning from 0 to 300 minutes for each genotype.
To compare activity post-molt, activity was analyzed two hours after the end of quiescence. If the molt occurred too late in our recording, activity could not be assessed. Activity was averaged across two hours.
To assess number of post-molt quiescent bins, we again assessed data two hours following the end of quiescence. Following those two hours, we determined how many ten-minute bins were spent quiescent as defined above. Total number of quiescent bins were quantified for a period of two hours.
Quiescence- neurite length Correlation
Larval stage 4 control and aptf-1(tm3287) were placed on a WorMotel set-up (above). The following day, males were recovered from the WorMotel and individually imaged following confocal microscopy methodology (see above). DVB neuron morphology was traced using FIJI and number of quiescent bins for each individual C. elegans was calculated as described. Data were plotted in GraphPad Prism 9.
DVB neuron morphology analysis
DVB neuron morphology analysis was performed in FIJI image J, blinded to genotype. Analysis was performed using the Simple Neurite Tracer or updated SNT Neuroanatomy plugin. Neurites were traced starting at the center of the DVB soma and extending to the longest point of neurite extension through the Z-stacks.64,65 Branches were then traced with the start site connected to the existing trace.64,65 All posteriorly extending branches of DVB were traced to the anterior turn of DVB on the ventral side. Total neurite length was determined using the Analyze Skeleton function. Data were graphed in GraphPad Prism 9.
DVB cla-1::gfp pre-synaptic puncta analysis
Puncta in control, aptf-1, and npr-1 mutant day 1 males were analyzed using the FIJI Analyze Particle function, which allows for unbiased quantification of cla-1::gfp puncta. lim-6int4p::cla-1::gfp transgenic C. elegans were crossed with the otIs541 (lim-6int4p::wCherry) strain and imaged at 63X on the Leica SP8 point scanning Confocal Microscope with an additional zoom of 2.5x. Z-stacks were set as described above. Lightning was applied to enhance resolution. Lightning deconvolution applies an appropriate reconstruction strategy to individual voxels rather than using a single parameter on the whole image to extract detailed image information.115 Micrographs were converted to 8-bit and auto-thresholded. If auto-thresholding was not representative of cla-1::gfp puncta, minor manual adjustment was made to match original images most closely. A region of interest (ROI) was defined to restrict puncta counts to DVB neurites (exclusion of soma and PVT neuron). Analyze Particles generated cla-1::gfp counts and bare outline images for the ROI. Pixel size of cla-1::gfp puncta was restrained to 0.01 pixels to exclude noise and circularity was not restricted. Data were graphed in GraphPad Prism 9. Traces for representative images were made in Adobe Illustrator and bare outlines of cla-1::gfp puncta were super-imposed on traces.
Vibration sleep deprivation
L4: Mid Larval stage 4 males, which were identified based on tail-tip retraction and morphology, were transferred to new NGM plates coated with OP50 bacteria. All animals were picked between molts based on movement and pharyngeal pumping behaviors and lack of molting characteristics. Plates housing the worms were then placed inside the WormWatcher platform developed by Tau Scientifics.100 One-second-long 8,000-12,000 Hz vibration was applied every ten minutes for a duration of six hours across the L4 molt. Adults were imaged for DVB morphology as described above. For vibration experiments before the L4 molt, worms picked as L3s based on morphology and stimulus was applied four hours later. For vibration experiments after the L4 molt, worms were picked as early L4s and vibration was applied ten hours later when the animals were early adults.
L2/L3: Larval stage 2 or 3 C. elegans identified based on size in comparison with L2 and L3 hermaphrodite siblings (identified using size and vulva morphology) underwent vibration as described above. The following day, males from these plates were removed as L4s or adults, respectively. Both groups were imaged as day 1 adults. As we cannot distinguish between males and hermaphrodites at L2 and L3, some hermaphrodites were present during the vibration protocol; however, L2 males were transferred to a new plate at L4 prior to sexual maturation and therefore unable to mate.
