Abstract
Antimicrobial resistance (AMR) is responsible for the spread and persistence of bacterial infections. Surveillance of AMR in healthy individuals is usually not considered, though these individuals serve as reservoirs for continuous disease transmission. Therefore, it is essential to conduct epidemiological surveillance of AMR in healthy individuals to fully understand the dynamics of AMR transmission in Nigeria. Thirteen multidrug-resistant Citrobacter spp., Enterobacter spp., Klebsiella pneumoniae, and Escherichia coli isolated from stool samples of healthy children were subjected to whole genome sequencing (WGS) using Illumina and Oxford nanopore sequencing platforms. A bioinformatics analysis revealed antimicrobial resistance genes such as the pmrB_Y358N gene responsible for colistin resistance detected in E. coli ST219, virulence genes such as senB, and ybtP&Q, and plasmids in the isolates sequenced. All isolates harbored more than three plasmid replicons of either the Col and/or Inc type. Plasmid reconstruction revealed an integrated tetA gene, a toxin production caa gene in two E. coli isolates, and a cusC gene in K. quasivariicola ST3879, which induces neonatal meningitis. The global spread of AMR pathogenic enteric bacteria is of concern, and surveillance should be extended to healthy individuals, especially children. WGS for epidemiological surveillance will improve the detection of AMR pathogens for management and control.
Keywords: whole genome sequencing, multidrug resistance, AMR genes, plasmid, virulence, enterobacteriaceae, healthy children
1. Introduction
Enterobacteriaceae are inhabitants of the gastrointestinal tract, sometimes called enteric bacteria. Most are commensals, such as gut flora or microbiota, and some are known to be pathogenic, mainly when found in some specific areas of the human body [1]. Enteric pathogens significantly cause morbidity and mortality among young children, especially in low and middle-income countries (LMICs), as well as in adults. However, many enteric bacterial infections are either asymptomatic or result in only mild-to-moderate disease [2]. Some common enteric pathogens found in children are Citrobacter freundii, Klebsiella spp., Escherichia coli, Enterobacter spp., Salmonella typhi, and Shigella spp. [3,4].
The exact burden of diseases caused by enteric bacteria in Sub-Saharan Africa is vague due to poor healthcare services and lack of proper data documentation, to mention a few reasons. Recent reports have shown an increase in the prevalence of diseases caused by enteric bacteria, such as sepsis [5], typhoid [6], cholera [7], shigellosis [8], and diarrheal infection [9]. This is of great concern, especially regarding proposed disease elimination and meeting the third sustainable development goal (SDG3), good health and well-being. It is, therefore, of great importance to embark on active surveillance of these pathogens for management and control if this goal is to be met by 2030.
Antimicrobial resistance (AMR) has been established to be a significant driver for the persistence and spread of enteric diseases, particularly in cases where the causative agents are multidrug-resistant (MDR) or extensively drug-resistant (XDR) pathogens [10,11] as MDR pathogens are known to be non-susceptible to one or more drugs in three antibiotics classes, while XDR pathogens are non-susceptible to at least one drug in all but two or fewer antibiotics classes. The overuse and misuse of antibiotics have been identified as significant causes of AMR in enteric bacteria, as these create selective pressure and promote the emergence of resistance, leading to an alarming increase in the prevalence of MDR enteric bacteria among healthy individuals [12]. In addition, the ease of transfer of plasmids and transposons harboring genes associated with AMR amongst enteric bacteria through horizontal gene transfer enhances the persistence and spread of AMR [13,14].
Currently, most management and control efforts on infectious diseases are focused on symptomatic cases. It is essential to equally consider and investigate asymptomatic individuals, especially in LMIC, where enteric diseases are endemic and transmission is high due to poor hygiene. Furthermore, asymptomatic carriers of pathogenic enteric bacteria are rarely focused on in research studies, even though they are crucial in disease transmission and persistence, serving as reservoirs for antimicrobial-resistant genes [15,16]. A healthy human microbiome is one of the major reservoirs of antibiotic-resistant genes (ARGs) which are transmissible to other pathogenic bacteria [14,17]. Recent research reveals that ARGs contribute to the prevalence of MDR enteric bacteria in healthy humans, ensuring the continuous circulation of ARGs in the population [18]. Children constitute a significant population burdened by MDR enteric infections [19,20,21,22]. In addition, reports have shown AMR enteric bacteria to be a significant cause of mortality in neonatal sepsis in Nigeria, despite government interventions and policies [22,23,24,25]. Furthermore, the country has only a few in-depth genomic analyses of enteric bacteria that are of public health interest [26,27,28].
