Abstract
In vitro transcription was used to analyze the promoter specificity of the α-amanitin-resistant RNA polymerase that is induced late during infection of Autographa californica multicapsid nuclear polyhedrosis virus. By modifying the preparation of crude nuclear extracts, we have established an assay that permits differentiation between weak late and strong very late viral promoters. The virus-induced RNA polymerase initiates at a TAAG sequence motif in both late and very late promoters. Based on the sensitivity of our in vitro transcription system, we have investigated the sequences responsible for a functional TAAG motif and their putative role with respect to the strength of very late promoters. By constructing hybrid promoters between the early pe38 and the very late polyhedrin promoters, we demonstrated that the replacement of 7 nucleotides upstream of the nonfunctional TAAG sequences in the pe38 promoter with the corresponding sequences of the polyhedrin promoter was sufficient for recognition by the virus-induced RNA polymerase. The strength of the very late polyhedrin promoter was established after replacing the 5′ untranslated sequences of the pe38 promoter by those of the polyhedrin promoter in addition to the 7 nucleotides upstream of the TAAG motif.
The infection cycle of Autographa californica multicapsid nuclear polyhedrosis virus (AcMNPV) includes a complex temporally regulated gene expression cascade and results in the production of two different infectious forms. Although replication of this DNA virus takes place in the nucleus, AcMNPV infection induces a novel RNA polymerase activity that is resistant to α-amanitin (7, 12). It is not known whether the virus-induced RNA polymerase is virus encoded, comprising subunits that are divergent from known RNA polymerases, or is a host complex modified by virus-encoded subunits. The targets of the virus-induced RNA polymerase are late and very late viral promoters, while host RNA polymerase II recognizes early promoters (for reviews, see references 5 and 31). Late and very late promoters differ not only in the temporal appearance of their activity but also in promoter strength. The promoters of the very late genes polyhedrin and p10 are much stronger than those of late genes, resulting in abundant protein expression. How the virus-induced RNA polymerase contributes to the hyperexpression of the p10 and polyhedrin genes is still under investigation.
Transcriptional initiation at late and very late promoters occurs within a conserved TAAG sequence motif (35). Mutagenesis studies of the polyhedrin and p10 promoters demonstrate that upstream sequences have only a minor effect on activity whereas any mutations in the TAAG sequence abolish transcriptional initiation (26, 30, 32, 34, 43, 44). In addition, mutations in the adenine- and thymine-rich sequences downstream of the TAAG motif result in dramatic reduction of transcription. In contrast to strong very late promoter activity, weak late expression can be sufficiently directed by a TAAG sequence motif (13, 28).
The differential regulation of late and very late promoters has been analyzed in vivo and in vitro. Transient-expression studies indicate that 18 genes of AcMNPV are able to transactivate the late promoters vp39 and p6.9 (39, 40). An additional factor, vlf-1 (27), enhances activation only of the very late promoters p10 and polyhedrin, without affecting the activity of late promoters (40). It is not known how this factor contributes to the high transcriptional efficiency or whether additional factors are directly involved in the regulation of late and very late promoters.
As an alternative, in vitro transcription systems were established to identify the factors involved in baculovirus early- and late-gene expression. These in vitro systems are based on crude nuclear extracts (10, 45) or fractionated nuclear extracts (3) from infected Spodoptera frugiperda cells, which support the transcription of early, late, and very late genes. To accelerate the analysis of late and very late in vitro-transcribed RNAs, cytidine-free cassettes were introduced (45), although the high transcriptional efficiency of very late promoters observed in vivo could not be mimicked in vitro.
We have introduced a modification in the extract preparation that leads to crude nuclear protein extracts capable of faithful in vitro transcription of baculovirus very late genes, which allows the distinction between late and very late promoters based on promoter strength. Our assay was a prerequisite for determining the promoter specificity of the virus-induced RNA polymerase in more detail. Transcriptional initiation at TAAG sequences implies the recognition of a rather short consensus site by the virus-induced RNA polymerase, which in turn leads to the question of how AcMNPV accomplishes the recognition of functional versus nonfunctional TAAG motifs on its genome of 133,894 bp (1). One mechanism is selection against TAAG sequences; instead of 1,410 statistically expected motifs, only 397 are present (1). Indeed, additional mechanisms might account for the initiation at distinct TAAG motifs. The analysis of late-promoter selection in the AcMNPV gp64 gene suggests that the immediate context, rather than the position relative to active TAAG sequence motifs, determines whether the TAAG sequence serves as transcriptional initiation site (9). To analyze the role of the immediate context of TAAG sequences in transcriptional initiation, we have constructed hybrid promoters between the early pe38 (21) and the very late polyhedrin promoters. Close to the early transcriptional start, the pe38 promoter contains a TAAG motif that is not recognized by the virus-induced RNA polymerase. By replacing the pe38 flanking sequences of the TAAG element with those of the polyhedrin promoter, we demonstrated the conversion of a nonfunctional into a functional TAAG element. These observations led us to investigate which additional sequences specify a very late promoter.
MATERIALS AND METHODS
Cells and viruses.
S. frugiperda IPLB21 (41) cells were grown at 27°C in TC100 medium (8) supplemented with 10% fetal calf serum. For protein extract preparation, S. frugiperda cells were grown in suspension culture. Infection with AcMNPV plaque isolate E (38) was performed in monolayer culture at a multiplicity of infection of 10 PFU per cell. Time zero was defined as the time when the inoculum was added to the cells.
Plasmid constructions and oligonucleotides.
Plasmids pME53-CAT202 (19), pPE38-CAT82 (20), pHE65-CAT474 (2), and pAcp10-CAT (17) were described previously. The pPE38-CAT57 plasmid was generated by insertion of the oligonucleotide comprising 57 bp of the pe38 promoter into the SalI-XhoI sites of pBLCAT3 (23). This promoter construct contains 18 bp upstream and 39 bp downstream of the transcriptional start site. Plasmid pph-CAT101 was constructed by inserting the polyhedrin promoter into the PstI-BglII sites of pBLCAT3. The promoter fragment was generated by PCR amplification with plasmid pAc-EcoH (22) as the template and the following primers carrying BglII and PstI sites, respectively: 5′TTTTCTGCAGGTTGCTGATATC3′ and 5′GGGGAGATCTCCGGCATATTTATAG3′. The resulting fragment contains 101 nucleotides upstream of the polyhedrin initiation codon, which is in frame with the chloramphenicol acetyltransferase (CAT) gene.
