Abstract
The bluetongue virus (BTV) minor protein VP4, with molecular mass of 76 kDa, is one of the seven structural proteins and is located within the inner capsid of the virion. The protein has a putative leucine zipper near the carboxy terminus of the protein. In this study, we have investigated the functional activity of this putative leucine zipper by a number of approaches. The putative leucine zipper region (amino acids [aa] 523 to 551) was expressed initially as a fusion protein by using the pMAL vector of Escherichia coli, which expresses a maltose binding monomeric protein. The expressed fusion protein was purified by affinity chromatography, and its size was determined by gel filtration chromatography. Proteins of two sizes, 51 and 110 kDa, were recovered, one equivalent to the monomeric form and the other equivalent to the dimeric form of the fusion protein. To prove that the VP4-derived sequence was responsible for dimerization of this protein, a mutated fusion protein was created in which a VP4 leucine residue (at aa 537) within the zipper was replaced by a proline residue. Analyses of the mutated protein demonstrated that the single mutation indeed prevented dimerisation of the protein. The dimeric nature of VP4 was further confirmed by using purified full-length BTV-10 VP4 recovered from recombinant baculovirus-expressing BTV-10 VP4-infected insect cells. Using chemical cross-linking and gel filtration chromatography, we documented that the native VP4 indeed exists as a dimer in solution. Subsequently, Leu537 was replaced by either a proline or an alanine residue and the full-length mutated VP4 was expressed in the baculovirus system. By sucrose density gradient centrifugation and gel filtration chromatography, these mutant forms of VP4 were shown to lack the ability to form dimers. The biological significance of the dimeric forms of VP4 was examined by using a functional assay system, in which the encapsidation activity of VP4 into core-like particles (CLPs) was studied (H. LeBlois, T. French, P. P. C. Mertens, J. N. Burroughs, and P. Roy, Virology 189:757–761, 1992). We demonstrated conclusively that dimerization of VP4 was essential for encapsidation by CLPs.
Leucine zipper and leucine zipper-like domains in eukaryotic proteins have been demonstrated to play essential roles in inter- and intramolecular interactions. The leucine zipper motif was initially defined as a sequence of four or five leucine residues spaced 7 amino acids (aa) apart. It was first described for the DNA binding proteins c-Myc and c-Jun and the yeast gene regulatory protein GCN4 (4, 12). The secondary structure of a leucine zipper exhibits a coiled-coil conformation (16). In the case of DNA binding proteins, these motifs are responsible for protein dimerization and DNA binding activity (7, 18). The leucine zipper motif has also been identified in other types of proteins, including the viral envelope glycoproteins of many retroviruses (5, 17). For example, a leucine zipper-like heptad repeat for the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp41 is required for fusogenic processes and virus entry into host cells (2).
Bluetongue virus (BTV) VP4 protein is a minor nucleocapsid protein which, together with 2 other minor proteins (VP1 and VP6) and 10 double-stranded RNA (dsRNA) segments, form the inner virion core, which is enclosed by three protein layers made up of four major structural proteins. A subcore layer of VP3 molecules surrounds the inner core and forms a scaffold for the next protein layer, which contains 780 molecules of VP7. Together, they form a complex icosahedral core structure. The core is intimately in contact with an outer capsid composed of two proteins, VP2 and VP5. The core, consisting of five proteins (VP1, VP3, VP4, VP6, and VP7) is transcriptionally active, and the three minor proteins (VP1, VP4, and VP6) are believed to be responsible for transcription and replication of the BTV genome. The VP4 protein (76 kDa), which is encoded by BTV RNA segment M4, has a guanylyltransferase activity, an enzyme essential for the modification of the 5′ termini of the viral mRNAs, including the positive-sense strand of the dsRNA genome (13, 15).
With a total of 654 aa, VP4 has a putative leucine zipper motif near the carboxyl end of the molecule (8, 21). This domain, encompassing 29 residues from aa 523 to 551 (LRVESSVLRVRNPTLHETADELKAMG LDL), contains leucine residues (in bold) at every seventh position and between short stretches of charged and other amino acids, indicating that it could form a leucine zipper, although the presence of a proline in the middle of the arrangement may inhibit a regularly coiled structure. However, whether the domain actually forms a leucine zipper and/or whether it is required for the function of the protein has not been investigated to date.
The present investigation was undertaken to determine the importance of the putative leucine zipper region of VP4 in terms of its structural and biological activities. We used a variety of methods, including gel filtration chromatography, chemical cross-linking, and sucrose density gradient centrifugation, to demonstrate that a purified recombinant VP4 occurs in the form of a dimer in solution. Second, we examined the ability of the putative leucine zipper in a chimeric foreign protein to drive the monomeric form of the protein into a dimeric form. Further, we used site-specific mutagenesis to disrupt the zipper arrangement and to demonstrate its requirement for dimerization. The data were confirmed by producing mutants with similar mutations in VP4 and by examining the effects of such mutations on the overall structure and oligomerization of the protein. The mutant and wild-type VP4 molecules were compared to examine the functional properties of the dimeric molecule. The data obtained demonstrate the involvement of a leucine zipper in the dimerization and encapsidation of VP4 in the BTV assembly process.
MATERIALS AND METHODS
Viruses and cells.
The wild-type baculovirus Autographa californica nuclear polyhedrosis virus (AcNPV) and the recombinant BAcPAK6 virus containing the lacZ (β-galactosidase) gene under the control of the AcNPV polyhedrin promoter were used (9). Spodoptera frugiperda cells (Sf9) were used to grow BAcPAK6 and other recombinant forms of AcNPV. The cells were cultured in TC-100 medium (GIBCO BRL, Paisley, United Kingdom) supplemented with 5% fetal calf serum.
