Abstract
The expression of human immunodeficiency virus type 1 (HIV-1) structural proteins requires the action of the viral trans-regulatory protein Rev. Rev is a nuclear shuttle protein that directly binds to its cis-acting Rev response element (RRE) RNA target sequence. Subsequent oligomerization of Rev monomers on the RRE and interaction of Rev with a cellular cofactor(s) result in the cytoplasmic accumulation of RRE-containing viral mRNAs. Moreover, Rev by itself is exported from the nucleus to the cytoplasm. Although it has been demonstrated that Rev multimerization is critically required for Rev activity and hence for HIV-1 replication, the number of Rev monomers required to form a trans-activation-competent complex on the RRE is unknown. Here we report a systematic analysis of the putative multimerization domains within the Rev trans-activator protein. We identify the amino acid residues which are part of the proposed single hydrophobic surface patch in the Rev amino terminus that mediates intermolecular interactions. Furthermore, we show that the expression of a multimerization-deficient Rev mutant blocks HIV-1 replication in a trans-dominant (dominant-negative) fashion.
The replication of human immunodeficiency virus type 1 (HIV-1) is characterized by a temporal pattern of viral mRNA expression. HIV-1 uses alternative splicing of its primary ∼9-kb transcript to generate the various mRNAs that are necessary for virus production (55, 77). Initially, only fully spliced mRNAs of the ∼2-kb class, encoding the regulatory proteins Tat, Nef, and Rev, are expressed (31, 67, 88, 92). While the Tat protein acts as a transcriptional activator of the viral long terminal repeat (LTR) promoter, Nef appears to be a regulatory protein that directly affects viral pathogenesis in infected individuals (for reviews, see references 20 and 21). The subsequent expression of the ∼9-kb and partly spliced ∼4-kb classes of viral mRNAs, encoding the structural proteins Gag, Pol, and Env, depends on the action of the Rev trans-activator (28, 40, 58). In the absence of Rev, these transcripts are retained in the cell nucleus (15, 41, 62, 76, 85, 93) and are either spliced to completion or subjected to degradation. In the presence of Rev, the incompletely spliced mRNAs accumulate in the cytoplasm to serve as templates for protein synthesis or as viral genomes (32, 65, 68). Rev activity is therefore essential for virus replication (30, 91, 94, 99).
Rev itself is a small phosphoprotein of 116 amino acids (aa) which, at steady state, accumulates primarily in the nucleoli of the host cell (14, 16, 22, 27, 43) and has the capacity to shuttle between the nucleus and the cytoplasm (53, 74, 86, 108). Nuclear localization is mediated by a short stretch of basic amino acids characterized by eight arginine residues located near the amino terminus of Rev (11, 16, 42, 48, 63, 104). In addition, this same protein domain (positions 33 to 46) is responsible for the binding of Rev to its cis-acting RNA target sequence, the Rev response element (RRE) (11, 24, 42, 44, 57, 70, 98, 109, 110), although some more-amino-terminal residues appear to contribute to binding specificity (26, 50). The RRE itself is encoded by sequences of the env gene (39, 68, 89) and is therefore present in all unspliced and incompletely spliced viral mRNAs, making them subject to Rev regulation.
A second essential region within Rev is located near the carboxy terminus and has been mapped to residues 75 to 93 (47, 69, 73, 103, 105). Mutation of the four leucine residues within this domain results in nonfunctional Rev proteins that retain wild-type nuclear localization and RRE binding characteristics but also display a pronounced dominant-negative (trans-dominant) phenotype (6, 29, 63, 66). Therefore, this domain appears to mediate effector functions and has been defined as the activation domain. Recently, this domain was shown to contain a nuclear export signal which, when conjugated to heterologous proteins, is sufficient to mediate their transport from the nucleus to the cytoplasm in eukaryotic cells (33, 75, 106). These findings suggest that the signals for nuclear RNA export reside in proteins that act as carriers to transport RNA across the nuclear envelope. At least in Xenopus oocytes, this viral mRNA transport appears to be independent of any pre-mRNA splicing events (34).
It is therefore expected that the Rev activation domain provides the site of interaction with host cell proteins that are involved in nuclear-cytoplasmic translocation. Indeed, various cellular proteins which appear to be specific binding partners of the Rev activation domain have been described; these include nucleoporin-like protein hRIP/Rab (10, 36), eukaryotic initiation factor 5A (eIF-5A) (90), and nuclear export factor CRM1, which appears to be a general nuclear export signal receptor (35, 37, 81, 95). For eIF-5A, nonfunctional mutants with a block in Rev trans-activation and therefore HIV-1 replication in human T cells (7, 52) were recently described, demonstrating a novel way to interfere with the viral life cycle: by inhibition of Rev-mediated nuclear export.
In addition, an important and often overlooked aspect of Rev with respect to biological activity is its ability to form multimers. It has been suggested that this multimerization event is responsible for a certain threshold level of intracellular Rev proteins and that it is this threshold level which must be overcome in order to establish a productive HIV-1 infection, thereby regulating viral latency (83, 84). Rev has a very high tendency to aggregate in solution (24, 54, 79, 107, 109) and has been reported to form homomultimeric complexes in vivo even in the absence of RRE RNA (9, 49, 59, 80, 96, 110). However, with respect to Rev-mediated viral RNA export and hence Rev biological activity, the formation of a Rev multimer on the RRE has been shown to be more critical (17, 18, 25, 51, 61, 64, 102, 111). Random mutational analysis of the rev gene revealed that the residues within Rev that participate in these intermolecular interactions reside in the amino terminus of the protein, in regions flanking the basic domain responsible for RNA binding and nuclear localization (61, 64). However, little is known about their precise localization and how many Rev molecules form the biologically active complex on the RRE, since as many as 12 Rev proteins have been shown to bind to the RRE in vitro (25, 56, 71, 110, 111).
This study was undertaken to characterize the regions in HIV-1 Rev which mediate the formation of homo-oligomeric complexes on the RRE in more detail.
MATERIALS AND METHODS
Molecular clones.
Expression plasmids pcRev and pcTat encode cDNA copies of the rev and tat genes, respectively, of isolate HXB-3 (67). Expression vectors encoding Rev mutants RevM4, RevM5, RevM6, RevM8, RevM10, RevM16, RevM23, and RevTC5 have been described in detail elsewhere (4, 63, 100). In transfection experiments, the vector pBC12/CMV was used to maintain constant input DNA levels and pBC12/CMV/IL-2 served as a negative control plasmid (19). Constructs pBC12/CMV/βGal (90) and pBC12/RSV/SEAP (5) have been used for internal control of transfection efficiencies. pDM128/CMV is a well-established Rev-responsive reporter construct encoding the bacterial chloramphenicol acetyltransferase (CAT) gene (48, 69). Construct pSLIIB/CAT contains an HIV-1 LTR promoter in which the trans-activation response element (TAR) sequence has been replaced by the RRE-derived stem-loop IIB (SLIIB) high-affinity Rev binding site (101). Plasmids in which the tat gene is fused to the rev gene (pcTat/Rev), encoding the fusion proteins Tat-Rev, Tat-RevM4, Tat-RevM5, and Tat-RevM10, have been described elsewhere (61). Plasmid pGEX-Rev is a bacterial expression plasmid that expresses Rev fused to the carboxy terminus of glutathione S-transferase (GST) (7). pGEM/RRE is an in vitro transcription vector that contains the RRE (64).
Oligonucleotide-directed mutagenesis with a bacteriophage M13 mutagenesis system (United States Biochemicals, Cleveland, Ohio) was used to introduce targeted nucleotide substitutions encoding amino acid alterations into the rev gene of pcRev. All introduced mutations were confirmed by DNA sequence determination with Sequenase 2.0 (United States Biochemicals). Variants of pGEX-Rev and pcTat/Rev possessing mutated rev genes were generated by exchange of the wild-type gene for the respective mutated gene by PCR technology. Finally, the coding regions of the constructs were confirmed by DNA sequence determination as described above. Protein expression of all mutants that were generated in the course of this study was confirmed by specific immunoprecipitation analysis with radiolabelled protein extracts from transiently transfected COS cells.
