Abstract
Redox-responsive hydropersulfide prodrugs are designed to enable a more controllable and efficient hydropersulfide (RSSH) supply and to thoroughly explore their biological and therapeutic applications in oxidative damage. To obtain novel activation patterns triggered by redox signaling, we focused on NAD(P)H: quinone acceptor oxidoreductase 1 (NQO1), a canonical antioxidant enzyme, and designed NQO1-activated RSSH prodrugs. We also performed a head-to-head comparison of two mainstream structural scaffolds with solid quantitative analysis of prodrugs, RSSH, and metabolic by-products by LC-MS/MS, confirming that the perthiocarbamate scaffold was more effective in intracellular prodrug uptake and RSSH production. The prodrug was highly potent in oxidative stress management against cisplatin-induced nephrotoxicity. Strikingly, this prodrug possessed potential feedback activation properties by which the delivered RSSH can further escalate the prodrug activation via NQO1 upregulation. Our strategy pushed RSSH prodrugs one step further in the pursuit of efficient release in biological matrices and improved druggability against oxidative stress.
Keywords: Hydropersulfide, Hydropersulfide prodrug, Reactive sulfur species, Oxidative stress, Nephrotoxicity
Highlights
-
•
Prodrugs release persulfide via enzymatic redox reaction rather than chemical ones.
-
•
Two mainstream prodrug scaffolds are compared to optimize persulfide release.
-
•
Intracellular persulfide and endogenous sulfur species are quantified.
-
•
Prodrug 1e efficiently reversed the toxicity and ROS overload caused by cisplatin.
-
•
The releasing persulfide boosts prodrug activation via redox enzyme upregulation.
1. Introduction
Hydropersulfide (RSSH) is a class of molecules with Janus character, presenting concomitantly an electrophilic and a strongly nucleophilic sites [[1], [2], [3], [4]]. Such a unique property originates from its sulfane structure and anionic form under physiological conditions [2]. This puts RSSH at the heart of sulfur trafficking and metabolism [5]. The generation, transformation, and biological activity of RSSH are closely related to hydrogen sulfide (H2S) and other sulfur species and are closely involved in regulating sulfur species homeostasis [[6], [7], [8], [9], [10]]. Although the interaction between RSSH and other reactive species and its signal transduction need to be extensively explored, the biological activity of RSSH is already fascinating enough, especially in the resistance to oxidative stress or electrophilic stress [[11], [12], [13], [14], [15], [16]].
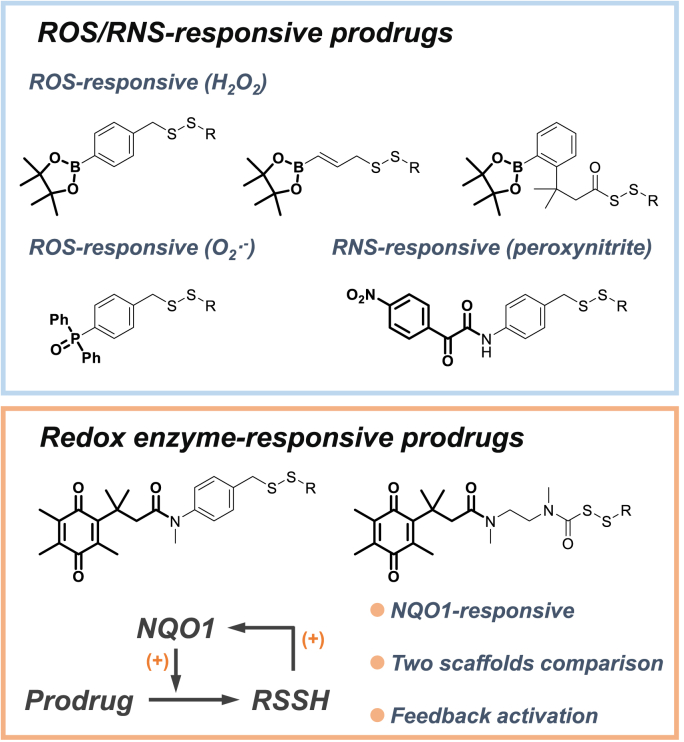
Oxidative stress is associated with a variety of toxicities and diseases involving two prime mechanisms, including the production of reactive species and the disturbance of redox signaling [[17], [18], [19], [20]]. Targeting both mechanisms, RSSH can effectively scavenge free radical species [[21], [22], [23]] as well as the biological oxidant H2O2 [24,25]. It can also be engaged in regulating redox signaling and protein persulfidation against oxidative injury [22,[26], [27], [28], [29]]. However, RSSH can be regarded as a reactive metabolite, and therefore its efficient delivery is more preferably achieved through prodrug strategies. In recent years, an array of RSSH prodrugs (also known as RSSH donors or RSSH precursors) have emerged [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54]], exhibiting ideal therapeutic effects against pathological processes such as ferroptosis [11,14], ischemia-reperfusion [12,15], paracetamol poisoning [37], doxorubicin-induced cardiotoxicity [13] and postoperative pain [36]. Furthermore, RSSH prodrugs are proven superior to their counterparts, such as H2S donors, in terms of antioxidant activity [36]. Therefore, developing novel RSSH prodrugs that can be triggered in disease states, especially under oxidative stress, could ignite a better understanding for the biological activity and therapeutic effects of RSSH. We noticed that the existing activation patterns of RSSH prodrugs were restricted to responding to reactive oxygen species (ROS) [42,43,45,48] or reactive nitrogen species (RNS) (Fig. 1, blue box) [37]. Compared with the chemical reaction between small molecules and oxidative species, we believe enzyme-catalytic reaction could be more efficient for prodrug activation [18]. To this end, we hope to extend prodrug types to those seizing prominent enzymes in redox signaling to achieve RSSH release.
Fig. 1.
ROS/RNS-responsive prodrugs (blue box) and redox enzyme-responsive prodrugs (orange box). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Human NAD(P)H quinone oxidoreductase 1 (NQO1) is an inducible antioxidant enzyme that acts as an effector of the Nrf2 (nuclear factor erythroid 2-related factor 2) system, a major cellular defense mechanism against oxidative and electrophilic stress [55,56]. NQO1 is a two-electron reductase with high kinetic rate constants [57]. Using NADH or NADPH as a reducing cofactor, the enzyme can rapidly reduce quinone to hydroquinone, thereby regenerating antioxidant forms of endogenous antioxidants, such as α-tocopherol [58]. In addition, we noticed that exogenous chemicals elevating the NQO1 levels or activating the Nrf2-NQO1 axis have proven potential for antioxidant therapy, some of which have been developed in clinical trials as Nrf2 activators [59,60]. Based on this, we assumed that once the prodrug is triggered by NQO1, the resulting RSSH might also act as an inducer of NQO1 upregulation, subsequently stimulating prodrug activation and extended RSSH release. To test this hypothesis of feedback activation, we designed two types of NQO1-responsive RSSH prodrugs, one releasing RSSH through 1,6-elimination and the other releasing RSSH through intramolecular cyclization of perthiocarbamate platform (Fig. 1, orange box). We also compared the RSSH release efficiency and biological activity of the two scaffolds.
2. Materials and methods
2.1. General information
Starting materials, solvents, and reagents were received from commercial sources (Adamas-beta, Bide Pharmatech, Aladdin, Energy Chemical and Sigma-Aldrich) unless otherwise noted and were used without purification. Recombinant human NQO1 (D1315) and NADPH were purchased from Sigma-Aldrich. HSip-1 (fluorescence probe for H2S detection) was purchased from Dojindo Laboratories (Kumamoto, Japan). N, N′-Dimethyl-2-Imidazolidinone (NMI, CAS: 80-73-9) and warfarin (CAS: 81-81-2) were purchased from Bide Pharmatech (Shanghai, China).
Analytical thin-layer chromatography (TLC) was carried out on Merck 60 F254 pre-coated silica gel plate (0.2 mm thickness). Visualization was accomplished with UV light (254 nm) or staining with phosphomolybdic acid followed by heating. Flash column chromatography was performed over silica gel (200–300 mesh).
1H NMR and 13C NMR spectra were recorded at ambient temperature using Bruker 300 M or 500 M spectrometers, chemical shifts (in ppm) were referenced to CDCl3 (δ = 7.26 ppm for 1H NMR and δ = 77.2 ppm for 13C NMR), DMSO-d6 (δ = 2.50 ppm for 1H NMR and δ = 39.5 ppm for 13C NMR) as internal standards. Deuterated solvents (Cambridge Isotope Laboratories and Adamas-beta) were used for NMR spectroscopic analyses. Data for 1H NMR are recorded as following abbreviations: multiplicity (s = singlet, d = doublet, t = triplet, q = quarter, m = multiplet), coupling constant (J, Hz).
High resolution mass spectrometry (HRMS) data were acquired using the electrospray ionization time-of-flight (ESI-TOF) method. The RSSH generation and prodrug stability were measured by a LC-MS/MS system containing a Shimadzu HPLC system (Kyoto, Japan) coupled with a SCIEX API 4000 triple-quadrupole mass spectrometer (Foster City, CA, USA) equipped with an electrospray ionization (ESI) interface in positive or negative ionization mode. MS analysis was performed using an AB Sciex 5600 + Triple TOF mass spectrometer (Concord, Ontario, Canada). Analyst software version 1.5.1 (SCIEX) was used for data acquisition and analysis. Excitation and emission measurements were recorded with a 96-well plate reader (BioTek SYNERGY-H1 multi-mode reader, Winooski, VT). Fluorescence imaging were acquired using an Olympus FV3000 laser scanning confocal microscope (Zeiss, Oberkochen, Germany).
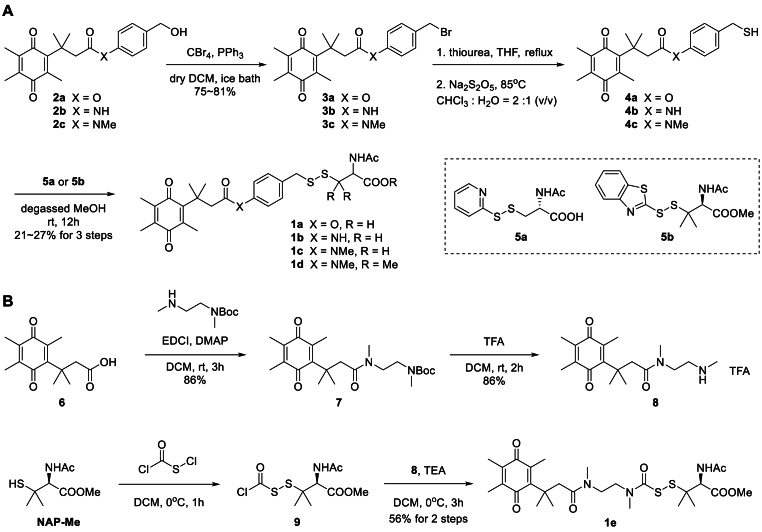
2.2. Chemical synthesis (see Scheme 1 and Scheme S1-S13)
Scheme 1.
Synthesis of prodrugs 1a-e.
