Abstract
Purpose
We hypothesize that lower grade gliomas (LGG) can be identified and classified into two distinct subtypes: radiologically circumscribed Lower-Grade Gliomas (cLGG) and infiltrating Lower-Grade Gliomas (iLGG) based on radiological parameters and that these two different subtypes behave differently in terms of clinical outcomes.
Methods
We conducted a retrospective cohort study on surgical patients diagnosed with lower grade glioma over five years. Patient records and MRIs were reviewed, and neurosurgeons classified tumors into cLGG and iLGG groups.
Results
From the 165 patients in our cohort, 30 (18.2%) patients were classified as cLGG and 135 (81.8%) patients were classified as iLGG Mean age in cLGG was 31.4 years while mean age in iLGG was 37.9 years (p = 0.004). There was significant difference in mean blood loss between cLGG and iLGG groups (270 and 411 ml respectively, p = 0.020). cLGG had a significantly higher proportion of grade II tumors (p < 0.001). The overall mean survival time for the iLGG group was 14.96 ± 1.23 months, and 18.77 ± 2.72 months for the cLGG group. In univariate cox regression, the survival difference between LGG groups was not significant (HR = 0.888, p = 0.581), however on multivariate regression cLGG showed a significant (aHZ = 0.443, p = 0.015) positive correlation with survival. Intense contrast enhancement (HZ = 41.468, p = 0.018), blood loss (HZ = 1.002, p = 0.049), and moderately high Ki-67 (HZ = 4.589, p = 0.032) were also significant on univariate analyses.
Conclusion: cLGG and iLGG are radiologically distinct groups with separate prognoses, surgical experience, and associations.
Keywords: Circumscribed, Infiltrating, Lower-grade, Glioma, Clinical outcomes, Radiological features
Abbreviations
- cLGG
Circumscribed Lower-Grade Gliomas
- iLGG
Infiltrating Lower-Grade Gliomas
- CNS
Central Nervous System
- IDH
Isocitrate dehydrogenase
- PFS
Progression-free survival
- OS
Overall survival
- AKUH
Aga Khan University Hospital
- KPS
Karnofsky Performance Score
1. Introduction
Gliomas are tumors originating from glial cells in the brain parenchyma and are the most common type of neoplasm in the central nervous system (CNS).1 There are well-described radiological features to help differentiate between low- and high-grade gliomas, such as contrast-enhancement and necrosis.2 However, the grade II and III categories are more ambiguous, termed as lower-grade glioma (LGG). This is more so seen when assessing morphological characteristics of the lesion; invasion, surround edema as seen on T2 and FLAIR images, and variable contrast-enhancing patterns make it difficult to accurately assess whether a certain glioma is likely to be aggressive or benign on imaging alone. Texture analysis on MRIs can also be a significant predictor of early malignant transformation of lower-grade gliomas and has potential implications in treatment.3,4
The World Health Organization (WHO) conducted major revisions to its classification of diffuse lower-grade gliomas (LGGs).5, 6, 7 LGGs include WHO grade II and III oligodendroglial and astrocytic tumors, which predominantly have a mutation in the isocitrate dehydrogenase (IDH).6,8 Several MRI features have been found to have prognostic value as they differentiate between grades, diffuse, and discrete tumors but there is insufficient data to evaluate them having any influence on resectability of tumor, histopathological characteristics, and overall survival. These tumors can either have very distinct borders or an infiltrating appearance with ill-defined boundaries. These two differently appearing gliomas on MRI can have the same histopathological diagnosis but present as unique surgical dilemmas with distinct onco-functional outcomes.
The authors intend on characterizing the morphological patterns of LGG into two separate groups: radiologically circumscribed Lower-Grade Gliomas (cLGG) AND infiltrating Lower-Grade Gliomas (iLGG), based on radio–morphological parameters. In this study, we assess the proposed classification as a robust method of capturing the two distinct morphologies and characterizing the demographic distribution, diagnostic characteristics, treatment modalities, and outcomes (recurrence, functionality, progression-free survival [PFS], and overall survival [OS]).