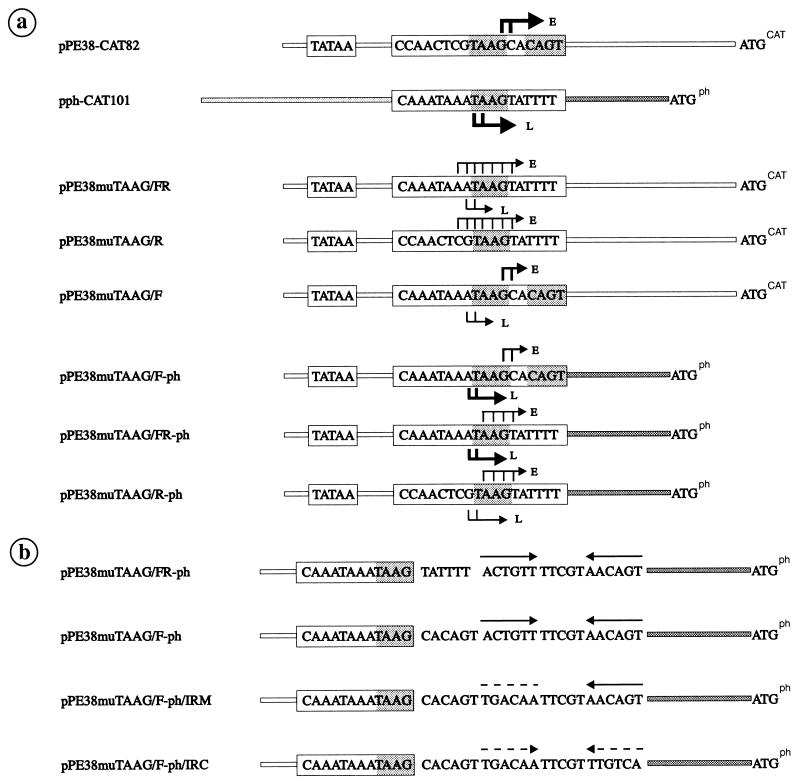
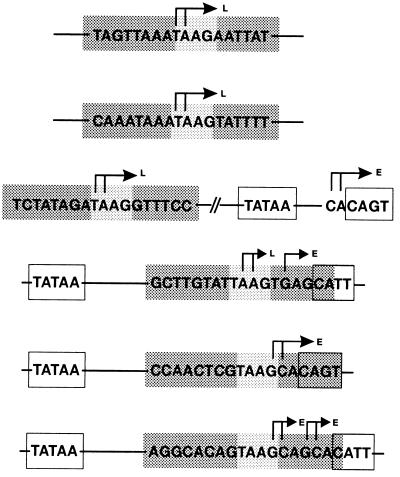
Plasmids containing pe38/polyhedrin hybrid promoters (see Fig. 4) were all constructed by inserting various oligonucleotides into the XbaI-XhoI sites of pBLCAT3. Oligonucleotides were synthesized on an Applied Biosystems 381 A DNA synthesizer. After oligonucleotides were heated at 95°C for 3 min, complementary pairs were annealed for 25 min each at 60 and 37°C and then inserted into the vector. The resulting plasmids, pPE38muTAAG/FR, pPE38muTAAG/R, and pPE38muTAAG/F, carry promoter fragments that are identical in their 5′ and 3′ sequences and correspond to those of plasmid pPE38-CAT82 (see Fig. 4). The promoter fragments in plasmids pPE38muTAAG/F-ph, pPE38muTAAG/FR-ph, pPE38muTAAG/R-ph, pPE38muTAAG/F-ph/IRM, and pPE38muTAAG/F-ph/IRC have the same 5′ sequences as the pe38 promoter in plasmid pPE38-CAT82 and the same 3′ sequences as the polyhedrin promoter in pph-CAT101 (see Fig. 4). The hybrid promoter constructs were named as follows: constructs are based on the pe38 promoter with mutations in the TAAG box of 18 bp (PE38muTAAG); 7 bp in front (F) and 5 bp in the rear (R) of the TAAG motif are replaced by those of the polyhedrin promoter; in addition to F and R mutations, the 5′ untranslated sequences of the pe38 promoter have been replaced by those of the polyhedrin promoter (ph); the inverted repeat in the 5′ untranslated region (UTR) of the polyhedrin promoter has been mutated (IRM) or the mutated inverted repeat has been restored by complementing mutations (IRC).
FIG. 4.
Schematic representation of the pe38/polyhedrin hybrid promoter constructs used for in vitro transcription. The putative TATA motifs and the sequences of the early and late transcriptional start sites are boxed. The hybrid promoters are followed by the translational initiation codon of the CAT gene (ATGCAT) or polyhedrin gene (ATGph). (a) Arrows indicate early (E) and late/very late (L) transcriptional start sites, and the thickness of the arrows is proportional to the efficiency of initiation. (b) Arrows represent the inverted repeats; broken lines indicate the mutated sequences.
The sequence of each promoter construct was verified by sequence analysis with an Applied Biosystems 373A DNA sequencer.
Preparation of nuclear extracts.
Crude nuclear extracts from uninfected S. frugiperda cells were prepared as described previously (20). To prepare nuclear extracts of infected S. frugiperda cells, nuclei were purified more extensively before their lysis, as described initially by Gorski et al. (11) for tissue-specific transcription from the mouse albumin promoter. We have adapted the method for nuclei of infected S. frugiperda cells as follows: 8 × 108 cells grown in spinner cultures were harvested at various times after infection and resuspended in a hypotonic buffer (10 mM HEPES [pH 8.3], 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, 2 M sucrose, 10% glycerol) in a twofold volume of the packed cells. The cells were broken by homogenization in a glass Dounce homogenizer (L-pestle). The homogenate was layered over a 0.5-ml cushion of the same buffer and centrifuged at 25,000 rpm for 30 min at 4°C in a Beckman SW60 rotor (11). The nuclei, which accumulated on top of the cushion, were resuspended in 1 ml of the hypotonic buffer, again in the Dounce homogenizer. This homogenate was centrifuged under the same conditions as described above. The nuclear fraction was resuspended in 4 volumes of nuclear extraction buffer [15 mM HEPES (pH 8.3), 105 mM KCl, 4.4 mM MgCl2, 0.01 mM EDTA, 0.01 mM dithioerythreitol, 1 mM phenylmethylsulfonyl fluoride, 2.9 KIU of aprotinin per ml, 340 mM (NH4)2SO4]. The nuclei were lysed by stirring for 30 min at 4°C, and the lysate was centrifuged at 40,000 rpm in a Beckman TLS-55 rotor for 60 min at 4°C. The supernatant was dialyzed against 40 mM KCl–10 mM Tris HCl (pH 8)–0.1 mM EDTA–1 mM dithiothreitol–10% glycerol for 4 h. The protein yield was usually 2.5 μg/μl. The aliquots were frozen in liquid N2 and stored at −70°C.