Cell cultures, transfections, and assays.
COS and HeLa cells were maintained and transfected with DEAE-dextran and chloroquine or calcium phosphate as previously described (101). Rev trans-activation was investigated by cotransfection of 2.5 × 105 COS cells with 375 ng of pDM128/CMV DNA and 75 ng of pBC12/CMV/βGal DNA, together with 250 ng of either pcREV (positive control), pBC12/CMV/IL-2 (negative control), or mutant Rev expression plasmid. At ∼60 h posttransfection, cell lysates were prepared and the levels of β-galactosidase activity were measured as described previously (101). These values were subsequently used to determine the amount of cell extract to be assayed for CAT by an enzyme-linked immunosorbent assay (ELISA) (Boehringer GmbH, Mannheim, Germany).
Dominant-negative inhibition of Rev function was investigated by cotransfection of 2.5 × 105 COS cells with 250 ng of HIV-1Δrev proviral DNA (87) and 250 ng of pBC12/RSV/SEAP DNA (transfection efficiency control), together with 125 ng of pcRev and 1.25 μg of mutant Rev expression plasmid. Total input DNA was kept constant in all transfections by inclusion of the parental vector pBC12/CMV. At ∼60 h posttransfection, cell supernatants were assayed for secreted alkaline phosphatase (SEAP) (5) and p24 Gag protein synthesis.
In vivo RNA binding of Tat-Rev fusion proteins was assessed by cotransfecting 2.5 × 105 HeLa cells with 2.5 μg of pSLIIB/CAT reporter plasmid, 1.5 μg of pBC12/CMV/βGal plasmid, and 2.5 μg of either pcTat/Rev (positive control), pcTat/RevM5 (negative control), or pcTat/mutant Rev expression vector. At 42 h posttransfection, cell lysates were prepared and the levels of β-galactosidase activity were determined. These values were then used to adjust the amount of protein extract to be assayed in a radioactive CAT assay (101).
The multimerization capacity of Rev proteins was assessed by cotransfection of 2.5 × 105 HeLa cells with 1.5 μg of pSLIIB/CAT reporter plasmid, 1.5 μg of internal control plasmid (pBC12/CMV/βGal), 1.5 μg of pcTat/RevM5, and 1.5 μg of either pcRev (positive control), pcRevM5 (negative control), or mutant Rev expression plasmid. At 42 h posttransfection, cell lysates were prepared for determination of β-galactosidase and CAT activities.
Purification of GST-Rev fusion proteins and in vitro RRE binding.
Wild-type Rev and mutant Rev were expressed as carboxy-terminal fusions to GST in Escherichia coli BL21. The fusion proteins were purified from crude cell lysates by affinity chromatography on glutathione-Sepharose 4B according to the manufacturer’s protocol (Pharmacia Biotech, Vienna, Austria). Eluted protein fractions were analyzed by Rev-specific Western analysis, pooled, concentrated by ultrafiltration with a PM10 filter device (Amicon Inc., Beverly, Mass.), and stored at −70°C. The final protein concentrations were determined by the method of Bradford (12). RNA binding assays with a radiolabelled 252-nucleotide RRE probe were performed as previously described (25), except that RNA-protein complexes were resolved on 6% polyacrylamide gels.
Retrovirus-mediated gene transfer and HIV-1 challenge experiments.
The retroviral vectors were constructed by inserting PCR-generated rev genes into the single XhoI site of retroviral vector pBC140 (6). The rev sequences were subsequently confirmed by DNA sequencing as described above. The amphotropic GP + envAm12 (Am12) packaging cell line (72) was transfected with the retroviral DNAs, and CD4+ human CEM cells were subsequently inoculated with virus-containing infectious Am12 supernatants and selected for neomycin-resistant populations as described previously (7, 8). RNA expression of the transgene in clonal populations of the transduced CEM cells was characterized as described previously (6).
CD4+ rev-transduced CEM cells (2.0 × 106) were infected with 2,000 tissue culture infective doses (TCID) of HIV-1 strain SF2 (60) at an ambient temperature for 2 h. Subsequently, the cells were washed three times, transferred into fresh RPMI 1640 medium containing 10% fetal bovine serum, and incubated at 37°C. On days 4, 7, and 11 of the incubation period, HIV-1 replication, measured by p24 Gag protein synthesis with an antigen capture assay (ELISA), and cell counts, measured with a Coulter device, were determined.
RESULTS
Biophysical measurements obtained with circular dichroism and fluorescence spectra provided the initial insights into the structure of the HIV-1 Rev protein (1). These data indicated that most of the amino-terminal half of Rev (residues 1 to 61–66), which contains the RNA binding-nuclear localization domain (98) as well as the putative multimerization domains (61, 64), is in an α-helical conformation. In particular, this region appears to form a helix-loop-helix motif via intramolecular hydrophobic contacts between two α helices that are separated by a stretch of proline-rich residues (1). Additional functional data obtained from mutagenesis studies (100) refined this structural model of the Rev amino terminus and allowed the identification of specific hydrophobic amino acids that are exposed on the surface of Rev and that form a surface for potential intermolecular interactions (Fig. 1A). Interestingly, the previously described nonfunctional Rev mutants RevM4 and RevM7 (63), which are unable to form multimers on their RRE RNA target in vitro (64), map to this region of the Rev protein (Fig. 1B). Using these data as a starting point, we set out to investigate the sequence requirements for HIV-1 Rev-specific multimer formation in more detail.
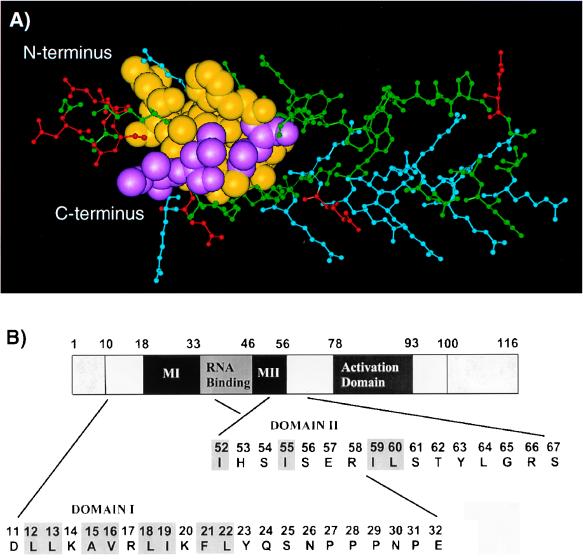
FIG. 1.
Location of the putative HIV-1 Rev multimerization domains. (A) Model of the Rev amino terminus, redrawn from Thomas et al. (100), shows the helix-loop-helix motif of Rev residues 1 to 60. It is evident that the two putative multimerization domains form a single exposed hydrophobic surface patch which presumably constitutes an oligomerization interface. Amino acids are colored as follows: basic residues (Arg, Lys), blue; acidic residues (Asp, Glu), red; residues analyzed in this study, yellow; and residues identified in this study as important for intermolecular interactions, purple. (B) Diagrammatic representation of the 116-aa Rev trans-activator protein showing the functional domains and the regions mutagenized. The two protein regions that form a putative multimerization surface are marked as domains MI and MII. The amino acid sequences of both regions systematically mutagenized in this study are shown as expanded sections. Residues potentially involved in multimerization and previously identified by examination of the refined structural model of Rev (100) are highlighted by shaded boxes.
Mutational analysis of the putative Rev multimerization domains.