Construction of a bacterial fusion protein.
pMal-p2 (New England Biolabs, Hitchin, Hertfordshire, United Kingdom) is an Escherichia coli bacterial expression vector that can be used to express and purify fusion proteins from cloned genes. A foreign sequence is inserted downstream from the MalE (maltose binding protein [MBP]) gene of E. coli. A recombinant plasmid to express a fusion protein containing a specific VP4 sequence was constructed by using PCR to amplify a portion of the cDNA of VP4 from plasmid pAcYM1VP4 (13) with oligonucleotide primers VP4LZF and VP4LZR (Table 1). The synthetic primers provided an upstream BamHI site and a downstream PstI site with a stop codon immediately preceding the PstI site. The PCR was performed with Vent polymerase (NEB). Following ligation into the BamHI and PstI sites of the bacterial expression plasmid pMal-p2, the products were transformed into E. coli XL-1 Blue.
TABLE 1.
Oligonucleotide primers used in PCR
| Primer | Sequencea |
|---|---|
| VP4LZF | GACGGGGATCCAGGTTCATCGGACTGCGCGTCGAA |
| VP4LZR | CGGCTGCAGTTACAAATCCAATCCCATCCGTTT |
| VP4LZP | CGGCTGCAGTTACAAATCCAATCCCATCCGTTTCAG TTCGTCAGCCGTCTCATGTGGTGTTGG |
| AvaII F(Pr) | TGTCTGAGGGTCCGAAACCCAACACCACATGAG |
| CD2VP4R | GATAATCTGCAGCTATTTCCGTTTGTATTCCCTT |
| BINF | AACTGCAGATGGACGCTTCCGTGACGAAAGTT |
| AvaII R | GGTTTCGGACCCTCAGCACGGAT |
| AvaII F(Al) | TGTCTGAGGGTCCGAAACCCAACAGCGCATGAG |
Restriction enzyme sites are underlined, and mutated sequences are shown in boldface type.
A point mutation for the central leucine of the putative leucine zipper was also created by PCR. Oligonucleotide primers VP4LZF and VP4LZP (Table 1) were used as forward and reverse primers, respectively. The reverse primer VP4LZP encodes a mutant sequence to convert the central leucine in the putative leucine zipper motif (VP4 residue Leu537) to a proline residue. The PCR product was ligated into the BamHI and PstI sites of the bacterial expression vector pMal-p2 and transformed into E. coli XL-1 Blue. The orientations of all the recombinant plasmids were verified by dideoxy sequencing (19).
Fusion protein expression and purification.
Bacterial expression plasmids containing VP4 derivatives were transformed into E. coli XL-1 Blue, which contains a lacIq inducible repressor. The bacteria were grown at 37°C in an orbital shaker to mid-logarithmic phase (optical density at 600 nm of 0.5 to 0.6) in Luria-Bertani LB medium containing 0.02% glucose and 100 μg of ampicillin per ml. Protein expression was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 3 mM followed by incubation for a further 2 h at 37°C. The cells were then pelleted and resuspended in column buffer (100 mM Tris-HCl [pH 7.4], 1 mM EDTA, 200 mM NaCl, 1 mM sodium azide, 10 mM β-mercaptoethanol) and frozen at −20°C overnight. The cells were thawed on ice, sonicated for 10 min with short pulses, and centrifuged at 9,000 × g for 30 min. The fusion proteins were purified by taking advantage of the affinity between MBP and amylose. To this end, the supernatant was passed through an amylose resin (NEB) column previously equilibrated with column buffer. The column was washed with 10 column volumes of column buffer to remove unbound proteins. The fusion protein was subsequently eluted with column buffer containing 10 mM maltose.
Construction of site-directed mutants and production of recombinant baculoviruses.
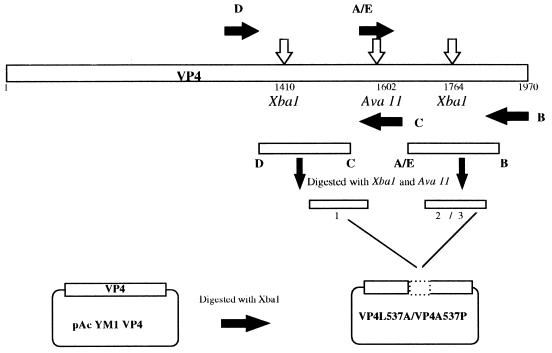
Mutations to the central leucine of the putative leucine zipper (VP4 residue Leu537) were also constructed in the full-length VP4 gene. The baculovirus transfer vector pAcYM1VP4 was digested with XbaI to remove an internal fragment between nucleotides 1410 and 1770 and to generate plasmid pAcYM1VP4 Xba. To create VP4 containing proline in place of leucine 537 (L537P) oligonucleotide primers AvaII F(Pr) (forward primer) and CD2VP4R (reverse primer) were used in a PCR (Table 1). The product was digested with XbaI and AvaII to give fragment 1 (Fig. 1). A second fragment was created with oligonucleotide primers BINF and AvaII R as the forward and reverse primers, respectively. This product was digested with XbaI and AvaII to give fragment 2. Fragments 1 and 2 were ligated into pAcYM1VP4 Xba and transformed in E. coli XL-1 Blue. To create mutant L537A (a VP4 mutant with alanine in place of leucine 537), a similar strategy was used, except that a fragment of DNA was produced with AvaII F(Al) and CD2VP4R as the forward and reverse primers, respectively (Table 1). This product was digested with XbaI and AvaII to give fragment 3. Fragments 2 and 3 were ligated into pAcYM1VP4 Xba and transformed into E. coli XL-1 Blue. All the DNA mutations and junction sites were confirmed by dideoxynucleotide sequencing (19). The template used for all these reactions was pAcYM1VP4. Recombinant baculoviruses expressing wild-type and mutant VP4 were obtained by cotransfection of Sf9 cells with Bsu36I-linearized AcNPV.PAC6 DNA (10) in the presence of recombinant transfer vectors and a Lipofectin reagent (GIBCO BRL). The viruses were plaque purified by conventional techniques.
FIG. 1.
Diagram of the cloning strategy used for creation of site-directed mutations in full-length VP4. A through E represent primers AvaII F(Pr), CD2VP4R, AvaII R, BINF, and AvaII F(Al) respectively. See Table 1 for primer sequences.
Expression and purification of recombinant baculovirus wild-type and mutant VP4.