In vitro transcription assay.
The conditions for in vitro reactions, including pH, temperature, and salt requirements, were as described previously (10). However, preincubation was not performed for early, or late, or very late promoters, since it did not influence the efficiency of transcription under our conditions. The optimal DNA template concentration was 4 nM. The optimal protein concentration was determined for each extract made, ranging between 0.6 and 1.2 μg/μl.
Transcriptional analysis.
The 5′ ends of the in vitro-transcribed RNAs were mapped by primer extension analyses as previously described (2) with the CAT-specific primer 5′GCCATTGGGATATATCAACGGTGGTATATCC3′. The extended products were analyzed on 8% polyacrylamide–7 M urea gels, and were quantified by a bioimaging analyzer system (FUJIX BAS 1000, version 3.0).
RESULTS
Experimental design.
We have addressed the question of how the virus-induced RNA polymerase distinguishes functional from nonfunctional TAAG motifs and subsequently differentiates between late and very late promoters by constructing hybrid promoters whose activity was tested in vitro. Although recent studies demonstrated that late and very late promoters can be transcribed in vitro by crude nuclear extracts (10, 45), the transcriptional activity detected did not reflect the strength of the very late promoters observed in vivo. Therefore, we have initially optimized the in vitro transcription system to differentiate between late and very late promoters.
In vitro transcription assay for late and very late promoters.
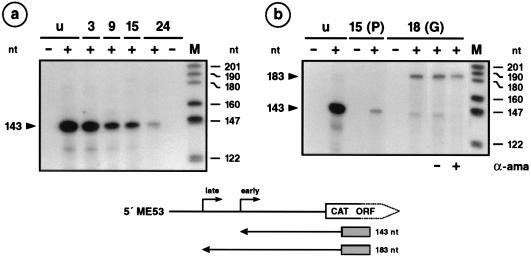
To optimize the in vitro assay for late transcription, we chose an early/late promoter unit such as the me53 promoter as the template for early and late transcription and the p10 promoter as the template for very late transcription. Previous experiments demonstrated both early and late me53 transcriptional start sites after infection of S. frugiperda and TN-368 cells (18), suggesting that the me53 promoter serves as a target for both host- and virus-induced RNA polymerases. In vitro transcription was performed with crude nuclear extracts of S. frugiperda cells and plasmids pME53-CAT202 and pAcp10-CAT containing the me53 and p10 promoters, respectively; this step was followed by primer extension analysis. The extended products of 143 and 183 nucleotides (nt) represent early and late me53 transcription, respectively (Fig. 1), and the 357-nt products reflect p10 very late transcription (Fig. 2).
FIG. 1.
In vitro transcription of the early/late me53 promoter. (a) Nuclear extracts of uninfected (lanes u) or AcMNPV-infected S. frugiperda cells prepared at 3, 9, 15, and 24 h p.i. (lanes 3, 9, 15, and 24) by the method of Krappa et al. (20) were used for in vitro transcription. Plasmid pME53-CAT202 was used as the template (lanes +). As negative control, the DNA template was omitted (lanes −). Transcripts were analyzed by primer extension with a 31-mer CAT-specific primer. The extended products of 143 and 183 nt represent the early and late me53 transcriptional start sites, respectively. (b) In vitro transcription was performed in extracts of uninfected cells (lanes u), in extracts prepared from cells at 15 h p.i. by the method of Krappa et al. (20) [lanes 15 (P)], or in extracts prepared from cells at 18 h p.i. by the method of Gorski et al. (11) [lanes 18(G)]. The absence (−) or presence (+) of 2.5 μg of α-amanitin (α-ama) per ml is indicated below the autoradiogram. Positions of DNA size markers (lanes M) are shown on the right. ORF, open reading frame. The localization of the extended products and the corresponding start sites are shown below.
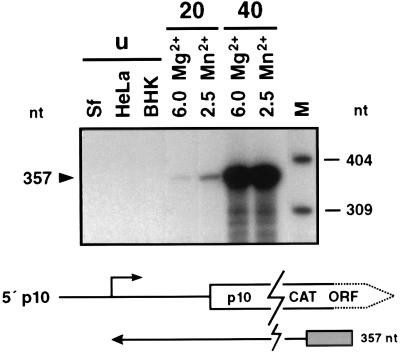
FIG. 2.
In vitro transcription of the very late p10 promoter. Plasmid pAcp10-CAT was used as template for in vitro transcription in nuclear extracts of the vertebrate cell lines HeLa and BHK and of uninfected (lanes u) or AcMNPV-infected S. frugiperda cells prepared at 20 h p.i. (lanes 20) and 40 h p.i. (lanes 40). Nuclear extracts of uninfected and AcMNPV-infected cells were prepared by the method of Gorski et al. (11). In vitro transcription with extracts of uninfected cells was performed in the presence of 6 mM Mg2+ and with extracts of infected cells in the presence of either 6 mM Mg2+ or 2.5 mM Mn2+; α-amanitin (2.5 μg/ml) was added to each reaction mixture. The extended products of 357 nt represent the very late p10 transcriptional start. Positions of DNA size markers (lane M) are shown on the right. ORF, open reading frame. The localization of the extended product and the corresponding start site are shown below.
Early me53 transcription was very efficiently supported by nuclear extracts of uninfected cells and by nuclear extracts prepared from cells at 3 h postinfection (p.i.) (Fig. 1a), as described by Krappa et al. (20). A decrease in transcription was observed in extracts prepared from cells at 9, 15, and 24 h p.i. (Fig. 1a), which corresponds to the decrease of early me53 transcription during the course of infection (18). However, late me53 transcription was not detectable in nuclear extracts prepared at 15 and 24 h p.i. (Fig. 1a). Since changing a variety of reaction conditions did not improve late transcription, we changed the procedure used to prepare the nuclear extracts. After addition of a step to purify the nuclei, crude nuclear extracts prepared at 18 h p.i. recognized the late me53 start site and still supported early transcription (Fig. 1b). Addition of α-amanitin abolished the accumulation of the early transcript but did not inhibit late transcription (Fig. 1b), which indicated that the late promoter is recognized by the virus-induced RNA polymerase in the optimized crude nuclear extracts.