General procedure A for synthesis of prodrugs 1a-1d: To a dried 50 mL round bottom flask was added 2a-c (4 mmol), CBr4 (2.6 g, 8 mmol) and dry DCM (15 mL). After PPh3 (2.1 g, 8 mmol) in dry DCM (15 mL) was added dropwise at 0 °C, the reaction solution was allowed to warm to room temperature with stirring. The reaction was always completed within 5 h. Then cold water (30 mL) was added, and the aqueous layer was extracted with DCM (3 × 20 mL). The combined organic layers were washed with brine and dried over Na2SO4, filtered, and concentrated on rotary evaporator. The residue was purified by trituration with diethyl ether or flash column chromatography (petroleum ether/ethyl acetate = 20/1–5/1) to obtain 3a-c.
General procedure B for synthesis of 1a-1d: A 20 mL sealed tube was charged with 3a-c (1 mmol), thiourea (312 mg, 4 mmol) and THF (5 mL). Then the mixture was stirred at 85 °C for 5 h. The resulting suspension liquid was concentrated on rotary evaporator, and used directly for the next step without purification. The residue was firstly suspended in degassed CHCl3 (4 mL), to which a solution of Na2S2O5 (380 mg, 2 mmol) in H2O (2 mL) was added in one portion. The reaction was violently stirred at 85 °C for 5 h in a sealed tube. The aqueous layer was extracted with DCM (3 × 10 mL). The combined organic layers were washed with brine and dried over Na2SO4, filtered, and concentrated on rotary evaporator. The residue (crude thiol 4a-c) was used directly for the next step without purification, avoiding the dimerization during column chromatography.
General procedure C for synthesis of 1a-1d: A double-neck round-bottom flask was charged with thiol 4a-e (1.0 equiv.), degassed MeOH (0.2 M), and a stir bar. Activated thiols 5a or 5b (for 1d) (1.1 equiv.) in degassed MeOH (0.2 M) was added dropwise under Ar atmosphere, resulting in a clear yellow solution. The reaction was stirred at room temperature overnight, monitoring reaction progress by TLC. The solvent was removed under vacuum, and the residue was extracted with DCM (3 × 10 mL). The combined organic layers were washed with brine and dried over Na2SO4, filtered, and concentrated on rotary evaporator. The crude product was purified by silica gel chromatography (DCM/MeOH = 100/1–10/1) to obtain 1a-d.
Compound 1a: Yellow solid, 126 mg, 27% yield for three steps, from 3a (419 mg, 1 mmol); m.p. 64–65 °C; 1H NMR (300 MHz, DMSO-d6) δ 8.33 (d, J = 7.8 Hz, 1H), 7.33 (d, J = 7.9 Hz, 2H), 6.96 (d, J = 7.8 Hz, 2H), 4.56-4.40 (m, 1H), 3.96 (s, 2H), 3.17 (s, 2H), 3.08-2.93 (m, 1H), 2.90–2.72 (m, 1H), 2.11 (s, 3H), 1.86 (s, 9H), 1.46 (s, 6H). 13C NMR (75 MHz, DMSO) δ 190.8, 187.2, 172.5, 171.2, 169.9, 152.1, 149.7, 142.9, 139.1, 138.5, 135.6, 130.8, 122.0, 51.6, 47.3, 41.4, 38.6, 28.9, 22.9, 14.5, 12.9, 12.3. HRMS (ESI) for C26H31NO7S2Na [M+Na]+ calcd 556.1434, found 556.1429.
Compound 1b: Yellow solid, 102 mg, 19% yield for three steps, from 3b (418 mg, 1 mmol); m.p. 101–102 °C; 1H NMR (300 MHz, DMSO-d6) δ 9.97 (s, 1H), 8.30 (d, J = 8.0 Hz, 1H), 7.44 (d, J = 8.2 Hz, 2H), 7.22 (d, J = 8.2 Hz, 2H), 4.53-4.41 (m, 1H), 3.91 (s, 2H), 3.05-2.97 (m, 1H), 2.95 (s, 2H), 2.85–2.76 (m, 1H), 2.05 (s, 3H), 1.90 (s, 6H), 1.86 (s, 3H), 1.39 (s, 6H). 13C NMR (75 MHz, DMSO) δ 190.8, 187.3, 172.5, 170.8, 169.9, 154.6, 144.0, 138.5, 137.1, 136.6, 132.3, 130.1, 119.6, 51.7, 49.3, 42.0, 38.1, 28.6, 22.8, 14.2, 13.1, 12.2. HRMS (ESI) for C26H32N2O6S2Na [M+Na]+ calcd 555.1594, found 555.1598.
Compound 1c: Yellow solid, 120 mg, 22% yield for three steps, from 3c (432 mg, 1 mmol); m.p. 75–76 °C; 1H NMR (300 MHz, DMSO-d6) δ 12.84 (s, 1H), 8.26 (d, J = 8.0 Hz, 1H), 7.41 (d, J = 7.7 Hz, 2H), 7.20 (d, J = 7.8 Hz, 2H), 4.52-4.38 (m, 1H), 4.00 (s, 2H), 3.03 (s, 3H), 2.86 (d, J = 11.6 Hz, 1H), 2.75 (d, J = 9.7 Hz, 1H), 2.66 (s, 2H), 1.99 (s, 3H), 1.88 (s, 6H), 1.84 (s, 3H), 1.21 (s, 6H). 13C NMR (75 MHz, DMSO) δ 190.8, 187.3, 172.5, 171.4, 169.9, 155.4, 143.8, 143.2, 137.5, 137.2, 135.6, 131.1, 127.8, 51.6, 47.3, 41.7, 38.2, 37.1, 28.5, 22.9, 14.2, 13.1, 12.2. HRMS (ESI) for C27H34N2O6S2Na [M+Na]+ calcd 569.1750, found 569.1741.
Compound 1d: Yellow solid, 73 mg, 12% yield for three steps, from 3c (432 mg, 1 mmol); m.p. 60–61 °C; 1H NMR (500 MHz, CDCl3) δ 7.38 (d, J = 7.7 Hz, 2H), 7.17 (d, J = 8.0 Hz, 2H), 6.32–6.18 (m, 1H), 4.79 (d, J = 9.0 Hz, 1H), 3.99 (s, 2H), 3.78 (s, 3H), 3.16 (s, 3H), 2.76 (s, 2H), 2.12 (s, 3H), 2.08 (s, 3H), 2.02 (s, 3H), 1.98 (s, 3H), 1.44 (s, 3H), 1.40 (s, 3H), 1.32 (s, 6H). 13C NMR (126 MHz, CDCl3) δ 191.2, 187.7, 172.0, 170.7, 169.8, 154.7, 143.6, 143.4, 137.8, 136.7, 136.3, 130.7, 127.7, 58.8, 52.5, 52.3, 47.7, 44.7, 38.1, 37.1, 28.5, 26.1, 24.8, 23.3, 14.1, 12.7, 12.1. HRMS (ESI) for C30H40N2O6S2Na [M+Na]+ calcd 611.2220, found 611.2212.
Synthesis of prodrugs 1e: To a dried double-neck flask was charged with 6 (498 mg, 2 mmol), tert-butyl methyl(2-(methylamino)ethyl)carbamate (404 mg, 2 mmol) and DMAP (163 mg, 1.34 mmol) in DCM (20 mL), and then EDCI (499 mg, 2.6 mmol) was added portionwise. The solution was stirred at room temperature for 3 h. After the complete consumption of 6, ice water (20 mL) was added, the resulting mixture extracted with DCM (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and concentrated. The crude product was purified by silica gel chromatography (dichloromethane/methanol = 50/1–10/1), giving a yellow oil 7 (724 mg, 86%). 1H NMR (500 MHz, Chloroform-d) δ 3.45-3.36 (m, 2H), 3.31-3.24 (m, 2H), 3.00 (s, 3H), 2.90 (s, 3H), 2.80 (s, 2H), 2.12 (s, 3H), 1.92 (s, 3H), 1.90 (s, 3H), 1.48 (s, 3H), 1.45 (s, 6H), 1.41 (s, 3H), 1.41 (s, 3H).
A 50 mL double-neck round bottom flask was charged with 7 (419 mg, 1 mmol) and DCM (24 mL). TFA (8 mL) was added in one portion. The reaction mixture was stirred at room temperature for 2 h. The resulting yellow solution was concentrated via rotary evaporation, followed by the cold NaHCO3 solution (50 mL) addition. The resulting mixture extracted with DCM (3 × 15 mL). The organic layers were combined, dried over Na2SO4, and concentrated. The crude product was purified by silica gel chromatography (dichloromethane/methanol = 50/1–10/1), affording a white solid 8 (275 mg, 63%). 1H NMR (500 MHz, Chloroform-d) δ 3.56 (t, J = 6.0 Hz, 2H), 3.09 (t, J = 5.7 Hz, 2H), 3.03 (s, 3H), 2.98 (s, 2H), 2.64 (s, 3H), 2.12 (s, 3H), 1.93 (s, 3H), 1.87 (s, 3H), 1.40 (s, 6H). TFA salt: 1H NMR (500 MHz, DMSO-d6) δ 8.43 (s, 1H), 3.45 (t, J = 6.4 Hz, 2H), 2.97 (s, 3H), 2.95-2.94 (m, 2H), 2.53 (s, 2H), 2.50 (d, J = 1.9 Hz, 3H), 2.04 (s, 3H), 1.87 (s, 3H), 1.84 (s, 3H), 1.36 (s, 6H).
A 25 mL two-neck flask was charged with N-acetyl-penicillamine methyl ester (246 mg, 1.2 mmol), dry DCM (5 mL) and equipped with a nitrogen balloon. After the gas replacement, to the flask was dropwise added a solution of chlorocarbonylsulfenyl chloride (0.11 mL, 1.26 mmol) in dry DCM (2 mL) at 0 °C. The resulting solution was stirred for 1 h and concentrated via rotary evaporation. The residue (crude 9) was used directly for the next step without purification.
A 25 mL two-neck flask was charged with 8 (419 mg, 1 mmol), trimethylamine (0.3 mL, 2.1 mmol). To the flask was subsequently added dry DCM (5 mL) and a solution of 9 in dry DCM (5 mL) at 0 °C under a nitrogen atmosphere. The resulting mixture was stirred for 3 h, quenched by ice water (10 mL) and extracted with DCM (3 × 10 mL). The organic layers were combined, washed with brine, dried over Na2SO4, and concentrated. The residue was purified by column chromatography (dichloromethane/methanol = 50/1–10/1) to give 1e as a yellow solid (325 mg, 56%), m.p. 82–83 °C; 1H NMR (500 MHz, Chloroform-d) δ 4.54 (dd, J = 24.2, 8.3 Hz, 1H), 3.72 (s, 3H), 3.57-3.50 (m, 2H), 3.47-3.44 (m, 2H), 3.06 (s, 3H), 3.01 (s, 3H), 2.97 (s, 2H), 2.12 (s, 3H), 2.05 (s, 3H), 1.93 (s, 3H), 1.90 (s, 3H), 1.45 (s, 3H), 1.41 (s, 6H), 1.33 (s, 3H). 13C NMR (126 MHz, CDCl3) δ 192.1, 187.9, 172.4, 170.7, 167.7, 154.7, 143.3, 138.5, 136.9, 59.5, 53.8, 52.9, 52.5, 48.4, 47.9, 45.2, 37.8, 36.5, 29.0, 29.0, 26.9, 26.3, 23.2, 14.5, 13.0, 12.4. HRMS (ESI) for C27H42N3O7S2 [M+H]+ calcd 584.2459, found 584.2461.