As outlined in Fig. 1B, the two regions of the Rev protein (here termed domains MI and MII) that create a potential multimerization surface were extensively mutagenized by introducing scanning missense mutations (Table 1). All mutant genes generated appeared to express protein at levels comparable to that of the wild-type rev gene, as tested by Rev-specific immunoprecipitation analysis with protein extracts of transiently transfected COS cells (data not shown). In order to test the functionality of these Rev mutants, we used the Rev-responsive reporter construct pDM128/CMV. This reporter plasmid contains the gene encoding CAT and the RRE target sequence of Rev, both of which are positioned between HIV-1 splice sites and are under the control of the cytomegalovirus immediate-early promoter (48, 69). RNA produced by pDM128/CMV therefore has a single intron containing the CAT gene and the RRE sequence, which is removed when the RNA is spliced. As HIV-1 splice sites are inefficiently recognized in primate cells (13), significant levels of both spliced and unspliced pDM128/CMV-specific mRNAs accumulate in the cell nucleus. However, in the presence of functional Rev protein, the unspliced message is exported to the cytoplasm, resulting in high levels of CAT expression. Therefore, by cotransfection of pDM128/CMV and the rev vector in question into COS cells, we were able to monitor Rev trans-activation.
TABLE 1.
Rev mutants generated by oligonucleotide-directed mutagenesisa
| Mutant | Position(s) (aa) | Domain | Mutation |
|---|---|---|---|
| RevSLT1 | 12, 13 | MI | LL to DD |
| RevSLT2 | 13, 14 | MI | LK to DD |
| RevSLT3 | 14, 15 | MI | KA to DD |
| RevSLT4 | 15, 16 | MI | AV to DD |
| RevSLT5 | 16, 17 | MI | VR to DD |
| RevSLT6 | 17, 18 | MI | RL to DD |
| RevSLT7 | 18, 19 | MI | LI to DD |
| RevSLT8 | 19, 20 | MI | IK to DD |
| RevSLT9 | 20, 21 | MI | KF to DD |
| RevSLT10 | 21, 22 | MI | FL to DD |
| RevSLT11 | 22, 23 | MI | LY to DD |
| RevSLT12 | 23, 24 | MI | YQ to DL |
| RevSLT13 | 24, 25 | MI | QS to LL |
| RevSLT14 | 25, 26 | MI | SN to LL |
| RevSLT15 | 26, 27 | MI | NP to LL |
| RevSLT16 | 27, 28 | MI | PP to DD |
| RevSLT17 | 28, 29 | MI | PP to DD |
| RevSLT18 | 29, 30 | MI | PN to DL |
| RevSLT19 | 30, 31 | MI | NP to LD |
| RevSLT20 | 31, 32 | MI | PE to DL |
| RevSLT21 | 16 | MI | V to D |
| RevSLT22 | 18 | MI | L to D |
| RevSLT23 | 19 | MI | I to D |
| RevSLT24 | 21 | MI | F to A |
| RevSLT25 | 22 | MI | L to A |
| RevSLT26 | 22 | MI | L to D |
| RevSLT27 | 23 | MI | Y to A |
| RevSLT28 | 23 | MI | Y to D |
| RevSLT29 | 21, 22, 23 | MI | FLY to AAA |
| RevSLT30 | 15, 16, 18, 19, 22 | MI | All to D |
| RevSLT31 | 15, 16, 18, 19, 22 | MI | 15 to L, rest to A |
| RevSLT32 | 51, 52 | MII | QI to LD |
| RevSLT33 | 52, 53 | MII | IH to DD |
| RevSLT34 | 53, 54 | MII | HS to DL |
| RevSLT35 | 54, 55 | MII | SI to DL |
| RevSLT36 | 55, 56 | MII | IS to DL |
| RevSLT37 | 56, 57 | MII | SE to LL |
| RevSLT38 | 57, 58 | MII | ER to LD |
| RevSLT39 | 58, 59 | MII | RI to DD |
| RevSLT40 | 59, 60 | MII | IL to DD |
| RevSLT41 | 60, 61 | MII | LS to DD |
| RevSLT42 | 62, 63 | MII | TY to AE |
| RevSLT43 | 63, 64 | MII | YL to ED |
| RevSLT44 | 64, 65 | MII | LG to DK |
| RevSLT45 | 65, 66 | MII | GR to KE |
| RevSLT46 | 66, 67 | MII | RS to EA |
| RevM16b | 11, 12 | MI | DL to ED |
| RevM4b | 23, 25, 26 | MI | YSN to DDL |
| RevM5b | 38, 39 | RNA-NLS | RR to DL |
| RevM6b | 41, 42, 43, 44 | RNA-NLS | RR to DL, rest deleted |
| RevM23b | 48, 49 | RNA | RQ to DL |
| RevTC5b | 52 | MII | I to D |
| RevM8b | 61, 62 | MII | ST to DL |
| RevM10b | 78, 79 | Activation | LE to DL |
The protein name, the position(s) mutated within the 116-aa Rev protein, the putative protein domain affected by the introduced mutation(s), and the amino acid changes introduced are shown. MI, multimerization domain I; MII, multimerization domain II; RNA-NLS, RNA binding-nuclear localization domain; Activation, protein effector domain.
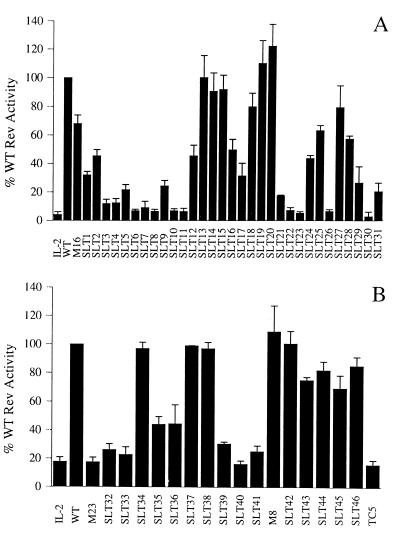
Figure 2 summarizes the results of the functional analysis of Rev domains MI and MII. These and all other transfection experiments in this study were internally controlled for various transfection efficiencies by inclusion of the constitutive control vector pBC12/CMV/βGal (90). All CAT values are expressed as a percentage of wild-type Rev activity (set arbitrarily to 100%) and were corrected for background (mock) activity. The scanning functional analysis of the two putative multimerization domains revealed a wide range of functional phenotypes, ranging from wild-type activity through partial activity to complete nonfunctionality (Fig. 2). The detection of mutants with partial activity indicated a moderately important functional role for the mutated residues. However, a series of mutants showed 10% or less wild-type trans-activation activity and were essentially completely nonfunctional. Evaluation of all mutant phenotypes obtained allowed us to identify amino acid residues Ala15, Val16, Leu18, Ile19, and Leu22 in domain MI as being critical for Rev function and therefore potentially involved in multimer formation (1, 100). Residues Ile52, Ile59, and Leu60 appear to be the functionally important residues in domain MII. The lack of biological activity seen with mutant RevM23, which was included as a control in these experiments, resulted from its inability to localize to the cell nucleus (4). Upon analysis of the data obtained, the important residues identified by functional dissection of domains MI and MII correlated well with the residues previously identified by examination of a refined Rev model (100) as components of the exposed hydrophobic patches which form the potential multimerization interfaces of Rev (Fig. 1B).
FIG. 2.
Biological activities of HIV-1 Rev mutant proteins. (A) Scanning mutagenesis of putative multimerization domain MI. Rev trans-activation capacity was determined by cotransfection of COS cell monolayers with the Rev-responsive reporter plasmid pDM128/CMV, the various mutant Rev expression plasmids (indicated on the x axis; see also Table 1), and the constitutive internal control vector pBC12/CMV/βGal. Data are expressed as a percentage of wild-type (WT) Rev activity (set to 100%), and the error bars represent the standard deviations of four independent experiments. All CAT values were adjusted for transfection efficiency by determining the level of β-galactosidase in each culture and were corrected for background (mock) activity. IL-2, negative control plasmid pBC12/CMV/IL-2; WT, wild-type Rev (pcRev). (B) Scanning mutagenesis of putative multimerization domain MII. Details are as for panel A.