Histamine silencing of RIS neurons
To silence RIS, transgenic worms expressing a histamine gated chloride channel in RIS (NQ1208, flp-11p::HisCl::SL2::mCherry) were used.99 Males with this transgene were identified based on expression of the co-injection marker myo-2p::mCherry and picked at larval stage 4. L4 C. elegans were placed on top of an individual NGM-filled well of a WorMotel76,100,. 1.5μL of 10mM histamine in water was applied to the surface of WorMotel wells filled with NGM agar. Following an incubation period of one hour to allow the histamine to diffuse into the agar, L4 males were transferred to the WorMotel with food (OP50) spread on each well. WorMotels were imaged using WormWatcher Platforms (Tau Scientific) with a picture taken every ten seconds for sixteen hours. Pixel subtraction between images was performed using MATLAB to determine activity and quiescence of worms. Day 1 adults were subjected to confocal microscopy to measure DVB morphology as described above.
Aldicarb spicule protraction assay
Larval stage 4 males of each tested genotype/condition were transferred to a new NGM plate coated with OP50. 8-15 worms were placed on each plate. For vibration sleep deprivation, worms were placed in a WormWatcher and subjected to the above vibration protocol. Males were tested the following day (day 1 adults) using an aldicarb spicule protraction assay.64 One hour prior to experiments, 130μL of 100mM aldicarb were applied to empty NGM plate, without OP50, and evenly spread across the plate. Aldicarb was allowed to absorb into the plate and dry for one hour. Following transfer to the aldicarb assay plate, we measured the latency to spicule protraction lasting at least five seconds, blinded to genotype. Males were removed from the plate after spicule protraction. Time to protraction was plotted in GraphPad Prism 9.
Quantification and statistical analysis
Number of worms used in each experiment were based on previous studies and effect size.64,65 Each experiment was performed with at least 3 independent replicates, with each trial performed on a different day alongside matched controls. Experimenter was blinded to genotype for all experiments. Animals were randomized by location on WorMotel device for sleep quantification. Data were excluded if males left the WorMotel during experiment. All data were plotted and analyzed in GraphPad Prism 9. To determine statistical significance, we used one-way ANOVA with Tukey’s post-hoc test. For comparisons of two data sets, a two-tailed unpaired t-test was used to compare significance. For the experiment in which neurite length and number of quiescent bins of individual animals were correlated, a line of best-fit was generated in Prism and R2 values were used to assess linearity. “n” represents the number of individual worms tested and is indicated in each figure. Error bars on figures represent standard error of the mean (SEM). P-values are shown in each figure. Bolded p-values indicate significance (P<0.05).
Acknowledgments
We thank the members of the Hart lab for technical assistance and intellectual input on this project. The authors thank all C. elegans labs at the University of Pennsylvania, including the labs of Meera Sundaram, John Murray, Chris Fang-Yen, and Colin Conine for providing feedback on this project. We express appreciation for Anthony D. Fouad from Tau Scientific for technical support on the WormWatcher Platforms. Figures were generated in part with BioRender.com. Some strains were provided by the CGC, funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by an SFARI Bridge to Independence Award (MPH), the Autism Spectrum Program of Excellence at the Perelman School of Medicine, the National Science Foundation Graduate Research Fellowship Program (M.H.C.), the 2020 Hearst Foundation Fellowship (MHC), and by NINDS (1R01NS129736(MPH), R01NS107969(D.M.R.), and R01NS122779(D.M.R.)) of the NIH.
Author contributions
M.H.C., D.M.R., and M.P.H. developed the experiments for this study. M.H.C. conducted all imaging and behavioral experiments apart from vibration stimulus performed by M.P.H. M.H.C. performed all analysis for this project and created all data figures. M.H.C., D.M.R., and M.P.H. wrote, reviewed, revised, and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 11, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109477.