The synthesis of reference compounds and trapping reagent HPE-IAM was summarized in Supplementary Materials.
2.3. LC-MS/MS assay for validating RSSH release
A total of three analytical methods were used. Analytical Method A and Analytical Method B was used to analyze the decomposition of prodrugs and the RSSH production in enzymatic reactions in buffers, and Analytical Method C was used to analyze the RSSH production and endogenous sulfides in cells. See Table S1∼S8 in Supplementary Materials for specific conditions and parameters.
2.3.1. HPLC condition
The study was carried out on a Shimadzu LC 20A HPLC system (Kyoto Japan). HPLC autosampler temperature: 4 °C. Injection volume: 5 μL. The chromatographic separation was performed on Waters XSelect-T3 column (3.5 μm, 4.6 × 150 mm, Ireland) with a flow rate of 0.8 mL/min (Method A and B) or on ZORBAX Eclipse XDB-C18 (150 mm × 4.6 mm, 5 μm) with a flow rate of 1.0 mL/min (Method C) at 40 °C.
2.3.2. MS condition
The compounds were analyzed by a LC-MS/MS system containing a Shimadzu HPLC system (Kyoto, Japan) coupled with a SCIEX API 4000 (Method A) and 6500 (Method B and C) triple-quadrupole mass spectrometer (Foster City, CA, USA) equipped with an electrospray ionization (ESI) interface. Analyst software version 1.5.1 (SCIEX) was used for data acquisition and analysis. The pressure of curtain gas, collision gas, ion source gas 1 and ion source gas 2 were 12, 30, 60 and 70 psi, respectively. The ion source temperature was maintained at 550 °C. The optimized ion spray voltage was 4500 V and dwell time of 100 ms for the analytes and IS applied. Selected Multiple Reaction Monitoring (MRM) was conducted for the quantification analysis in the mass analyzers.
2.3.3. Preparation of the stock and standard solutions
Stock solution of human NQO1 was prepared in PBS at 1 mg/mL; Stock solution of NADPH was prepared in PBS at 400 mM; stock solutions of all analytes and deuterated internal standards (IS) were prepared in DMSO at 10 mM; stock solution of FDNB and HPE-IAM was were prepared in acetonitrile at 10 mM; stock solution of warfarin was made at 1.0 mg/mL in DMSO; all the above stock solutions were stored at 4 °C.
2.3.4. Reaction solutions with FDNB or HPE-IAM
10 μL of each prodrug solution (1 mM) was transferred into a 1.5 mL EP tube, followed by the addition of FDNB solution (10 μL, 10 mM) or HPE-IAM solution (10 μL, 5, 10, or 50 mM), and 960 μL of PBS buffer solution. The mixture was vortexed for approximately 30 s prior to addition of 10 μL of NADPH solution (2, 4, or 8 mM in PBS), and then 10 μL of human NQO1 solution (0.08 mg/mL in PBS). After a second 30 s vortex, the reaction was incubated at 37 °C. At a series of time points (0, 5, 10, 15, 30, 60 min and 12, 24, 36, 48, 72, 96 h), 50 μL of the reaction solution was diluted by 250 μL working solution containing IS. The solution was centrifuged twice at 18,000 rpm at 4 °C for 5 min to remove invisible impurities. 80 μL of the resulting supernatant was analyzed by LC-MS/MS directly (with Analytical Method A; Table S2). In each control group, we replaced NQO1 with PBS. Above assays were repeated in triplicate and recorded as the mean ± SD from three experiments.
2.3.5. Reaction solutions without HPE-IAM
20 μL of 1e solution (1 mM) was transferred into a 4 mL EP tube, followed by 1940 μL of PBS buffer solution. The mixture was vortexed for approximately 30 s prior to addition of 20 μL of NADPH solution (8 mM in PBS), and then 20 μL of human NQO1 solution (0.08 mg/mL in PBS). After a second 30 s vortex, the reaction was incubated at 37 °C. At a series of time points (0, 5, 10, 15, 30, 60 min and 2, 4, 6 h), 50 μL of the reaction solution was diluted by 250 μL working solution containing IS. The solution was centrifuged twice at 18,000 rpm at 4 °C for 5 min to remove invisible impurities. 80 μL of the resulting supernatant was analyzed by LC-MS/MS directly (with Analytical Method B; Table S5). In each control group, we replaced NQO1 with PBS. Above assays were repeated in triplicate and recorded as the mean ± SD from three experiments.
2.3.6. Metabolism of prodrug in cells
For the quantification of endogenous RSSH levels by LC-MS/MS, HPE-IAM was chosen as the trapping agent. HK-2 cell line was seeded in 6-well cell culture plates (3.5 × 105 cells/well) and grown to confluence. Then 1e or 18 (0, 50, 100, or 200 μM) was added to each well with serum free MEM medium. After incubation at 37 °C for 0.5 h, drug-containing medium was removed, drug-free medium was treated for 0, 1, or 2 h, and cells were then washed with PBS three times and lysed in cold 70% methanol solution of HPE-IAM (1 mM, 300 μL for each well). The mixture was vortexed for 5 min for more complete analyte abstract and protein precipitation, and then centrifuged at 18,000 rpm for 5 min. An aliquot (100 μL) of the supernatant was transferred to a new tube, diluted with acetonitrile solution containing IS (100 μL, IS working solution), and recentrifuged at 18,000 rpm before LC-MS/MS analysis (with Analytical Method C; Table S7). The protein content was measured using BCA colorimetric protein kit.
2.4. H2S detection with fluorescence probe
A solution of 1e (10 μL, 1 mM) or 18 (10 μL, 1 mM) was added to individual wells on a 96 well plate followed by the addition of PBS buffer (60 μL), NADPH solution (10 μL, 8 mM), and hNQO1 solution (10 μL, 80 μg/mL) in sequence. The reaction system was incubated for 0.5 h at 37 °C. Then the solution of NAP-Me (10 μL, 4 mM) or PBS (as control group, 50 mM, pH 7.4) was added. Thus, each well contained final concentration of 100 μM for 1e and 18 with a total volume of 100 μL. The plate was incubated in dark for 2 h at 37 °C, and agitated to ensure homogeneity. 10 mM solution of HSip-1 (fluorescence probe) was prepared in freshly degassed PBS buffer (50 mM, pH 7.4). After the addition of HSip-1 (10 μL, 200 μM), fluorescence measurements were recorded.
The fluorescence experiments were carried out according to the manufacture's recommended protocol provided with appropriate modification. Fluorescence spectroscopic studies were performed on a 96-well plate reader (BioTek SYNERGY-H1 multi-mode reader, Winooski, VT). Excitation wavelength was 470 nm. Emission wavelength was at 550 nm. This assay was repeated in triplicate and recorded as the mean ± SD from three experiments.
2.5. Cell culture
The HK-2 human proximal tubular cell line was acquired from the Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were maintained in complete high-glucose Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F-12; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Genetimes Technology Inc., Shanghai, China) and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an incubator containing 95% humidified air and 5% CO2 at 37 °C.
Transient over-expression of NQO1: Small interfering RNA (siRNA) was used to determine the effect of NQO1 knockdown on RSSH formation in HK-2 cells. HK-2 cells were seeded in 12-well cell culture plates (0.6 × 105 cells/well) and incubated with scrambled siControl or siRNA targeting human NQO1 (5′-GAAAGGACAUCACAGGUAA-3′) (Genomeditech, Shanghai) together with transfection reagent (Lipofectamine™ RNAiMAX, ThermoFisher) and made the final concentration of siControl or siRNA in each well to 50 nM. After incubation for 8 h, cells were treated with fresh MEM medium for another 40 h. The cells were then used to perform the prodrug metabolism assays described in 2.3.6.
Dicoumarol-mediated inhibition of NQO1: Dicoumarol (DIC) was used to determine the effect of NQO1 inhibition on RSSH formation in HK-2 cells. HK-2 cell line was seeded in 6-well cell culture plates (2 × 105 cells/well) and grown to confluence. Then DIC (25, 50, and 100 μM) was added to each well with serum free MEM medium. After incubation at 37 °C for 24 h, drug-containing medium was removed, and cells were washed with PBS three times. The cells were then used to perform the prodrug metabolism assays described in 2.3.6.
2.6. Cellular protection
The intracellular antioxidant activity of the studied prodrugs was evaluated in HK-2 cells. Briefly, cells were seeded in a 96-well plate at 5 × 103 cells/well, incubated for 24 h and subsequently incubated with NAP-Me (100 and 200 μM), 1d (100 and 200 μM), 1e (25, 50, 100, 200 μM), or 18 (100 and 200 μM) for 30 min at 37 °C in 5% CO2. At the end of incubation, the treatment medium was removed, and medium containing Cis (40 μM) was added to each well. After 24 h of incubation at room temperature, CCK-8 reagent was added to the cells, which were incubated for 1 h at 37 °C. The absorbance was measured at 450 nm using a microplate (Synergy H1 Hybrid Multi-Mode Reader, BioTek Instruments, Inc., Winooski, Vermont, USA). The results were expressed as the percentage of cell viability (%) with respect to the control (medium treated cells). The cell viability was tested in four independent experiments performed in triplicate.
2.7. Cellular imaging assays
Cellular imaging for ROS: HK-2 cells were treated with 100 μM of 1e for 30 min, followed by the treatment with Cis (40 μM) for 6 h. Then the cells were incubated with 5.0 μM of DCFH-DA (Dojindo Laboratories, Kumamoto, Japan) for 40 min and Hoechst 33342 solution (1:1000 diluted) for 15 min before imaging. Fluorescence imaging at 488 nm were acquired using an Olympus FV3000 laser scanning confocal microscope (Zeiss, Oberkochen, Germany). Fluorescence variations were calculated with the definition and measurement of regions of interest (ROIs) using ImageJ software and expressed as relative Medium Fluorescence Index (MFI) compared to control.
Immunofluorescence imaging for NQO1: HK-2 cells were pretreated with 1e for 0.5 h followed by exposure to Cis (40 μM) for 24 h. HK-2 Cells growing on cover slides were washed in PBS, fixed with 4% para-formaldehyde on ice for 15 min and washed once with PBS before blocking with block-sperm buffer (1% BSA in PBS) for 20 min. After washing once with PBS, cells were incubated with the primary antibody (rabbit anti-NQO1, Abcam ab80588) for 2 h in antibody buffer (1% BSA in PBS). Cells were washed three times with PBS and incubated with the Fluorescein Isothiocyanate (FITC)-conjugated secondary antibody (Jackson ImmunoResearch, 111-095-003, USA) (1:200) in antibody buffer for 1 h. Cells were washed three times with PBS and incubated with the DAPI for 5 min. Cells were imaged using a confocal laser scanning microscope (OLYMPUS FV300).