To test whether the strong increase in transcription of very late promoters can also be supported by the optimized nuclear extracts, the p10 and polyhedrin promoters were transcribed in vitro. The transcriptional activity of the p10 gene increases dramatically in vivo from 24 to 48 h p.i. (30, 37). We have analyzed the increase in transcriptional efficiency in vitro by comparing nuclear extracts prepared from cells at 20 and 40 h p.i. As expected, p10 transcription was not supported by extracts from uninfected insect (S. frugiperda) and mammalian (HeLa and BHK) cell lines (Fig. 2). Weak p10 transcription was observed in extracts prepared from cells at 20 h p.i., while extracts isolated at 40 h p.i. gave rise to very high p10 transcriptional activity, showing a more than 50-fold stimulation (Fig. 2). Similar results were obtained for the polyhedrin promoter (Fig. 3). As a control, late me53 transcription was tested in extracts prepared from cells at 20 and 40 h p.i., demonstrating no increase of transcriptional activity in extracts isolated at 40 h p.i. (data not shown). Therefore, our in vitro system reflects the transcriptional burst of the p10 and polyhedrin promoters which is observed in vivo during the very late phase of infection. We also investigated the requirement of the virus-induced RNA polymerase for divalent cations, since differences from the requirements of the host polymerase have been observed in vivo (7) and in vitro (10). Our results confirmed the preference of the virus-induced RNA polymerase for low concentrations of Mg2+ or Mn2+ in vitro (Fig. 2), indicating that the nature of the divalent cations does not seem to be particularly important (data not shown).
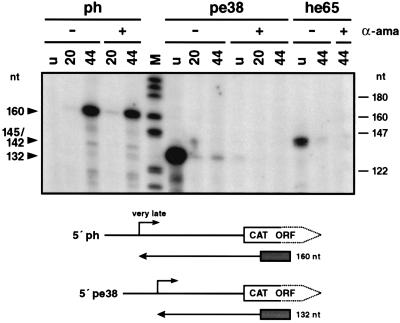
FIG. 3.
In vitro transcription of the very late polyhedrin promoter in comparison to early pe38 and he65 transcription. Plasmids pph-CAT101, pPE38-CAT82, and pHE65-CAT474 were used as templates for in vitro transcription in nuclear extracts of uninfected S. frugiperda cells (lanes u) or AcMNPV-infected S. frugiperda cells prepared at 20 h p.i. (lanes 20) and 44 h p.i. (lanes 44) by the method of Gorski et al. (11). In vitro transcription reactions were performed in the presence (lanes +) or absence (lanes −) of α-amanitin (α-ama) (2.5 μg/ml). The extended products of 160 nt represent the very late polyhedrin transcriptional start site, and 132 nt and 145/142 nt represent the early pe38 and he65 start sites, respectively. Positions of DNA size markers (lane M) are shown on the right. ORF, open reading frame. The localization of the extended product and the corresponding start site are shown below.
Functional and nonfunctional TAAG sequence motifs in transcriptional initiation by the virus-induced RNA polymerase.
In late and very late promoters, TAAG sequence motifs from the core of transcriptional start sites recognized by the virus-induced RNA polymerase. In addition to early transcriptional start sites, some promoters such as the me53 (19) and p35 (29) promoters carry TAAG sequences which can be recognized by the virus-induced RNA polymerase while others such as the ie2 (6), pe38 (21), and he65 (2) promoters include TAAG sequences that are nonfunctional during the late phase of infection (Table 1). To analyze whether these TAAG sequences indeed do not serve as recognition sites of the virus-induced RNA polymerase, we performed in vitro transcription assays with our optimized nuclear extracts and with plasmids pPE38-CAT82 and pHE65-CAT474 as templates, followed by primer extension analysis. The extended products of 132 and 142 nt represent the early pe38 and he65 transcripts, respectively, which were abundant when extracts were prepared from uninfected cells and weakly present in extracts prepared from cells at 20 or 44 h p.i. (Fig. 3). In the presence of α-amanitin, the extended products disappeared (Fig. 3), confirming that the pe38 and he65 promoters were not recognized by the virus-induced RNA polymerase. As a control, strong transcription of the polyhedrin promoter in extracts prepared from cells at 44 h p.i. is shown in the presence of α-amanitin (Fig. 3).
TABLE 1.
Functional and nonfunctional TAAG motifs in AcMNPV promoters
These observations prompt the question of how the virus-induced RNA polymerase differentiates between functional and nonfunctional TAAG sequence motifs. Previous results demonstrate that a synthetic promoter of 18 bp containing the TAAG sequence motif is sufficient to direct late transcription (28). Since these results suggest that only the adjacent flanking sequences of the TAAG motif are essential for recognition by the virus-induced RNA polymerase, we compared sequences flanking functional and nonfunctional TAAG sequences. The remarkable difference in AT richness led us to calculate the helix stability of the flanking 8 and 6 nt (36), which revealed a correlation between this thermodynamic parameter and the recognition by the virus-induced RNA polymerase. The helix stability was lowest for the flanking sequences of the TAAG motif in very late promoters whereas the corresponding sequences of nonfunctional TAAG motifs were characterized by a higher helix stability (Table 1). These findings support the view that recognition by the virus-induced RNA polymerase depends on a region that facilitates strand separation. The presence of such a region is described for all RNA polymerases (15, 16, 24, 25), except for RNA polymerase II, which uses the high energy β-γ bond of ATP to open the initiation site (42).
Conversion of the early pe38 promoter into a late promoter.