Our proposed classification of LGGs into radiologically circumscribed and infiltrating is subjective. It may be prone to errors due to difference in experience and expertise of the observers. To make this process more objective, we also trained machine learning classifiers using various shape-based features on FLAIR MRI volume scans. Statistical shape analysis is an analysis of the geometrical properties of a set of shapes by statistical methods to model shape variations. Shape-based classification in brain tumor MRI, stems from the ability to run shape analysis and identify morphological abnormalities of the neuroanatomy. The shape features are extracted by the application of Spherical Harmonics (SPHARM). It is an extension of the fourier analysis, creating a parametric surface description by using an arbitrary shape function and expanding it onto a sphere using a set of orthogonal spherical functions as a basis functions.9 Earlier shape-based features have been used to classify brain tumors into regular vs irregular tumor and benign vs malignant tumor and showed reasonable accuracies with random forest classifiers.10,11
2. Methodology
We conducted a retrospective cohort study at the Aga Khan University Hospital (AKUH), in Karachi, Pakistan. Institutional Board approval was obtained from the Ethics Review Committee (ERC: 2021-6333-18753) at AKUH and patient records were obtained from January 2017 to December 2021. Data were extracted from medical records of patients diagnosed with lower-grade (grade II and grade III) glioma in the last five years (2017–2021). These patients had a confirmed histopathological diagnosis of a glioma brain tumor, had a surgical resection, and had Magnetic Resonance Imaging (MRI) available. Patients below the age of 18 and those who refused to give consent were excluded from the study. Patients with outsourced MRI were also excluded due to unavailability of imaging records. Histopathological diagnosis was made using the WHO 2016 criteria available at the time.
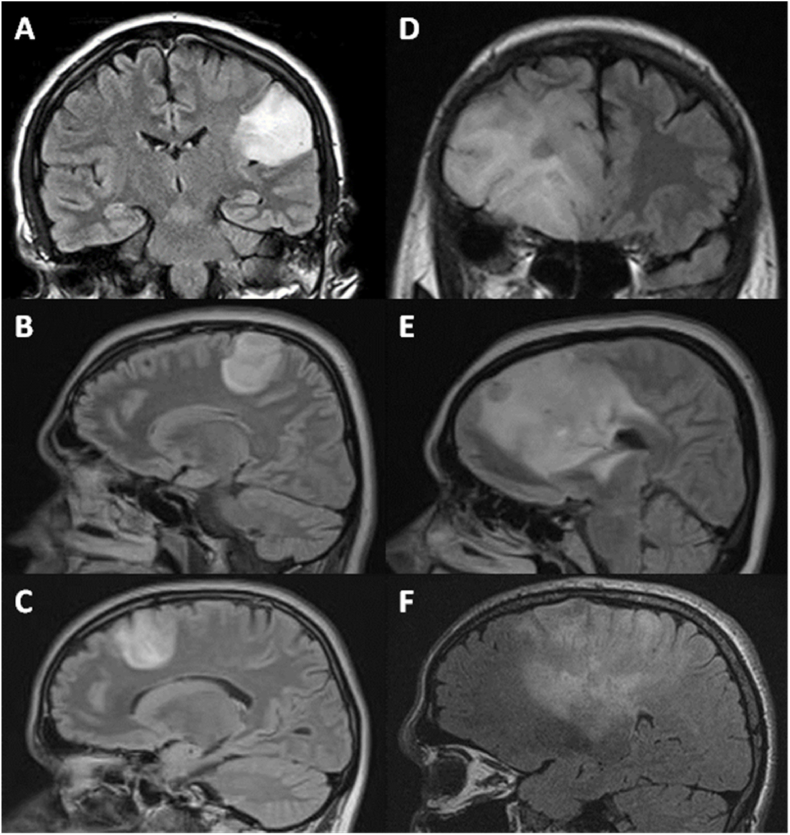
Patients were divided into the cLGG and iLGG group based on tumor morphology and appearance. Tumors with diffuse infiltration and indistinct edges were classified as iLGG, while tumors with discrete, well-defined edges and involving only one lobe were classified as cLGG. A neurosurgeon divided these tumors in two groups after reading T1, contrast enhanced T1, T2, and FLAIR sequences of each patient. Following are the examples of cLGG and iLGG (Fig. 1). Specific criteria established during the training and validation portion of data assessment were as follows: assessment of tumor boundaries, extent of infiltration into surrounding tissue, and tumor morphology/homogeneity. This was independently reviewed by the senior author.
Fig. 1.
MRI scans of cases showing radiologically circumscribed LGG (A, B, C) and diffuse LGG (D, E, F).
We investigated interobserver reliability by introducing a neurosurgical resident and informing him of the definition of the groups. 10 randomized MR numbers from each LGG group were chosen and he was asked to classify them, blinded to original results. The results were compared with the initial classification by our neurosurgical faculty. Inter observer reliability was 70% for our group.