Supplemental information
References
- 1.Maquet P. The Role of Sleep in Learning and Memory. Science. 2001;294:1048–1052. doi: 10.1126/science.1062856. [DOI] [PubMed] [Google Scholar]
- 2.Xie L., Kang H., Xu Q., Chen M.J., Liao Y., Thiyagarajan M., O’Donnell J., Christensen D.J., Nicholson C., Iliff J.J., et al. Sleep Drives Metabolite Clearance from the Adult Brain. Science. 2013;342:373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobson J.A. Sleep is of the brain, by the brain and for the brain. Nature. 2005;437:1254–1256. doi: 10.1038/nature04283. [DOI] [PubMed] [Google Scholar]
- 4.Nollet M., Wisden W., Franks N.P. Sleep deprivation and stress: a reciprocal relationship. Interface Focus. 2020;10 doi: 10.1098/rsfs.2019.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Killgore W.D.S. In: Progress in Brain Research. Kerkhof G.A., van Dongen H.P.A., editors. Elsevier; 2010. Effects of sleep deprivation on cognition; pp. 105–129. [DOI] [PubMed] [Google Scholar]
- 6.Fifel K., Videnovic A. Circadian and Sleep Dysfunctions in Neurodegenerative Disorders—An Update. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.627330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra R.K. Neurodegenerative Disorders and Sleep. Sleep Med. Clin. 2018;13:63–70. doi: 10.1016/j.jsmc.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Mims K.N., Kirsch D. Sleep and Stroke. Sleep Med. Clin. 2016;11:39–51. doi: 10.1016/j.jsmc.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Hermann D.M., Bassetti C.L. Role of sleep-disordered breathing and sleep-wake disturbances for stroke and stroke recovery. Neurology. 2016;87:1407–1416. doi: 10.1212/WNL.0000000000003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musiek E.S., Holtzman D.M. Mechanisms Linking Circadian Clocks, Sleep, and Neurodegeneration. Science. 2016;354:1004–1008. doi: 10.1126/science.aah4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrest M.P., Parnell E., Penzes P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 2018;19:215–234. doi: 10.1038/nrn.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baglioni C., Nanovska S., Regen W., Spiegelhalder K., Feige B., Nissen C., Reynolds C.F., Riemann D. Sleep and mental disorders: A meta-analysis of polysomnographic research. Psychol. Bull. 2016;142:969–990. doi: 10.1037/bul0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernardinelli Y., Nikonenko I., Muller D. Structural plasticity: mechanisms and contribution to developmental psychiatric disorders. Front. Neuroanat. 2014;8:123. doi: 10.3389/fnana.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheetham C., Finnerty G. Plasticity and its Role in Neurological Diseases of the Adult Nervous System. Adv. Clin. Neurosci. Rehabil. 2007;7:8–9. [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey A.G., Murray G., Chandler R.A., Soehner A. Sleep Disturbance as Transdiagnostic: Consideration of Neurobiological Mechanisms. Clin. Psychol. Rev. 2011;31:225–235. doi: 10.1016/j.cpr.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson K.P., Zarrinnegar P. Autism Spectrum Disorder and Sleep. Child Adolesc. Psychiatr. Clin. N. Am. 2021;30:195–208. doi: 10.1016/j.chc.2020.08.012. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds A.M., Malow B.A. Sleep and Autism Spectrum Disorders. Pediatr. Clin. 2011;58:685–698. doi: 10.1016/j.pcl.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Accardo J.A., Malow B.A. Sleep, epilepsy, and autism. Epilepsy Behav. 2015;47:202–206. doi: 10.1016/j.yebeh.2014.09.081. [DOI] [PubMed] [Google Scholar]
- 19.Ballester P., Richdale A.L., Baker E.K., Peiró A.M. Sleep in autism: A biomolecular approach to aetiology and treatment. Sleep Med. Rev. 2020;54 doi: 10.1016/j.smrv.2020.101357. [DOI] [PubMed] [Google Scholar]
- 20.Cuomo B.M., Vaz S., Lee E.A.L., Thompson C., Rogerson J.M., Falkmer T. Effectiveness of Sleep-Based Interventions for Children with Autism Spectrum Disorder: A Meta-Synthesis. Pharmacotherapy. 2017;37:555–578. doi: 10.1002/phar.1920. [DOI] [PubMed] [Google Scholar]
- 21.Elkhatib Smidt S.D., Ghorai A., Taylor S.C., Gehringer B.N., Dow H.C., Langer A., Rawot E., Zhang J., Mitchell J.A., Rader D.J., et al. The relationship between autism spectrum and sleep–wake traits. Autism Res. 2022;15:641–652. doi: 10.1002/aur.2660. [DOI] [PubMed] [Google Scholar]
