Abstract
Adenovirus type 5 E4 open reading frame 4 (E4orf4) protein has been previously shown to counteract transactivation of the junB and c-fos genes by cyclic AMP plus E1A protein and to interact with protein phosphatase 2A (PP2A). Here, we show that the wild-type E4orf4 protein induces apoptosis in the E1A-expressing 293 cells, in NIH 3T3 cells transformed with v-Ras, and in the lung carcinoma cell line H1299. The induction of apoptosis is not accompanied by enhanced levels of p53 in 293 cells and occurs in the absence of p53 in H1299 cells, indicating involvement of a p53-independent pathway. A mutant E4orf4 protein that had lost the ability to induce apoptosis also lost its ability to bind PP2A. We suggest that E4orf4 antagonizes continuous signals to proliferate, like those given by E1A or v-Ras, and that the conflicting signals lead to the induction of cell death.
Apoptosis, or programmed cell death, is a genetically controlled cellular process, which leads cells to commit suicide in response to a variety of stimuli. Regulation of cell death is essential for normal development and is an important defense against viral infection and the emergence of cancer. Increased cell death can lead to impaired development and degenerative diseases, whereas decreased cell death can lead to cancer and persistent viral infection. Apoptosis is accompanied by morphological changes, including loss of cell-cell contact, cell rounding, loss of cell volume, and condensation of the nucleus with subsequent nuclear fragmentation and DNA degradation (4). The process of apoptosis is controlled through the expression of a large number of genes, many of which are conserved from nematodes to mammals and viruses (reviewed in reference 60).
Adenoviruses (Ads) encode proteins that function as inducers of apoptosis, as well as proteins that inhibit apoptosis (50, 53). Ad E1A protein increases p53 levels and induces p53-dependent apoptosis (10, 29, 46). During Ad infection of p53-negative cells, E1A can induce apoptosis in a p53-independent fashion, a process which may involve products of early region 4 (E4) of Ad (30, 54).
The E4 gene region was shown to be required for lytic virus growth, and it provides functions that facilitate viral DNA replication, accumulation of nuclear and cytoplasmic RNAs derived from the major late transcription unit, and host cell shutoff (16, 57). Furthermore, it has recently been demonstrated that the E4 region participates in cellular transformation of nonpermissive cells (24, 36, 42). The Ad E4 transcription unit is complex and encodes at least seven different protein products from seven open reading frames (13, 56). To date, we know of several biological functions that are performed by five of the E4 open reading frames (E4orfs). E4orf3 and E4orf6 play a role in the control of alternative splicing of the major late tripartite leader during lytic viral growth (43, 44). The products of E4orf3 and E4orf6 appear to have redundant activities during Ad infection, and expression of either one is sufficient to support wild-type levels of virus production (6, 21). E4orf6 binds the E1B 55-kDa protein, and the complex is involved in nucleocytoplasmic mRNA transport (7, 28). Recently, E4orf6 was shown to bind p53, to block p53-mediated transcriptional activation, to block p53-mediated apoptosis, and to enhance transformation by the E1 region of Ad (11, 36, 42). The E4orf6/7 protein enhances E2 gene transcription by facilitating cooperative binding of transcription factor E2F to the E2 promoter (22, 31, 41). The E4orf1 gene encodes a transforming protein, which may be distantly related to dUTP pyrophosphatase enzymes and which interacts with a mammalian homolog of the Drosophila discs large tumor suppressor protein (27, 58). Ad9 E4orf1 induces mammary tumors in rats (24).
The Ad5 E4orf4 protein plays a role in down-regulation of virally induced signal transduction. Previous work has shown that the Ad E1A proteins and cyclic AMP (cAMP) cooperate to induce the accumulation of AP-1 transcription factor by activating the transcription of the cellular c-fos and junB genes (encoding the AP-1 components). The induced AP-1 activates the transcription of early Ad genes through Ap-1 and activating transcription factor sites in Ad promoters (12, 39). The induction of AP-1 by E1A plus cAMP is counterbalanced by the 14-kDa Ad E4orf4 protein, whose levels rise upon stimulation by E1A and cAMP (38). The phenotype of two mutant Ads, which lack the E4orf4 coding region, indicates that the E4orf4 protein inhibits transcription of the cellular junB and c-fos genes and depresses translation of c-fos (38). It appears, therefore, that as E4orf4 accumulates, it causes AP-1 DNA binding to return to its basal levels. E4orf4 activities also result in hypophosphorylation of E1A and c-fos proteins (38), and as a consequence of the cumulative E4orf4 effects, its own promoter is down-regulated as well (5, 59).
While investigating the mechanisms underlying E4orf4-induced down-regulation of stimulated gene expression, we have found that the E4orf4 protein binds several cellular proteins, one of which has been identified as protein phosphatase 2A (PP2A) (26). We and others have further shown that the E4orf4-PP2A interaction plays a role in down-regulation of stimulated transcription (5, 26).
The down-regulation of environmentally triggered signals is of great importance to maintaining a normal cell, since constitutive activation of signal transduction pathways that stimulate transcription is known to result in cancer. Examples include pathways stimulated by tyrosine kinase receptors and c-Ras, which eventually activate AP-1 transcription factor (25). Mutated genes for the receptors or Ras encode constitutively active proteins that are responsible for triggering their signal transduction pathways in the absence of extracellular signals, thus enhancing cell proliferation and contributing to tumorigenesis (1, 23, 33). Therefore, genes that are down-regulators of signal transduction may act to inhibit growth. If continuous contradictory signals are given to the cell, i.e., to grow and to arrest growth, apoptosis may ensue.