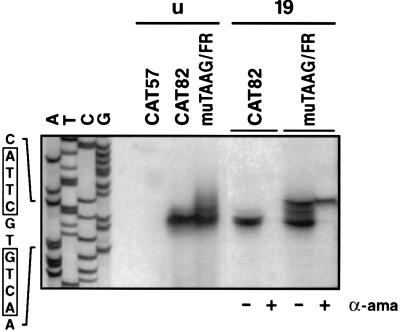
To confirm that sequences adjacent to the TAAG motif determine whether TAAG sequences are recognized by the virus-induced RNA polymerase, we replaced the flanking sequences of the nonfunctional TAAG motif in the pe38 promoter construct pPE38-CAT82 by those of the functional motif in the polyhedrin promoter. We chose pPE38-CAT82 as the shortest pe38 promoter construct for the mutation studies, since deletion of the TATA sequences in plasmid pPE38-CAT57 abolished promoter activity (see Fig. 5). The promoter construct pPE38muTAAG/FR (Fig. 4) was tested by in vitro transcription and analyzed by primer extension. In extracts prepared from cells at 19 h p.i., the hybrid pe38 promoter was recognized by the virus-induced RNA polymerase (Fig. 5), indicating that the sequence replacement converted the nonfunctional TAAG motif into a functional TAAG motif. Transcriptional initiation occurred accurately at the 5′ end of the TAAG sequence motif (Fig. 5). Although the sequence replacement affected the early transcriptional start site in the pe38 promoter (Fig. 4), construct pPE38muTAAG/FR still served as a target for the host RNA polymerase II in nuclear extracts of uninfected cells and in nuclear extracts prepared from cells at 19 h p.i. (Fig. 4 and 5). In contrast to the wild-type initiation site in construct pPE38-CAT82 (Fig. 4), transcriptional initiation at the mutated early start site in pPE38muTAAG/FR was dispersed and 5- to 10-fold less efficient (Fig. 4 and 5). The addition of α-amanitin to nuclear extracts prepared from cells at 19 h p.i. abolished recognition by the host RNA polymerase II and illustrates that the hybrid pe38 promoter was recognized by the virus-induced RNA polymerase (Fig. 5). Recognition of the hybrid promoter by both the host and the virus-induced RNA polymerases might be the result of either simultaneous recognition or competition for the same template.
FIG. 5.
In vitro transcription of the hybrid promoter pPE38muTAAG/FR in comparison to pe38 transcription. Plasmids pPE38-CAT57 and pPE38-CAT82, containing wild-type pe38 promoter sequences, and plasmid pPE38muTAAG/FR, containing a pe38/polyhedrin promoter (Fig. 4), were used as templates for in vitro transcription in nuclear extracts of uninfected (lanes u) or AcMNPV-infected S. frugiperda cells prepared at 19 h p.i. (lanes 19) by the method of Gorski et al. (11). In vitro transcription reactions were performed in the presence (lanes +) or absence (lanes −) of α-amanitin (α-ama) (2.5 μg/ml). The extended products were analyzed on a 6% polyacrylamide sequencing gel, and the results of the precise transcriptional mapping are depicted in Fig. 4. The sequencing ladder of the bottom strand of the pe38 promoter is shown on the left.
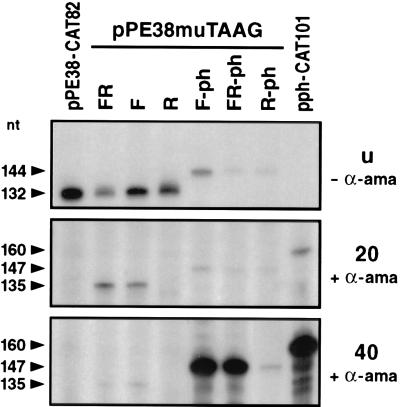
The importance of the upstream and downstream flanking sequences of the TAAG motif was further investigated by independently replacing each of the flanking sequences in the pe38 promoter by those of the polyhedrin promoter. The resulting plasmid pPE38muTAAG/F contains a 7-bp polyhedrin sequence upstream of the TAAG motif, whereas construct pPE38muTAAG/R contains a 5-bp polyhedrin sequence downstream of the TAAG motif (Fig. 4). Both constructs were transcribed in vitro by nuclear extracts of uninfected cells (Fig. 6). In extracts prepared from cells at 20 h p.i., only plasmid pPE38muTAAG/F was transcribed in the presence of α-amanitin, indicating that the flanking sequences upstream of the TAAG motif are responsible for the activity of the virus-induced RNA polymerase (Fig. 6).
FIG. 6.
In vitro transcription of pe38/polyhedrin hybrid promoters. In vitro transcription of the hybrid promoters pPE38muTAAG/FR, pPE38muTAAG/F, pPE38muTAAG/R, pPE38muTAAG/F-ph, and pPE38muTAAG/R-ph was compared to transcription of the wild-type pe38 (pPE38-CAT82) and polyhedrin (pph-CAT101) promoters in extracts of uninfected cells (u) in the absence of α-amanitin (− α-ama) or in extracts of AcMNPV-infected cells prepared at 20 and 40 h p.i. by the method of Gorski et al. (11), in the presence of α-amanitin (+ α-ama). The extended products are indicated by arrowheads and detailed in Fig. 4.
The accuracy of early transcriptional initiation was retained in plasmid pPE38muTAAG/F, which contains the authentic early transcriptional start site. Accordingly, initiation was dispersed in the constructs pPE38muTAAG/R and pPE38-muTAAG/FR (Fig. 4).
Conversion of the early pe38 promoter into a very late promoter.
The transcriptional activity of the constructs pPE38muTAAG/FR and pPE38muTAAG/F was tested in extracts prepared from cells at 40 h p.i. to analyze whether the flanking sequences of the TAAG motif have any influence on the transcriptional burst observed for very late promoters. As shown in Fig. 6, no increase in transcription was observed when the activity in nuclear extracts prepared at 20 and 40 h p.i. was compared. These results confirm previous mutagenesis studies which suggest that additional promoter elements such as the 5′ UTR are needed along with the TAAG sequence motif for very late expression (34, 35, 37). Hence, we have replaced the 5′ UTR of the pe38 promoter in constructs pPE38muTAAG/FR and pPE38muTAAG/R by the corresponding sequences of the polyhedrin promoter, generating plasmids pPE38muTAAG/FR-ph and pPE38muTAAG/R-ph (Fig. 4). Both constructs differ only in 7 bp upstream of the TAAG motif. In plasmid pPE38muTAAG/FR-ph, this 7 bp originates from the polyhedrin promoter, and in construct pPE38muTAAG/R-ph, it originates from the pe38 promoter. Therefore, the hybrid promoter in pPE38muTAAG/FR-ph consists of a functional TAAG motif and that in pPE38muTAAG/R-ph consists of a nonfunctional TAAG motif.