RNA binding and multimerization characteristics of nonfunctional Rev mutants.
Use of the pDM128/CMV-based trans-activation assay allowed nonfunctional Rev proteins to be identified but provided no insight into the reason for their aberrant function. Therefore, the ability of selected mutants identified above to bind to and multimerize on the RRE target RNA was investigated.
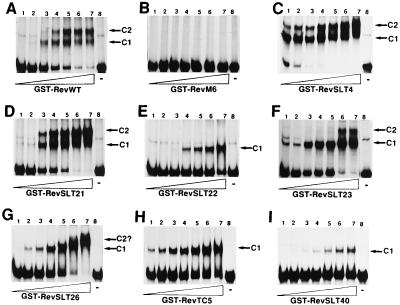
The RRE binding characteristics of Rev mutants RevSLT4, RevSLT21 to RevSLT23, RevSLT26, RevSLT40, and RevTC5 (Table 1) were first assessed in vitro by RNA gel retardation analysis. For this, the respective proteins were expressed and purified in the context of fusions to GST and then analyzed in combination with an in vitro-transcribed RRE RNA probe (42) (Fig. 3). Despite the fact that GST may itself form homodimers, GST-Rev fusion proteins have been used to successfully identify in vitro multimerization-deficient Rev mutants (64), provided that the addition of increasing amounts of GST-Rev to a preformed initial complex does not cause higher-order complexes. Control experiments confirmed that the addition of increasing amounts of Rev wild-type protein to the binding reaction mixture resulted in the successive appearance of RNA-protein complexes with slower mobilities in nondenaturing gel electrophoresis (C1 and C2 in Fig. 3A). These complexes were not detected when mutant RevM6 was used (Fig. 3B); this mutant is characterized by an internal deletion in the RNA binding domain and therefore serves as a negative control for RRE recognition (63, 64). Inspection of the data revealed that nonfunctional Rev mutants RevSLT22, RevTC5, and RevSLT40 retained their in vitro RRE binding capacity (Fig. 3E, H, and I, respectively). However, all of these proteins lacked the ability to bind to the RRE in a cooperative manner, a result which is indicated by the formation of a single retarded complex (C1), even at the highest protein concentrations tested. An intermediate result was observed with RevSLT26 protein in these experiments (Fig. 3G). RevSLT26 clearly bound to the RRE and formed the initial C1 complex. However, at higher protein concentrations, no distinct multimeric complexes comparable to those seen with the wild-type protein were formed. This result may indicate that RevSLT26 is severely impaired in this in vitro multimerization assay.
FIG. 3.
In vitro RRE binding analysis of wild-type and mutant GST-Rev proteins. A constant level of a 32P-labelled RRE RNA probe was incubated with increasing amounts (30 ng to 2 μg, lanes 1 to 7) of wild-type Rev (RevWT) (A) or the indicated Rev mutant fusion proteins (B to I) and then subjected to RNA gel mobility shift analysis. In each case, lane 8 contained free (unbound) RRE RNA. The various distinct complexes formed upon Rev binding to the RRE RNA probe (C1 and C2) are indicated to the right of each panel.
It was recently shown that Rev mutants that are multimerization deficient in vitro do in fact form multimers in vivo (61, 64). The reason for this obvious discrepancy is not yet known. Nevertheless, in order to characterize the selected nonfunctional Rev mutant proteins as fully as possible with respect to RRE binding and multimerization capabilities, we also tested them in vivo.
The primary high-affinity Rev binding site within the RRE was previously assigned to a region known as SLIIB (3, 18, 51, 82, 102). The identification of this site allowed the development of a reporter gene-based in vivo system to measure the binding capacity (101) and, moreover, the multimerization capacity of Rev on its SLIIB RNA target site (61).
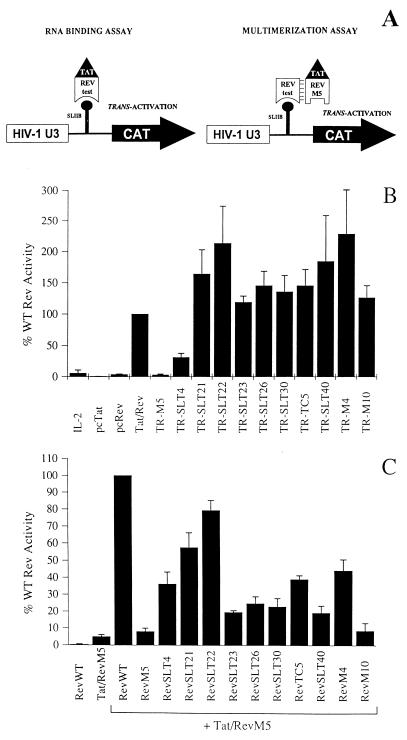
As depicted in Fig. 4A, the pSLIIB/CAT reporter construct used for these assays contains the CAT gene under the transcriptional control of the HIV-1 LTR promoter. The wild-type TAR element, which is the promoter-proximal RNA target sequence of the HIV-1 Tat transcriptional trans-activator, is replaced by a 29-nucleotide RNA sequence encoding SLIIB of the RRE (101). This promoter is only activated by Tat-Rev fusion proteins and is not responsive to Tat or Rev alone. Thus, the quantity of CAT produced gives an indication of the RNA binding ability of a given Rev mutant tested in the context of a Tat-Rev fusion protein.
FIG. 4.
RNA binding and multimerization activities of nonfunctional Rev mutant proteins in a mammalian cell culture system. (A) Diagrammatic representation of the pSLIIB/CAT reporter system. Activation of a chimeric HIV-1 LTR promoter in which the TAR sequence was replaced by the RRE SLIIB element (101) allowed the monitoring of in vivo RRE SLIIB RNA binding and multimer formation by Rev (see the text for further details). (B) In vivo RNA binding activity of nonfunctional Rev mutants. HeLa cells were cotransfected with reporter construct pSLIIB/CAT, the various Tat-Rev (TR) expression plasmids (indicated on the x axis), and constitutive internal control plasmid pBC12/CMV/βGal. pBC12/CMV/IL-2 (IL-2), pcTat, pcRev, and pcTat/Rev (Tat/Rev) were included as controls. At 42 h posttransfection, protein extracts were prepared to assay for β-galactosidase and CAT levels. Data are expressed as a percentage of wild-type (WT) Rev activity (set to 100%) and were corrected for background (mock) activity. (C) Analysis of Rev multimerization in vivo. The ability of the selected nonfunctional Rev mutants to form multimers was determined by cotransfection of HeLa cells with reporter construct pSLIIB/CAT, pcTat/RevM5 (an RNA binding-negative mutant), the various mutant Rev expression plasmids (indicated on the x axis), and constitutive internal control plasmid pBC12/CMV/βGal. pcRev (RevWT) and pcTat/RevM5 (Tat/RevM5) alone were included as negative controls. At 42 h posttransfection, levels of β-galactosidase and CAT were determined as described above. Data are expressed as a percentage of wild-type (WT) Rev activity (set to 100%). Error bars represent the standard deviations of four independent experiments.
As expected, cotransfection of HeLa cells with an unrelated control plasmid expressing human interleukin-2 (IL-2) or with expression vectors encoding HIV-1 Tat or Rev did not result in trans-activation of the pSLIIB/CAT reporter construct (Fig. 4B). However, significant CAT activity was detected when an expression plasmid encoding a Tat-Rev fusion protein was included in the transfection. This promoter activation was not detected when the SLIIB binding-incompetent Tat-RevM5 mutant protein (61) was used in these experiments. The data obtained clearly demonstrated that all of our nonfunctional Rev mutant proteins bound to RRE SLIIB in vivo, although mutant RevSLT4 appeared to have impaired activity in this assay (Fig. 4B). In general, the RNA binding data obtained in this in vivo assay correlated well with the data from the RNA gel retardation assays presented above.