Study variables included demographic information, such as age, gender, and marital status, duration of hospital stay, past medical and surgical history, family history. Details of the pathology i.e., tumor type, genetic/molecular analysis, KPS score pre-op and post-op, lesion location and size, and grade of tumor were recorded as well. Information about the surgical course i.e., extent of tumor resection, operating time, ASA levels, and blood loss was collected. Finally, post-surgical details such as recurrence, and last known status were recorded from the files.
Patients (who were not reported as expired) were called telephonically. Three attempts at contact were made with a gap of 20 min and 1 day respectively. Patients or attendants were asked for informed verbal consent and administered a questionnaire. The variables assessed were mortality status, and evidence of recurrence or progression on recent MRIs.
2.1. Plan of analysis
STATA version 15 and R was used for statistical analysis. Significance was assessed by independent t-test/Mann Whitney U test (for 2 groups) and ANOVA/Kruskal Walis test (for 3 groups) as appropriate. The qualitative variables have been reported as frequency and percentages and were assessed by chi-square/fisher exact test as appropriate.
Logistic regression was used to investigate correlations between variables and LGG group. Variables include in the univariate model were age, gender, grade, contrast enhancement, KPS pre- and post-op, blood loss, length of stay, surgery type, Ki-67, and progression/recurrence. Variables that had p < 0.25 or clinically relevant were carried over to the multivariate model. These included age, gender, grade, contrast enhancement, KPS pre- and post-op, blood loss, surgery type, and Ki-67.
The mean survival time of the participants was reported by Kaplan Meier curves and assessed by the log-rank test. The factors associated with overall survival have been determined by multivariate cox regression analysis and an unadjusted and adjusted hazard ratio with 95% CI has been reported. Variables of interest were age, gender, KPS at presentation, grade, blood loss, length of hospital stay, and cellularity. Univariate cox was reported with Hazard Ratio (HR), p-value, and confidence interval (CI), while multivariate was reported with Adjusted Hazard Ratio (HR), p-value, and confidence interval (CI). A p-value of <0.05 was considered significant throughout the study.
2.2. Machine learning classifier
Grade 2 and 3 gliomas with a pre-operative FLAIR volume MRI scan available were included (n = 118). These were annotated for glioma lesions by the clinical experts and classified into cLGG (n = 26) and iLGG (n = 92) cases using the 3D Slicer software. Handcrafted shape features were extracted by the application of Spherical Harmonics (SPHARM) and fed into machine learning algorithms. We used classical machine learning approaches including decision trees, random forests and support vector machine and compared their performance parameters (Fig. 2).
Fig. 2.
Pipeline showing development of machine learning classifier.
3. Results
From January 2017 through December 2021, 746 patients presented to our center with a diagnosis of glioma. Among these, 288 were grade 2 and grade 3 patients. MRI and clinical records were available for 165 patients and comprised the sample population for data analysis. The best accuracy for machine learning classification was achieved at 96.4% using Random Forest followed by 82.1% for both Decision Tree and Support Vector Machine.
3.1. Patient characteristics
From the 165 patients in our cohort, 135 (81.8%) patients were from the iLGG group, and 30 (18.2%) patients were from the cLGG group. Of these 165 patients, 118 (71.5%) patients were male, and 47 (28.5%) patients were female. Gender of the patient was not significantly associated with their respective LGG groups (p = 0.114).
The mean age was 36.7 ± 11.3 years overall, with the youngest patient being 18 years of age and the oldest 70 years. In the cLGG group the mean age was 31.4 ± 7.9 while the mean age in iLGG was 37.9 ± 11.6. Younger patients were more likely to be diagnosed with cLGG than with iLGG (p = 0.004). The demographic features of our population are summarized in Table 1. Presenting complaints are stratified by LGG group and demonstrated in Fig. 3 (supplement).
Table 1.