In extracts of uninfected cells or extracts prepared from cells at 20 h p.i., construct pPE38muTAAG/FR-ph was weakly transcribed in vitro (Fig. 6). A dramatic increase in transcription was observed in nuclear extracts prepared at 40 h p.i. indicating that the hybrid promoter in construct pPE38muTAAG/FR-ph functions as a very late promoter which can still be recognized rather weakly by the host RNA polymerase II. The lack of the polyhedrin sequences upstream of the TAAG motif in construct pPE38muTAAG/R-ph abolished the characteristics of a very late promoter. The observation that this construct still served as a target of the virus-induced RNA polymerase indicates that a nonfunctional TAAG motif can be recognized by the virus-induced RNA polymerase if the 5′ UTR of a very late promoter is present (Fig. 6). To support very late transcription, both the upstream sequences of a functional TAAG motif and the 5′ UTR are essential.
Since we observed that the upstream sequences of a functional TAAG motif were sufficient for recognition by the virus-induced RNA polymerase, we investigated whether mutation of the 5 bp downstream of the TAAG motif also had no influence on the transcriptional burst. To answer this question, we constructed a hybrid promoter which was identical to the one in plasmid pPE38muTAAG/FR-ph except for the 5 bp downstream of the TAAG motif, which was replaced by the sequence from the pe38 promoter. Accordingly, the resulting plasmid, pPE38muTAAG/F-ph, carries a mutation of 5 bp in the 5′ UTR of the polyhedrin promoter (Fig. 4). In vitro transcription assays revealed that the activity of plasmid pPE38muTAAG/F-ph was comparable to that of pPE38muTAAG/FR-ph, which indicates that the 5 bp downstream of the TAAG motif is not essential for the transcriptional burst (Fig. 6). The recognition by the host RNA polymerase II was less efficient for plasmid pPE38muTAAG/FR-ph than for pPE38muTAAG/F-ph, which carries the authentic early start site of the pe38 promoter (Fig. 4 and 5).
In comparison to the transcriptional activity of the wild-type polyhedrin promoter, both hybrid promoter constructs pPE38muTAAG/F-ph and pPE38muTAAG/FR-ph showed a three- to fivefold decrease in transcription (Fig. 6 and 7), which might be due to the absence of upstream polyhedrin sequences that slightly enhance the activity of the promoter (30, 34).
FIG. 7.
In vitro transcription of pe38/polyhedrin hybrid promoters in comparison to constructs with mutations in the polyhedrin 5′ UTR. Plasmids pPE38-muTAAG/F-ph/IRM and pPE38muTAAG/F-ph/IRC with mutations in the polyhedrin 5′ UTR, the parental construct pPE38muTAAG/F-ph, pPE38muTAAG/FR-ph, and pph-CAT101 (Fig. 4) were used as templates for in vitro transcription in nuclear extracts of AcMNPV-infected cells prepared at 44 h p.i. by the method of Gorski et al. (11), in the presence of α-amanitin (+ α-ama). The extended products are indicated on the left and detailed in Fig. 4. Positions of DNA size markers (lane M) are shown on the right.
Role of mutations in the 5′ UTR of the polyhedrin promoter in very late transcription.
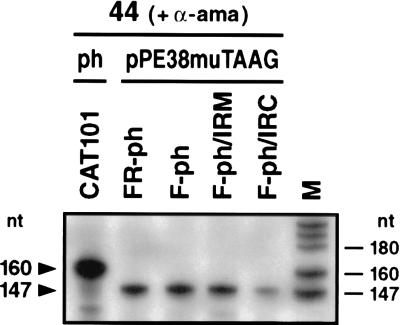
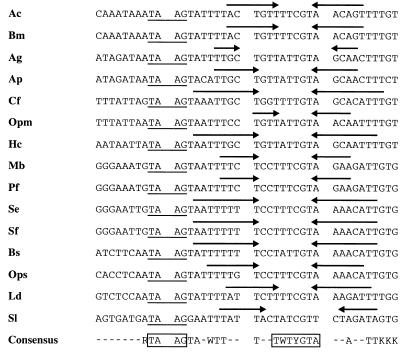
Previous mutagenesis studies of the p10 (43) and polyhedrin (30) promoters suggest that integrity of the 5′ UTR is essential for very late expression. Hence, we have not progressively mutated the 5′ UTR but instead have investigated whether 5′ UTRs of known very late promoters have common sequence elements or motifs which might play a role in the transcriptional burst. A previous comparison of polyhedrin promoters among several baculoviruses showed limited sequence homology and a structural frame that was reminiscent of RNA polymerase III promoters (46). Our updated sequence comparison revealed a perfect or imperfect inverted repeat in the 5′ UTR of all known polyhedrin promoters, which is localized downstream of the TAAG sequence motif (Fig. 8). The spacing sequence of the inverted repeats is highly conserved, with the general consensus motif WTYRT always located 12 bp downstream of the TAAG motif (Fig. 8). To investigate the significance of the inverted repeat for the transcriptional burst, plasmids pPE38muTAAG/F-ph/IRM and pPE38muTAAG/F-ph/IRC were generated. Both constructs are based on pPE38muTAAG/F-ph; one contains six mutations that abolish half of the inverted repeat (pPE38muTAAG/F-ph/IRM), and in the second, the inverted repeat is restored by complementary mutations (pPE38muTAAG/F-ph/IRC) (Fig. 4).
FIG. 8.
Alignment of baculovirus polyhedrin sequences. Sequences downstream of the TAAG motif in the polyhedrin were compared among baculoviruses. The conserved inverted repeat in the polyhedrin 5′ UTR of AcMNPV (46) (Ac), Bombyx mori NPV (46) (Bm), Anticarsia gemmatalis MNPV (46) (Ag), Antheraea pernyi MNPV (M57666) (Ap), Choristoneura fumiferana NPV (Cf) (33), Orgyia pseudotsugata MNPV (46) (Opm), Hyphantria cunea NPV (D14573) (Hc), Mamestra brassicae MNPV (46) (Mb), Panolis flammea MNPV (46) (Pf), Spodoptera exigua MNPV (46) (Se), Spodoptera frugiperda MNPV (46) (Sf), Buzura suppressaria SNPV (14) (Bs), Orgyia pseudotsugata SNPV (Ops), Lymantria dispar MNPV (46) (Ld), and Spodoptera littoralis NPV (D01017) (Sl) is indicated by arrows.