Sf9 cells were infected with recombinant baculoviruses at a multiplicity of infection of 5 and harvested 72 h postinfection by centrifugation at 3,500 × g for 10 min. The pellet was washed with phosphate-buffered saline and resuspended in HNN lysis buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 0.5% Nonidet P-40) containing the protease inhibitors leupeptin, APMSF (10 mM), E-64 (10 mM), pepstatin A (1 mM), and dithiothreitol (1 mM). The cells were lysed with 10 strokes of a Dounce homogenizer and centrifuged as above. Most of the expressed wild-type VP4 pelleted with the nuclei and cellular debris. The protein was subsequently solubilized from the pellet with 50 mM HEPES (pH 7.4) containing 1 M NaCl. This high-salt buffer also released nucleic acids from the cell debris. To remove the nucleic acids, polyethylenimine (final concentration, 0.1% [vol/vol]) was added to the high-salt extract and the precipitated nucleic acids were removed by centrifugation. To remove excess polyethyleneimine, the supernatant was passed through an S-Sepharose column, and the protein was eluted by washing the column with 50 mM HEPES (pH 7.4) containing 1 M NaCl. The eluted protein was equilibrated in 50 mM HEPES–100 mM NaCl (pH 7.4) (HN buffer) by being passed through a PD10 Sephadex G-25 column (Pharmacia). For further purification, the sample was bound to a poly(U) column and VP4 was eluted with 50 mM HEPES–500 mM NaCl.
In contrast to the wild-type VP4 and as estimated by gel electrophoresis, some 90% of the mutant VP4L537A and VP4L537P proteins were soluble in HNN lysis buffer and were recovered in the supernatant. These mutant proteins were further purified by precipitating the solution first with 30% ammonium sulfate and then with 90% ammonium sulfate. The latter pellets were then dissolved in HN buffer. The final preparations were passed through a PD10 Sephadex G-25 column to equilibrate the protein in 50 mM HEPES–100 mM NaCl (pH 7.4).
Chemical cross-linking.
Purified VP4 (200 ng) core-like particles (CLPs) encapsidating VP4 in HN buffer were incubated with freshly prepared glutaraldehyde solution (final concentrations, 0.0001 to 0.01% [vol/vol] in H2O) in a 20-μl reaction mixture at 25°C for 30 min. The reaction was then stopped by addition of 2 μl of 1 M Tris-HCl (pH 8.0) followed by incubation on ice for 5 min, and the cross-linked products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The identity of the VP4 species was confirmed by Western blotting with a VP4-specific guinea pig antiserum, and the identity of VP3, VP7, and VP4 proteins was confirmed by Western blotting with a BTV-specific rabbit antiserum as described below.
Gel filtration chromatography.
Gel filtration chromatography was performed with Zorbax GF250 (Dupont, Wilmington, Del.) and Superose 12 (Pharmacia) columns. Protein samples at 1 mg/ml were passed through the column and eluted with 50 mM Tris–200 mM NaCl or 50 mM HEPES–100 mM NaCl at a flow rate of 0.7 or 0.1 ml/min, respectively. The protein content of each eluted fraction was measured spectrophotometrically at a wavelength of 214 nm, and the presence of VP4 was verified by SDS-PAGE and Western blotting as described below. The columns were calibrated with known molecular weight standards (Sigma), and a graph was drawn by plotting the partition coefficient Kav on the x axis and log molecular weight on the y axis. Kav was calculated from the formula Kav = (Ve −V0)/(Vt − V0), where Ve is the eluted volume, V0 is the void volume, and Vt is the total volume. From this calibration graph, the molecular weights of the eluted proteins were calculated. An oligomer-to-monomer ratio was then calculated by measuring the regions under both the oligomer and monomer A214 peaks, and the percent oligomerization was determined from this ratio.
SDS-PAGE and Western blot analysis.
Protein samples were analyzed by SDS-PAGE with either a 10 or 8% polyacrylamide resolving gel (11). The gels were either stained with Coomassie brilliant blue or electroblotted onto polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore) for immunodetection. The membranes were blocked with 5% fat-free milk powder in phosphate-buffered saline for 1 h and probed with a polyclonal guinea pig anti-VP4 antiserum (dilution of 1:1,000), an anti-MBP monoclonal anti-serum (NEB), or a polyclonal rabbit anti-BTV antiserum for 60 min at room temperature. The membranes were washed and treated with a 1:40,000 dilution of alkaline phosphatase-conjugated goat anti-guinea pig immunoglobulin G or anti-rabbit immunoglobulin G antiserum or a 1:1,000 dilution of peroxidase-conjugated anti-rabbit immunoglobulin G antiserum (Sigma, Poole, United Kingdom). When alkaline phosphatase conjugate was used, the membranes were developed with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate (GIBCO), whereas luminescence substrate solution A and starting solution B (Boehringer Mannheim) were used as specified by the manufacturer when peroxidase-conjugated anti-rabbit immunoglobulin G antiserum was used.
Sucrose density gradient centrifugation.
Sucrose gradients were used to examine the oligomeric forms of the proteins. Samples (1 ml) containing approximately 1 mg of purified or partially purified VP4 and mutant VP4 proteins were layered onto linear gradients of 25 to 10% (wt/vol) sucrose dissolved in HN buffer and centrifuged at 30,000 × g at 4°C for 18 h in an SW41 centrifuge. Fractions of 0.5 ml were collected from the top of the gradients and numbered with fraction 1 at the top and fraction 24 at the bottom. To monitor the sizes of the protein samples, the molecular mass protein markers β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (68 kDa), and chicken ovalbumin (41 kDa) were similarly fractionated. A 20-μl sample of each fraction was mixed with 10 μl of SDS-PAGE sample buffer, heated to 90°C for 2 min, and analyzed by SDS-PAGE. The gels were electroblotted onto PVDF membranes, and proteins were detected by probing with guinea pig anti-VP4 antiserum as described above.
RESULTS
The putative leucine zipper motif of VP4 forms an oligomer when introduced into a carrier protein.