Demographic characteristics, surgical features, type of tumor, contrast enhancement and histopathological features of patients in the cLGG and iLGG groups.
| Demographic Characteristics | ||||||||
|---|---|---|---|---|---|---|---|---|
| cLGG |
iLGG |
p-value | ||||||
| Grade 2 |
Grade 3 |
Total |
Grade 2 |
Grade 3 |
Total |
|||
| n = 27 | n = 3 | n = 30 | n = 73 | n = 62 | n = 135 | |||
| Age (mean ± SD) | 31.1 ± 8.1 | 34.2 ± 4.9 | 31.4 ± 7.9 | 36.1 ± 9.9 | 39.7 ± 13.2 | 37.9 ± 11.6 | 0.004 | |
| Gender | Male | 22 | 3 | 25 (83.3%) | 53 | 39 | 93 (68.9%) | 0.443 |
| Female | 5 | 0 | 5 (16.7%) | 20 | 22 | 42 (31.1%) | ||
|
Average Length of Stay (days) |
1.7 |
1 |
1.6 |
2.2 |
2.1 |
2.2 |
0.207 |
|
| Surgical characteristics | ||||||||
| cLGG | iLGG | |||||||
| Grade 2 | Grade 3 | Total | Grade 2 | Grade 3 | Total | |||
| N (% within grade) |
N (% within grade) |
N (% within cLGG) |
N (% within grade) |
N (% within grade) |
N (% within iLGG) |
|||
| Gross total resection | 7 (43.8) | 3 (100.0) | 10 (52.6) | 22 (37.9) | 20 (46.5) | 42 (41.6) | ||
| Sub-total resection | 8 (50.0) | 0 (0.0) | 8 (42.1) | 34 (58.6) | 22 (51.2) | 56 (55.4) | ||
| Biopsy | 1 (6.3) | 0 (0.0) | 1 (5.3) | 2 (3.4) | 1 (2.3) | 3 (3.0) | ||
|
Blood loss (mean, in ml) |
263.8 ± 130.2 |
370 |
270 ± 128 |
404.4 ± 270.1 |
419.6 ± 210.3 |
410.6 ± 246.0 |
||
| Tumor type | ||||||||
| cLGG | iLGG | |||||||
| Grade 2 | Grade 3 | Total | Grade 2 | Grade 3 | Total | |||
| N (% within grade) |
N (% within grade) |
N (% within cLGG) |
N (% within grade) |
N (% within grade) |
N (% within iLGG) |
|||
| Oligodendroglioma | 18 (66.7) | 2 (66.7) | 20 (70) | 56 (80.0) | 34 (56.7) | 90 (69) | ||
|
Astrocytoma |
9 (33.3) |
1 (33.3) |
10 (30) |
14 (20.0) |
26 (43.3) |
40 (31) |
||
| Contrast enhancement | ||||||||
| cLGG | iLGG | |||||||
| Grade 2 | Grade 3 | Total | Grade 2 | Grade 3 | Total | |||
| N (%) |
N (%) |
N |
N (%) |
N (%) |
N |
|||
| None | 13 (54.2) | 1 (33.3) | 14 | 17 (24.6) | 11 (18.6) | 28 | ||
| Subtle | 10 (41.7) | 1 (33.3) | 11 | 45 (65.2) | 33 (55.9) | 78 | ||
| Definite | 1 (4.2) | 1 (33.3) | 2 | 3 (4.3) | 11 (18.6) | 14 | ||
|
Intense |
0 (0.0) |
0 (0.0) |
0 |
4 (5.8) |
4 (6.8) |
8 |
||
| Histopathological features | ||||||||
| cLGG | iLGG | |||||||
| Grade 2 | Grade 3 | Total | Grade 2 | Grade 3 | Total | |||
| N (% within grade) |
N (% within grade) |
N |
N (% within grade) |
N (% within grade) |
N |
|||
| High Ki67 | 1 (100.0) | 0 (0.0) | 1 | 8 (44.4) | 10 (55.6) | 18 | ||
| Nuclear atypia | 4 (80.0) | 1 (20.0) | 5 | 22 (48.9) | 23 (51.1) | 45 | ||
| Necrosis | 0 (0.0) | 0 (0.0) | 0 | 2 (18.2) | 9 (81.8) | 11 | ||
| Reactive gliosis | 4 (100.0) | 0 (0.0) | 4 | 20 (62.5) | 12 (37.5) | 32 | ||
| Vascular/endothelial proliferation | 2 (66.7) | 1 (33.3) | 3 | 10 (40.0) | 15 (60.0) | 25 | ||
| Increased Cellularity | 3 (50.0) | 3 (50.0) | 6 | 19 (30.2) | 44 (69.8) | 63 | ||
3.2. Surgical characteristics
The mean length of in-hospital stay was 1.9 days for our patients, it was lower for cLGG at 1.63 days as opposed to 2.15 days for iLGG. The mean KPS score at presentation was 78.3 ± 12.5 overall while the mean postoperative KPS was 76.3 ± 13.6. The average change in KPS was not significant in either of the groups.