2.8. Western blot assay
HK-2 cells were pretreated with compounds for 0.5 h followed by exposure to Cis (40 μM) for 6 h. Cell samples were lysed with RIPA buffer containing phosphatase and protease inhibitors. The protein content was measured using BCA colorimetric protein kit. Equal amount of protein were separated with 12% SDS-PAGE and transferred onto a PVDF membrane. After blocking with 10% nonfat milk, the membranes were incubated with primary antibody overnight with mild shake at 4 °C. Then the membrane was washed for 3 times with TBST buffer followed by 1 h incubation with horseradish peroxidase-conjugated secondary antibody. The immunoblots were visualized with ECL Western blotting substrate. Protein bands were normalized with GAPDH, beta-actin or Histone H3.
The reagents used as follows were purchased from Keygen Biotech (Nanjing, China): Total Protein Extraction Kit (KGP250), Protein BCA Protein Assay Kit (KGA902), Western Blot Assay Kit (KGP1201), anti-GAPDH (KGAA002), and beta-actin antibody (KGC6106). Anti-Nrf2 (ab62352) and anti-NQO1 (ab80588) were obtained from Abcam (Cambridge, UK). Histone H3 antibody (D1H2) were obtained from Cell Signaling Technology (Boston, USA).
2.9. Gene expression analysis (qPCR assay)
HK-2 cells were pretreated with compounds for 0.5 h followed by exposure to Cis (40 μM) for 6 h. Total RNA from HK-2 cells was extracted using TRIzol (Invitrogen), and 2 μg of total RNA was used for reverse transcription using the One Step TB Green™ PrimeScript™ RT-PCR Kit II (Takara, Kusatsu, Japan). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using a SYBR Green Supermix kit (Takara). Relative expression was normalized to the expression levels of GAPDH or actin. The following primer sequences were used. Reactions were performed in triplicate for each sample and gene expression was normalized to the mRNA expression of GAPDH.
2.10. Flow cytometry analysis
HK-2 cells were seeded in a 6-well plate at 5 × 105 cells/mL, incubated for 24 h and subsequently incubated with 1e (50, 100, 200 μM), or 18 (200 μM) for 1 h at 37 °C in 5% CO2. At the end of incubation, the treatment medium was removed, and medium containing Cis (40 μM) was added to each well. After 24 h of incubation at room temperature, cells were collected and washed with cold sterile PBS. For DCFH-DA-based flow cytometric detection of cellular ROS, cells were incubated with 10 μM DCFH-DA (KEYGEN, KGT010-1, Nanjing) for 20 min at 37 °C and washed. Then, cells were analyzed at excitation/emission wavelengths at 488/530 nm on a Beckman Coulter Cytoflex (Indianapolis, USA). Mean channel fluorescence was converted to absolute fluorescence using an inverse log transformation and normalized to the fluorescence in untreated cells. Flow cytometry data were analyzed using Summit Software (Beckman Coulter). See Fig. S8.
2.11. Statistical analysis
GraphPad Prism 9.0 software (GraphPad Software) was used for statistical analysis. One-way ANOVA was used appropriately where needs to quantified the statistical differences. Data were expressed as mean ± SD where applicable. p < 0.05 was regarded as significant.
3. Results and discussion
3.1. Synthesis of prodrugs 1a-1e
Two mainstream prodrug scaffolds with distinct mechanisms for RSSH release were selected for our study. The first scaffold carries asymmetric disulfides containing stimulant-cleavable groups that trigger 1, 6-elimination to release RSSH [42,45,51]. This is represented by the prodrugs BDP-NAC [51], SOPD-NAC [45], and NDP-NAC [42], which achieve RSSH release in response to hydrogen peroxide, superoxide, and nitroreductase, respectively. Our group also designed RSSH prodrugs in response to RNS against paracetamol poisoning using the above-described scaffold [37]. The second scaffold contains acyl persulfide moieties that can undergo intramolecular acyl migration to release RSSH [40,41,44]. This class of prodrugs includes an alkylamine-substituted perthiocarbamate [44] or alkylsulfenyl thiocarbonate scaffold [41], which recently focused on pH-controlled spontaneous hydrolysis to achieve RSSH release. This class of prodrugs has successfully demonstrated the therapeutic potential of RSSH, effectively alleviating many pathological processes such as ferroptosis [11,14] and ischemia-reperfusion [12,15]. Inspired by these work, we adopted two mainstream scaffolds of prodrugs and designed two series of prodrugs, namely series A (prodrug 1a-d, Scheme 1A) and series B (prodrug 1e, Scheme 1B). These prodrugs 1a-e were synthesized according to Scheme 1. Benzyl alcohol 2a-c were converted to the corresponding benzyl bromide 3a-c under Appel condition. With thiourea as a nucleophilic sulfur source, the resulting bromides 3a-c formed intermediate isothiouronium salt, which underwent Na2S2O5-mediated hydrolysis to thiols 4a-c. The reaction of thiols with activated disulfides (5a and 5b) afforded the disulfides 1a-d. The above three steps were carried out without isolating the intermediates with overall yields of 12∼27%. In the synthesis of prodrug 1e, the acid 6 was converted to the amine 8 after amide coupling and deprotection with good yield. The target RSSH prodrug 1e was obtained by acylation of the amine 8 with the intermediate 9, which can be prepared by the substitution reaction between N-acetyl-penicillamine methyl ester (NAP-Me) and chlorocarbonylsulfenyl chloride.
3.2. Prodrug activation and RSSH release
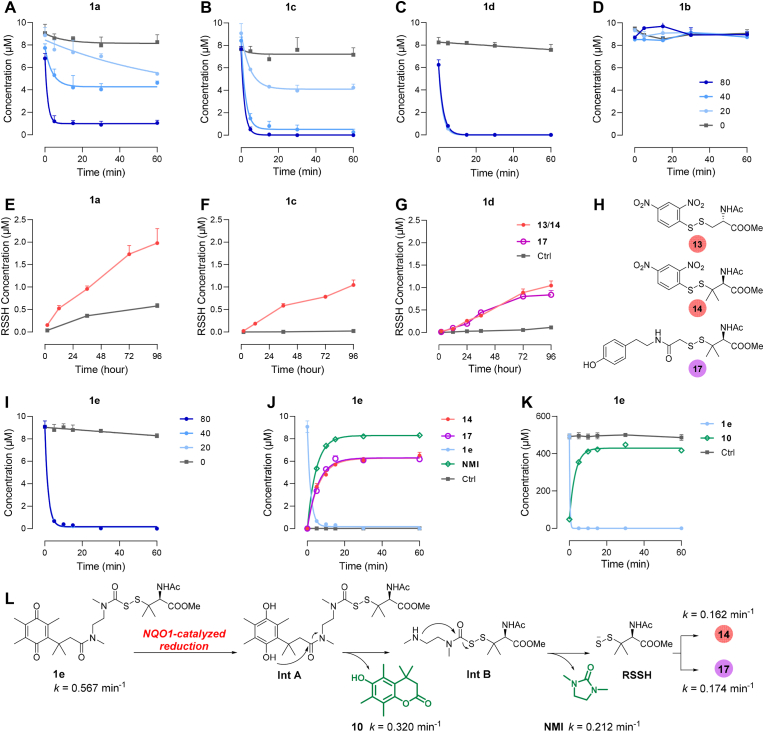
After obtaining the two structural types of prodrugs, we investigated their characteristics during activation, containing prodrug degradation and RSSH release. Prodrugs 1a, 1c, and 1d degraded under the catalysis of NQO1, except for prodrug 1b (Fig. 2A–D). We speculate that 1b containing the NH structure are not good substrates for the enzyme, even though it differ from 1c by only the N–CH3 group. This is supported by articles [61,62] reporting that the activity of NQO1 activated prodrugs containing the NH structure is much weaker than that of those with the N–CH3 group, possibly due to the tendency of the former to generate a class of spirolactone [63] intermediates that interfere with enzyme recognition. With the increase of the NADPH concentration, 1a and 1c showed a gradual NADPH-dependent degradation. While the more rapid and complete degradation of 1d occurred at any of the three NADPH concentrations. To validate the RSSH release, we added 1-fluoro-2, 4-dinitrobenzene (FDNB) as a trapping agent to the NQO1-catalytic reaction system. The levels of RSSH were reflected by quantifying the generated RSSH derivatives RSS-DNB (13 or 14; Fig. 2H). As shown in the figures, 13 or 14 were detected in all reactions treated with 1a, 1c or 1d. However, it should be pointed out that the production of 13 or 14 is significantly delayed compared to the rapid degradation of the prodrugs. The yields of these compounds within 96 h are only 10%–20% (Fig. 2E–G). This gradual release at the hourly level was also observed in our previous quantification using LC-MS/MS [37] and in Matson's NMR experiments for quantitative analysis [51]. Moreover, in the 1a group, 13 was also detected in blank reactions without enzyme (Fig. 2E), suggesting that prodrugs containing aryl ester moieties are metastable upon prolonged incubation. In the 1d group, as the rate of the RSSH alkylation step is crucial [[64], [65], [66], [67]], we replaced β-(4-hydroxyphenyl) ethyl iodoacetamide (HPE-IAM) as the trapping agent and observed the formation of RSS-HPE-AM (17) (Fig. 2G, H and S3). It was found that the generation curves of 14 and 17 were almost overlapping, which indicated that the two reported trapping agents, FDNB and HPE-IAM, possessed a similar trapping efficiency for RSSH.
Fig. 2.
NQO1-triggered prodrug activation and RSSH release. (A–D) Degradation of prodrugs 1a-1d with different concentrations of NADPH. (E–G) Generation of RSSH with NADPH (80 μM), characterized by the RSSH derivatives 13 (NAC-SS-DNB), 14 (RSS-DNB), or 17 (RSS-HPE-AM). (H) The structures of trapped RSSH 13, 14, and 17. (I) Degradation of 1e with different concentrations of NADPH (0/20/40/80 μM). (J) Generation of RSSH and NMI after 1e activation with NADPH (20 μM); 14 (k = 0.162 min−1, t1/2 = 4.29 min), 17 (k = 0.174 min−1, t1/2 = 3.98 min), NMI (k = 0.212 min−1, t1/2 = 3.27 min). (K) Generation of 10 and degradation of 1e; 10 (k = 0.320 min−1, t1/2 = 2.17 min). (L) Mechanism for the RSSH release from 1e in response to NQO1-catalyzed reduction. Enzymatic reaction conditions: (A–J) prodrugs (10 μM), NQO1 (0.8 μg/mL), NADPH (0/20/40/80 μM), FDNB/HPE-IAM (100 μM) in PBS (with 1% DMSO, pH 7.4, 37 °C), followed by LC-MS/MS quantification; (K) 1e (0.5 mM), NQO1 (40 μg/mL), NADPH (4 mM) in PBS (with 1% DMSO, pH 7.4, 37 °C), followed by HPLC-UV quantification. Data represent the average ± SD (n = 3).