- 22.Hodge D., Carollo T.M., Lewin M., Hoffman C.D., Sweeney D.P. Sleep patterns in children with and without autism spectrum disorders: Developmental comparisons. Res. Dev. Disabil. 2014;35:1631–1638. doi: 10.1016/j.ridd.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Doldur-Balli F., Imamura T., Veatch O.J., Gong N.N., Lim D.C., Hart M.P., Abel T., Kayser M.S., Brodkin E.S., Pack A.I. Synaptic dysfunction connects autism spectrum disorder and sleep disturbances: A perspective from studies in model organisms. Sleep Med. Rev. 2022;62 doi: 10.1016/j.smrv.2022.101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tordjman S., Najjar I., Bellissant E., Anderson G.M., Barburoth M., Cohen D., Jaafari N., Schischmanoff O., Fagard R., Lagdas E., et al. Advances in the research of melatonin in autism spectrum disorders: literature review and new perspectives. Int. J. Mol. Sci. 2013;14:20508–20542. doi: 10.3390/ijms141020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seibt J., Frank M.G. Primed to Sleep: The Dynamics of Synaptic Plasticity Across Brain States. Front. Syst. Neurosci. 2019;13 doi: 10.3389/fnsys.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raven F., Van Der Zee E.A., Meerlo P., Havekes R. The role of sleep in regulating structural plasticity and synaptic strength: Implications for memory and cognitive function. Sleep Med. Rev. 2018;39:3–11. doi: 10.1016/j.smrv.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Areal C.C., Warby S.C., Mongrain V. Sleep loss and structural plasticity. Curr. Opin. Neurobiol. 2017;44:1–7. doi: 10.1016/j.conb.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Appelbaum L., Wang G., Yokogawa T., Skariah G.M., Smith S.J., Mourrain P., Mignot E. Circadian and Homeostatic Regulation of Structural Synaptic Plasticity in Hypocretin Neurons. Neuron. 2010;68:87–98. doi: 10.1016/j.neuron.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandra R., Farah F., Muñoz-Lobato F., Bokka A., Benedetti K.L., Brueggemann C., Saifuddin M.F.A., Miller J.M., Li J., Chang E., et al. Sleep is required to consolidate odor memory and remodel olfactory synapses. Cell. 2023;186:2911–2928.e20. doi: 10.1016/j.cell.2023.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin A., Chen M.-Y., Kirszenblat L., Reinhard J., van Swinderen B., Claudianos C. Neurexin-1 regulates sleep and synaptic plasticity in Drosophila melanogaster. Eur. J. Neurosci. 2015;42:2455–2466. doi: 10.1111/ejn.13023. [DOI] [PubMed] [Google Scholar]
- 31.Kayser M.S., Mainwaring B., Yue Z., Sehgal A. Sleep deprivation suppresses aggression in Drosophila. Elife. 2015;4 doi: 10.7554/eLife.07643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donlea J.M., Ramanan N., Shaw P.J. Use-Dependent Plasticity in Clock Neurons Regulates Sleep Need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donlea J.M., Pimentel D., Miesenböck G. Neuronal Machinery of Sleep Homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss J.T., Donlea J.M. Sleep deprivation results in diverse patterns of synaptic scaling across the Drosophila mushroom bodies. Curr. Biol. 2021;31:3248–3261.e3. doi: 10.1016/j.cub.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altenhofen S., Bonan C.D. Zebrafish as a Tool in the Study of Sleep and Memory-related Disorders. Curr. Neuropharmacol. 2022;20:540–549. doi: 10.2174/1570159X19666210712141041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro-da-Silva J., Tran S., Luchiari A.C. Sleep deprivation impairs cognitive performance in zebrafish: A matter of fact? Behav. Process. 2018;157:656–663. doi: 10.1016/j.beproc.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Elbaz I., Zada D., Tovin A., Braun T., Lerer-Goldshtein T., Wang G., Mourrain P., Appelbaum L. Sleep-Dependent Structural Synaptic Plasticity of Inhibitory Synapses in the Dendrites of Hypocretin/Orexin Neurons. Mol. Neurobiol. 2017;54:6581–6597. doi: 10.1007/s12035-016-0175-x. [DOI] [PubMed] [Google Scholar]