Next, the ability of the mutants to multimerize in vivo was tested with a similar assay that also used the pSLIIB/CAT reporter construct. The principle of this assay is explained diagrammatically in Fig. 4A. The assay relies on the ability of the test mutant to rescue the nonbinding phenotype of another mutant, Tat-RevM5, by the formation of a Rev-Rev multimer (61). The Tat portion of the nonbinding RNA fusion protein is thus brought into close proximity with the LTR promoter, resulting in trans-activation. Figure 4C shows the activities of the nonfunctional Rev mutants in this assay. Negative controls of wild-type Rev or Tat-RevM5 alone were inactive in these experiments. Likewise, when Tat-RevM5 was assayed with RevM5, only background levels of activity were measured. Control mutants RevM4 and RevM10 (63) gave the phenotypes that had been previously described (61), while the test mutants resulted in a variety of different phenotypes. RevSLT22 multimerized with essentially wild-type efficiency. RevSLT4, RevSLT21, and RevTC5 showed a partial phenotype. Rev mutants RevSLT23, RevSLT26, and RevSLT40 as well as the control RevSLT30 all multimerized with approximately 20% wild-type activity, and we therefore considered these mutants multimerization defective in this artificial mammalian cell culture system. Interestingly, testing of multimerization capacity resulted in contradictory data for some of these nonfunctional Rev mutants. For example, no in vitro multimer formation was detected with RevSLT22 and RevTC5 (Fig. 3E and H). In contrast, both proteins clearly displayed multimerization activity in the mammalian cell culture system (Fig. 4C). With RevSLT23, multimerization activity was detected in vitro but not in vivo (compare Fig. 3F and Fig. 4C). However, the same result was already reported in earlier studies. In particular, mutant RevM4 was originally proposed to be a prototype multimerization-deficient Rev protein due to its multimerization deficiency in RNA gel retardation experiments (64). A subsequent study, however, showed that RevM4 multimerizes about 40% as effectively as wild-type Rev in the in vivo assay (61), although there is no good explanation for this discrepancy. One explanation might be that the interaction of mutant proteins is measured in the in vitro assay, while the in vivo assay reflects the ability of a mutant Rev protein to interact with the wild-type protein. Nevertheless, considering both in vitro and in vivo data generated in this study, two mutant proteins, namely, RevSLT26 and RevSLT40, appeared to be inactive due to an inherent inability to form functional multimeric complexes.
trans-Dominant inhibition of HIV-1 replication.
It was previously shown that Rev multimerization is required for Rev activity (17, 18, 25, 51, 61, 64, 102, 111). Therefore, true multimerization-deficient mutants should be able to block Rev function in a dominant-negative manner. In theory, these Rev mutants, when present in trans, should act as competitive inhibitors of the wild-type protein by sequestering RRE-containing viral RNAs.
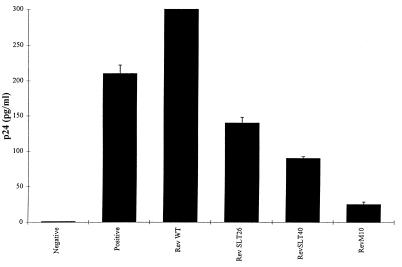
To test this hypothesis, we first investigated the effects of RevSLT26 and RevSLT40 on Rev trans-activation using a provirus rescue assay. As described before (87), cotransfection of COS cells with an HXB-2-derived Rev-deficient proviral DNA (HIV-1Δrev) and pcRev resulted in the Rev-dependent accumulation of p24 Gag antigen in COS cell supernatants (Fig. 5). As expected, the additional coexpression (10-fold excess) of wild-type Rev protein increased the release of p24 Gag into the supernatants. However, the expression of RevSLT26 or RevSLT40 protein clearly correlated with the inhibition of wild-type Rev function. Obviously, the strong inhibitory effect seen with the RevM10 protein, which is characterized by a mutation in the Rev activation domain and has been described so far as the most powerful trans-dominant Rev protein (69), could not be reached by the RevSLT26 or RevSLT40 protein in this transient assay. Notwithstanding this result, these experiments indicated that both RevSLT26 and RevSLT40 might have a moderate dominant-negative potential, with the inhibitory effect of RevSLT40 appearing to be more pronounced (Fig. 5).
FIG. 5.
Competitive inhibition of Rev function. COS cell cultures were cotransfected with the HXB-2-derived Rev-deficient proviral DNA HIV-1Δrev, pBC12/RSV/SEAP (internal control vector), and pBC12/CMV/IL-2 (negative), pcRev (positive), or pcRev plus a 10-fold excess of expression vector encoding wild-type (WT) Rev, RevSLT26, RevSLT40, or RevM10. Total input DNA was maintained at a constant level by inclusion of parental vector pBC12/CMV. All p24 Gag antigen values were adjusted for transfection efficiency by determining the level of SEAP (5) in each culture and were corrected for background (mock) activity. Error bars represent the standard deviations of three independent experiments.
To test the RevSLT40 phenotype in a more rigorous way, we investigated the ability of RevSLT40 to inhibit HIV-1 replication. For this, retroviral vector pBC140 (6) was used for retrovirus-mediated gene transfer of various rev genes into human CEM T cells. Transduced, neomycin-resistant CD4+ CEM cell clones were characterized for transgene expression by Northern analysis (data not shown) and subsequently inoculated with 2,000 TCID of replication-competent HIV-1 strain SF2 (60). The numbers of cells in the cultures were determined on days 0, 4, 7, and 11 postinfection, and p24 Gag protein levels were assayed as a measure of HIV-1 replication (Table 2). In all cases, infected cells proliferated at the same rate as uninfected control cells (data not shown), indicating that constitutive expression of the mutant rev genes was not toxic for the cells. As shown in Table 2, CEM cells transduced with pBC140 alone provided no protection against HIV-1 infection. Likewise, significant levels of p24 Gag antigen were measured in the culture containing CEM clone RevM4, a mutant previously described to be multimerization defective in vitro (61, 64) and serving as a control in these experiments. The two CEM cell lines expressing Rev mutant RevSLT40 (RevSLT40/1 and RevSLT40/2; Table 2), which were considered to be multimerization deficient in vitro and in vivo, gave low levels of p24 Gag antigen when supernatants were assayed at day 4, 7, or 11 postinfection. RevM10, serving as a positive control for the trans-dominant phenotype, strongly inhibited virus replication, as has been reported previously (6, 63, 66).
TABLE 2.
trans-Dominant inhibition of HIV-1 replicationa
| CEM clone | 106 Cells/ml (p24 Gag protein level, pg/ml) on day:
|
|||
|---|---|---|---|---|
| 0 | 4 | 7 | 11 | |
| Vector | 0.2 (120) | 2.6 (1,141) | 2.4 (1,951) | 2.0 (>12,300) |
| RevM4 | 0.2 (125) | 3.8 (1,230) | 3.4 (386) | 2.4 (>12,300) |
| RevSLT40/1 | 0.2 (118) | 3.4 (824) | 3.0 (72) | 2.6 (1,860) |
| RevSLT40/2 | 0.2 (122) | 3.8 (411) | 3.4 (60) | 2.8 (2,373) |
| RevM10 | 0.2 (110) | 2.4 (62) | 3.0 (25) | 2.5 (617) |
Transduced CEM cell lines were infected with 2,000 TCID of HIV-1 strain SF2 (60) on day 0. After 2 h at room temperature, the cells were washed three times and transferred into fresh medium, and p24 levels were determined; these data represent day 0. On days 4 and 7 postinfection, cells were counted and p24 levels in the medium were measured. Subsequently, the cells were washed three times, and 0.2 × 106 infected cells were reseeded into fresh medium. On day 11 postinfection, cells were counted and the levels of p24 in cell supernatants were measured by an ELISA. Vector, retrovirus vector pBC140 (6) from which all rev transgenes were expressed.