Subsequently, we investigated the activation of prodrug 1e. Like 1d, 1e also achieved rapid and complete degradation at various NADPH concentrations (k = 0.567 min−1, t1/2 = 1.2 min) (Fig. 2I). However, unlike 1c and 1d, 1e exhibited substantial RSSH production with a yield of up to 60%, which was initiated concurrently with self-degradation and swiftly reached a steady state within 30 min (Fig. 2J, S1, and S2). Furthermore, the derivatization efficiencies of the two trapping agents, FDNB and HPE-IAM, were almost equal (14 formation: k = 0.162 min−1, t1/2 = 4.3 min; 17 formation: k = 0.174 min−1, t1/2 = 4.0 min). We also tracked NMI, a by-product during the prodrug activation, in more than 80% yield with an observed rate constant of 0.212 min−1 (t1/2 = 3.3 min) (Fig. 2J). In the HPLC-UV experiment, the kinetics of lactone 10 were analogously measured (k = 0.320 min−1, t1/2 = 2.2 min) (Fig. 2K and S4). The rate of 10 generation can be considered to be close to the rate at which 1e undergoes enzyme-catalyzed reduction and spontaneous lactonization, and this rate is slightly lower than the reported rate of NMI generation from Int B [44], suggesting that the enzyme-catalyzed reduction-lactonization tandem reaction is rate-limiting. The above results all confirmed that 1e could launch a lactonization-lactamization ring closure cascade reaction during its activation and enable the high-efficiency releasing of RSSH (Fig. 2L, Scheme S14).
Taken together, all prodrugs were rapidly degraded under the catalysis of NQO1, suggesting that both types of structures could be better recognized and transformed by enzymes. In contrast, the release behavior of RSSH was significantly different. We believe that such prodrugs involving 1, 6-elimination are unfit for RSSH release, at least in our incubation system, because the production of RSSH is delayed and low-level. Whereas 1e has a more defined RSSH release and can serve as a preferential prodrug.
3.3. Intermolecular interactions of sulfur series
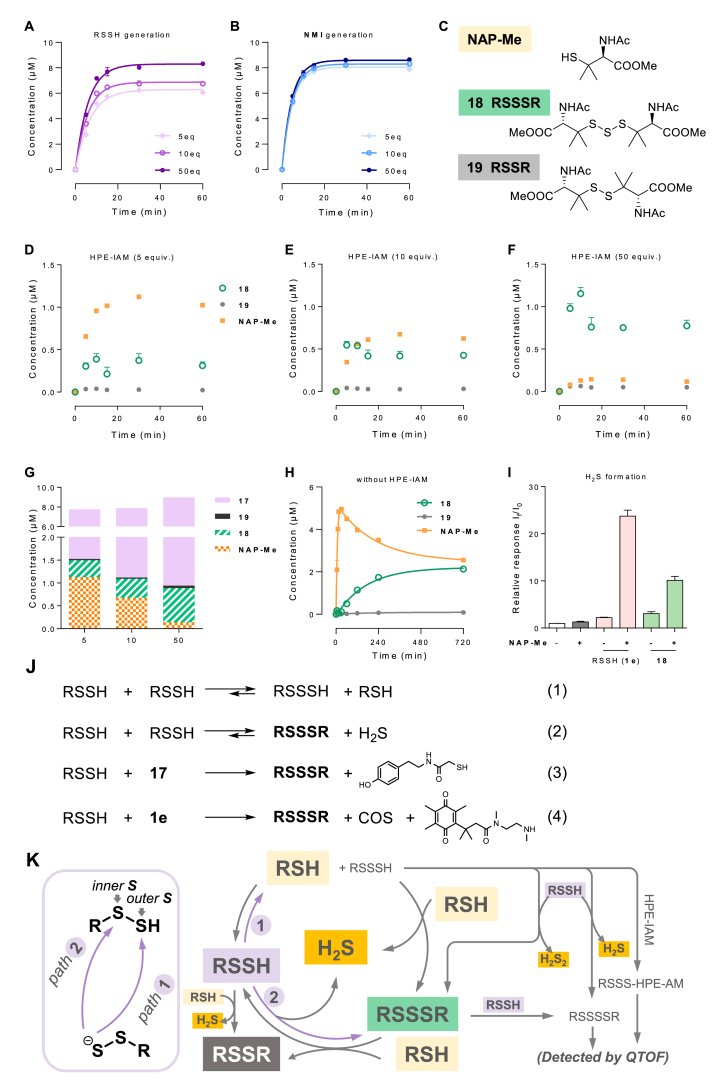
We proposed that the deviation between the kinetics of 17 and NMI (Fig. 2J) was due to the alkylation reaction of RSSH; and the span between the steady-state concentrations of the two might originate from the intermolecular interactions of sulfur series. Therefore, the study investigated the impact of varying trapping agent concentrations on the production of RSSH and related sulfur species.
The yield of RSSH increased significantly from 61% to 83% when the degree of HPE-IAM excess was raised from 5 to 50 equivalents (Fig. 3A). In contrast, the amount and rate of NMI generation remained almost unaffected by the HPE-IAM concentration, with a yield of 80∼86% (Fig. 3B). The yield of RSSH produced by 1e within our reaction system was at least 80%, as the generation of NMI and the release of RSSH were mechanically synchronized. This indicates that increasing the concentration of trapping agent can more accurately reflect the actual RSSH production. We also examined other sulfur species within the reaction system, including RSSSR (18), RSSR (19), and RSH (NAP-Me) (Fig. 3C). The amount of 18 increased as the concentration of the trapping agent increased, while the amount of NAP-Me decreased gradually and the amount of 19 was negligible (Fig. 3D–F). Since the amount of each species stabilized at 30 min, we selected this moment and plotted the accumulation of the target sulfur species. It was observed that increasing the concentration of HPE-IAM resulted in an increase in the total sulfur species content. Additionally, the proportion of RSSH in the total sulfur species also increased (Fig. 3G). At lower concentrations of the trapping agent, the untrapped RSSH may undergo intermolecular disproportionation reactions, resulting in the production of sulfur-containing mixtures, with NAP-Me being the major product (Fig. 3D and J, eq 1). In contrast, at higher concentrations of the trapping agent, RSSH reacts more readily with 17, resulting in an increase in the proportion of trisulfide 18 produced (Fig. 3J, eq 3). As the enzyme-catalyzed degradation of 1e is very rapid (t1/2 = 1.2 min), it is believed that RSSH accounts for less of the RSSSR generated with 1e (Fig. 3J, eq 4).
Fig. 3.
The intermolecular interactions of RSSH and related sulfur species. (A, B) Detectable RSSH (A) and NMI (B) at different concentrations of trapping agents HPE-IAM (5, 10, and 50 equiv.) (C) The structures of RSH (NAP-Me), RSSSR (18), and RSSR (19). (D–F) Kinetics of NAP-Me, 18, and 19 at different concentrations of HPE-IAM (5, 10, and 50 equiv.). (G) Concentrations of sulfur species (17, NAP-Me, 18, and 19) at 30 min with concentrations of HPE-IAM (5, 10, and 50 equiv.). (H) Production of NAP-Me, 18, and 19 generated after 1e activation without HPE-IAM. Reaction condition: 1e (10 μM), NQO1 (0.8 μg/mL), HPE-IAM (0, 50, 100, and 500 μM), NADPH (20 μM) in PBS (with 1% DMSO, pH 7.4, 37 °C), followed by LC-MS/MS quantification. (I) Production of H2S after 1e (100 μM) or 18 (100 μM) was activated by NQO1 (8 μg/mL) and NADPH (800 μM) for 30 min with or without additional NAP-Me (400 μM). (J, K) Putative transformation of sulfur species. Data represent the average ± SD (n = 3).
It was attractive for us to further explore the behavior of the reactive RSSH in the NQO1-catalyzed reaction without trapping agents. In the absence of the shielding effect of the trapping agents, RSSSR (18), RSSR (19), and RSH (NAP-Me) were traced out (Fig. 3H). Following the prodrug degradation and the RSSH release, NAP-Me was produced sharply, reaching a peak concentration of 5 μM approximately 15 min after the addition of NQO1. It then slowly degraded, showing interesting biphasic kinetics. In contrast, 18 gradually accumulated in this system, while the amount of 19 remained low. Finally, 18 and NAP-Me reached a steady coexistence state (Fig. 3H). At physiological pH, RSSH can be deprotonated, resulting in an anion (persulfide, RSS−) that is nucleophilic compared to structurally analogous thiol (RSH) and H2S [[1], [2], [3], [4]]. The neutral form of RSSH, also known as hydropersulfide, is electrophilic, and both inner- and outer-sulfur can be attacked by the RSSH anion [2,3]. The distinct behaviors of the NAP-Me and 18 stemmed from which of the two sulfur atoms of RSSH participated in the electrophilic reaction (Fig. 3J, eq 1 and eq 2; Fig. 3K, path 1 and path 2). RSH and intermediate RSSSH were preferentially generated when the outer sulfur acted as an electrophilic site (Fig. 3K, path 1). The resulting RSSSH can continue to react with RSH or RSSH to provide RSSSR or tetrasulfide RSSSSR. By high-resolution mass spectrometry (QTOF), we detected the existence of RSSSSR in the above reaction system (Fig. S5) and identified derived RSSSH when HPE-IAM was added (Fig. S6). On the other hand, when the intermolecular reaction site was located at the inner sulfur, RSSSR and H2S was generated (Fig. 3K, path 2). It is worth mentioning that RSSSR can also be regarded as a RSSH donor, which can react with RSH and reverse back to RSSH. Therefore, the cyclic interconversion between RSH (NAP-Me) and RSSSR (18) tended to be balanced, as observed in Fig. 3H. Additionally, both RSSH and RSSSR can react with RSH to form disulfide RSSR. However, the results indicate that the proportion of such transformations is small. The intermolecular interaction of RSSH and related sulfur species was briefly summarized in Fig. 3K.
H2S formation is consistently intertwined with sulfide series interactions [5]. In the above process, H2S was another important sulfur species besides RSH and RSSSR. We investigated the generation of H2S with the aid of the fluorescent probe HSip-1 (Fig. 3I) [68]. The addition of RSH (NAP-Me) resulted in the production of significant amounts of H2S, with a 15-fold increase in the amount of H2S in the 1e group and a 4-fold increase in the RSSSR (18) group, compared to the group treated without RSH. This suggests that the conversion of RSSH to H2S is more efficient compared to RSSSR. These results confirm that RSH drives the conversion flow of sulfane-containing species to H2S through thiol-polysulfide exchange (Fig. 3K).