Table 1 summarizes the surgical characteristics of the patients by grade and iLGG/cLGG group. There was a significant difference in mean blood loss between cLGG and iLGG patients. The overall mean intraoperative blood loss for both cohorts was 389 ml ± 237 ml. For the cLGG group, the mean blood loss was 270 ± 128 ml; for the iLGG group, it was 411 ± 246 ml (p = 0.020). In the type of surgical procedure performed, 11 patients from the cLGG group and 34 patients from the iLGG group were excluded from the analysis due to unavailable data.
3.3. Tumor characteristics
In the iLGG group 90 (69%) tumors were oligodendroglioma and 40 (31%) were astrocytoma, while the cLGG group had 20 (70%) Oligodendroglioma and 10 (30%) astrocytoma. 5 tumors from the iLGG group were not characterized because of missing data. There was no significant difference in the tumor type between the two LGG groups (p = 0.785). There was a significant difference in grade distribution amongst our proposed LGG groups, with cLGG having a higher proportion of grade II tumors (p < 0.001). Table 1 demonstrates the tumor type within each category.
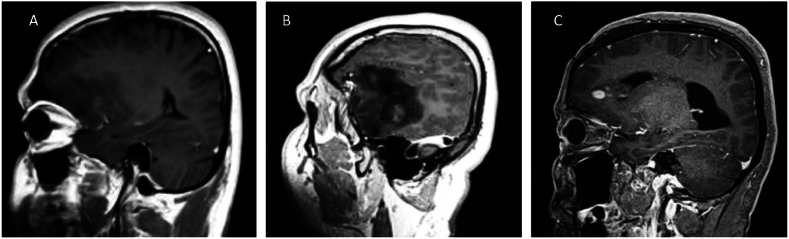
Contrast enhancement was assessed in 155 patients. Only iLGG tumors had intense contrast enhancement (p = 0.011), cLGG tumors were most likely to have no contrast enhancement (p = 0.013), while iLGG tumors were most likely to have subtle contrast enhancement (p < 0.001). Fig. 3 shows the different patterns of contrast enhancement. Table 1 demonstrates contrast enhancement stratified by grade and cLGG/iLGG group and Fig. 5 (supplement) compares the percentage of patients with contrast enhancement in each category according to LGG group. Histopathological features are also categorized according to grade and LGG group and summarized in Table 1.
Fig. 3.
Pattern of contrast enhancements. A: Subtle enhancement, B: Definite enhancement, C: Intense enhancement.
3.4. Regression analysis
In the univariate model, age had a significant correlation with lower grade glioma group (B coeff:-6.493, p = 0.004). Similarly grade 3 tumors also had a significant negative correlation with the cLGG group (OR = −0.223, p < 0.001). In terms of contrast enhancement, when compared to no enhancement, subtle enhancement (OR = −0.209, p = 0.003) and intense enhancement (OR = −0.333, p = 0.014) had a significant negative correlation with cLGG. Blood loss also had a strong correlation with the type of lower grade glioma. CLGG were likely to have much lower intraoperative blood loss than iLGG (B coeff:-140.652, p = 0.024). Similarly, iLGG were significantly more likely to have both definite and intense contrast enhancement when compared to cLGG (OR = −0.179, p = 0.008). Gender, KPS pre or post-op, length of stay, type of surgical intervention, and progression had no significant correlation with LGG group.
Variables that were significant (p < 0.05), that were clinically relevant, or that had a (p < 0.25) were carried forward to the multivariate analysis. These included age, gender, grade, contrast enhancement, pre-op and post-op KPS score, blood loss, length of stay, surgery type and Ki-67 levels. In the multivariate model subtle contrast enhancement (OR = −0.175, p = 0.035), subtotal resection (OR = −0.605, p = 0.028), and moderately high Ki-67 (OR = −0.213, p = 0.028) all had significant negative correlations with the LGG group (cLGG compared to iLGG). Our findings are summarized in Table 2 (supplement).