DISCUSSION
The rev gene of HIV-1 encodes a key activity for the complex regulation of viral gene expression. Rev acts early in the HIV-1 replication cycle to bring about the export from the nucleus to the cytoplasm of incompletely spliced and unspliced viral mRNAs encoding the viral structural proteins (reviewed in references 23 and 46). Numerous independent studies have determined a number of specific requirements for full Rev activity, resulting in a frequently updated model of Rev trans-activation. The protein must be localized in the correct subcellular compartment (the nucleus), must be able to bind directly and specifically to its RRE RNA target sequence, and, after multimerization by cooperative binding of further Rev molecules, must interact via its activation domain with one or more cellular cofactors. These cofactors, including nucleoporin-like proteins such as hRIP/Rab (10, 36, 97), CRM1 (35, 37, 81, 95), and nuclear eIF-5A (7, 90), are thought to mediate the subsequent nuclear export of Rev (reviewed in reference 38). Rev was recently shown to be a shuttling protein (53, 74, 86, 108); therefore, it is thought that after Rev enters the cytoplasm and dissociates from the RRE, it then returns to the nucleus to transport further viral messages.
To date, these aspects of Rev function have been addressed mostly separately and not in a complete model system. Therefore, a number of important questions still remain unanswered. For example, the amino acid residues in Rev which mediate Rev multimer formation have not been fully characterized, and no true multimerization-deficient Rev mutants (defined as inactive in the in vitro and in vivo assays) have been identified.
However, the capacity of Rev to form multimers and the clear requirement of multimer formation for Rev function have placed this viral trans-regulatory protein at the center of models which describe the regulation of viral latency. It has been shown that some cell lines which are nonproductively infected with HIV-1 are characterized by low Rev protein levels (84). Reaching a critical intracellular threshold level of Rev protein obviously correlates with Rev function and allows virus production (83). Thus, the transition from a quiescent (latent) state to a productive state of infection appears to be controlled in these cells at least in part by the ability of Rev to multimerize (64).
Advances in the determination of the structure of Rev have allowed the development of a refined structural model of the amino terminus of the protein (1, 100). This model has provided the first indication that the Rev multimerization interface, although generated by two separate protein regions, may actually form a single interaction area in the functional protein structure (Fig. 1A). Interestingly, the previously described Rev mutants RevM4 and RevM7, which appear to be defective in their ability to multimerize in vitro (64), map to these regions. The detailed functional analysis of these regions carried out in this study has allowed the amino acid residues that are critical for Rev activity to be identified (Fig. 2). Of note, these experimentally defined residues coincide perfectly with the amino acids which have been suggested by the structural model of the Rev amino terminus to form the Rev dimerization interface (1, 100).
Testing the nonfunctional Rev mutants generated in this study in both in vitro and in vivo multimerization assay systems revealed conflicting multimerization activities (e.g., RevSLT22 and RevSLT23 [Fig. 3 and 4]). Such inconsistency among phenotypes obtained with two different assay systems was previously reported for the RevM4 protein (61, 64) and led to some confusion as to what the precise requirements for Rev multimer formation are. However, these data also emphasized that a combination of both in vitro and in vivo assays for Rev multimer formation is needed in order to identify true multimerization-deficient Rev mutants.
Analysis of mutants generated in this study allowed dissection of the RevM4 mutant (63) (YSN to DDL at aa positions 23, 25, and 26) phenotype. Individual mutations in the component residues revealed no functional importance for these residues with respect to multimerization. It therefore seems likely that the partial multimerization deficiency phenotype seen for RevM4 is the result of a localized structural change which is due to the introduction of three mutations in close proximity and which consequently affects the α helix in this region. Likewise, it appears that the isoleucine residue at position 55, rather than the residues on either side (Ser54 and Ser56) that are deleted in RevM7 (63), is the functionally important residue which produces the multimerization deficiency phenotype and that it is the highly localized structural disruption caused by the mutation in this region which is responsible for the complete loss of Rev function.
Despite efficiently binding to its RNA target in vitro, nonfunctional mutant RevSLT4 appeared to be at least partially active in RNA binding and multimerization in vivo. Thus, some defect other than one in the capacity to multimerize might have been responsible for its nonfunctionality. We were, however, able to attribute the functional inactivity of mutants RevSLT26 and RevSLT40 to their impaired ability to form Rev multimers. The phenotypes of these two mutants identified amino acid residues Leu22, Ile59, and Leu60 as being essential for multimerization. Obviously, this result is in agreement with the structural models of the Rev amino terminus which suggested that these hydrophobic residues (Leu22, Ile59, and Leu60) are exposed and presumably create a single intermolecular oligomerization surface (1, 100).
A number of trans-dominant Rev mutants have been identified, but so far the best characterized appears to be RevM10 (2, 6, 29, 63, 66, 69). The basis of the trans-dominant phenotype of an activation domain mutant such as RevM10 is thought to be an inherent inability to interact with a cellular cofactor. Therefore, by extension of the same idea, a multimerization-deficient mutant such as RevSLT26 or RevSLT40 should also show a dominant-negative phenotype. This was indeed found to be the case in a transient transfection assay carried out to investigate the trans-dominant phenotypes of RevSLT26 and RevSLT40 (Fig. 5). Dominant-negative phenotypes were observed for RevSLT40 and, to a lesser degree, for RevSLT26, although the inhibitory effects were not as great as those seen for RevM10. The molecular basis for the trans-dominant phenotypes of multimerization-deficient Rev mutants such as RevSLT26 and RevSLT40 is likely to be an ability to bind to the RRE but not form the homomultimeric complex which is required either to allow Rev to assume the correct conformation for interaction of one or more cellular cofactors with the activation domain or to allow the formation of a multicomponent binding site to which these cofactors then bind. Hence, by sequestering all available RRE-containing RNA, multimerization-deficient Rev mutants should be able to competitively inhibit wild-type Rev function.
The single most trans-dominant multimerization-deficient mutant identified (RevSLT40) in the transient assay was then tested for its ability to inhibit virus replication. Upon challenge of an RevSLT40-transduced human T-lymphocyte cell line with high doses of replication-competent HIV-1 strain SF2, significant inhibition of virus replication was seen during the experiments (up to 11 days postinfection). Of note, these virus challenge experiments were designed to achieve maximal virus replication. The virus titers used (2,000 TCID) are comparable to those typically seen in the peripheral blood mononuclear cells of patients suffering from full-blown AIDS (45). Clearly, the HIV-1 inhibitory level seen with multimerization-deficient mutant RevSLT40 did not reach the level seen with activation domain mutant RevM10 (Table 2). However, this result is almost to be expected since, as mentioned above, a multimerization-deficient mutant is only able to compete with wild-type Rev for RRE binding, while an activation domain mutant should, at least in theory, also be able to enter preexisting Rev complexes, thereby rendering them inactive. Therefore, activation domain mutants such as RevM10 are still the trans-dominant reagents of choice for the clinical development of anti-HIV somatic gene therapies (78). Although the trans-dominant multimerization-deficient mutant RevSLT40 will, for this reason, most likely not be used in these types of gene intervention strategies, the detailed characterization in this study of the regions in Rev which are required for protein dimerization and the generation of true multimerization-deficient mutants such as RevSLT40 will provide the tools to allow further analysis of the biologically active multimeric Rev complex in the future. It is obvious that the detailed determination of how many Rev monomers form the trans-activation-competent complex will have consequences for cofactor interaction and therefore Rev-mediated nuclear export.
ACKNOWLEDGMENTS
We thank Lotte Hofer and Johannes Pertl for excellent technical assistance and Thomas Baumruker for helpful discussions during the course of this study.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft (to M.O. and P.H.; SFB 466).