- 38.Havekes R., Park A.J., Tudor J.C., Luczak V.G., Hansen R.T., Ferri S.L., Bruinenberg V.M., Poplawski S.G., Day J.P., Aton S.J., et al. Sleep deprivation causes memory deficits by negatively impacting neuronal connectivity in hippocampal area CA1. Elife. 2016;5 doi: 10.7554/eLife.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havekes R., Aton S.J. Impacts of Sleep Loss versus Waking Experience on Brain Plasticity: Parallel or Orthogonal? Trends Neurosci. 2020;43:385–393. doi: 10.1016/j.tins.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L., Zhou H., Cichon J., Yang G. Experience and sleep-dependent synaptic plasticity: from structure to activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375 doi: 10.1098/rstb.2019.0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spano G.M., Banningh S.W., Marshall W., de Vivo L., Bellesi M., Loschky S.S., Tononi G., Cirelli C. Sleep Deprivation by Exposure to Novel Objects Increases Synapse Density and Axon–Spine Interface in the Hippocampal CA1 Region of Adolescent Mice. J. Neurosci. 2019;39:6613–6625. doi: 10.1523/JNEUROSCI.0380-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang G., Gan W.-B. Sleep contributes to dendritic spine formation and elimination in the developing mouse somatosensory cortex. Dev. Neurobiol. 2012;72:1391–1398. doi: 10.1002/dneu.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G., Lai C.S.W., Cichon J., Ma L., Li W., Gan W.-B. Sleep promotes branch-specific formation of dendritic spines after learning. Science. 2014;344:1173–1178. doi: 10.1126/science.1249098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vecsey C.G., Baillie G.S., Jaganath D., Havekes R., Daniels A., Wimmer M., Huang T., Brown K.M., Li X.-Y., Descalzi G., et al. Sleep deprivation impairs cAMP signalling in the hippocampus. Nature. 2009;461:1122–1125. doi: 10.1038/nature08488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Ma L., Yang G., Gan W.-B. REM sleep selectively prunes and maintains new synapses in development and learning. Nat. Neurosci. 2017;20:427–437. doi: 10.1038/nn.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamoto H., Hensch T.K. Bidirectional interaction of sleep and synaptic plasticity: A view from visual cortex. Sleep Biol. Rhythm. 2006;4:35–43. doi: 10.1111/j.1479-8425.2006.00204.x. [DOI] [Google Scholar]
- 47.Mohmed Nor N.S., Nawi A., Wan Ahmad W.A.N., Noordin L. Sleep Deprivation Models in Rodents. J. Sains Kesihatan Malaysia. 2021;19:29–34. doi: 10.17576/jskm-2021-1902-04. [DOI] [Google Scholar]
- 48.Frank M.G., Cantera R. Sleep, clocks, and synaptic plasticity. Trends Neurosci. 2014;37:491–501. doi: 10.1016/j.tins.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Helou J., Bélanger-Nelson E., Freyburger M., Dorsaz S., Curie T., La Spada F., Gaudreault P.-O., Beaumont É., Pouliot P., Lesage F., et al. Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. USA. 2013;110:9974–9979. doi: 10.1073/pnas.1221381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freyburger M., Pierre A., Paquette G., Bélanger-Nelson E., Bedont J., Gaudreault P.-O., Drolet G., Laforest S., Blackshaw S., Cermakian N., et al. EphA4 is Involved in Sleep Regulation but Not in the Electrophysiological Response to Sleep Deprivation. Sleep. 2016;39:613–624. doi: 10.5665/sleep.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Callaghan E.K., Ballester Roig M.N., Mongrain V. Cell adhesion molecules and sleep. Neurosci. Res. 2017;116:29–38. doi: 10.1016/j.neures.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Porter N.M., Bohannon J.H., Curran-Rauhut M., Buechel H.M., Dowling A.L.S., Brewer L.D., Popovic J., Thibault V., Kraner S.D., Chen K.C., Blalock E.M. Hippocampal CA1 Transcriptional Profile of Sleep Deprivation: Relation to Aging and Stress. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massart R., Freyburger M., Suderman M., Paquet J., El Helou J., Belanger-Nelson E., Rachalski A., Koumar O.C., Carrier J., Szyf M., Mongrain V. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl. Psychiatry. 2014;4:e347. doi: 10.1038/tp.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betancur C., Sakurai T., Buxbaum J.D. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.SFARI . SFARI; 2017. SFARI Gene.https://www.sfari.org/resource/sfari-gene/ [Google Scholar]