3.4. Cellular RSSH production
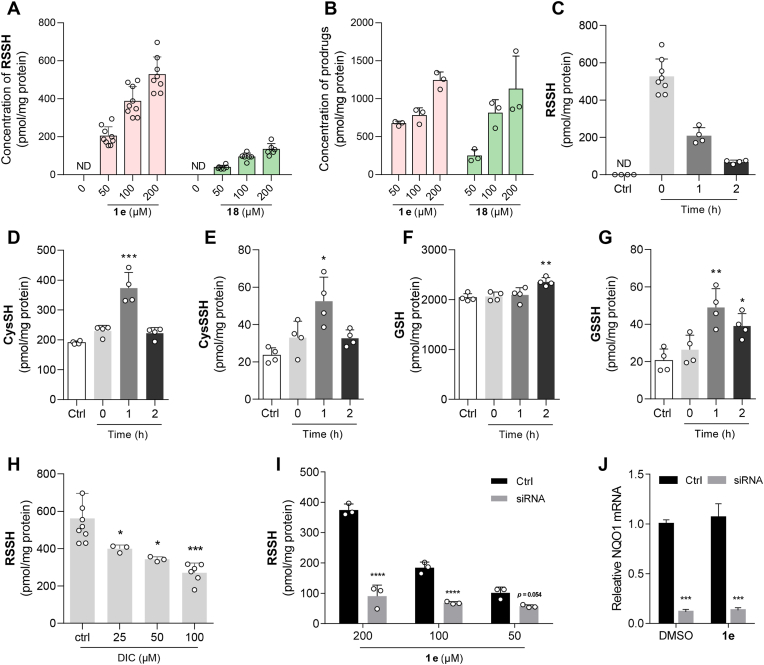
Next, we attempted to probe the intracellular release of RSSH. The intracellular release of RSSH has been previously confirmed using fluorescent probe [41]. In this study, we quantified the intracellular RSSH release from the prodrugs using LC-MS/MS with isotopic internal standard. After incubating 1e or 18 with HK-2 cells, we used methanol containing HPE-IAM to simultaneously extract and protect the RSSH from the samples. The results showed a concentration-dependent increase in intracellular RSSH after cells were treated with 1e for 30 min (Fig. 4A), suggesting that 1e can be activated and release RSSH in cells. Likewise, when 18 was co-incubated with cells, intracellular RSSH release was also achieved, although to a much lesser extent than in the 1e group (Fig. 4A). The undegraded 1e and 18 was also detected (Fig. 4B). This indicated that they possessed appropriate stability, which is conducive for their cellular uptake and metabolic activation. Since the cellular uptake of 18 was not inferior to that of 1e, we believed that RSSH prodrugs can increase the intracellular delivery of RSSH more directly and effectively than RSSSR, the traditional RSSH donor.
Fig. 4.
NQO1-triggered intracellular RSSH release and the resulting switch of endogenous sulfur species. HK-2 cells were pretreated with prodrugs (1e or 18) for 30 min and extracted using methanol containing HPE-IAM. (A, B) The cellular production of RSSH (A) and the cellular concentration of prodrugs 1e and 18 (B). (C–G) Intracellular content of RSSH (C), CysSH (D), CysSSH (E), GSH (F), and GSSH (G) at different time points (0, 1, and 2 h) after pretreatment with 1e for 30 min. (H–J) DIC-mediated inhibition of NQO1 (H) and reduced NQO1 expression (I, J) leads to decrease in intracellular RSSH release. The resulting samples were analyzed using LC-MS/MS. Data represent the average ± SD (n = 3); ND = not detected. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 versus control group.
We investigated the impact of the exogenous RSSH provided by the prodrug on the homeostasis of endogenous sulfur species. HK-2 cells were treated with 1e for 0.5 h, and the prodrug was subsequently withdrawn. The intracellular RSSH content was then measured before, 1 h, and 2 h after the withdrawal of 1e. The highest intracellular levels of RSSH were observed after 0.5 h of 1e treatment, but decreased by 60% after 1 h and by 87% after 2 h of prodrug withdrawal. These results suggest a cellular disposition of RSSH and a possible concomitant transformation of sulfur species (Fig. 4C). Deuterated endogenous thiols were synthesized as reference compounds [[69], [70], [71]] and used to demonstrate this transformation. The experimental results indicate that prodrug 1e, although delivering a structure of RSSH (penicillamine analog) different from that of endogenous thiols, still causes a change in the content of endogenous sulfur species over time (Fig. 4D–G). Specifically, the levels of cysteine (CysSH) and cysteine persulfide (CysSSH) significantly increased after 1 h of prodrug withdrawal (Fig. 4D and E). In contrast to the delayed changes in glutathione (GSH) content, which only became significant after 2 h of prodrug withdrawal, glutathione persulfide (GSSH) showed a significant accumulation at 1 and 2 h (Fig. 4F and G). It is important to note that the concentration of exogenous RSSH consistently exceeded that of endogenous RSSH at all time points.
The release of intracellular RSSH from 1e is also dependent on NQO1 activity. To confirm this, HK-2 cells were pretreated with dicoumarol (DIC), a known NQO1 inhibitor, followed by treatment with 1e. A dose-dependent reduction in the amount of intracellular RSSH was observed, with a reduction of over 50% when the DIC concentration was increased to 100 μM (Fig. 4H). In HK-2 cells with low NQO1 expression, a nearly 5-fold decrease in intracellular RSSH content was observed (Fig. 4I and J). This decrease was attributed to the reduced intracellular NQO1 levels, which affected the activation of the prodrug and subsequent release of RSSH. These results suggest that a prodrug has been designed, which is dependent on NQO1 for intracellular activation and RSSH release.
3.5. Cellular protective effects of prodrugs against cisplatin-induced oxidative stress
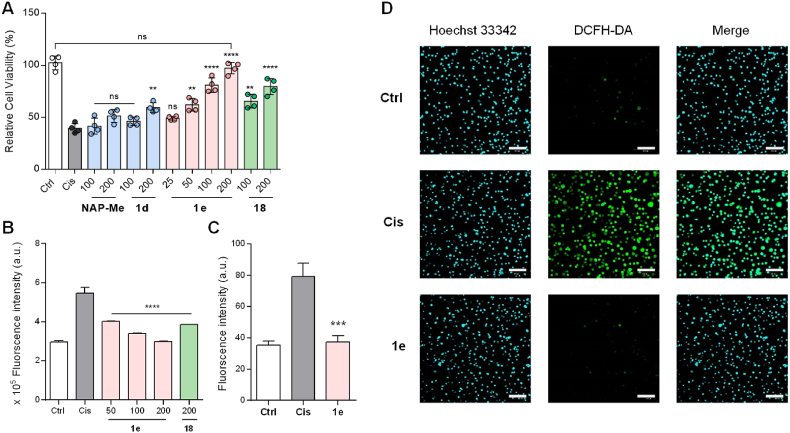
Cisplatin (Cis) is an effective chemotherapeutic agent widely used in the treatment of various solid tumors. It can promote intracellular ROS levels to induce cancer cell death [72]. However, due to the double-edged sword role of ROS, cisplatin also results in nephrotoxicity, in particular in the proximal tubule, which lingers as a major dose-limiting factor in its clinical application [73]. Studies have suggested that the excessive generation of ROS is a hallmark of cisplatin-induced acute kidney injury [74]. Therefore, we hypothesized that RSSH prodrugs could modulate cisplatin-induced oxidative damage in HK-2 cells (human renal proximal tubule cells) via the inhibition of ROS generation. Initially, we investigated the antioxidant activity of 1e at the cellular level. We induced damage in HK-2 cells by incubation with cisplatin (Fig. S7) and determined the effect of the prodrugs against cytotoxicity. As Fig. 5A shown, the protective effect of 1e was significant, and the cell viability continued to increase with the concentration elevation. Strikingly, high concentration (200 μM) of 1e almost completely reversed the damage. High concentrations of 18 also moderately attenuated cisplatin-induced damage, whereas the protective effects of 1d and NAP-Me were not obvious. The above results suggested that the release levels of RSSH has positive effects on the cellular protection.
Fig. 5.
Cellular protective effects of prodrugs against cisplatin-induced oxidative stress. (A) Protection for HK-2 cells pretreated with NAP-Me (100, 200 μM), 1d (100, 200 μM), 1e (25, 50, 100, 200 μM), and 18 (100, 200 μM) for 0.5 h followed by exposure to cisplatin (Cis, 40 μM) for 24 h. (B) ROS quantification (DCFH-DA assay) by flow cytometry with pretreat of 1e (50, 100, 200 μM), and 18 (200 μM) for 1 h and cisplatin (40 μM) damage for 24 h. (C, D) Confocal fluorescence imaging for ROS detection in cisplatin-damaged HK-2 cells. HK-2 cells were pretreated with 1e (100 μM) for 0.5 h followed by exposure to Cis (40 μM) for 6 h. Scale bar = 100 μm. The results are expressed as the mean ± SD (n = 3). **p < 0.01, ***p < 0.001, and ****p < 0.0001 vs. cisplatin group.
To determine the role of 1e in alleviating oxidative stress, we quantitatively analyzed the intracellular ROS levels by flow cytometry. As shown in Fig. 5B, a remarkable increase in intracellular ROS was observed in HK-2 cells under cisplatin-induced conditions compared with the control group. Pretreatment of cells with 1e and 18 significantly inhibited the accumulation of intracellular ROS under the same conditions (Fig. 5B and S8). Furthermore, we more intuitively observed that cisplatin significantly caused intracellular ROS accumulation through cell imaging studies using a ROS fluorescent probe (DCFH-DA) (Fig. 5C and D). However, after 1e pretreatment, cisplatin-induced ROS was significantly reduced. Taken together, 1e can protect cells by resisting oxidative damage.