3.5. Survival analysis
Our patients were followed up for a mean of 69 months (5.8 years). At point of last follow-up, 71 patients (46.1%) had a progression or recurrence of the disease, while 82 patients (53.8%) had no reported recurrence or progression. From the patients with recurrence or progression, 55 (44% of the iLGG cohort) patients were from the iLGG group and 16 (55.2% of the cLGG cohort) were from the cLGG group. There was no significant association between LGG group and progression of disease. Twelve patients had no follow-up data available and were excluded from the analysis. From patients with known outcomes, 12 (8.3%) patients had passed away, and 132 (91.7%) patients were still alive. Of the deceased patients 2 (6.7% of the cLGG group) were from the cLGG group and 10 (7.4% of the iLGG group) were from the iLGG group. Twenty-one patients (12.7%) had no follow-up data available and were excluded from survival analysis.
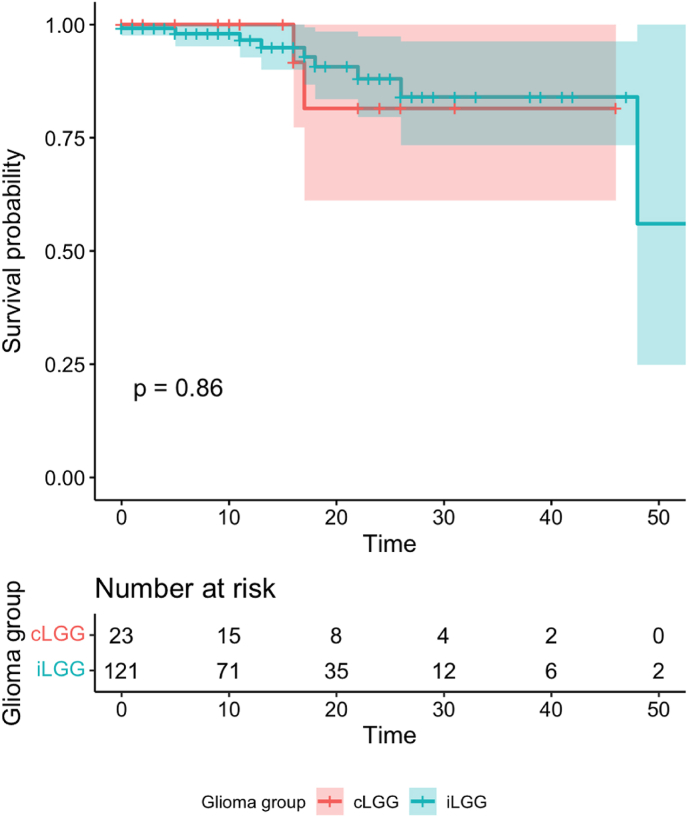
Survival analysis was performed using Kaplan Meier analysis with Log-rank test and Cox regression analysis for crude and adjusted hazard ratios. The results are depicted in Table 3 (supplement) and Fig. 4. The survival rate at 1 year was 94.6% with all 3 mortalities being from the iLGG group. Survival rate at 3 years was 92.9% for iLGG, and 90.5% for cLGG. At 5 years the survival rate for cLGG remained the same but for iLGG fell to 91.7%. The overall mean survival time for the iLGG group was 14.96 ± 1.23 months, and 18.77 ± 2.72 months for the cLGG group. The difference in survival between the LGG groups was not significant when stratified by type of surgery as shown in Fig. 7 (supplement). On univariate cox regression analysis, the survival difference between cLGG and iLGG group was not significant (HR = 0.888, p = 0.581), however on multivariate regression, cLGG showed a significant (aHZ = 0.443, p = 0.015) positive correlation with survival. Other positive variables on univariate cox regression were intense contrast enhancement (HZ = 41.468, p = 0.018), blood loss (HZ = 1.002, p = 0.049), and moderately high Ki-67 (HZ = 4.589, p = 0.032). However, the significance did not carry over to the multivariate model.
Fig. 4.
Kaplan Meier plot for survival analysis according to LGG group. (Time given in months).