In this report, we demonstrate that E4orf4 induces apoptosis in the E1A-expressing 293 cells, in NIH 3T3 cells transformed by v-Ras, and in H1299 cells, which lack an intact p53 gene. The induction of apoptosis is not accompanied by enhanced levels of p53 expression in 293 cells and occurs in the absence of p53 expression in H1299 cells, suggesting that a p53-independent pathway is used. Since E4orf4 is a down-regulator of virally induced signal transduction, we suggest that E4orf4 antagonizes continuous signals to proliferate, like the ones given by E1A or by v-Ras, and that the conflicting signals lead to induction of cell death.
MATERIALS AND METHODS
Plasmids and cells.
The plasmids expressing E4orf4 either as a glutathione S-transferase (GST)-fusion protein (GST-E4orf4) or from the cytomegalovirus (CMV) immediate-early promoter (pCMV-E4orf4), have been previously described (26), as were the CMV vector itself (19) and the plasmid used for in vitro expression of the PP2A B subunit (45). pMAMneo-E4orf4 expresses E4orf4 from a dexamethasone-inducible mouse mammary tumor virus promoter. pMAMneo and pEGFP-C1, expressing a red-shifted green fluorescent protein (GFP), are from Clontech. pBabe-puro expresses the puromycin resistance gene from the early simian virus 40 (SV40) promoter (37). The E4orf4 A3 mutant was created by random PCR mutagenesis, and it contains proline instead of serine at position 95. The mutated sequence was recloned into the CMV vector and into the GST fusion vector pGEX-2TK (Pharmacia). pCH110 expresses the β-galactosidase gene from the early SV40 promoter.
Human 293 cells were derived from human embryonic kidney cells and express Ad5 E1A and E1B proteins (14). NIH 3T3 cells transformed by v-Ras (NIH 3T3-vRas) were a gift from A. Levitzki. The two cell lines were cultured in Dulbecco’s minimal essential medium supplemented with 10% calf serum. The human non-small-cell lung carcinoma H1299 cells (34) were cultured in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum. A stable cell line expressing E4orf4 (293-15-DM) was obtained by cotransfection of pBabe-puro and the E4orf4-expressing plasmid, pMAMneo-E4orf4, followed by selection with 2.5 μg of puromycin per ml. E4orf4 expression in 293-15-DM cells was induced by the addition of 1 μM dexamethasone.
Transfections, Western blot analysis, and protein binding assays.
Cells were plated in 60-mm culture dishes, and transfections were carried out by the standard method of calcium phosphate precipitation of DNA (15). At 48 h after transfection, the cells were harvested, washed with phosphate-buffered saline (PBS), and lysed with lysis buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.5% Nonidet P-40, 2 μg of leupeptin per ml, 2 μg of aprotinin per ml, and 0.5 mM phenylmethylsulfonyl fluoride. The levels of E4orf4 and p53 proteins were analyzed by Western blot analysis. Expression of E4orf4 was detected with a rabbit polyclonal antibody raised against E4orf4 expressed as a histidine-fusion protein in bacteria. The Ab421 monoclonal antibody against p53 (a generous gift of A. J. Levine) was used to detect p53. A polyclonal antibody recognizing the PP2A C subunit was raised against the C-terminal peptide of PP2A C, coupled to hemocyanin. Immune complexes were detected by chemiluminescence.
GST fusion protein binding assays and coimmunoprecipitations were carried out as previously described (26). Bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide gels for PP2A and 15% polyacrylamide gels for E4orf4) and detected either by autoradiography or by Western blot analysis, as indicated.
β-Galactosidase assays.
Cells in 60-mm-diameter culture dishes were transfected with 5 μg each of pCH110 and the CMV vector expressing wild-type or mutant E4orf4 proteins. Cell lysates were prepared and enzymatic reactions were carried out as previously described (47).
In vitro translation.
Supercoiled plasmid DNA was transcribed and translated with the TNT system from Promega.
Fluorescence microscopy.
Cells were plated in 60-mm culture dishes and transfected with 5 μg each of pEGFP-C1 and the CMV vector expressing wild-type or mutant E4orf4 proteins. At 48 h posttransfection, the cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, washed again, and stained with 0.1 μg of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma) per ml. A coverslip was mounted on the plates with Fluoromount-G (Southern Biotechnology Associates, Birmingham, Ala.). The fluorescent cells were visualized under ×600 magnification with a Zeiss Axioskop microscope.
Flow cytometry.
Cells were harvested, washed once with PBS, and fixed with methanol for at least 30 min at −20°C. The cells were then washed twice with PBS, resuspended in PBS containing 50 μg of RNase A per ml, and incubated for 30 min at 37°C. To stain the DNA, propidium iodide (Sigma) was added to a final concentration of 25 μg per ml. Samples were analyzed in a cell sorter (FACSCalibur; Becton Dickinson).
DNA fragmentation.
Low-molecular-weight DNA was isolated from cells by a modified Hirt extraction procedure (20). The cells were harvested and resuspended in proteinase K lysis buffer (10 mM Tris-HCl [pH 8], 5 mM EDTA, 100 mM NaCl, 1 mg of proteinase K per ml). SDS was added to the cells at a final concentration of 0.5%. Cell lysates were incubated at 37°C for 2 h, and NaCl was added to a final concentration of 1 M. The samples were incubated at 4°C overnight and were then centrifuged at 15,000 × g for 30 min. The supernatants, containing the low-molecular-weight DNA, were extracted with phenol-chloroform and precipitated with ethanol. The nucleic acids were incubated with RNase A for 30 min at 37°C, and concentrations of DNA were determined. Equal amounts of DNA were resolved on a 1% agarose gel containing ethidium bromide.
RESULTS
Transfection with wild-type E4orf4, but not with a mutant that had lost the ability to bind PP2A, reduces colony formation by 293 cells.