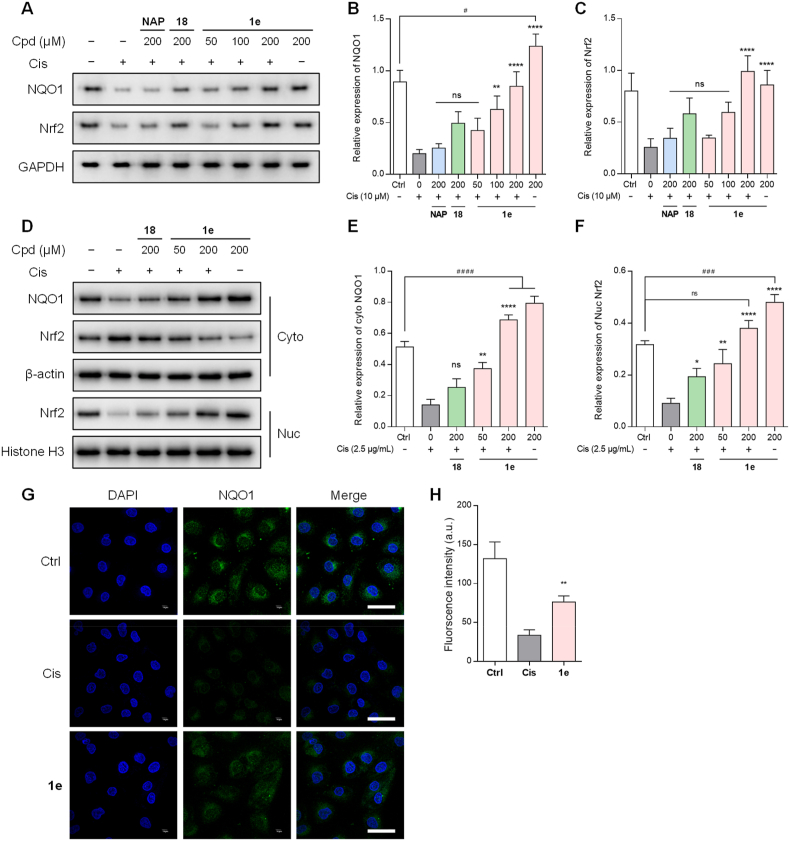
Cisplatin-induced overwhelming ROS is a major challenge for HK-2 cell survival. Therefore, while RSSH directly quenches ROS, upregulating genes/proteins with ROS scavenging ability seems to be a more necessary and reasonable choice to improve cell survival. Nrf2, a transcription factor involved in the antioxidant response, is a fundamental activator of genes encoding antioxidant enzymes and detoxification enzymes (such as NQO1), contributing to ROS and RNS reduction [75]. Previous studies showed that pharmacologic boost of the Nrf2/NQO1 signaling could neutralize the elevated ROS levels during cisplatin nephrotoxicity [76]. Furthermore, a recent work has revealed that RSSH fended off the doxorubicin-induced cardiotoxicity [13], and our previous work also found that RSSH prodrug alleviated acetaminophen hepatoxicity [37]. Therefore, we attempted to determine whether 1e mediated the activity of Nrf2-NQO1 pathway, especially under oxidative stress. We tested the protein expression of NQO1 and Nrf2 by Western blot. In whole-cell extracts, the total protein expression of NQO1 and Nrf2 were significantly inhibited after Cis stimulation for 6 h (Fig. 6A–C). Pretreatment of 1e effectively reversed this inhibition, with a dose-dependent increase in the expression of the two proteins (P < 0.0001). Furthermore, 1e treatment led to significant upregulation of the mRNA levels of Nrf2 and NQO1, in a concentration-dependent manner (Fig. S9). In contrast, 18 and NAP-Me (shown as NAP in Fig. 6A–C)) showed no upregulation of the target proteins even at the highest concentration (200 μM). Notably, 1e also increased NQO1 expression in the absence of cisplatin induction. In subcellular extracts, 1e also significantly increased cytoplasmic NQO1 and intranuclear Nrf2 expression more than 18 in a dose-dependent manner (Fig. 6D–F). This study demonstrated that 1e enhanced nuclear translocation of Nrf2, which induces gene expression for counteracting oxidative stress. In addition, the immunofluorescence results were consistent with the Western blot results, in which 1e restored the NQO1 levels reduced by Cis in HK-2 cells (Fig. 6G and H). Our findings indicated that 1e can promote the Nrf2-NQO1 signaling cascade in HK-2 cells, exerting a renoprotective effect. More importantly, we believe that a positive feedback loop exists whereby the upregulation of NQO1 induced by produced RSSH, which in turn benefits the prodrug activation mediated by NQO1 (Fig. 1).
Fig. 6.
Prodrug 1e enhances the nuclear translocation of Nrf2 and the resulting activation of NQO1. HK-2 cells were pretreated with 1e (100 μM) for 0.5 h followed by exposure to Cis (40 μM) for 6 h. (A–C) The impact of RSH (NAP, 200 μM), RSSSR (18) (200 μM), prodrug 1e (50, 100, 200 μM) on the expression levels of whole cell NQO1 (A, B) and Nrf2 (A, C) induced by cisplatin in HK-2 cells. (D–F) The impact of RSSSR (18) (200 μM) and prodrug 1e (50, 200 μM) on the expression levels of cytosolic NQO1 (D, E), cytosolic Nrf2 (D), and nuclear Nrf2 (D, F) induced by cisplatin in HK-2 cells. (G, H) The expression levels of NQO1 was measured by immunofluorescence (scale bar, 50 μm). The results are expressed as the mean ± SD (n = 3). **p < 0.01, and ****p < 0.0001 vs. cisplatin group; #p < 0.05, ###p < 0.001, and ####p < 0.0001 vs. control group.
4. Conclusion
In summary, we designed novel NQO1-triggered RSSH prodrugs based on two representative scaffolds and verified their properties under the same conditions. To our knowledge, this constitutes the first comparison of their structure-property relationships. We found that rapid prodrug degradation did not guarantee sufficient RSSH generation and that self-immolative elimination of linkers played a critical role. By detecting RSSH derivatives (14 and 17) and metabolic by-products (NAP-Me and 10), we confirmed that prodrug 1e can release RSSH smoothly. The release of intracellular RSSH and the resulting effects on endogenous thiol homeostasis were also carefully described. Corresponding to its rapid and high-yield release of RSSH, 1e also possesses the expected excellent antioxidant activity. It reversed the cytotoxicity and ROS overload caused by cisplatin, a potent inducer of oxidative stress. More importantly, we have demonstrated that this prodrug strategy has potential feedback activation features. Following uptake into the cell, 1e was metabolized by NQO1 with RSSH release. The resulting RSSH could upregulate the expression of NQO1, further stimulating the prodrug activation and amplifying the RSSH release. Moreover, our findings provided more references for selecting prodrug scaffolds, activation patterns, and even trapping agents for future research. We will use 1e to further explore the pharmacological activity and mechanism of RSSH in the following research.
CRediT authorship contribution statement
Bi-Xin Xu: Methodology, Formal analysis, Data curation. Tian-Yu Hu: Methodology, Investigation, Formal analysis, Data curation. Jin-Biao Du: Data curation. Tao Xie: Data curation. Ya-Wen Xu: Methodology, Data curation. Xin Jin: Data curation. Si-Tao Xu: Methodology. Hao-Wen Jin: Methodology. Guangji Wang: Supervision, Resources, Funding acquisition. Jiankun Wang: Writing – original draft, Validation, Formal analysis, Conceptualization. Le Zhen: Writing – review & editing, Writing – original draft, Supervision, Resources, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos. 82173686 and 81973186), Outstanding Youth Foundation of Jiangsu Province of China (No. BK20200081), Leading Technology Foundation Research Project of Jiangsu Province (BK20192005), and Haihe Laboratory of Cell Ecosystem Innovation Fund (22HHXBSS00005).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103130.
Contributor Information
Guangji Wang, Email: guangjiwang@hotmail.com.
Jiankun Wang, Email: wangjiankun789@126.com.
Le Zhen, Email: i_m_zhenle@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Iciek M., Bilska-Wilkosz A., Kozdrowicki M., Górny M. Biosci. Rep. 2022;42 doi: 10.1042/BSR20221006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipovic M.R., Zivanovic J., Alvarez B., Banerjee R. vol. 118. 2018. p. 377. (Chem. Rev.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasamatsu S., Nishimura A., Morita M., Matsunaga T., Hamid H.A., Akaike T. Molecules. 2016;21:1721. doi: 10.3390/molecules21121721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park C.-M., Weerasinghe L., Day J.J., Fukuto J.M., Xian M. Mol. Biosyst. 2015;11:1775. doi: 10.1039/c5mb00216h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller E.G. Nat. Chem. Biol. 2006;2:185. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 6.Bora P., Manna S., Nair M.A., Sathe R.R.M., Singh S., Adury V.S.S., Gupta K., Mukherjee A., Saini D.K., Kamat S.S., Hazra A.B., Chakrapani H. Chem. Sci. 2021;12 doi: 10.1039/d1sc03828a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau N., Pluth M.D. Curr. Opin. Chem. Biol. 2019;49:1. doi: 10.1016/j.cbpa.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Yuan S., Shen X., Kevil C.G. Antioxid. Redox Signaling. 2017;27:634. doi: 10.1089/ars.2017.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuevasanta E., Lange M., Bonanata J., Laura Coitino E., Ferrer-Sueta G., Filipovic M.R., Alvarez B. J. Biol. Chem. 2015;290 doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ono K., Akaike T., Sawa T., Kumagai Y., Wink D.A., Tantillo D.J., Hobbs A.J., Nagy P., Xian M., Lin J., Fukuto J.M. Free Radic. Biol. Med. 2014;77:82. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barayeu U., Schilling D., Eid M., da Silva T.N.X., Schlicker L., Mitreska N., Zapp C., Grater F., Miller A.K., Kappl R., Schulze A., Angeli J.P.F., Dick T.P. Nat. Chem. Biol. 2023;19:28. doi: 10.1038/s41589-022-01145-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths K., Ida T., Morita M., Lamb R.J., Lee J.J., Frenneaux M.P., Fukuto J.M., Akaike T., Feelisch M., Madhani M. Redox Biol. 2023;60 doi: 10.1016/j.redox.2023.102605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pharoah B.M., Zhang C., Khodade V.S., Keceli G., McGinity C., Paolocci N., Toscano J.P. Redox Biol. 2023;60 doi: 10.1016/j.redox.2023.102625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z., Khodade V.S., Chauvin J.-P.R., Rodriguez D., Toscano J.P., Pratt D.A. J. Am. Chem. Soc. 2022;144 doi: 10.1021/jacs.2c06804. [DOI] [PubMed] [Google Scholar]
- 15.Pharoah B.M., Khodade V.S., Eremiev A., Bao E., Liu T., O'Rourke B., Paolocci N., Toscano J.P. Antioxidants. 2022;11:1010. doi: 10.3390/antiox11051010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang T., Tsutsuki H., Ono K., Akaike T., Sawa T. J. Clin. Biochem. Nutr. 2021;68:5. doi: 10.3164/jcbn.20-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sies H., Belousov V.V., Chandel N.S., Davies M.J., Jones D.P., Mann G.E., Murphy M.P., Yamamoto M., Winterbourn C. Nat. Rev. Mol. Cell Biol. 2022;23:499. doi: 10.1038/s41580-022-00456-z. [DOI] [PubMed] [Google Scholar]
- 18.Forman H.J., Zhang H. Nat. Rev. Drug Discov. 2021;20:689. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schieber M., Chandel N.S. Curr. Biol. 2014;24:R453. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray P.D., Huang B.W., Tsuji Y. Cell. Signal. 2012;24:981. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko T., Mita Y., Nozawa-Kumada K., Yazaki M., Arisawa M., Niki E., Noguchi N., Saito Y. Free Radic. Res. 2022;56:677. doi: 10.1080/10715762.2023.2165918. [DOI] [PubMed] [Google Scholar]