- 56.Südhof T.C. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L.Y., Jiang M., Zhang B., Gokce O., Südhof T.C. Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins. Neuron. 2017;94:611–625.e4. doi: 10.1016/j.neuron.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai J., Aoto J., Südhof T.C. Alternative Splicing of Presynaptic Neurexins Differentially Controls Postsynaptic NMDA and AMPA Receptor Responses. Neuron. 2019;102:993–1008.e5. doi: 10.1016/j.neuron.2019.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flaherty E., Zhu S., Barretto N., Cheng E., Deans P.J.M., Fernando M.B., Schrode N., Francoeur N., Antoine A., Alganem K., et al. Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat. Genet. 2019;51:1679–1690. doi: 10.1038/s41588-019-0539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuccillo M.V., Földy C., Gökce Ö., Rothwell P.E., Sun G.L., Malenka R.C., Südhof T.C. Single-Cell mRNA Profiling Reveals Cell-Type Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuccillo M.V., Pak C. Copy number variants in neurexin genes: phenotypes and mechanisms. Curr. Opin. Genet. Dev. 2021;68:64–70. doi: 10.1016/j.gde.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu B., Ho Y., Fasolino M., Medina J., O’Brien W.T., Lamonica J.M., Nugent E., Brodkin E.S., Fuccillo M.V., Bucan M., Zhou Z. Allelic contribution of Nrxn1α to autism-relevant behavioral phenotypes in mice. PLoS Genet. 2023;19 doi: 10.1371/journal.pgen.1010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu Y., Huang Z.J. Differential dynamics and activity-dependent regulation of α- and β-neurexins at developing GABAergic synapses. Proc. Natl. Acad. Sci. USA. 2010;107:22699–22704. doi: 10.1073/pnas.1011233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hart M.P., Hobert O. Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature. 2018;553:165–170. doi: 10.1038/nature25192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hart M.P. Stress-Induced Neuron Remodeling Reveals Differential Interplay Between Neurexin and Environmental Factors in Caenorhabditis elegans. Genetics. 2019;213:1415–1430. doi: 10.1534/genetics.119.302415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brenner S. The Genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cook S.J., Jarrell T.A., Brittin C.A., Wang Y., Bloniarz A.E., Yakovlev M.A., Nguyen K.C.Q., Tang L.T.-H., Bayer E.A., Duerr J.S., et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571:63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang H., Singh K., Hart A.C. Measuring Caenorhabditis elegans Sleep During the Transition to Adulthood Using a Microfluidics-based System. Bio. Protoc. 2017;7 doi: 10.21769/BioProtoc.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lawler D.E., Albrecht D.R. Monitoring neural activity during sleep/wake events in adult C. elegans by automated sleep detection and stimulation. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawler D.E., Chew Y.L., Hawk J.D., Aljobeh A., Schafer W.R., Albrecht D.R. Sleep Analysis in Adult C. elegans Reveals State-Dependent Alteration of Neural and Behavioral Responses. J. Neurosci. 2021;41:1892–1907. doi: 10.1523/JNEUROSCI.1701-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urmersbach B., Besseling J., Spies J.-P., Bringmann H. Automated analysis of sleep control via a single neuron active at sleep onset in C. elegans. Genesis. 2016;54:212–219. doi: 10.1002/dvg.22924. [DOI] [PubMed] [Google Scholar]
- 73.Spies J., Bringmann H. Automated detection and manipulation of sleep in C. elegans reveals depolarization of a sleep-active neuron during mechanical stimulation-induced sleep deprivation. Sci. Rep. 2018;8:9732. doi: 10.1038/s41598-018-28095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cho J.Y., Sternberg P.W. Multilevel Modulation of a Sensory Motor Circuit during C. elegans Sleep and Arousal. Cell. 2014;156:249–260. doi: 10.1016/j.cell.2013.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyazaki S., Kawano T., Yanagisawa M., Hayashi Y. Intracellular Ca2+ dynamics in the ALA neuron reflect sleep pressure and regulate sleep in Caenorhabditis elegans. iScience. 2022;25 doi: 10.1016/j.isci.2022.104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Churgin M.A., Szuperak M., Davis K.C., Raizen D.M., Fang-Yen C., Kayser M.S. Quantitative Imaging of Sleep Behavior in Caenorhabditis elegans and larval Drosophila melanogaster. Nat. Protoc. 2019;14:1455–1488. doi: 10.1038/s41596-019-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang H., Zhu C.-T., Skuja L.L., Hayden D.J., Hart A.C. Genome-Wide Screen for Genes Involved in Caenorhabditis elegans Developmentally Timed Sleep. G3 (Bethesda) 2017;7:2907–2917. doi: 10.1534/g3.117.300071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belfer S.J., Chuang H.