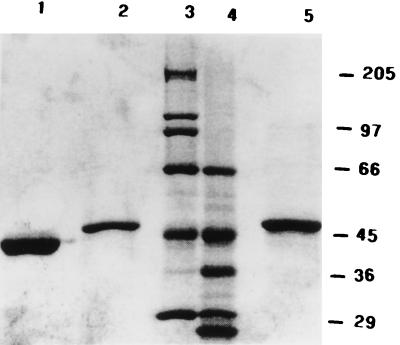
To investigate the ability of the putative leucine zipper region of VP4 to oligomerize a monomeric protein and to facilitate the purification of VP4 derivatives, we used a bacterial expression vector, pMal-p2, which encodes MBP, a monomeric protein that has an affinity to amylose resins. The BTV VP4 gene fragment (encoding aa 523 to 551 and encompassing the putative leucine zipper region) was expressed as a fusion protein with MBP and purified to homogeneity as described in Materials and Methods. The synthesis of the fusion protein MBP/VP4LZ was examined by SDS-PAGE (Fig. 2). The fusion protein MBP/VP4LZ had a molecular mass of 51 kDa, equivalent to its predicted size (Fig. 2, lane 2) and larger than the carrier 41-kDa MBP (lane 1).
FIG. 2.
SDS-PAGE of MBP-VP4 fusion proteins. The samples of MBP fusion protein containing either wild-type or mutant forms of the leucine zipper region of VP4 were analyzed by SDS-PAGE (10% polyacrylamide). Lanes: 1, purified MBP; 2, purified fusion protein derived from plasmid MBP/VP4LZ expressing the wild-type leucine zipper region; 3 and 4, standard molecular mass markers; 5, purified fusion protein derived from plasmid MBP/VP4LZP expressing the mutant (L537P) leucine zipper region.
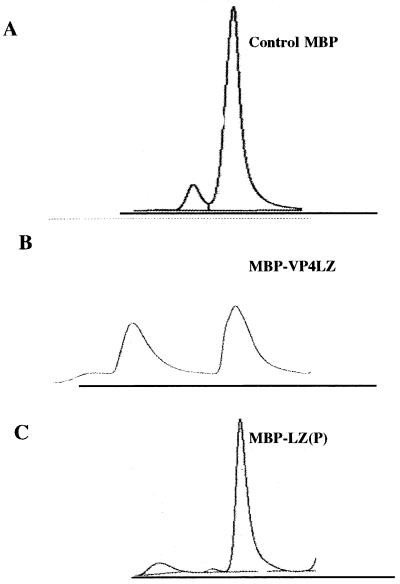
The oligomeric nature of the fusion protein was examined by gel filtration chromatography. As shown in Fig. 3A, the control MBP eluted as a single sharp peak at an elution time of 14 min, corresponding, as expected, to a single monomer of 41 kDa. By contrast, the fusion protein MBP/VP4LZ eluted as two sharp peaks, one at an elution time of 9.5 min and the second at an elution time of 14 min (Fig. 3B), indicating that two forms of the MBP/VP4LZ protein were present. The low-molecular-mass peak eluted close to that expected for a 51-kDa monomeric form of the fusion protein. The earlier-eluting high-molecular-mass peak corresponded to a protein with a molecular mass of 110 kDa, as expected for the dimeric form of MBP/VP4LZ. An aliquot of each peak was examined by Western blot analysis with a monoclonal anti-MBP antibody to confirm the presence of the monomeric and dimeric species. The presence of dimers in the chimeric but not the parent MBP suggests that the leucine zipper region of VP4 was responsible for the observed dimerization.
FIG. 3.
Gel filtration analysis of MBP fusion proteins. Aliquots of purified MBP, MBP/VP4LZ, and MBP/VP4LZP were loaded onto a GF250 column. Samples were eluted at a flow rate of 0.5 ml/min with 50 mM Tris–200 mM NaCl as the eluent, and the UV absorption at 214 nm was measured. (A) MBP eluted as a single peak; (B) MBP/VP4LZ eluted as two peaks of approximately the same size; (C) MBP/VP4LZP (L537P) eluted mainly as a single peak.
A single mutation in the center of the putative leucine zipper motif prevents oligomerization of the fusion protein.
To confirm that the putative leucine zipper motif was responsible for oligomerization of the fusion protein, a mutant form of the fusion protein, in which the central leucine residue of the heptad leucine residue was replaced with a proline residue (L537P), was generated. The mutant protein (MBP/VP4LZP) was expressed and purified as described above. Analysis by gel filtration chromatography showed that the profiles of the eluted protein peaks differed significantly from that of the parent fusion protein. The dimeric form was significantly reduced (Fig. 3C). When calculated, the ratio of monomer to dimer was 1:0.006; i.e., only 0.6% of the total protein was in the form of a dimer. By contrast, the MBP/VP4L2 protein exhibited a ratio of 1:1. These data demonstrate that a single mutation in a key leucine residue perturbed the oligomerization function of the domain, although it did not abrogate it completely.
VP4 exists as a dimer.
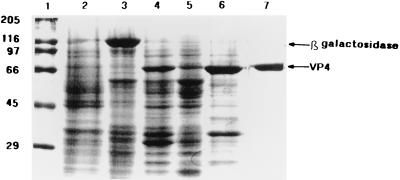
Although the fragment encompassing the putative leucine zipper motif exhibited an ability to oligomerize a foreign protein, the issue of whether it also is involved in oligomerization of VP4 remained to be determined. For this purpose, a baculovirus vector that expresses a functional form of VP4 was used for comparative analyses with baculovirus-expressed mutant forms of VP4 (13). The level of VP4 in insect cell cultures is very high, albeit present predominantly as an insoluble form (13). Since the use of purified VP4 was essential to investigate the dimeric nature of the protein, a procedure was developed to obtain a soluble and homogeneous form of VP4. Solubilization of VP4 was initially achieved by the addition of a high salt concentration (to 1 M NaCl). We also took advantage of the nonspecific RNA binding property of VP4 to obtain a final purification, by binding the protein onto a poly(U) column and separating it from residual cellular proteins. The purity of the 76-kDa protein was analyzed by SDS-PAGE and Western blotting (Fig. 4).