4. Discussion
The current study proposes a novel approach to lower-grade gliomas in terms of radiological appearance translating to surgical, histological, and survival outcomes. Overall, cLGG tumors were more amenable to gross total resection compared to iLGG (42% vs. 36%). Our cohort indicates a trend for cLGG to be more likely present within younger patients. Radiologically, iLGG within our cohort were more likely to show significant (definite or intense) contrast enhancement compared to cLGG tumors. Intraoperatively, cLGG group tumors had a significantly lower mean blood loss, possibly indicating favorable surgical outcomes. Our histopathological analysis showed statistically significant higher Ki67 indices within iLGG (13.8% vs. 3.4%, p=0.003) and cellularity (56.25% vs. 24%, p=0.004). iLGG were also more likely to have higher nuclear atypia, necrosis, reactive gliosis and microvascular proliferation. These factors correlate with the significant difference in WHO grade, with cLGG tumors more likely to be reported as grade II glioma. Survival analysis yielded no significant difference (p = 0.257); however, median survival at 1 year and 5 years was higher for cLGG within our cohort (1 year: 9 vs. 7 months, 5 years: 75 vs. 63 months). Cox multivariate regression analysis indicates significant correlation of LGG type with survival. Proportional regression analysis showed significant associations of contrast enhancement, Ki67, and extent of resection in predicting for LGG subtype.
Lower-grade glioma are a heterogeneous group of tumors. Although grade III gliomas are thought to be more aggressive and require postoperative chemoradiotherapy, recent data has shown that morphological appearances of glioma correlate with distinct survival patterns and molecular signatures.12 Diffuse infiltration of glioma makes complete resection difficult, and this aggressive phenotype is associated with worse outcomes. On the other hand, radiologically circumscribed gliomas are easily accessible and completely resected. These two morphological patterns have been shown in previous studies to depict distinct molecular signatures.13 Our study applied a more stringent radiological criteria for differentiating grade II/III gliomas and validated this through evaluating key histopathological, surgical, and survival characteristics of our cohort. Within the literature, machine learning models have shown value in predicting subgroups of LGG according to CT and MRI findings with prediction of molecular subtypes as well.14 These models focus on features such as T2/FLAIR mismatch, contrast enhancement patterns, and clinical data. Similarly, our proposed model, although subjective, provides a nuanced and practicable approach for surgeons preoperatively evaluating LGGs for resectability and determining the patient's clinical course.
We see that patients had no significant difference in presenting complaints, with seizures as the most common symptom. Mean length of stay was higher for iLGG (2.2 vs. 1.6 days). Surgically, grade II cLGG and iLGG showed similar rates of resection – however, grade III cLGG were more likely to be completely resected compared to iLGG. A recent study in 2019 conducted on 172 diffuse LGG showed an association between greater surgical resection and overall survival, with a stronger impact with astrocytomas over oligondrogliomas.15 Similarly, a large cohort from 2018 evaluating postoperative glioma volume for grade II diffuse gliomas showed worse survival with greater postoperative tumor volume, with a similar strong association with astrocytomas over oligodendroglioma.16 Our findings show similar trends, with greater median 5-year survival for cLGG and higher rates of gross total resection in the cohort.
Darvishi et al discovered that preoperative MRI metrics of LGGs patients can offer prognostic information within molecularly defined classes. They found that contrast enhancement was associated with WHO grade III among IDHwt and IDHmut-Noncodel LGGs, but not IDHmut-Codel LGGs.17 This study highlights that MRIs are an exceptional imaging modality to identify brain tumors, but there is no existing classification to recognize the type or grade of lower-grade gliomas, such that disease progression and outcomes can be foretold on imaging alone.
Our study is limited by sample size – although we were able to report significance for a few key characteristics, greater differentiation may be seen with larger cohorts and the inclusion of a T1 post-contrast sequence. Our findings are also based on subjective assessment of the imaging. Lastly, we were unable to evaluate the significance of molecular markers for classifying LGG according to the 2021 WHO Classification of CNS Tumors, due to many of these cases having occurred before the change in criteria.18
5. Conclusion
The importance of prognosticating and evaluating the morphology of LGG will help optimize patient care and postsurgical outcomes. The application of our radiological criteria to a cohort of patients shows distinct patterns in terms of histology, surgical resection, and survival. It might also be practical to add cLGG and iLGG classifiers in WHO classification to better help classify in multidisciplinary discussions regarding patient management. Future research into validating these findings and developing radiological biomarkers may improve preoperative planning and prognostication.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Ethics approval
Institutional Board approval was obtained from the Ethics Review Committee (ERC: 2021-6333-18753) at AKUH.