Previous studies have shown that Ad E4orf4 protein, an early Ad protein that is induced by E1A and cAMP, down-regulates E1A-enhanced gene expression (26, 38). Thus, it is possible that E4orf4 interferes with E1A-induced cell proliferation. The effect of E4orf4 on cell growth was therefore investigated.
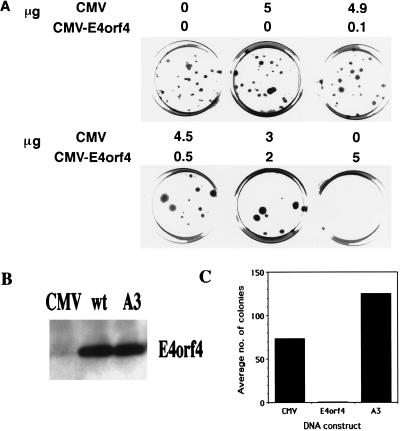
In the assays, 293 cells, expressing the Ad5 E1 genes, were cotransfected with a gene encoding puromycin resistance and a combination consisting of a plasmid in which E4orf4 expression is driven by the CMV immediate-early promoter and of the empty CMV vector. The total amount of plasmids containing the CMV promoter was kept constant. The number of puromycin-resistant colonies was determined after 2 weeks of growth in selective medium. The results of a representative experiment are shown in Fig. 1A. Increasing the amount of CMV-E4orf4 reduced the number of puromycin-resistant colonies. At maximal levels of E4orf4 transfection, colony formation was reduced to zero. The fact that the amount of CMV promoter was not varied in these experiments precludes the possibility that the reduction in the number of colonies resulted from promoter competition that reduced the expression of the puromycin resistance gene.
FIG. 1.
Wild-type E4orf4 reduces colony formation by 293 cells. (A) 293 cells were transfected with 5 μg of pBabe-puro and 5 μg (total) of pCMV plus pCMV-E4orf4. The ratios between pCMV and pCMV-E4orf4 were varied, as indicated. Stable colonies were selected in 2.5 μg of puromycin per ml for 2 weeks. (B) 293 cells were transfected with 5 μg of the CMV vector, wild-type E4orf4 (wt), or mutant A3. Proteins were harvested 40 h posttransfection and subjected to Western blot analysis with an antibody specific to E4orf4. (C) 293 cells were transfected with 5 μg of pBabe-puro and 5 μg of effector plasmid (CMV, wild-type E4orf4, or mutant A3). Selection was carried out as described for panel A. The results are the mean of three experiments.
In the course of a functional analysis of E4orf4, mutations were introduced into the protein by random PCR mutagenesis. One mutant, containing a point mutation that changes serine to proline at position 95 (mutant A3), lost the ability to reduce the number of puromycin-resistant colonies. This mutant is expressed and is recognized by an antiserum raised against E4orf4 (Fig. 1B). However, in several experiments, mutant A3 increased the number of colonies by 1.25- to 2.5-fold, (average, 1.7-fold) whereas E4orf4-wt reduced this number by 10- to 100-fold (average, 73-fold) (Fig. 1C). The product of the wild-type E4orf4 gene is thus required to inhibit colony formation by 293 cells.
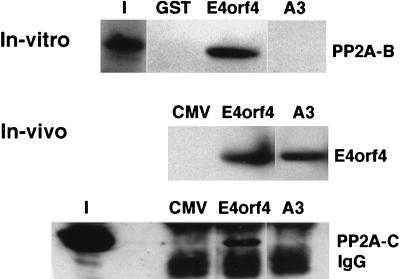
Since wild-type E4orf4 protein is known to bind PP2A (26), mutant A3 was further characterized by testing its ability to bind PP2A both in vitro and in vivo. GST fusion constructs containing wt E4orf4 protein or mutant A3 were prepared. Equal amounts of the fusion proteins were used in binding assays containing the in vitro-translated B subunit of PP2A, as previously described (26). As seen in Fig. 2, wild-type E4orf4, but not mutant A3, bound the PP2A B subunit in vitro. E4orf4 and mutant A3 were also expressed in 293 cells and immunoprecipitated from the extracts by the specific anti-E4orf4 antiserum. Although a Western blot analysis of the immune complexes demonstrated that equal amounts of the E4orf4 and A3 proteins were immunoprecipitated, the PP2A C subunit was detected only in the E4orf4 immune complex, not in the A3 immunoprecipitation (Fig. 2). Thus, mutant A3 had lost the ability both to reduce 293 colony formation and to bind PP2A.
FIG. 2.
Mutant A3 does not bind PP2A. In the in vitro experiments, GST alone or GST fused to E4orf4 or mutant A3 was incubated with in vitro-translated, 35S-labeled PP2A B subunit. Bound PP2A B was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and detected by autoradiography. Lane I contains 7% of the input PP2A B. In the in vivo experiments, the empty CMV vector, E4orf4, or mutant A3 was transfected into 293 cells and the cell extracts were immunoprecipitated with the antiserum recognizing E4orf4. The immune complexes were subjected to Western blot analysis, and E4orf4 or the PP2A C subunits were detected with the appropriate antibodies. Lane I contains 20% of the input extract used for immunoprecipitations.