FIG. 4.
Expression and purification of full-length VP4 examined by SDS-PAGE (10% polyacrylamide). Lanes: 1, molecular mass markers; 2, uninfected Sf9 cell lysate; 3, Sf9 cells infected with PAK6 expressing β-galactosidase; 4, Sf9 cells infected with recombinant baculovirus expressing VP4; 5, supernatant of Sf9 cells infected with recombinant baculovirus expressing VP4 following lysis with HNN buffer; 6, solubilization of VP4 in 1 M NaCl; 7, purified VP4 after passage through the PD10 column and the poly(U) column.
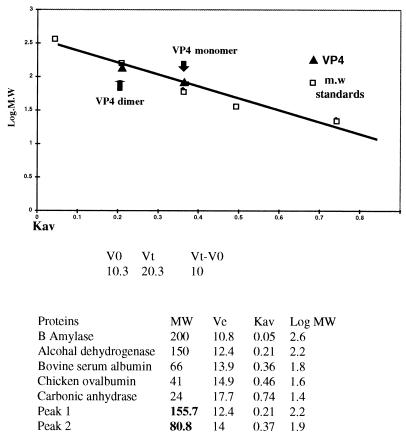
To determine whether recombinant VP4 exists as monomers or as dimers, we first analyzed the purified VP4 by gel filtration chromatography in the presence of a low salt concentration, as described in Materials and Methods. The presence of VP4 in each fraction was initially monitored by SDS-PAGE followed by Western blot analysis (data not shown). VP4 was eluted as two sharp peaks, one between fractions 120 and 125 and the second between fractions 138 and 142. The molecular masses of the protein samples in each peak were determined in relation to the known molecular mass markers as described in Materials and Methods. As shown in Fig. 5, the molecular mass of the protein in one peak was 158 kDa, equivalent to a dimeric form of VP4, and that of the second, 80 kDa, was equivalent to monomeric VP4. When calculated, the ratio of monomer to dimer was 1:1 (data not shown). These results clearly demonstrate that under these conditions VP4 exists as monomers and dimers.
FIG. 5.
Molecular mass determination of the purified full-length VP4 by gel filtration chromatography. A molecular mass calibration curve was obtained from the elution profiles of the protein molecular weight (m.w.) standards shown in the bottom panel. Apparent molecular masses of different forms of VP4 (monomer and dimer as indicated) were determined by comparing their average Ve/V0 ratios (obtained from three independent determinations) with the calibration curve shown in the upper panel.
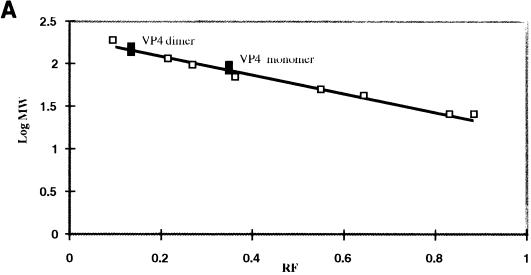
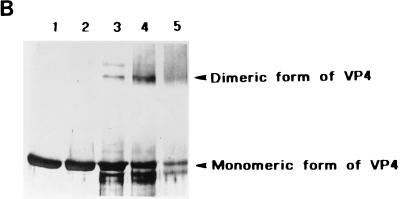
To analyze VP4 further, we used glutaraldehyde to cross-link the dimer and to produce a stable form of such dimers. Initially, and to examine the effects of glutaraldehyde on VP4, the protein was treated with various concentrations of glutaraldehyde (final concentration, 0.0001 to 0.1% [vol/vol]) and the protein complexes were analyzed by SDS-PAGE gel (data not shown). Following glutaraldehyde treatment, a significant portion of the VP4 protein migrated as a dimer (20 to 25% [data not shown]), although larger aggregates of protein were also observed at the top of the gel. The molecular masses of the monomers and dimers were estimated more accurately by generating a graph, drawn by plotting Rf values on the x axis and log molecular masses on the y axis (Fig. 6A). The VP4 dimers had molecular masses of 162 kDa, whereas the VP4 monomers had apparent molecular masses of 79 kDa. Since the calculated molecular mass of VP4 is 75 kDa, the latter value is within the range of the expected size (±10%) (Fig. 6A). The identities of the bands were confirmed by Western blotting with an anti-VP4 polyclonal guinea pig antiserum (Fig. 6B). The data clearly showed that VP4 forms dimers, since the protein bands corresponding to both dimeric and monomeric forms of VP4 were detected in the presence of 0.001 and 0.01% glutaraldehyde (Fig. 6B, lanes 3 and 4). At a higher concentration of glutaraldehyde (0.1%) (lane 5), the amount of monomeric protein decreased and a smear was observed in the position of the dimers and higher up the gel. When treated with a low concentration of glutaraldehyde (0.0001%), the VP4 monomer remained unaffected. In addition to dimeric and monomeric bands, a second high-molecular-mass band was identified (lane 3) in PAGE. This is probably due to either partially cross-linked polymers of VP4 or multimeric forms of a lower-molecular-mass degradation product of VP4.
FIG. 6.
Determinations of the molecular mass oligomeric status of baculovirus-expressed VP4 after glutaraldehyde cross-linking. (A) After SDS-PAGE (10% polyacrylamide), log molecular masses of protein standards (y axis) (□) were plotted against Rf values (distance moved by the protein/distance moved by the dye front) (x axis) as described in Materials and Methods. Size estimates of VP4 monomers and dimers (▪) were extrapolated from the graph. (B) Purified VP4 was cross-linked with glutaraldehyde, subjected to SDS-PAGE (10% polyacrylamide), and transferred to a PVDF membrane for Western blot analyses. The blot was reacted with a polyclonal anti-VP4 guinea pig antiserum. Lanes: 1, VP4 without glutaraldehyde; 2 to 5, VP4 cross-linked with 0.0001, 0.001, 0.01, or 0.1% glutaraldehyde, respectively. (C) Purified CLPs encapsidating VP4 were cross-linked with glutaraldehyde, subjected to SDS-PAGE (8% polyacrylamide), and transferred to a PVDF membrane for Western analyses. The blot was reacted with a polyclonal anti-BTV rabbit antiserum. Lanes: 1, CLPs encapsidating VP4, cross-linked with 0.001% (final concentration) glutaraldehyde; 2, CLPs encapsidating VP4 without glutaraldehyde.