-S., Freedman B.L., Yuan J., Norton M., Bau H.H., Raizen D.M. Caenorhabditis-in-Drop Array for Monitoring C. elegans Quiescent Behavior. Sleep. 2013;36:689–698. doi: 10.5665/sleep.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trojanowski N.F., Nelson M.D., Flavell S.W., Fang-Yen C., Raizen D.M. Distinct Mechanisms Underlie Quiescence during Two Caenorhabditis elegans Sleep-Like States. J. Neurosci. 2015;35:14571–14584. doi: 10.1523/JNEUROSCI.1369-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trojanowski N.F., Raizen D.M. Call it Worm Sleep. Trends Neurosci. 2016;39:54–62. doi: 10.1016/j.tins.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nelson M.D., Raizen D.M. A sleep state during C. elegans development. Curr. Opin. Neurobiol. 2013;23:824–830. doi: 10.1016/j.conb.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raizen D.M., Zimmerman J.E., Maycock M.H., Ta U.D., You Y.j., Sundaram M.V., Pack A.I. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 83.Singh K., Huang H., Hart A.C. Do C. elegans Sleep? A Closer Look. Sleep. 2013;36:307–308. doi: 10.5665/sleep.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busack I., Bringmann H. A sleep-active neuron can promote survival while sleep behavior is disturbed. PLoS Genet. 2023;19 doi: 10.1371/journal.pgen.1010665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nichols A.L.A., Eichler T., Latham R., Zimmer M. A global brain state underlies C. elegans sleep behavior. Science. 2017;356 doi: 10.1126/science.aam6851. [DOI] [PubMed] [Google Scholar]
- 86.Schwarz J., Lewandrowski I., Bringmann H. Reduced activity of a sensory neuron during a sleep-like state in Caenorhabditis elegans. Curr. Biol. 2011;21:R983–R984. doi: 10.1016/j.cub.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 87.Bringmann H. Sleep-Active Neurons: Conserved Motors of Sleep. Genetics. 2018;208:1279–1289. doi: 10.1534/genetics.117.300521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turek M., Besseling J., Spies J.-P., König S., Bringmann H. Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife. 2016;5 doi: 10.7554/eLife.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson B., Goetting D.L., Cisneros Desir J., Van Buskirk C. aptf-1 mutants are primarily defective in head movement quiescence during C. elegans sleep. microPubl. Biol. 2019;2019 doi: 10.17912/micropub.biology.000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turek M., Lewandrowski I., Bringmann H. An AP2 Transcription Factor Is Required for a Sleep-Active Neuron to Induce Sleep-like Quiescence in C. elegans. Curr. Biol. 2013;23:2215–2223. doi: 10.1016/j.cub.2013.09.028. [DOI] [PubMed] [Google Scholar]
- 91.Dabbish N.S., Raizen D.M. GABAergic Synaptic Plasticity during a Developmentally Regulated Sleep-Like State in C. elegans. J. Neurosci. 2011;31:15932–15943. doi: 10.1523/JNEUROSCI.0742-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choi S., Chatzigeorgiou M., Taylor K.P., Schafer W.R., Kaplan J.M. Analysis of NPR-1 Reveals a Circuit Mechanism for Behavioral Quiescence in C. elegans. Neuron. 2013;78:869–880. doi: 10.1016/j.neuron.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mani A., Radhakrishnan J., Farhi A., Carew K.S., Warnes C.A., Nelson-Williams C., Day R.W., Pober B., State M.W., Lifton R.P. Syndromic patent ductus arteriosus: Evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc. Natl. Acad. Sci. USA. 2005;102:2975–2979. doi: 10.1073/pnas.0409852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kucherenko M.M., Ilangovan V., Herzig B., Shcherbata H.R., Bringmann H. TfAP-2 is required for night sleep in Drosophila. BMC Neurosci. 2016;17:72. doi: 10.1186/s12868-016-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu Y., Bringmann H. Tfap2b acts in GABAergic neurons to control sleep in mice. Sci. Rep. 2023;13:8026. doi: 10.1038/s41598-023-34772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soto R., Goetting D.L., Van Buskirk C. NPR-1 Modulates Plasticity in C. elegans Stress-Induced Sleep. iScience. 2019;19:1037–1047. doi: 10.1016/j.isci.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xuan Z., Manning L., Nelson J., Richmond J.E., Colón-Ramos D.A., Shen K., Kurshan P.T. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. Elife. 2017;6 doi: 10.7554/eLife.29276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoon S., Ju J., Kwon S., Jeon T.-J., Kim S.M. Growth and Activity of Caenorhabditis elegans Exposed to Mechanical Vibration During the Embryonic Period. Biotechnol. Bioproc. Eng. 2020;25:126–131. doi: 10.1007/s12257-019-0433-7. [DOI] [Google Scholar]