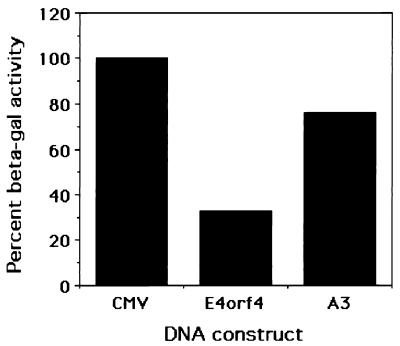
E4orf4 protein has been shown to down-regulate transcription from both cellular and viral promoters (5, 26, 38), raising the possibility that the reduction in the number of puromycin-resistant colonies resulted from down-regulation of the early SV40 promoter driving the puromycin resistance gene. β-Galactosidase assays were carried out to test the effect of E4orf4 on the SV40 early promoter at early times after transfection. A reporter construct containing the β-galactosidase gene, driven by the SV40 early promoter, was cotransfected into 293 cells with E4orf4 or the A3 mutant, and β-galactosidase assays were performed 2 days after transfection. Figure 3 shows that E4orf4 reduced β-galactosidase activity threefold. The maximal decrease in this activity did not surpass fourfold in several experiments. Similar results were obtained when β-galactosidase activity was assayed 1 day after transfection. Mutant A3 reduced β-galactosidase activity to a lesser extent, by 20 to 40% compared with the empty vector. Thus, although E4orf4 may reduce the expression of the puromycin resistance gene by up to fourfold, this does not explain the 10- to 100-fold reduction in the number of puromycin-resistant colonies.
FIG. 3.
E4orf4 reduces β-galactosidase levels in 293 cells. 293 cells were transfected with 5 μg of pCH110 and 5 μg of the CMV vector, wild-type E4orf4, or mutant A3. Proteins were harvested 40 h posttransfection, and β-galactosidase assays were carried out with equal amounts of proteins. The activity of β-galactosidase in the cells transfected with the empty vector was defined as 100%.
E4orf4 induces apoptosis in 293 cells.
In some of the puromycin selection experiments, the stable colonies were stained with neutral red, a dye that stains viable cells. Some of the colonies obtained after cotransfection of the puromycin resistance gene and E4orf4 were not stained red, and visualization under a phase-contrast microscope revealed the cells to be dead, indicating that the cells started growing and then died (results not shown). This finding suggested that E4orf4 may inhibit colony formation by inducing apoptosis in the cells.
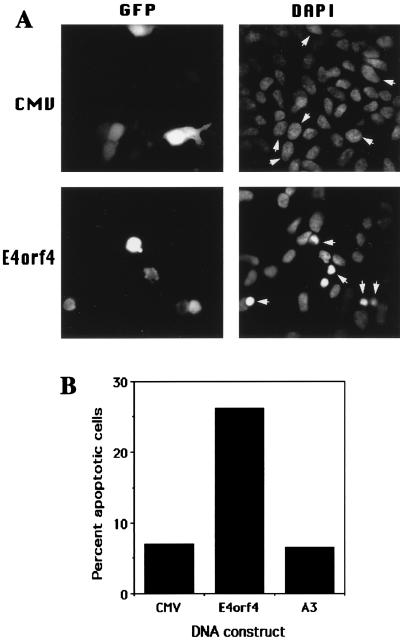
To test whether E4orf4 induces apoptosis in 293 cells, the cells were transiently cotransfected with E4orf4 or an empty vector and GFP, which was used as a marker for transfection. Two days later, the cells were stained with DAPI and subjected to fluorescence microscopy. Transfected cells were detected by their green fluorescence, and the apoptotic state of transfected nuclei was determined by the DAPI staining. Nuclei from cells transfected with the empty vector were stained uniformly and weakly with DAPI, indicating that they were intact, whereas nuclei from cells transfected with E4orf4 appear condensed or fragmented, a hallmark of apoptotic nuclei (Fig. 4A). The apoptotic cells were loosely adherent to the tissue culture dishes, and thus their nuclei appeared in a slightly different focal plane from nonapoptotic nuclei. Five similar transfection experiments were carried out to determine the levels of apoptosis induced by E4orf4 and by the mutant E4orf4 protein A3 (Fig. 4B). E4orf4 increased apoptosis in 293 cells by three- to sixfold (averaging fourfold) compared with the empty vector, whereas mutant A3 did not significantly increase apoptosis. These results agree with the previous results concerning the effect of the E4orf4 proteins on the growth of puromycin-resistant colonies. E4orf4 decreased the growth of puromycin-resistant colonies and increased apoptosis in 293 cells, whereas mutant A3 did not decrease the colony growth and did not enhance apoptosis in the cells compared with the empty vector.
FIG. 4.
E4orf4 induces apoptosis in transfected 293 cells. (A) 293 cells were transfected with 5 μg of pEGFP-C1 and 5 μg of either the CMV vector or the CMV vector expressing wild-type E4orf4. The cells were fixed 48 h posttransfection, stained with DAPI, and subjected to fluorescence microscopy at a magnification of ×600. Each field was photographed twice with filters suitable for detection of the GFP and DAPI staining. The arrowheads mark the nuclei of transfected cells, detected by the presence of the GFP. (B) Experiments were carried out as for panel A. For each experiment, at least 100 transfected cells were counted, and the percentage of nuclei manifesting apoptotic phenotypes, as seen in panel A, was calculated. The graph shows the mean of five experiments.