REFERENCES
- 1.Auer M, Gremlich H-U, Seifert J-M, Daly T J, Parslow T G, Casari G, Gstach H. Helix-loop-helix motif in HIV-1 Rev. Biochem. 1994;33:2988–2996. doi: 10.1021/bi00176a031. [DOI] [PubMed] [Google Scholar]
- 2.Bahner I, Zhou C, Yu X-J, Hao Q-L, Guatelli J C, Kohn D B. Comparison of trans-dominant inhibitory mutant human immunodeficiency virus type 1 genes expressed by retroviral vectors in human T lymphocytes. J Virol. 1993;67:3199–3207. doi: 10.1128/jvi.67.6.3199-3207.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 4.Berger J, Aepinus C, Dobrovnik M, Fleckenstein B, Hauber J, Böhnlein E. Mutational analysis of functional domains in the HIV-1 Rev trans-regulatory protein. Virology. 1991;183:630–635. doi: 10.1016/0042-6822(91)90992-k. [DOI] [PubMed] [Google Scholar]
- 5.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 6.Bevec D, Dobrovnik M, Hauber J, Böhnlein E. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc Natl Acad Sci USA. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bevec D, Jaksche H, Oft M, Wöhl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, Lottspeich F, Hauber J. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science (Washington, DC) 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 8.Bevec D, Volc-Platzer B, Zimmermann K, Dobrovnik M, Hauber J, Veres G, Böhnlein E. Constitutive expression of chimeric neo-Rev response element transcripts suppresses HIV-1 replication in human CD4+ T lymphocytes. Hum Gene Ther. 1994;5:193–201. doi: 10.1089/hum.1994.5.2-193. [DOI] [PubMed] [Google Scholar]
- 9.Bogerd H, Greene W C. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J Virol. 1993;67:2496–2502. doi: 10.1128/jvi.67.5.2496-2502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 11.Böhnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 13.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 14.Cochrane A, Kramer R, Ruben S, Levine J, Rosen C A. The human immunodeficiency virus rev protein is a nuclear phosphoprotein. Virology. 1989;171:264–266. doi: 10.1016/0042-6822(89)90535-7. [DOI] [PubMed] [Google Scholar]
- 15.Cochrane A W, Jones K S, Beidas S, Dillon P J, Skalka A M, Rosen C A. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J Virol. 1991;65:5305–5313. doi: 10.1128/jvi.65.10.5305-5313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochrane A W, Perkins A, Rosen C A. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol. 1990;64:881–885. doi: 10.1128/jvi.64.2.881-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole J L, Gehman J D, Shafer J A, Kuo L C. Solution oligomerization of the Rev protein of HIV-1: implications for function. Biochemistry. 1993;32:11769–11775. doi: 10.1021/bi00095a004. [DOI] [PubMed] [Google Scholar]
- 18.Cook K S, Fisk G J, Hauber J, Usman N, Daly T J, Rusche J R. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991;19:1577–1583. doi: 10.1093/nar/19.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 20.Cullen B R. Does HIV-1 Tat induce a change in viral initiation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- 21.Cullen B R. The role of Nef in the replication cycle of the human and simian immunodeficiency viruses. Virology. 1994;205:1–6. doi: 10.1006/viro.1994.1613. [DOI] [PubMed] [Google Scholar]
- 22.Cullen B R, Hauber J, Campbell K, Sodroski J G, Haseltine W A, Rosen C A. Subcellular localization of the human immunodeficiency virus trans-acting art gene product. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cullen B R, Malim M H. The HIV-1 Rev protein: prototype of a novel class of eukaryotic post-transcriptional regulators. Trends Biochem Sci. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 24.Daly T J, Cook K S, Gray G S, Maione T E, Rusche J R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature (London) 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 25.Daly T J, Doten R C, Rennert P, Auer M, Jaksche H, Donner A, Fisk G, Rusche J R. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev responsive element. Biochemistry. 1993;32:10497–10505. doi: 10.1021/bi00090a028. [DOI] [PubMed] [Google Scholar]
- 26.Daly T J, Doten R C, Rusche J R, Auer M. The amino terminal domain of HIV-1 Rev is required for discrimination of the RRE from nonspecific RNA. J Mol Biol. 1995;253:243–258. doi: 10.1006/jmbi.1995.0549. [DOI] [PubMed] [Google Scholar]
- 27.Dundr M, Leno G H, Hammarskjöld M-L, Rekosh D, Helga-Maria C, Olson M O J. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- 28.Emerman M, Vazeux R, Peden K. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 29.Escaich S, Kalfoglou C, Plavec I, Kaushal S, Mosca J D, Böhnlein E. RevM10-mediated inhibition of HIV-1 replication in chronically infected T cells. Hum Gene Ther. 1995;6:625–634. doi: 10.1089/hum.1995.6.5-625. [DOI] [PubMed] [Google Scholar]
- 30.Feinberg M B, Jarret R F, Aldovini A, Gallo R C, Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 31.Felber B K, Drysdale C M, Pavlakis G N. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 34.Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 36.Fritz C C, Zapp M L, Green M R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 38.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 39.Hadzopoulou-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammarskjöld M-L, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammarskjöld M-L, Li H, Rekosh D, Prasad S. Human immunodeficiency virus env expression becomes Rev independent if the env region is not defined as an intron. J Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammerschmid M, Palmeri D, Ruhl M, Jaksche H, Weichselbraun I, Böhnlein E, Malim M H, Hauber J. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans activator. J Virol. 1994;68:7329–7335. doi: 10.1128/jvi.68.11.7329-7335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hauber J, Bouvier M, Malim M H, Cullen B R. Phosphorylation of the rev gene product of human immunodeficiency virus type 1. J Virol. 1988;62:4801–4808. doi: 10.1128/jvi.62.12.4801-4804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 45.Ho D D, Moudgil T, Alam M. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 46.Hope T, Pomerantz R J. The human immunodeficiency virus type 1 Rev protein: a pivotal protein in the viral life cycle. Curr Top Microbiol Immunol. 1995;193:91–105. doi: 10.1007/978-3-642-78929-8_5. [DOI] [PubMed] [Google Scholar]
- 47.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hope T J, Huang X, McDonald D, Parslow T G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hope T J, Klein N P, Elder M E, Parslow T G. trans-Dominant inhibition of human immunodeficiency virus type 1 Rev occurs through formation of inactive protein complexes. J Virol. 1992;66:1849–1855. doi: 10.1128/jvi.66.4.1849-1855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hope T J, McDonald D, Huang X, Low J, Parslow T G. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J Virol. 1990;64:5360–5366. doi: 10.1128/jvi.64.11.5360-5366.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwai S, Pritchard C, Mann D A, Karn J, Gait M J. Recognition of the high affinity binding site in rev-response element RNA by the human immunodeficiency virus type-1 rev protein. Nucleic Acids Res. 1992;20:6465–6472. doi: 10.1093/nar/20.24.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Junker U, Bevec D, Barske C, Kalfoglou C, Escaich S, Dobrovnik M, Hauber J, Böhnlein E. Intracellular expression of cellular eIF-5A mutants inhibits HIV-1 replication in human T cells: a feasibility study. Hum Gene Ther. 1996;7:1861–1869. doi: 10.1089/hum.1996.7.15-1861. [DOI] [PubMed] [Google Scholar]
- 53.Kalland K-H, Szilvay A M, Brokstad K A, Sætrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karn J, Dingwall C, Finch J T, Heaphy S, Gait M J. RNA binding by the tat and rev proteins of HIV-1. Biochimie. 1991;73:9–16. doi: 10.1016/0300-9084(91)90068-c. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, Byrn R, Groopman J, Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989;63:3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kjems J, Brown M, Chang D D, Sharp P A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kjems J, Calnan B J, Frankel A D, Sharp P A. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knight D M, Flomerfelt F A, Ghrayeb J. Expression of the art/trs protein of HIV and study of its role in viral envelope synthesis. Science (Washington, DC) 1987;236:837–840. doi: 10.1126/science.3033827. [DOI] [PubMed] [Google Scholar]