- 99.Grubbs J.J., Lopes L.E., van der Linden A.M., Raizen D.M. A salt-induced kinase is required for the metabolic regulation of sleep. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Churgin M.A., Jung S.-K., Yu C.-C., Chen X., Raizen D.M., Fang-Yen C. Longitudinal imaging of Caenorhabditis elegans in a microfabricated device reveals variation in behavioral decline during aging. Elife. 2017;6 doi: 10.7554/eLife.26652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kurshan P.T., Merrill S.A., Dong Y., Ding C., Hammarlund M., Bai J., Jorgensen E.M., Shen K. γ-Neurexin and Frizzled mediate parallel synapse assembly pathways antagonized by receptor endocytosis. Neuron. 2018;100:150–166.e4. doi: 10.1016/j.neuron.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Garcia L.R., Mehta P., Sternberg P.W. Regulation of Distinct Muscle Behaviors Controls the C. elegans Male’s Copulatory Spicules during Mating. Cell. 2001;107:777–788. doi: 10.1016/S0092-8674(01)00600-6. [DOI] [PubMed] [Google Scholar]
- 103.Caylor R.C., Jin Y., Ackley B.D. The Caenorhabditis elegans voltage-gated calcium channel subunits UNC-2 and UNC-36 and the calcium-dependent kinase UNC-43/CaMKII regulate neuromuscular junction morphology. Neural Dev. 2013;8:10. doi: 10.1186/1749-8104-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morizawa Y.M., Matsumoto M., Nakashima Y., Endo N., Aida T., Ishikane H., Beppu K., Moritoh S., Inada H., Osumi N., et al. Synaptic pruning through glial synapse engulfment upon motor learning. Nat. Neurosci. 2022;25:1458–1469. doi: 10.1038/s41593-022-01184-5. [DOI] [PubMed] [Google Scholar]
- 105.Berger R.J., Phillips N.H. Energy conservation and sleep. Behav. Brain Res. 1995;69:65–73. doi: 10.1016/0166-4328(95)00002-B. [DOI] [PubMed] [Google Scholar]
- 106.Siegel J.M. Sleep viewed as a state of adaptive inactivity. Nat. Rev. Neurosci. 2009;10:747–753. doi: 10.1038/nrn2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmidt M.H. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci. Biobehav. Rev. 2014;47:122–153. doi: 10.1016/j.neubiorev.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 108.Schmidt M.H., Swang T.W., Hamilton I.M., Best J.A. State-dependent metabolic partitioning and energy conservation: A theoretical framework for understanding the function of sleep. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bringmann H. Genetic sleep deprivation: using sleep mutants to study sleep functions. EMBO Rep. 2019;20 doi: 10.15252/embr.201846807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu S., Liu Q., Tabuchi M., Wu M.N. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell. 2016;165:1347–1360. doi: 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abel T., Havekes R., Saletin J.M., Walker M.P. Sleep, Plasticity and Memory from Molecules to Whole-Brain Networks. Curr. Biol. 2013;23:R774–R788. doi: 10.1016/j.cub.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tononi G., Cirelli C. Sleep and the Price of Plasticity: From Synaptic and Cellular Homeostasis to Memory Consolidation and Integration. Neuron. 2014;81:12–34. doi: 10.1016/j.neuron.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Corbett B.A., Simon D. Adolescence, Stress and Cortisol in Autism Spectrum Disorders. OA Autism. 2014;1:2. [PMC free article] [PubMed] [Google Scholar]
- 114.Ishizuka K., Yoshida T., Kawabata T., Imai A., Mori H., Kimura H., Inada T., Okahisa Y., Egawa J., Usami M., et al. Functional characterization of rare NRXN1 variants identified in autism spectrum disorders and schizophrenia. J. Neurodev. Disord. 2020;12:25. doi: 10.1186/s11689-020-09325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Corporate Communications Obtain maximum information from your specimen with LIGHTNING. 2020. https://www.leica-microsystems.com/science-lab/life-science/obtain-maximum-information-from-your-specimen-with-lightning/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data have been deposited in Mendeley and are publicly available as of the date of publication. Mendeley Data: https://doi.org/10.17632/d74xzc9kz7.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.