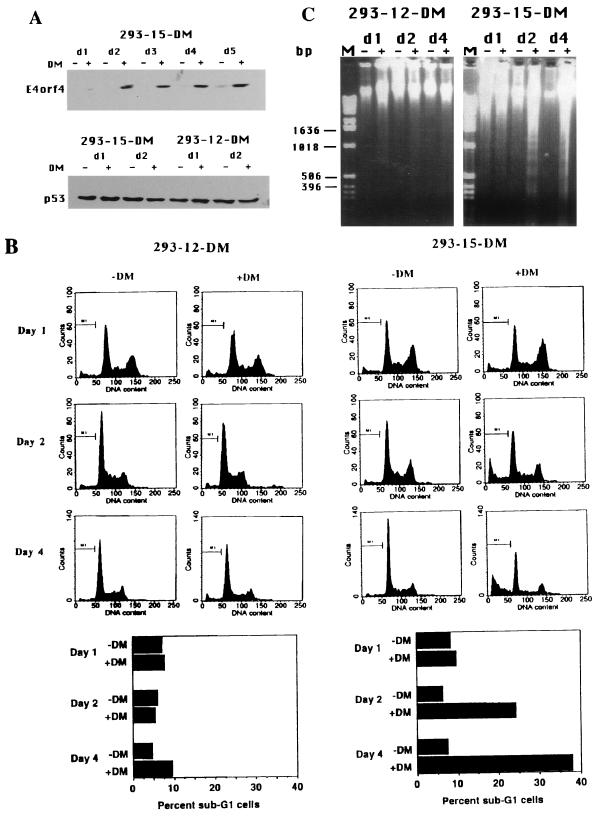
To further confirm the ability of E4orf4 to induce apoptosis, a stable 293 cell line, expressing E4orf4 under the influence of an inducible promoter, was established (293-15-DM). E4orf4 expression was induced by dexamethasone, and high levels of expression were detected by day 2 after induction and persisted until at least day 5 (Fig. 5A). Another cell line (293-12-DM), originating from the same screen, was chosen to serve as a negative control. No E4orf4 expression was detected in this cell line in the absence or presence of the inducer (data not shown). Accumulation of E4orf4 protein in 293-15-DM cells was not accompanied by significant alterations in p53 levels (Fig. 5A). Induced and uninduced cells were stained with propidium iodide to determine their DNA content and subjected to a flow cytometric analysis. When 293-12-DM cells were induced with dexamethasone, there was no significant increase in the sub-G1 cell population (Fig. 5B). Sub-G1 cells are cells containing less than 2 N DNA, which is indicative of apoptotic death (9, 18). No increase in the number of sub-G1 cells was observed on day 1 after induction in 293-15-DM cells, before the accumulation of high levels of E4orf4 protein. However, once E4orf4 expression was enhanced (starting on day 2 postinduction), increasing numbers of sub-G1 cells were observed (24% on day 2 and 38% on day 4 postinduction [Fig. 5B]). Furthermore, DNA of low molecular weight was extracted from induced and uninduced cells of both cell lines and separated on an agarose gel. Figure 5C shows that high levels of nucleosome-sized DNA fragments were present in samples of DNA extracted from induced 293-15-DM cells on days 2 and 4 after induction but not in the samples from uninduced cells. Degradation of DNA to nucleosome-sized fragments is another hallmark of apoptotic cell death (3). The apoptotic DNA degradation patterns were not observed in DNA samples extracted from 293-15-DM cells on day 1 postinduction, or from induced 293-12-DM cells. These results demonstrate that increased DNA degradation occurs upon induction of E4orf4 expression in the 293 cells, confirming that E4orf4 induces apoptosis in these cells.
FIG. 5.
A stable 293 cell line, expressing inducible wild-type E4orf4, undergoes apoptosis upon induction of E4orf4 expression. (A) 293-15-DM and 293-12-DM cells were induced by dexamethasone (DM), and proteins from uninduced (−) and induced (+) cells were harvested at the indicated times (d1 = day 1, etc.) and subjected to Western blot analysis. The blots were stained either with an antiserum raised against E4orf4 protein or with an antibody recognizing the p53 protein. (B) Cells were induced as in panel A, and at each time point a sample was taken for flow cytometric analysis. Cell numbers (marked as counts) are plotted against DNA content, measured by staining with propidium iodide. The percentage of cells in the sub-G1 region (marked M1) was calculated and plotted. (C) DNAs were prepared from uninduced (−) and induced (+) cells at each time point (d1 = day1, etc.), resolved on a 1% agarose gel, and stained with ethidium bromide. Lane M contains molecular size markers.
Induction of apoptosis by E4orf4 is not confined to 293 cells.
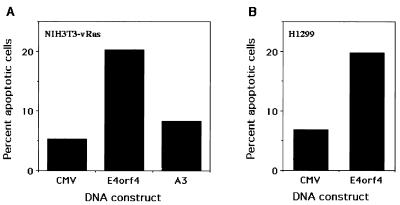
To test whether E4orf4 can induce apoptosis in cells that do not express any Ad proteins but express an oncogenic protein that sends constitutive signals to enhance cell proliferation, we tested the ability of E4orf4 to induce apoptosis in NIH 3T3 cells transformed with v-Ras (NIH 3T3-vRas). Cells were cotransfected with E4orf4 and GFP, and apoptotic nuclei were counted in the transfected cell population. Figure 6A shows the results of these experiments. Wild-type E4orf4 induced three- to fourfold more apoptosis than did the empty vector and mutant A3. Thus, E4orf4 can induce apoptosis in both human and murine transformed cells, and the presence of other Ad proteins is not required.
FIG. 6.
E4orf4 protein induces apoptosis in NIH 3T3 cells transformed with v-Ras and in H1299 cells. The experiment described in the legend to Fig. 4 was carried out by using NIH 3T3 cells transformed with v-Ras (A) and by using H1299 cells (B). Numerical analysis was performed as described in the legend to Fig. 4, and the results are means of three experiments.
The results shown in Fig. 5A demonstrate that E4orf4 expression does not lead to accumulation of the p53 protein and thus may induce apoptosis by a p53-independent pathway. To confirm this possibility, we repeated the experiments shown in Fig. 4 and 6A with the human lung carcinoma cell line H1299, which lacks p53 (34). Figure 6B shows the mean of three independent experiments, in which E4orf4 induced apoptosis 2.4- to 4-fold in comparison with the empty vector as measured by DAPI staining of transfected cells. Thus, p53 is not required for the induction of apoptosis by E4orf4.