- 22.Ida T., Sawa T., Ihara H., Tsuchiya Y., Watanabe Y., Kumagai Y., Suematsu M., Motohashi H., Fujii S., Matsunaga T., Yamamoto M., Ono K., Devarie-Baez N.O., Xian M., Fukuto J.M., Akaike T. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7606. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Everett S.A., Folkes L.K., Wardman P., Asmus K.D. Free Radic. Res. 1994;20:387. doi: 10.3109/10715769409145638. [DOI] [PubMed] [Google Scholar]
- 24.Benchoam D., Semelak J.A., Cuevasanta E., Mastrogiovanni M., Grassano J.S., Ferrer-Sueta G., Zeida A., Trujillo M., Moller M.N., Estrin D.A., Alvarez B. J. Biol. Chem. 2020;295 doi: 10.1074/jbc.RA120.014728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H.J., Liu H.W., Chen Z.G., Zhao R., Wang Q.D., Ran M.X., Xia Y.Z., Hu X., Liu J.H., Xian M., Xun L.Y. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vignane T., Filipovic M.R. Antioxid. Redox Signaling. 2023;39:19. doi: 10.1089/ars.2023.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura H. Br. J. Pharmacol. 2020;177:720. doi: 10.1111/bph.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C.-t., Devarie-Baez N.O., Hamsath A., Fu X.-d., Xian M. Antioxid. Redox Signaling. 2020;33:1092. doi: 10.1089/ars.2019.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju Y., Fu M., Stokes E., Wu L., Yang G. Molecules. 2017;22:1334. doi: 10.3390/molecules22081334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu B., Yang X., Yuan Z., Wang B. Curr. Opin. Chem. Biol. 2023;75 doi: 10.1016/j.cbpa.2023.102329. [DOI] [PubMed] [Google Scholar]
- 31.Khodade V.S., Aggarwal S.C., Eremiev A., Bao E., Porche S., Toscano J.P. Antioxid. Redox Signaling. 2022;36:309. doi: 10.1089/ars.2021.0149. [DOI] [PubMed] [Google Scholar]
- 32.Dillon K.M., Matson J.B. ACS Chem. Biol. 2021;16:1128. doi: 10.1021/acschembio.1c00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y., Wang J., Zhen L., Wang G. Chin. J. Org. Chem. 2021;41:2601. [Google Scholar]
- 34.Yu B., Yuan Z., Yang X., Wang B. Antioxid. Redox Signaling. 2020;33:1046. doi: 10.1089/ars.2019.7880. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Z.-N., Zheng Y.-Q., Wang B.-H. Chin. J. Nat. Med. 2020;18:296. doi: 10.1016/S1875-5364(20)30037-6. [DOI] [PubMed] [Google Scholar]
- 36.Yu B., Kang T., Xu Y., Liu Y., Ma Y., Ke B. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202201668. [DOI] [PubMed] [Google Scholar]
- 37.Xu Y.-W., Xu B.-X., Wang J., Jin H.-W., Xu S.-T., Wang G., Zhen L. Chem. Eur J. 2022;28 [Google Scholar]
- 38.Fosnacht K.G., Cerda M.M., Mullen E.J., Pigg H.C., Pluth M.D. ACS Chem. Biol. 2022;17:331. doi: 10.1021/acschembio.1c00805. [DOI] [PubMed] [Google Scholar]
- 39.Bora P., Sathian M.B., Chakrapani H. Chem. Commun. 2022;58:2987. doi: 10.1039/d1cc07162a. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal S.C., Khodade V.S., Porche S., Pharoah B.M., Toscano J.P. J. Org. Chem. 2022;87 doi: 10.1021/acs.joc.2c01049. [DOI] [PubMed] [Google Scholar]
- 41.Khodade V.S., Aggarwal S.C., Pharoah B.M., Paolocci N., Toscano J.P. Chem. Sci. 2021;12:8252. doi: 10.1039/d1sc01550h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dillon K.M., Morrison H.A., Powell C.R., Carrazzone R.J., Ringel-Scaia V.M., Winckler E.W., Council-Troche R.M., Allen I.C., Matson J.B. Angew. Chem. Int. Ed. 2021;60:6061. doi: 10.1002/anie.202014052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hankins R.A., Suarez S.I., Kalk M.A., Green N.M., Harty M.N., Lukesh J.C., 3rd Angew. Chem. Int. Ed. 2020;59 doi: 10.1002/anie.202010530. [DOI] [PubMed] [Google Scholar]
- 44.Khodade V.S., Pharoah B.M., Paolocci N., Toscano J.P. J. Am. Chem. Soc. 2020;142:4309. doi: 10.1021/jacs.9b12180. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Dillon K.M., Li Z., Winckler E.W., Matson J.B. Angew. Chem. Int. Ed. 2020;59 doi: 10.1002/anie.202006656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon K.M., Carrazzone R.J., Wang Y., Powell C.R., Matson J.B. ACS Macro Lett. 2020;9:606. doi: 10.1021/acsmacrolett.0c00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhuri A., Venkatesh Y., Das J., Gangopadhyay M., Maiti T.K., Singh N.D.P. J. Org. Chem. 2019;84 doi: 10.1021/acs.joc.9b01224. [DOI] [PubMed] [Google Scholar]
- 48.Bora P., Chauhan P., Manna S., Chakrapani H. Org. Lett. 2018;20:7916. doi: 10.1021/acs.orglett.8b03471. [DOI] [PubMed] [Google Scholar]
- 49.Khodade V.S., Toscano J.P. J. Am. Chem. Soc. 2018;140 doi: 10.1021/jacs.8b08469. [DOI] [PubMed] [Google Scholar]
- 50.Kang J., Xu S., Radford M.N., Zhang W., Kelly S.S., Day J.J., Xian M. Angew. Chem. Int. Ed. 2018;57:5893. doi: 10.1002/anie.201802845. [DOI] [PubMed] [Google Scholar]
- 51.Powell C.R., Dillon K.M., Wang Y., Carrazzone R.J., Matson J.B. Angew. Chem. Int. Ed. 2018;57:6324. doi: 10.1002/anie.201803087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan Z., Zheng Y., Yu B., Wang S., Yang X., Wang B. Org. Lett. 2018;20:6364. doi: 10.1021/acs.orglett.8b02611. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Y., Yu B., Li Z., Yuan Z., Organ C.L., Trivedi R.K., Wang S., Lefer D.J., Wang B. Angew. Chem. Int. Ed. 2017;56 doi: 10.1002/anie.201704117. [DOI] [PubMed] [Google Scholar]
- 54.Artaud I., Galardon E. Chembiochem. 2014;15:2361. doi: 10.1002/cbic.201402312. [DOI] [PubMed] [Google Scholar]
- 55.Ross D., Siegel D. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Q. Annu. Rev. Pharmacol. Toxicol. 2013;53:401. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anoz-Carbonell E., Timson D.J., Pey A.L., Medina M. Antioxidants. 2020;9:772. doi: 10.3390/antiox9090772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegel D., Bolton E.M., Burr J.A., Liebler D.C., Ross D. Mol. Pharmacol. 1997;52:300. doi: 10.1124/mol.52.2.300. [DOI] [PubMed] [Google Scholar]
- 59.Pisoschi A.M., Pop A., Iordache F., Stanca L., Predoi G., Serban A.I. Eur. J. Med. Chem. 2021;209 doi: 10.1016/j.ejmech.2020.112891. [DOI] [PubMed] [Google Scholar]
- 60.Valko M., Leibfritz D., Moncol J., Cronin M.T.D., Mazur M., Telser J. Int. J. Biochem. Cell Biol. 2007;39:44. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 61.Zhang C., Qu Y., Ma X., Li M., Li S., Li Y., Wu L. Bioorg. Chem. 2020;103 doi: 10.1016/j.bioorg.2020.104200. [DOI] [PubMed] [Google Scholar]
- 62.Qu Y., Zhang C., Ma X., Gao Y., Liu J., Wu L. Bioorg. Med. Chem. 2020;28 doi: 10.1016/j.bmc.2020.115821. [DOI] [PubMed] [Google Scholar]
- 63.Volpato M., Abou-Zeid N., Tanner R.W., Glassbrook L.T., Taylor J., Stratford I., Loadman P.M., Jaffar M., Phillips R.M. Mol. Cancer Therapeut. 2007;6:3122–3130. doi: 10.1158/1535-7163.MCT-07-0519. [DOI] [PubMed] [Google Scholar]
- 64.Schilling D., Barayeu U., Steimbach R.R., Talwar D., Miller A.K., Dick T.P. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202203684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dillon K.M., Matson J.B. ACS Chem. Biol. 2021;16:1128. doi: 10.1021/acschembio.1c00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bogdandi V., Ida T., Sutton T.R., Bianco C., Ditroi T., Koster G., Henthorn H.A., Minnion M., Toscano J.P., van der Vliet A., Pluth M.D., Feelisch M., Fukuto J.M., Akaike T., Nagy P. Br. J. Pharmacol. 2019;176:646. doi: 10.1111/bph.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamid H.A., Tanaka A., Ida T., Nishimura A., Matsunaga T., Fujii S., Morita M., Sawa T., Fukuto J.M., Nagy P., Tsutsumi R., Motohashi H., Ihara H., Akaike T. Redox Biol. 2019;21 doi: 10.1016/j.redox.2019.101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasakura K., Hanaoka K., Shibuya N., Mikami Y., Kimura Y., Komatsu T., Ueno T., Terai T., Kimura H., Naganot T. J. Am. Chem. Soc. 2011;133 doi: 10.1021/ja207851s. [DOI] [PubMed] [Google Scholar]
- 69.Matsunaga T., Sano H., Takita K., Morita M., Yamanaka S., Ichikawa T., Numakura T., Ida T., Jung M., Ogata S., Yoon S., Fujino N., Kyogoku Y., Sasaki Y., Koarai A., Tamada T., Toyama A., Nakabayashi T., Kageyama L., Kyuwa S., Inaba K., Watanabe S., Nagy P., Sawa T., Oshiumi H., Ichinose M., Yamada M., Sugiura H., Wei F.-Y., Motohashi H., Akaike T. Nat. Commun. 2023;14:4476. doi: 10.1038/s41467-023-40182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takata T., Jung M., Matsunaga T., Ida T., Morita M., Motohashi H., Shen X., Kevil C.G., Fukuto J.M., Akaike T. Nitric Oxide. 2021;116:47–64. doi: 10.1016/j.niox.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akaike T., Ida T., Wei F.-Y., Nishida M., Kumagai Y., Alam M.M., Ihara H., Sawa T., Matsunaga T., Kasamatsu S., Nishimura A., Morita M., Tomizawa K., Nishimura A., Watanabe S., Inaba K., Shima H., Tanuma N., Jung M., Fujii S., Watanabe Y., Ohmuraya M., Nagy P., Feelisch M., Fukuto J.M., Motohashi H. Nat. Commun. 2017;8:1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mirzaei S., Mohammadi A.T., Gholami M.H., Hashemi F., Zarrabi A., Zabolian A., Hushmandi K., Makvandi P., Samec M., Liskova A., Kubatka P., Nabavi N., Aref A.R., Ashrafizadeh M., Khan H., Najafi M. Pharmacol. Res. 2021;167 doi: 10.1016/j.phrs.2021.105575. [DOI] [PubMed] [Google Scholar]
- 73.Tang C., Livingston M.J., Safirstein R., Dong Z. Nat. Rev. Nephrol. 2023;19:53. doi: 10.1038/s41581-022-00631-7. [DOI] [PubMed] [Google Scholar]
- 74.Guan J., Tong X., Zhang Y., Xu F., Zhang Y., Liang X., Jin J., Jing H., Guo L., Ni X., Fu J. Chem. Biol. Interact. 2021;349 doi: 10.1016/j.cbi.2021.109662. [DOI] [PubMed] [Google Scholar]
- 75.Moldogazieva N.T., Mokhosoev I.M., Feldman N.B., Lutsenko S.V. Free Radic. Res. 2018;52:507. doi: 10.1080/10715762.2018.1457217. [DOI] [PubMed] [Google Scholar]
- 76.Oh G.-S., Kim H.-J., Choi J.-H., Shen A., Choe S.-K., Karna A., Lee S.H., Jo H.-J., Yang S.-H., Kwak T.H., Lee C.-H., Park R., So H.-S. Kidney Int. 2014;85:547. doi: 10.1038/ki.2013.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.