In extracts prepared from cells at 44 h p.i., pPE38muTAAG/F-ph/IRM was as strongly transcribed in vitro as were the constructs without mutations in the 5′ UTR whereas the transcriptional activity of pPE38muTAAG/F-ph/IRC was severely reduced (Fig. 7). These results indicate that the mutation of the inverted repeat comprising 12 bp immediately downstream of the TAAG sequences (Fig. 4) does not influence transcription in vitro. Only after mutation of the second half of the inverted repeat is the transcriptional burst lost, demonstrating that an inverted repeat per se is not responsible for the strong transcriptional activity of very late promoters.
DISCUSSION
In beginning to understand the mechanism of promoter recognition by the virus-induced RNA polymerase, we have determined promoter elements that are sufficient for transcriptional initiation. Transcription in vitro is advantageous for these studies because it allows the definition of the minimal sequences that are necessary to form the transcriptional complex leading to accurate initiation independently of sequences that influence proper mRNA polyadenylation, stability, and transport. By replacing sequences of the pe38 promoter by those of the very late polyhedrin promoter in a stepwise manner, we have converted the early pe38 promoter, which serves only as a target for the host RNA polymerase II, into a target promoter of the virus-induced RNA polymerase. The replacement of 7 nt upstream of the nonfunctional TAAG sequence motif in the pe38 promoter by those of the polyhedrin promoter was sufficient for recognition by the virus-induced RNA polymerase. These results support our assumption that thermodynamic stability rather than sequence specificity of nucleotides located upstream of the TAAG motif might be the deciding factor for its functionality. Reduced helix stability was found to correlate with functional TAAG motifs (Table 1), indicating that easier strand separation might be essential for transcriptional initiation by the virus-induced RNA polymerase. In addition, a nonfunctional TAAG motif could be compensated for if the polyhedrin 5′ UTR was present. The finding that recognition by the virus-induced RNA polymerase was dependent either on 7 bp upstream of the TAAG sequences or on the presence of the 5′ UTR of a very late promoter might explain previous observations which led to the prediction of an initiator element of different lengths (30, 34, 35). Based on mutagenesis studies of the very late p10 and polyhedrin promoters, an initiator of 12 bp (5′AATAAGTATTTT3′) (35) and 8 bp (5′TAAGTATT3′) was postulated (30, 34). Furthermore, the use of linker-scanning mutations of the late vp39 promoter led to the conclusion that an 18-bp promoter, including 8 bp upstream and 6 bp downstream of the TAAG motif, is sufficient for late expression (28). Taken together with the absence of conserved nucleotides upstream of the TAAG in known very late promoters (Fig. 8), our results derived from the analysis of hybrid promoters in vitro suggest that recognition is due simply to a TAAG motif in a sequence background of low helix stability. This rather simple mode of recognition implies that the virus-induced RNA polymerase needs less stringent promoter conditions than known nuclear RNA polymerases.
Transcriptional regulation that leads to hyperexpression of baculovirus very late genes is still an enigma. Previous studies of the polyhedrin promoter suggest that the transcriptional burst depends on the entire 5′ UTR of 50 bp (30). Here, we have demonstrated that deletion of 12 bp immediately downstream of the TAAG motif did not interfere with transcription in vitro. To further dissect the 5′ UTR into functional domains, we searched for common sequence elements among known baculovirus very late genes and identified an inverted repeat of different lengths whose spacing sequences are always at the same position (Fig. 8). Although the sequences of the inverted repeats showed no significant homology, an inverted repeat per se is not involved in the regulation of the polyhedrin promoter. The replacement of the inverted repeat by the complementary sequence resulted in loss of the transcriptional burst, which was retained if only the first half of the inverted repeat was mutated. Based on the sensitivity of our improved in vitro transcription assay, we have provided first evidence that strong very late transcription is the result of cooperative action between a functional TAAG motif and only part of the polyhedrin 5′ UTR.
ACKNOWLEDGMENTS
We thank Christiane Zock for providing protein extracts of BHK21 and HeLa cells and Andrew Barker for critical reading of the manuscript.
This research was supported by the EC Biotechnology program BIOTECH (BIO2-CT930457).
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of the Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Becker D, Knebel-Mörsdorf D. Sequence and temporal appearance of the early transcribed baculovirus gene HE65. J Virol. 1993;67:5867–5872. doi: 10.1128/jvi.67.10.5867-5872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beniya H, Funk C J, Rohrmann G F, Weaver R F. Purification of a virus-induced RNA polymerase from the Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells that accurately initiates late and very late transcription in vitro. Virology. 1996;216:12–19. doi: 10.1006/viro.1996.0029. [DOI] [PubMed] [Google Scholar]
- 4.Blissard G W, Kogan P H, Wei R, Rohrmann G F. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology. 1992;190:783–793. doi: 10.1016/0042-6822(92)90916-d. [DOI] [PubMed] [Google Scholar]
- 5.Blissard G W, Rohrmann G F. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 6.Carson D D, Summers M D, Guarino L A. Molecular analysis of a baculovirus regulatory gene. Virology. 1991;182:279–286. doi: 10.1016/0042-6822(91)90671-w. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs L Y, Woods M S, Weaver R F. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J Virol. 1983;48:641–646. doi: 10.1128/jvi.48.3.641-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardiner G R, Stockdale H. Two tissue culture media for production of lepidopteran cells and nuclear polyhedrosis viruses. J Invertebr Pathol. 1975;25:363–370. [Google Scholar]
- 9.Garrity D B, Chang M-J, Blissard G W. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology. 1997;231:167–181. doi: 10.1006/viro.1997.8540. [DOI] [PubMed] [Google Scholar]