DISCUSSION
In this report, we show that an active E4orf4 protein can induce apoptosis in cells that receive a constant signal to proliferate by oncogenes, such as E1A or v-Ras. Initially, two cell types were used in this study. The E1A-expressing 293 cells were used since E4orf4 was specifically shown to counteract E1A transactivation, raising the possibility that E4orf4 can modulate the effects of E1A on cell growth. NIH 3T3 cells transformed with v-Ras were used as another example of a transformed cell line, which expresses a constitutively activated enhancer of cell proliferation. The interaction of E4orf4 with PP2A (26) and the ability of PP2A to inactivate mitogen-activated protein (MAP) kinase and MAP kinase kinase (2, 52) suggested the possibility that E4orf4 counteracts activation by the Ras-MAP kinase pathway. For example, it has recently been demonstrated that the residues in the E1A protein that are hypophosphorylated when E4orf4 is expressed (38) may be phosphorylated by MAP kinase (59).
Our results demonstrate that the E4orf4 protein induces apoptosis in both 293 cells and NIH 3T3 cells transformed with v-Ras. This conclusion is supported by several lines of evidence.
First, overexpression of E4orf4 in transfected cells leads to the appearance of condensed and fragmented nuclei, which are a hallmark of apoptosis (Fig. 4A). The percentage of apoptotic cells in the transfected-cell population was 20 to 30% on day 2 posttransfection. It is not known if all the 293 cells that express E4orf4 are destined to die or if the induction of apoptosis depends on additional unidentified factors. However, the ability of E4orf4 to prevent colony growth (Fig. 1) suggests that eventually E4orf4 may induce cell death in all E4orf4-expressing 293 cells. A mutant E4orf4 protein does not induce apoptosis under the same conditions (mutant A3 [Fig. 4B and 6A]) and does not reduce colony formation (Fig. 1C), despite being expressed to high levels (Fig. 1B). These results suggest that a defined, as yet unknown function performed by E4orf4 is required for the induction of apoptosis. Such a function may depend on the interaction between E4orf4 and PP2A. We showed that mutant A3 lost the ability to induce apoptosis, as well as the ability to bind PP2A (Fig. 2). However, a more comprehensive mutational analysis should be carried out to determine whether the interaction between E4orf4 and PP2A correlates with the ability of E4orf4 to induce apoptosis.
The observation that mutant A3 consistently increases the number of puromycin-resistant colonies (Fig. 1C) is reminiscent of the ability of mutant p53 to transform cells while the wild-type protein is inhibitory to growth. However, a further investigation is required to understand the differences between the activities of wild-type and mutant E4orf4 proteins.
The reduction observed in the number of puromycin-resistant colonies in the presence of E4orf4 is most probably a consequence of E4orf4-mediated cell death rather than down-regulation of the SV40 promoter driving the expression of the puromycin resistance gene (determined by the β-galactosidase assays [Fig. 3]). This conclusion is based on the observation that colony formation was reduced by up to 100-fold whereas β-galactosidase activity at early times posttransfection was reduced by no more than 4-fold.
A second line of evidence demonstrates that in a cell line expressing E4orf4 from an inducible promoter, induction of E4orf4 expression is followed by the appearance of a sub-G1 cell population (Fig. 5B) and by degradation of DNA into nucleosome-sized fragments (Fig. 5C). Both these events are indicative of apoptosis in the cell population (4, 9). Surprisingly, even as late as day 4 postinduction, only 38% of the cells displayed an apoptotic phenotype, indicated by the presence of sub-G1 cells. However, E4orf4 expression in the 293-15-DM cell line was leaky and some E4orf4 expression was seen in the absence of induction (Fig. 5A, d5). Since E4orf4 induces apoptosis in the cells, selection against the expression of wild-type E4orf4 in this cell line may have occurred. Indeed, when this cell line was passaged a few more times, induction of apoptosis was no longer observed (data not shown). When E4orf4 DNA from the late passage was amplified and subjected to sequencing, several mutations in the E4orf4 gene were detected (data not shown). Thus, during our experiments, an increasing fraction of the population became resistant to apoptosis, explaining why only 38% of the cells underwent apoptosis 4 days postinduction.
E4orf4-induced apoptosis is not accompanied by changes in p53 protein levels in 293 cells (Fig. 5A). Furthermore, E4orf4 induced apoptosis in H1299 cells, which lack p53 (Fig. 6B). These results suggest that E4orf4 induces apoptosis by a p53-independent route. This conclusion is supported by a previous report suggesting that the E4 gene region is involved in p53-independent, E1A-induced apoptosis at late times of viral infection (30). During viral infection, E1A transactivates the early viral genes, including E4 (51), thus allowing the expression of E4orf4 and creating the potential for induction of apoptosis by this protein.
Ad expresses another apoptosis-inducing protein, E1A (46), as well as an additional protein that is implicated in Ad-induced cell death (the E3-11.6K gene product [55]), although it is still unclear whether cell death induced by the E3 protein is due to apoptosis. Other viruses also encode gene products that induce apoptosis (reviewed in references 50 and 53), and evidence has accumulated showing that such virally induced apoptosis may contribute directly to the cytopathogenic effects of the viruses. It has been suggested that cell lysis, the final stage in virus production, is associated with apoptosis in some viruses and can be blocked by the bcl-2 gene product (50, 53). At this stage, it is not clear if apoptosis induction by E4orf4 is useful to the virus under some circumstances or whether it is a by-product of other processes induced by E4orf4. However, E4orf4 probably does not induce apoptosis in the course of a normal viral infection, since Ad encodes at least one gene product, the E1B 19-kDa protein, which inhibits p53-independent apoptosis (60).