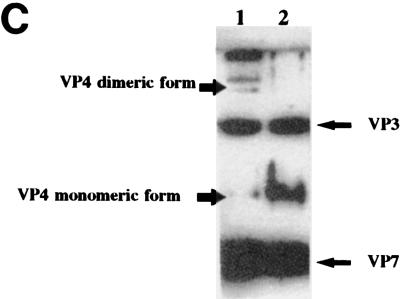
To evaluate the biological relevance of the dimeric form of VP4, it was necessary to investigate the oligomeric nature of VP4 within the BTV-derived particle. To facilitate the identification of the VP4 dimer within the virion and to avoid formation of oligomers of VP4 with VP1 and/or VP6 upon chemical cross-linking, we used CLPs containing only VP4 but not the other two minor proteins, VP1 and VP6. The CLPs were treated with glutaraldehyde in a similar way to that described above. After cross-linking the CLPs encapsidating VP4 with glutaraldehyde (0.001%, final concentration), the products were analyzed by SDS-PAGE (8% polyacrylamide) followed by Western blotting with anti-BTV polyclonal rabbit antiserum. Figure 6C (lane 1) shows the cross-linked product of CLPs encapsidating VP4, in which both VP4 monomers and dimeric bands were visualized clearly. As expected, both VP3 and VP7 monomeric bands were also present. In addition, a large band migrating slightly slower (∼200 kDa) than that of VP4 dimer was recognized by BTV antibody. This 200-kDa band was most probably VP3 dimer, since the VP3 monomeric size is 103 kDa. Alternatively, it could be a heterodimer of VP3 and VP4. However, this band was not detected by VP4-specific antisera, and so it is highly unlikely that it is a heterodimer of VP3 and VP4. Some aggregates of the proteins which could not enter the gel were also detected above this band. Lane 2 shows non-cross-linked product, where only monomeric bands of VP3, VP7, and VP4 were seen. The identity of the VP4 bands was reconfirmed by using anti-VP4 polyclonal guinea pig antiserum (data not shown). The results clearly demonstrate that VP4 indeed exists as a dimer in CLPs and forms homodimers but does not oligomerize with VP3 and VP7.
Leucine at aa 537 drives the dimerization of VP4.
To confirm the data obtained from the chimeras analyzed above and to determine the importance of the central leucine residue in the putative leucine zipper, two different substitution mutations were created. The first involved proline in lieu of leucine 537, and the second involved alanine in lieu of leucine 537. As described in Materials and Methods, both mutant forms of VP4, VP4L537A and VP4L537P, were generated with recombinant baculovirus expression systems. As expected, SDS-PAGE and Western blot analyses confirmed the presence and size of the mutant VP4 proteins (Fig. 7).
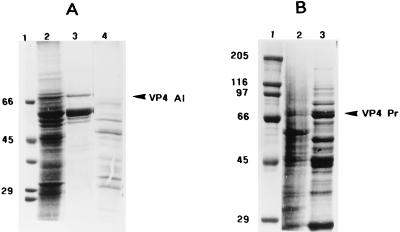
FIG. 7.
SDS-PAGE (10% polyacrylamide) of recombinant baculovirus-expressed full-length mutant VP4 proteins. (A) Lanes: 1, molecular mass markers; 2, Sf9 cells infected with recombinant baculovirus expressing VP4L537A; 3, supernatant Sf9 cells infected with recombinant baculovirus expressing VP4L537A following lysis with HNN buffer; 4, pellet of Sf9 cells infected with recombinant baculovirus expressing VP4L537A following lysis with HNN buffer (see Materials and Methods). (B) Lanes: 1, molecular mass markers; 2, pellet of Sf9 cells infected with recombinant baculovirus expressing VP4L537P following lysis with HNN buffer (see Materials and Methods); 3, supernatant Sf9 cells infected with recombinant baculovirus expressing VP4L537P following lysis with HNN buffer.
As with wild-type recombinant protein, the expression levels of both mutants were very high. However, surprisingly, and unlike wild-type VP4, both mutant proteins were soluble in low-salt buffer (see Materials and Methods). Figure 7A (lane 3) and Fig. 7B (lane 3) show samples of the supernatants after solubilization with HNN buffer (50 mM HEPES, 100 mM NaCl, 0.5% Nonidet P-40 [pH 7.4]) and centrifugation at 3,000 × g. Very little mutant protein was present in the pellet (Fig. 7A, lane 4). Despite the solubility and high expression levels, these proteins could not be purified by the usual column chromatography procedures (such as ion-exchange, hydrophobic, or blue Sepharose columns). The mutant proteins also failed to attach to the poly(U) column (data not shown), indicating that they had lost their RNA binding ability. As a consequence, only partially purified mutant VP4 proteins were used for further studies.