- 10.Glocker B, Hoopes R R, Rohrmann G F. In vitro transcription from baculovirus late gene promoter: accurate mRNA initiation by nuclear extracts prepared from infected Spodoptera frugiperda cells. J Virol. 1993;67:3771–3776. doi: 10.1128/jvi.67.7.3771-3776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 12.Grula M A, Buller P L, Weaver R F. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J Virol. 1981;38:916–921. doi: 10.1128/jvi.38.3.916-921.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino L A, Smith M. Regulation of delayed-early gene transcription by dual TATA boxes. J Virol. 1992;66:3733–3739. doi: 10.1128/jvi.66.6.3733-3739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z H, Liu M F, Jin F, Wang Z X, Liu X Y, Li M J, Liang B F, Xie T E. Nucleotide sequence of the Buzura suppressaria single nucleocapsid nuclear polyhedrosis virus polyhedrin gene. J Gen Virol. 1993;74:1617–1620. doi: 10.1099/0022-1317-74-8-1617. [DOI] [PubMed] [Google Scholar]
- 15.Jaehning J A. Mitochondrial transcription: is a pattern emerging? Mol Microbiol. 1993;8:1–4. doi: 10.1111/j.1365-2958.1993.tb01197.x. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis G A, Blanco J A, Johnson T E, Geiduschek E P. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol. 1992;226:47–58. doi: 10.1016/0022-2836(92)90123-2. [DOI] [PubMed] [Google Scholar]
- 17.Knebel D, Lübbert H, Doerfler W. The promoter of the late p10 gene in the insect nuclear polyhedrosis virus Autographa californica: activation by viral gene products and sensitivity to DNA methylation. EMBO J. 1985;4:1301–1306. doi: 10.1002/j.1460-2075.1985.tb03776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knebel-Mörsdorf D, Flipsen J T M, Roncarati R, Jahnel F, Kleefsman A W F, Vlak J M. Baculovirus infection of Spodoptera exigua larvae: lacZ expression driven by promoters of early genes pe38 and me53 in larval tissue. J Gen Virol. 1996;77:815–824. doi: 10.1099/0022-1317-77-5-815. [DOI] [PubMed] [Google Scholar]
- 19.Knebel-Mörsdorf D, Kremer A, Jahnel F. Baculovirus gene ME53, which contains a putative zinc finger motif, is one of the major early-transcribed genes. J Virol. 1993;67:753–758. doi: 10.1128/jvi.67.2.753-758.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krappa R, Behn-Krappa A, Jahnel F, Doerfler W, Knebel-Mörsdorf D. Differential factor binding at the promoter of early baculovirus gene PE38 during viral infection: GATA motif is recognized by an insect protein. J Virol. 1992;66:3494–3503. doi: 10.1128/jvi.66.6.3494-3503.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krappa R, Knebel-Mörsdorf D. Identification of the very early transcribed baculovirus gene PE-38. J Virol. 1991;65:805–812. doi: 10.1128/jvi.65.2.805-812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lübbert H, Kruczek I, Tjia S, Doerfler W. The cloned EcoRI fragments of Autographa californica nuclear polyhedrosis virus. Gene. 1981;16:343–345. doi: 10.1016/0378-1119(81)90092-5. [DOI] [PubMed] [Google Scholar]
- 23.Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margalit H, Shapiro B A, Nussinov R, Owens J, Jernigan R L. Helix stability in prokaryotic promoter regions. Biochemistry. 1988;27:5179–5188. doi: 10.1021/bi00414a035. [DOI] [PubMed] [Google Scholar]
- 25.Marilley M, Pasero P. Common DNA structural features exhibited by eukaryotic ribosomal gene promoters. Nucleic Acids Res. 1996;24:2204–2211. doi: 10.1093/nar/24.12.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuura Y, Possee R D, Overton H A, Bishop D H L. Baculovirus expression vectors: the requirements for high level expression of proteins, including glycoproteins. J Gen Virol. 1987;68:1233–1250. doi: 10.1099/0022-1317-68-5-1233. [DOI] [PubMed] [Google Scholar]
- 27.McLachlin J L, Miller L K. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris T D, Miller L K. Mutational analysis of a baculovirus major late promoter. Gene. 1994;140:147–153. doi: 10.1016/0378-1119(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 29.Nissen M S, Friesen P D. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J Virol. 1989;63:493–503. doi: 10.1128/jvi.63.2.493-503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ooi B G, Rankin C, Miller L K. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J Mol Biol. 1989;210:721–736. doi: 10.1016/0022-2836(89)90105-8. [DOI] [PubMed] [Google Scholar]
- 31.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors—a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1994. [Google Scholar]
- 32.Possee R D, Howard S C. Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1987;15:10233–10248. doi: 10.1093/nar/15.24.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qui W, Liu J J, Carstens E B. Studies of Choristoneura fumiferana nuclear polyhedrosis virus gene expression in insect cells. Virology. 1996;217:564–572. doi: 10.1006/viro.1996.0151. [DOI] [PubMed] [Google Scholar]
- 34.Rankin C, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene. 1988;70:39–49. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 35.Rohrmann G F. Polyhedrin structure. J Gen Virol. 1986;67:1499–1513. doi: 10.1099/0022-1317-67-8-1499. [DOI] [PubMed] [Google Scholar]
- 36.SantaLucia J, Allawi H T, Seneviratne P A. Improved nearest neighbour parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 37.Smith G E, Vlak J M, Summers M D. Physical analysis of Autographa californica nuclear polyhedrosis virus transcripts for polyhedrin and 10,000-molecular-weight protein. J Virol. 1983;45:215–225. doi: 10.1128/jvi.45.1.215-225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tjia S T, Carstens E B, Doerfler W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus. Virology. 1979;180:492–508. doi: 10.1016/0042-6822(79)90017-5. [DOI] [PubMed] [Google Scholar]
- 39.Todd J W, Passarelli A L, Miller L K. Eighteen baculovirus genes, including lef-11, p35, 39K, and p47, support late-gene expression. J Virol. 1995;69:968–974. doi: 10.1128/jvi.69.2.968-974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd J W, Passarelli A L, Lu A, Miller L K. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J Virol. 1996;70:2307–2317. doi: 10.1128/jvi.70.4.2307-2317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda. In Vitro (Rockville) 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 42.Wang W, Carey M, Gralla J D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 43.Weyer U, Possee R D. Functional analysis of the p10 gene 5′ leader sequence of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 1988;16:3635–3653. doi: 10.1093/nar/16.9.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weyer U, Possee R D. Analysis of the promoter of the Autographa californica nuclear polyhedrosis virus p10 gene. J Gen Virol. 1989;70:203–208. doi: 10.1099/0022-1317-70-1-203. [DOI] [PubMed] [Google Scholar]
- 45.Xu B, Yoo S, Guarino L A. Differential transcription of baculovirus late and very late promoters: fractionation of nuclear extracts by phosphocellulose chromatography. J Virol. 1995;69:2912–2917. doi: 10.1128/jvi.69.5.2912-2917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanotto P M, Sampaio M J A, Johnson D W, Rocha T L, Maruniak J E. The Anticarsia gemmatalis nuclear polyhedrosis virus polyhedrin gene region: sequence analysis, gene product and structural comparisons. J Gen Virol. 1992;73:1049–1056. doi: 10.1099/0022-1317-73-5-1049. [DOI] [PubMed] [Google Scholar]