It was previously reported that a mutant Ad, dl358, which does not express the E4orf4 protein (16), reduces the ability of infected CREF cells to form colonies, compared with a wild-type infection (38). These results are in apparent contradiction to the present study. However, it is not known whether the reduced number of CREF cell colonies resulted from enhanced apoptosis or from other growth-inhibitory events. Furthermore, experiments involving whole-virus infections are more difficult to interpret. In these experiments, dl366, which lacks the entire E4 region (16), did not reduce cell viability compared to a wild-type virus infection (data not shown). These results indicate that another E4 gene may have an inhibitory effect on CREF cell growth and that only when both E4 genes are removed does viral infection lose some of its cytotoxicity. In a wild-type virus infection of CREF cells, when both genes are present, the E4orf4 gene may reduce the expression of the other gene, thus reducing its inhibitory effect. It remains to be seen if E4orf4 does not induce apoptosis in the untransformed CREF cells.
The specific mechanisms by which E4orf4 induces apoptosis are unknown. E4orf4 can down-regulate gene expression on both the transcriptional and translational levels (38), and down-regulation of genes involved in control of apoptosis may play a role in the induction of apoptosis. In the case of the p53 protein, it was suggested that p53-induced repression may be involved in induction of apoptosis (49), and, indeed, p53 inhibits endogenous bcl-2 expression at the same time that it induces apoptosis (35, 48). It is also possible that E4orf4, through its association with PP2A, affects the phosphorylation of apoptosis-related proteins, resulting in modulation of apoptosis induction pathways. Phosphorylation has been shown to alter the apoptosis-inducing potential of the death agonist Bad (63), and changes in Bcl-2 phosphorylation were suggested to affect its antiapoptotic activity (8, 17, 32). Interfering with various MAP kinase pathways may also contribute to the induction of apoptosis (62).
E4orf4 induces apoptosis, and not growth arrest, in the transformed cells that were examined. This finding may suggest that a conflict between the proliferation signals given by E1A or v-Ras and the constant down-regulation of these signals by E4orf4 leads to cell death. In another example, E1A proteins stimulate DNA synthesis and cell proliferation of primary baby rat kidney cells but also cause accumulation of the apoptosis-inducing p53 protein (10, 29, 46, 61). Induction of apoptosis in this case occurs when cell growth is inhibited by cell-cell contact or by serum withdrawal and not when the cells are actively growing (40). The mechanisms involved in E4orf4-induced apoptosis are currently under investigation.
ACKNOWLEDGMENTS
We are grateful to A. J. Levine for the antibody to p53, to A. Levitzky for the NIH 3T3-vRas cells, and to T. Shenk, in whose laboratory the polyclonal antibodies were prepared. We thank D. Kornitzer, E. Bengal, and H. Barr for helpful comments on the manuscript.
This work was supported in part by The Israel Cancer Research Fund and by The Israel Cancer Association.
REFERENCES
- 1.Aaronson S A, Tronick S R. Constitutive activation of growth factor signalling pathways in cancer cells. In: Broder S, editor. Molecular foundations of oncology. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 41–56. [Google Scholar]
- 2.Anderson N G, Maller J L, Tonks N K, Sturgill T W. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinases. Nature. 1990;343:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 3.Arends M J, Morris R G, Wyllie A H. Apoptosis: the role of endonuclease. Am J Pathol. 1990;136:593–608. [PMC free article] [PubMed] [Google Scholar]
- 4.Bellamy C O C, Malcomson R D G, Harrison D J, Wyllie A H. Cell death in health and disease: the biology and regulation of apoptosis. Semin Cancer Biol. 1995;6:3–16. doi: 10.1006/scbi.1995.0002. [DOI] [PubMed] [Google Scholar]
- 5.Bondesson M, Ohman K, Mannervik M, Fan S, Akusjarvi G. Adenovirus E4 open reading 4 protein autoregulates E4 transcription by inhibiting E1A transactivation of the E4 promoter. J Virol. 1996;70:3844–3851. doi: 10.1128/jvi.70.6.3844-3851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridge E, Ketner G. Redundant control of adenovirus late gene expression by early region 4. J Virol. 1989;63:631–638. doi: 10.1128/jvi.63.2.631-638.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridge E, Ketner G. Interaction of adenoviral E4 and E1B products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Faller D V. Phosphorylation of Bcl-2 protein and association with p21Ras in Ras-induced apoptosis. J Biol Chem. 1996;271:2376–2379. doi: 10.1074/jbc.271.5.2376. [DOI] [PubMed] [Google Scholar]
- 9.Darzynkiewicz Z, Bruno S, Delbino G, Gorczyca W, Hotz M A, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 10.Debbas M, White E. Wild type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 11.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 12.Engel D A, Hardy S, Shenk T. cAMP acts in synergy with E1A protein to activate transcription of the adenovirus early genes E4 and E1A. Genes Dev. 1988;2:1517–1528. doi: 10.1101/gad.2.12a.1517. [DOI] [PubMed] [Google Scholar]
- 13.Freyer G A, Katoh Y, Roberts R J. Characterization of the major mRNAs from adenovirus 2 early region 4 by cDNA cloning and sequencing. Nucleic Acids Res. 1984;12:3503–3519. doi: 10.1093/nar/12.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham F L, Smiley J, Russel W C, Nairn R. Characteristics of a human cell line transformed by human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 16.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldar S, Jena N, Croce C M. Inactivation of Bcl-2 by phosphorylation. Proc Natl Acad Sci USA. 1995;92:4507–4511. doi: 10.1073/pnas.92.10.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 19.Hinds P, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 20.Hirt B. Selective extraction of polyoma DNA from infected mouse cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 21.Huang M M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang P, Hearing M M. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor E2F through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 23.Hunter T. Cooperation between oncogenes. Cell. 1991;64:249–270. doi: 10.1016/0092-8674(91)90637-e. [DOI] [PubMed] [Google Scholar]