- 59.Kubota S, Furuta R, Maki M, Hatanaka M. Inhibition of human immunodeficiency virus type 1 Rev function by a Rev mutant which interferes with nuclear/nucleolar localization of Rev. J Virol. 1992;66:2510–2513. doi: 10.1128/jvi.66.4.2510-2513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy J A, Hoffman A D, Kramer S M, Landis J A, Shimabukuro J M, Oshiro L S. Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science (Washington, DC) 1984;225:840–842. doi: 10.1126/science.6206563. [DOI] [PubMed] [Google Scholar]
- 61.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 62.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malim M H, Böhnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 64.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 65.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. Stable expression of transdominant Rev protein in human T cells inhibits human immunodeficiency virus replication. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malim M H, Hauber J, Fenrick R, Cullen B R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature (London) 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 68.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 69.Malim M H, McCarn D F, Tiley L S, Cullen B R. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 71.Mann D A, Mikaélian I, Zemmel R W, Green S M, Lowe A D, Kimura T, Singh M, Butler P J G, Gait M J, Karn J. A molecular rheostat. Co-operative Rev binding to stem I of the Rev-response element modulates human immunodeficiency virus type-1 late gene expression. J Mol Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 72.Markowitz D, Goff S, Bank A. Construction and use of a safe and efficient amphotropic packaging cell line. Virology. 1988;167:400–406. [PubMed] [Google Scholar]
- 73.Mermer B, Felber B K, Campbell M, Pavlakis G N. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 1990;18:2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 75.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mikaelian I, Krieg M, Gait M J, Karn J. Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNAs. J Mol Biol. 1996;257:246–264. doi: 10.1006/jmbi.1996.0160. [DOI] [PubMed] [Google Scholar]
- 77.Muesing M A, Smith D H, Cabradilla C D, Benton C V, Lasky L A, Capon D J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature (London) 1985;313:450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 78.Nabel G J, Fox B A, Post L, Thompson C B, Woffendin C. A molecular genetic intervention for AIDS—effects of a transdominant negative form of Rev. Hum Gene Ther. 1994;5:79–92. doi: 10.1089/hum.1994.5.1-79. [DOI] [PubMed] [Google Scholar]
- 79.Nalin C M, Purcell R D, Antelman D, Mueller D, Tomchak L, Wegrzynski B, McCarney E, Toome V, Kramer R, Hsu M C. Purification and characterization of recombinant Rev protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7593–7597. doi: 10.1073/pnas.87.19.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olsen H S, Cochrane A W, Dillon P J, Nalin C M, Rosen C A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990;4:1357–1364. doi: 10.1101/gad.4.8.1357. [DOI] [PubMed] [Google Scholar]
- 81.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science (Washington, DC) 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 82.Peterson R D, Bartel D P, Szostak J W, Horvath S J, Feigon J. 1H NMR studies of the high-affinity Rev binding site of the Rev responsive element of HIV-1 mRNA: base pairing in the core binding element. Biochemistry. 1994;33:5357–5366. doi: 10.1021/bi00184a001. [DOI] [PubMed] [Google Scholar]
- 83.Pomerantz R J, Seshamma T, Trono D. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J Virol. 1992;66:1809–1813. doi: 10.1128/jvi.66.3.1809-1813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pomerantz R J, Trono D, Feinberg M B, Baltimore D. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model of latency. Cell. 1990;61:1271–1276. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 85.Reddy T R, Kraus G, Suhasini M, Leavitt M C, Wong-Staal F. Identification and mapping of inhibitory sequences in the human immunodeficiency virus type 2 vif gene. J Virol. 1995;69:5167–5170. doi: 10.1128/jvi.69.8.5167-5170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richard N, Iacampo S, Cochrane A. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology. 1994;204:123–131. doi: 10.1006/viro.1994.1516. [DOI] [PubMed] [Google Scholar]
- 87.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Functional replacement of the HIV-1 rev protein by the HTLV-I rex protein. Nature (London) 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 88.Robert-Guroff M, Popovic M, Gartner S, Markham P, Gallo R C, Reitz M S. Structure and expression of tat-, rev-, and nef-specific transcripts of human immunodeficiency virus type 1 in infected lymphocytes and macrophages. J Virol. 1990;64:3391–3398. doi: 10.1128/jvi.64.7.3391-3398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosen C A, Terwilliger E, Dayton A, Sodroski J G, Haseltine W A. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, Hauber J. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sadaie M R, Benter T, Wong-Staal F. Site-directed mutagenesis of two trans-regulatory genes (tat-III, trs) of HIV-1. Science (Washington, DC) 1988;239:910–914. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- 92.Schwartz S, Felber B K, Benko D M, Fenyö E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwartz S, Felber B K, Pavlakis G N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of the Rev protein. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sodroski J, Goh W C, Rosen C, Dayton A, Terwilliger E, Haseltine W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature (London) 1986;321:412–417. doi: 10.1038/321412a0. [DOI] [PubMed] [Google Scholar]
- 95.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 96.Stauber R, Gaitanaris G A, Pavlakis G N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 97.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 98.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. RNA recognition by an isolated α helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 99.Terwilliger E, Burghoff R, Sia R, Sodroski J, Haseltine W, Rosen C. The art gene product of human immunodeficiency virus is required for replication. J Virol. 1988;62:655–658. doi: 10.1128/jvi.62.2.655-658.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas S L, Hauber J, Casari G. Probing the structure of the HIV-1 Rev trans-activator protein by functional analysis. Protein Eng. 1997;10:103–107. doi: 10.1093/protein/10.2.103. [DOI] [PubMed] [Google Scholar]
- 101.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 102.Tiley L S, Malim M H, Tewary H K, Stockley P G, Cullen B R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci USA. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Venkatesh L K, Chinnadurai G. Mutants in a conserved region near the carboxy-terminus of HIV-1 Rev identify functionally important residues and exhibit a dominant negative phenotype. Virology. 1990;178:327–330. doi: 10.1016/0042-6822(90)90414-m. [DOI] [PubMed] [Google Scholar]
- 104.Venkatesh L K, Mohammed S, Chinnadurai G. Functional domains of the HIV-1 rev gene required for trans-regulation and subcellular localization. Virology. 1990;176:39–47. doi: 10.1016/0042-6822(90)90228-j. [DOI] [PubMed] [Google Scholar]
- 105.Weichselbraun I, Farrington G K, Rusche J R, Böhnlein E, Hauber J. Definition of the human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex protein activation domain by functional exchange. J Virol. 1992;66:2583–2587. doi: 10.1128/jvi.66.4.2583-2587.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 107.Wingfield P T, Stahl S J, Payton M A, Venkatesan S, Misra M, Steven A J. HIV-1 rev expressed in recombinant Escherichia coli: purification, polymerization and conformational properties. Biochemistry. 1991;30:7527–7534. doi: 10.1021/bi00244a023. [DOI] [PubMed] [Google Scholar]
- 108.Wolff B, Cohen G, Hauber J, Meshcheryakova D, Rabeck C. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp Cell Res. 1995;217:31–41. doi: 10.1006/excr.1995.1060. [DOI] [PubMed] [Google Scholar]
- 109.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature (London) 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 110.Zapp M L, Hope T J, Parslow T G, Green M R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci USA. 1991;88:7734–7738. doi: 10.1073/pnas.88.17.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zemmel R W, Kelley A C, Karn J, Butler P J G. Flexible regions of RNA structure facilitate co-operative Rev assembly on the Rev-response element. J Mol Biol. 1996;258:763–777. doi: 10.1006/jmbi.1996.0285. [DOI] [PubMed] [Google Scholar]