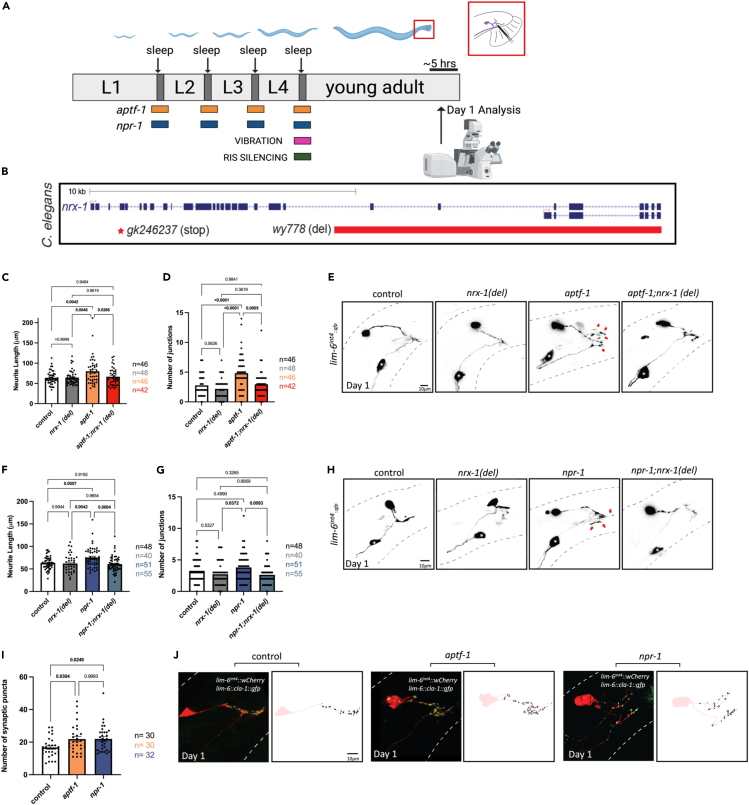
Figure 1.
Genetic disruption of developmental sleep (via npr-1 and aptf-1) increases DVB neurite outgrowth dependent on nrx-1
(A) Schematic of C. elegans developmental sleep stages between larval stages with timing of each sleep deprivation method indicated. aptf-1 (orange) and npr-1 (blue) animals experience impaired sleep during all larval sleep periods, while vibration (pink) and RIS silencing (green) are applied specifically during the L4 to adult transition/molt. Cartoon zoom in of the adult male tail with DVB shown.
(B) Schematic of C. elegans nrx-1 gene coding locus, red box indicates deletion in nrx-1(wy778) referred to as nrx-1(del), red star indicates premature stop in nrx-1(gk246237) referred to as nrx-1(stop). Quantification of DVB (C) total neurite length and (D) junctions in controls, nrx-1(del), aptf-1, and aptf-1; nrx-1(del) males at day 1.
(E) Representative images of DVB neuron in day 1 males using lim-6p::gfp. Red arrows indicate neurite branches. Black dashed lines represent body outline. White asterisk marks PVT neuron (not traced, see64). Quantification of DVB (F) total neurite length and (G) junctions in controls, nrx-1(del), npr-1, and npr-1; nrx-1(del) day 1 males.
(H) Representative images of DVB at day 1 in npr-1 mutant males and all controls.
(I) Quantification of number of cla-1::gfp puncta in DVB in controls and sleep deprived aptf-1 and npr-1 males at day 1.
(J) Representative images of DVB (red) and cla-1::gfp puncta (green) in controls, aptf-1, and npr-1 day 1 males. Outlines of synaptic puncta generated from FIJI particle analysis are shown (black) superimposed on a trace of the DVB neuron (light red). Number of individual animals is indicated by “n”, p values from one-way ANOVA with Tukey’s post-hoc test shown, error bars represent SEM, scale bar size as labelled.