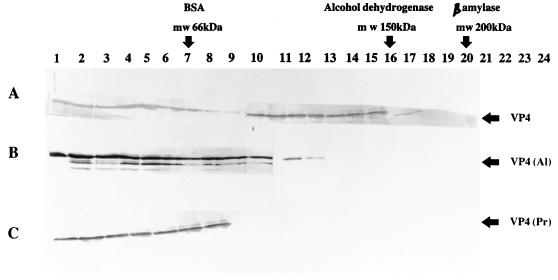
To monitor the oligomeric forms of the mutant proteins, we used an alternative sucrose density gradient centrifugation assay system in which the different sizes of VP4, such as monomer, dimer, and other multimers, could be separated. The gradients were fractionated, the presence of VP4 was monitored by Western blot analysis, and the molecular masses were determined as described in Materials and Methods. As shown in Fig. 8A, in the gradients the monomeric forms of VP4 were identified between fractions 6 and 10 (i.e., toward the top of the gradients), equivalent to the fractions that contain 68-kDa bovine serum albumin. The dimeric forms, on the other hand, and as expected, migrated in the middle of the gradient (fractions 10 to 15), similar to a 150-kDa protein, alcohol dehydrogenase. These results correspond to the data obtained from gel filtration chromatography and chemical cross-linking. The data obtained with two mutant forms were different. For the VP4L537A mutant protein, both monomeric (lanes 1 to 10) and dimeric (lanes 10 to 12) forms of the protein were present (Fig. 8B). However, there was less dimeric mutant protein than that obtained for the wild-type VP4. In contrast, the VP4L537P mutant was recovered only as a monomeric form (Fig. 8C) and not as a dimeric form, indicating that the substitution prevented dimerization of the protein. By way of comparison, the MBP/VP4LZ fusion protein was analyzed similarly. As expected, similar monomeric and dimeric forms of the fusion proteins were identified for MBP/VP4LZ. For the mutant fusion protein MBP/VP4LZP, only the monomeric form was identified (data not shown). These results not only confirmed the ability of the leucine zipper region to drive a monomeric protein into a dimeric form but also demonstrated that certain mutations (such as proline substitution) in the central leucine of the putative leucine zipper region of BTV-10 VP4 can abolish dimerization of the protein.
FIG. 8.
Western analysis of baculovirus-expressed wild-type and mutant forms of VP4 after fractionation by sucrose gradient centrifugation. Lanes 1 to 24 represent fractions collected from the sucrose gradient; fraction 1 represents the top of the gradient, and fraction 24 represents the bottom. After SDS-PAGE (10% polyacrylamide), all the fractions were subjected to Western analysis with anti-VP4 antisera. The positions of the standard marker proteins bovine serum albumin (BSA) (68 kDa), alcohol dehydrogenase (150 kDa), and β-amylase (200 kDa) are indicated at the top. (A) Baculovirus-expressed wild-type VP4; (B) VP4L537A, showing monomer and dimers (predominantly monomer, and a small portion as dimer); (C) VP4L537P mutant, showing only monomeric forms.
The dimeric form of VP4 is essential for encapsidation of the molecules within CLPs.
To investigate the biological significance of the leucine zipper motif and the dimeric forms of VP4, we examined the encapsidation activity of native and mutant forms of VP4 molecule by using an assay system in which proficient assembly of CLPs can be achieved (6).
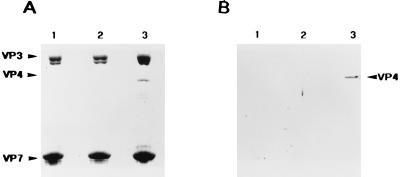
CLPs were prepared in the presence of wild-type or mutated forms of VP4, and the presence of VP4 in each preparation was analyzed by SDS-PAGE and Western blotting with an anti-VP4 antiserum. As shown in Fig. 9, recombinant wild-type VP4 was encapsidated efficiently within CLPs consisting of VP3 and VP7 (see lane 3). Surprisingly, not only the proline substitution mutant, VP4L537P (lane 2) but also the alanine substitution mutant VP4L537A (lane 1) failed to be encapsidated by CLPs, even though VP4L537A produced some dimeric forms of VP4 in isolation.
FIG. 9.
Encapsidation of the wild-type and mutant VP4 molecules by CLPs. Sf9 cells were coinfected with a dual recombinant baculovirus expressing recombinant proteins VP3 and VP7 and either with a recombinant virus expressing wild-type VP4 or with one of the recombinant viruses expressing mutant VP4. (A) Coomassie blue-stained gel. (B) Western blot with guinea pig anti-VP4 antiserum. Lanes: 1, gradient-purified CLPs obtained from coinfection of Sf9 cells with a dual-recombinant baculovirus expressing VP3 and VP7 and a recombinant baculovirus expressing VP4L537A; 2, gradient-purified CLPs obtained from coinfection of Sf9 cells with a dual-recombinant baculovirus expressing VP3 and VP7 and a recombinant baculovirus expressing VP4L537P; 3, gradient-purified CLPs obtained from coinfection of Sf9 cells with a dual-recombinant baculovirus expressing VP3 and VP7 and a recombinant virus expressing wild-type VP4.
DISCUSSION
Our initial investigation involving the putative leucine zipper region of VP4 was carried out by generating a bacterial fusion protein expressing the heptad repeat of leucine residues within the leucine zipper region of VP4, together with a bacterial MBP. The rationale behind this was to examine whether the monomeric form of MBP could be converted into a dimeric form by these VP4 sequences. The data clearly demonstrated that this region of VP4 can drive the dimerization of at least this monomeric protein. It is noteworthy in this context that when the hydrophobic heptad repeat of the HIV-1 transmembrane protein was similarly fused with monomeric proteins, such as MBP or protein A, the fusion proteins also formed dimers (2, 20). We have obtained direct proof of the involvement of a central leucine residue in the chimeric protein dimerization from analyses of a proline substitution mutant. Similar results have been reported for proline and aspartic acid substitution of the central isoleucine of the heptad repeat of HIV-1 gp41 (2).
Purified VP4, when passed through a gel filtration column, eluted as two major peaks, confirming the existence of both monomeric and dimeric forms of the protein. When we exposed purified VP4 to glutaraldehyde cross-linking, two sizes of VP4 molecules were identified in PAGE, one with an apparent molecular mass of 79 kDa and one with a mass of 162 kDa, i.e., equivalent to the monomeric and dimeric forms of VP4, respectively. Dimer formation by cross-linking occurred at 0.001 and 0.1% (vol/vol) glutaraldehyde. At 0.0001% glutaraldehyde, dimers were not stabilized. At high concentrations (e.g., 0.1%) most of the protein was present in the position of the dimeric forms but as a smear on SDS-PAGE. In addition, on SDS-PAGE, a second high-molecular-mass band was identified. Since this band reacted with VP4 polyclonal antiserum, it is likely that it consists of either cross-linked aggregates of VP4 or multimeric forms of a lower-molecular-mass degradation product of VP4. The existence of both monomeric and dimeric forms of VP4 under native conditions was confirmed by sucrose density gradient centrifugation of purified VP4.