CRediT authorship contribution statement
Ahsan Ali Khan: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Muhammad Usman Khalid: Writing – review & editing, Writing – original draft, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation. Mohammad Hamza Bajwa: Writing – review & editing, Writing – original draft, Validation, Software, Resources, Project administration, Methodology, Investigation. Faiza Urooj: Writing – original draft, Investigation, Data curation. Izza Tahir: Validation, Methodology, Investigation, Data curation. Meher Angez: Validation, Methodology, Investigation, Data curation. Fahad Zahid: Writing – review & editing, Data curation. Muhammad Waqas Saeed Baqai: Validation, Data curation. Kiran Aftab: Writing – review & editing, Writing – original draft, Validation, Investigation, Data curation. Shahabuddin Ansari: Validation, Data curation. Ummul Wara Khan: Validation, Data curation. Ali Azan Ahmed: Validation, Data curation. Syed Ather Enam: Writing – review & editing, Writing – original draft, Validation, Project administration, Methodology, Investigation, Conceptualization.
Declaration of competing interest
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.wnsx.2024.100356.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Mesfin F.B., Al-Dhahir M.A. StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan–. PMID: 28722904. 2023. Gliomas. StatPearls. [PubMed] [Google Scholar]
- 2.G C., M S., I G., et al. Cognitive functions in Repeated glioma surgery. Cancers. 2020;12 doi: 10.3390/CANCERS12051077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.BieÅ„kowski M., Wöhrer A., Moser P., et al. Molecular diagnostic testing of diffuse gliomas in the real-life setting: a practical approach. Clin Neuropathol. 2018;37:166–177. doi: 10.5414/NP301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capellades J., Teixidor P., Villalba G., et al. Results of a multicenter survey showing interindividual variability among neurosurgeons when deciding on the radicality of surgical resection in glioblastoma highlight the need for more objective guidelines. Clin Transl Oncol. 2017;19:727–734. doi: 10.1007/S12094-016-1598-6/TABLES/4. [DOI] [PubMed] [Google Scholar]
- 5.Louis D.N., Perry A., Reifenberger G., et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/S00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 6.Eckel-Passow J.E., Lachance D.H., Molinaro A.M., et al. Glioma groups based on 1p/19q, IDH, and TERT Promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMOA1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dj B., Rg V., Kd A., et al. Comprehensive, Integrative Genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMOA1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H., Parsons D.W., Jin G., et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMOA0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen L., Cong S., Inlow M. 2017. Statistical Shape Analysis for Brain Structures. Statistical Shape and Deformation Analysis: Methods, Implementation and Applications; pp. 351–378. [DOI] [Google Scholar]
- 10.Wu P., Xie K., Zheng Y., Wu C. Brain tumors classification based on 3D shape. Advances in Intelligent and Soft Computing. 2012;160 doi: 10.1007/978-3-642-29390-0_45/COVER. AISC:277–83. [DOI] [Google Scholar]
- 11.Asodekar B.H., Gore S.A., Thakare A.D. 2019 5th International Conference on Computing, Communication, Control and Automation. ICCUBEA; 2019. Brain tumor analysis based on shape features of MRI using machine learning. [DOI] [Google Scholar]
- 12.Olar A., Sulman E.P. Molecular markers in low-grade glioma-Toward tumor Reclassification. Semin Radiat Oncol. 2015;25:155–163. doi: 10.1016/J.SEMRADONC.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan J., Karsy M., Brock A.A., et al. Impact of a more restrictive overlapping surgery policy: an analysis of pre- and postimplementation complication rates, resident involvement, and surgical wait times at a high-volume neurosurgical department. J Neurosurg. 2018;129:515–523. doi: 10.3171/2017.5.JNS17183. [DOI] [PubMed] [Google Scholar]
- 14.Matsui Y., Maruyama T., Nitta M., et al. Prediction of lower-grade glioma molecular subtypes using deep learning. J Neuro Oncol. 2020;146:321–327. doi: 10.1007/S11060-019-03376-9. [DOI] [PubMed] [Google Scholar]
- 15.Patel S.H., Bansal A.G., Young E.B., et al. Extent of surgical resection in lower-grade gliomas: Differential impact based on molecular subtype. AJNR Am J Neuroradiol. 2019;40:1149–1155. doi: 10.3174/AJNR.A6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijnenga M.M.J., French P.J., Dubbink H.J., et al. The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol. 2018;20:103–112. doi: 10.1093/NEUONC/NOX176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darvishi P., Batchala P.P., Patrie J.T., et al. Prognostic value of preoperative MRI metrics for diffuse lower-grade glioma molecular subtypes. AJNR Am J Neuroradiol. 2020;41:815–821. doi: 10.3174/AJNR.A6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis D.N., Perry A., Wesseling P., et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/NEUONC/NOAB106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.