- 24.Javier R T. Adenovirus type 9 E4 open reading frame 1 encodes a transforming protein required for the production of mammary tumors in rats. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin M, Smeal T. Control of transcription factors by signal transduction pathways: the beginning of the end. Trends Biochem Sci. 1992;17:418–422. doi: 10.1016/0968-0004(92)90012-x. [DOI] [PubMed] [Google Scholar]
- 26.Kleinberger T, Shenk T. Adenovirus E4orf4 protein binds to protein phosphatase 2A, and the complex down regulates E1A-enhanced junB transcription. J Virol. 1993;67:7556–7560. doi: 10.1128/jvi.67.12.7556-7560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hD1g/SAP97, a mammalian homolog of the drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leppard K N, Shenk T. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe S, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus-5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 30.Marcellus R C, Teodoro J G, Wu T, Brough D E, Ketner G, Shore G, Branton P E. Adenovirus type 5 early region 4 is responsible for E1A-induced p53-independent apoptosis. J Virol. 1996;70:6207–6215. doi: 10.1128/jvi.70.9.6207-6215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marton M J, Baim S B, Ornelles D A, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May W S, Tyler P G, Ito T, Armstrong D K, Qatsha K A, Davidson N E. Interleukin-3 and bryostatin-1 mediate hyperphosphorylation of BCL2α in association with suppression of apoptosis. J Biol Chem. 1994;269:26865–26870. [PubMed] [Google Scholar]
- 33.McCormick F. ras oncogenes. In: Weinberg R A, editor. Oncogenes and the molecular origins of cancer. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 125–145. [Google Scholar]
- 34.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, D’Amico D, Bodner S, Oie H K, Linnoila R I, Mulshine J L, Minna J D, Gazdar A F. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 35.Miyashita T, Krajevski S, Krajevska M, Wang H G, Lin H K, Liebermann D A, Hoffman B, Reed J C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 36.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller U, Kleinberger T, Shenk T. Adenovirus E4orf4 protein reduces phosphorylation of c-fos and E1A proteins while simultaneously reducing the level of AP-1. J Virol. 1992;66:5867–5878. doi: 10.1128/jvi.66.10.5867-5878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller U, Roberts M P, Engel D A, Doerfler W, Shenk T. Induction of transcription factor AP-1 by adenovirus E1A protein and cAMP. Genes Dev. 1989;3:1991–2002. doi: 10.1101/gad.3.12a.1991. [DOI] [PubMed] [Google Scholar]
- 40.Mymryk J S, Shire K, Bayley S T. Induction of apoptosis by adenovirus type 5 E1A in rat cells requires a proliferation block. Oncogene. 1994;9:1187–1193. [PubMed] [Google Scholar]
- 41.Neill S D, Hemstrom C, Virtanen A, Nevins J R. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nordqvist K, Ohman K, Akusjarvi G. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–445. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohman K, Nordqvist K, Akusjarvi G. Two adenovirus proteins with redundant activities in virus growth facilitate tripartite leader mRNA accumulation. Virology. 1993;194:50–58. doi: 10.1006/viro.1993.1234. [DOI] [PubMed] [Google Scholar]
- 45.Pallas D C, Weller W, Jaspers S, Miller T B, Lane W S, Roberts T M. The third subunit of protein phosphatase 2A (PP2A), a 55-kilodalton protein which is apparently substituted for by T antigens in complexes with the 36- and 63-kilodalton PP2A subunits, bears little resemblance to T antigens. J Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao L M, Debbas M, Sabbatini P, Hockenberry D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis which is inhibited by the E1B-19k and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 16.59–16.67. [Google Scholar]
- 48.Selvakumaran M, Lin H-K, Miyashita T, Wang H G, Krajewski S, Reed J C, Hoffman B, Liebermann D. Immediate early up-regulation of bax expression by p53 but not TGFβ1: a paradigm for distinct apoptotic pathways. Oncogene. 1994;9:1791–1798. [PubMed] [Google Scholar]
- 49.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19k protein or the cellular bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Shenk T. Viruses and apoptosis. Curr Opin Genet Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 51.Shenk T. Adenoviridae, the viruses and their replication. In: Fields B, Howley P, Knipe D, editors. Virology. New York, N.Y: Raven Press; 1995. pp. 2111–2148. [Google Scholar]
- 52.Sontag E, Fedorov S, Kaminayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the Map kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 53.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teodoro J G, Shore G C, Branton P E. Adenovirus E1A proteins induce apoptosis by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:467–474. [PubMed] [Google Scholar]
- 55.Tollefson A E, Scaria A, Hermiston T W, Ryerse J S, Wold L J, Wold W S M. The adenovirus death protein (E3-11.6K) is required at very late stages in infection for efficient cell lysis and release of adenovirus from infected cells. J Virol. 1996;70:2296–2306. doi: 10.1128/jvi.70.4.2296-2306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virtanen A, Gilardi P, Naslund A, LeMoullec J M, Petterson U, Perricaudet M. mRNAs from human adenovirus 2 early region 4. J Virol. 1984;51:822–831. doi: 10.1128/jvi.51.3.822-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinberg D H, Ketner G. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J Virol. 1986;57:833–838. doi: 10.1128/jvi.57.3.833-838.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss R S, Lee S S, Prasad B V B, Javier R T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whalen S G, Marcellus R C, Whalen A, Ahn N G, Ricciardi R P, Branton P E. Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J Virol. 1997;71:3545–3553. doi: 10.1128/jvi.71.5.3545-3553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White E. Life, death and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 63.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]