The predicted leucine zipper motif in BTV VP4 has an unusual structure with a proline residue in the middle of the structure. Prolines often lead to an unacceptable disruption of helix hydrogen bonding, for example by dictating a kink in the structure and disturbing the packing of the helix side chains (3, 14). Generally, it is considered that proline residues act as α-helix breakers. Nevertheless, despite the presence of a proline residue, our data indicate that this region of VP4 functions as a driving force for dimeric forms of VP4.
Barlow and Thornton (1) have reported a survey of 291 helices in 57 proteins. Ten of these helices contained an internal proline, and Barlow and Thornton showed that conserved prolines in the α-helices played a definite structural and functional role in such proteins and suggested that the kink created by proline residues in each helix provides a necessary, rather than undesirable, distortion for such sequences.
The functional and structural importance of the leucine zipper of VP4 was confirmed by creating two substitution mutants with recombinant baculovirus expression systems. Our data demonstrate that replacement of leucine by alanine reduced the dimeric form of the protein but did not abolish it completely. In contrast, the replacement by proline essentially abolished dimer formation. Interestingly, both mutant proteins were highly soluble in low-salt buffer, unlike native VP4. The reason why these properties of the protein were altered is not known.
In the virion, VP4 resides within the inner core together with 2 other minor proteins VP1 and VP6 and 10 dsRNA segments. VP4 can be incorporated in the CLPs by coexpression of VP4 and the major core proteins VP3 and VP7, using recombinant baculovirus expression systems. By using this functional assay system, we have demonstrated that the mutant VP4 proteins that have lost the capability to form the VP4 dimer were not incorporated into CLPs, indicating that VP4 has to be in dimeric form to be encapsidated into the BTV core.
In summary, a putative leucine zipper has been shown to be a functional domain that drives the dimerization of VP4, and it has been demonstrated that the dimeric VP4 is essential for correct assembly into BTV cores.
ACKNOWLEDGMENTS
We thank Stephanie Price for typing the manuscript, Chris Hatton for doing photography, and Adele Peak for helping with in cell culture.
This work was partially funded by BBSRC Link grant AO2643 and NIH grant A126879.
REFERENCES
- 1.Barlow D J, Thornton J M. Helix geometry in proteins. J Mol Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein H B, Simon P T, Sambit R K, Sylvia A M, David T M, John W D, Jacob L, Richard W C, Eric H. Oligomerization of hydrophobic heptad repeat of gp41. J Virol. 1995;69:2745–2750. doi: 10.1128/jvi.69.5.2745-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou C P Y, Fasman G D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 4.Dang C V, McGuire M, Buckmire M, Lee W M F. Involvement of the “leucine zipper” region in the oligomerization and transforming activity of C-Myc protein. Nature. 1989;337:664–660. doi: 10.1038/337664a0. [DOI] [PubMed] [Google Scholar]
- 5.Delwart E L, Mosialos G. Retroviral envelope glycoproteins contain a “leucine zipper”-like repeat. AIDS Res Hum Retroviruses. 1990;6:703–706. doi: 10.1089/aid.1990.6.703. [DOI] [PubMed] [Google Scholar]
- 6.French T J, Roy P. Synthesis of bluetongue virus (BTV) core-like particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J Virol. 1990;64:1530–1536. doi: 10.1128/jvi.64.4.1530-1536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu J C, O’Shea E K, Kim P S, Sauer R T. Sequence requirements for coiled coil: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 8.Huang I J, Hayama E, Jeong Y J, Li J K K. Conservation of the segment 4 gene sequence and of a leucine zipper motif in VP4 among five US bluetongue viruses. Virology. 1993;195:772–779. doi: 10.1006/viro.1993.1429. [DOI] [PubMed] [Google Scholar]
- 9.Kitts P A, Possee R D. A method for producing recombinant baculovirus expression vectors at high frequency. BioTechniques. 1993;14:810–816. [PubMed] [Google Scholar]
- 10.Kitts P A, Ayres M D, Possee R D. The linearisation of baculovirus DNA enhances the recovery of recombinant expression vectors. Nucleic Acids Res. 1990;18:5667–5671. doi: 10.1093/nar/18.19.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 13.LeBlois H, French T, Mertens P P C, Burroughs J N, Roy P. The expressed VP4 protein of BTV binds GTP and is the candidate guanylyltransferase of the virus. Virology. 1992;189:757–761. doi: 10.1016/0042-6822(92)90600-t. [DOI] [PubMed] [Google Scholar]
- 14.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 15.Mertens P P C, Burroughs J N, Wade-Evans A M, LeBlois H, Oldfield S, Basak A, Loudon P, Roy P. Bluetongue, African horsesickness, and related orbiviruses. Proceedings of the Second International Symposium. 1992. Analysis of guanylyltransferase and transmethylase activities associated with bluetongue virus core and recombinant baculovirus-expressed core like particles; pp. 404–415. [Google Scholar]
- 16.O’Shea E K, Klemm J K, Alber T. X-ray structure of the GCN4 leucine zipper, a two stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 17.Ramsdale E E, Kingsman S M, Kingsman A J. The putative leucine zipper region of murine leukemia virus transmembrane protein (p150) is essential for viral infectivity. J Virol. 1996;220:100–108. doi: 10.1006/viro.1996.0290. [DOI] [PubMed] [Google Scholar]
- 18.Ransone L J, Visvader P, Sassonecorsi P, Verma I M. Fos-jun interaction: mutational analysis of the leucine zipper domain of both proteins. Genes Dev. 1989;3:770–781. doi: 10.1101/gad.3.6.770. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shugars D C, Carl T W, Teresa K G, Thomas J M. Biophysical characterization of recombinant proteins expressing the leucine zipper-like domain in human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1996;70:2982–2991. doi: 10.1128/jvi.70.5.2982-2991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Fukusho A, Roy P. Nucleotide sequence of the VP4 core protein gene (M4 RNA) of US bluetongue virus serotype 10. Nucleic Acids Res. 1987;15:7206. doi: 10.1093/nar/15.17.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]