Genomic surveillance of AMR enteric bacteria that are of public health importance, such as Escherichia coli, Klebsiella pneumoniae, Salmonella typhi, Vibrio cholera, and Shigella spp., not only informs treatment guidelines for diseases caused by these human pathogens but is necessary for the design and implementation of AMR interventions and control measures [29]. Whole genome sequencing (WGS) provides a high-resolution method capable of providing in-depth genomic characterization and epidemiological information about a pathogen. This further sheds more light on genomic diversity, transmission dynamics, evolution, AMR dynamics, and spread, aiding in better control measures [30]. This has been implemented in studies of asymptomatic carriers to characterize the pathogens further, understand the actual dynamics of AMR, and investigate continuous pathogen development within the natural host [14,31,32].
Thus, this study utilized a whole genome sequencing (WGS) approach to characterize AMR genes and mobile genetic elements in isolates obtained from fecal samples to understand the dynamics of AMR enteric pathogens in healthy children from Osun State, Nigeria.
2. Methods
2.1. Ethical Approval
Ethical clearance for this study was obtained from the research ethics committee of the Ladoke Akintola University of Technology Teaching Hospital (LAUTECH) (LTH/EC/2019/09/431) and the Ministry of Health (OSHREC/PRS/569T/164), Osogbo, Osun State, Nigeria.
2.2. Bacteria Isolation, Identification, and Selection
A total of 147 fecal samples were collected from presumptively healthy children between 1 and 15 years of age between 2019 and 2020 in a study focused on typhoid fever’s seroprevalence and the genomic characterization of enteric pathogens in healthy children. Serological data for typhoid fever prevalence have been previously published [33]. Fecal samples were cultured on Salmonella/Shigella and MacConkey agar. Initial identification was performed with an API 20E test kit (BioMeriuxe, Marcy-l’Étoile, France) for 52 positive enteric cultures. Thirteen (13) enteric isolates were selected based on their antibiotic resistance profile for genomic characterization.
2.3. Antimicrobial Susceptibility Test of the Isolates
Antimicrobial susceptibility testing was performed using the disk diffusion technique. The antibiotics disc used were Gentamicin (30 µg), Tetracycline (30 µg), Ciprofloxacin (5 µg), Chloramphenicol (30 µg), Trimethoprim/sulfamethoxazole (25 µg), Ceftazidime (30 µg), Ceftriaxone (30 µg), Cefotaxime (30 µg), and Aztreonam (30 µg) as described previously by [34]. Results were interpreted according to the 2017 guidelines provided by the Clinical and Laboratory Standards Institute (CLSI). Results were analyzed and interpreted using the ABIS online software v12 (http://www.tgw1916.net/bacteria_logare_desktop.html) accessed on 22 May 2021 [35].
2.4. Whole Genome Sequencing
The isolates were subcultured before DNA extraction. DNA was extracted using a Qiagen DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Extracted samples were quantified using a dsDNA high-sensitivity assay kit on a Qubit fluorometer (ThermoFisher Scientific, Waltham MA, USA)). Sequencing libraries were prepared using a Nextera DNA flex preparation kit (Illumina, San Diego, CA, USA). Library preparation protocol was adopted from the CDC PulseNet Nextera DNA Flex Standard operating protocol and sequenced using a Illumina Miseq platform and NextSeq 1000/2000 at the African Center of Excellence for Genomics of Infectious Diseases (ACEGID), Redeemer’s University, Nigeria.
To improve the plasmid assembly, we performed a single run on a GridION x5 to generate long reads. Library preparation and sequencing were performed using a Rapid PCR Barcoding kit (SQK-RPB004) (Oxford Nanopore Technologies, Oxford, UK), following the manufacturer’s recommendations. We used a GridION MK1 sequencer, FLO-MIN10 6D R9 flow cell, and MinKNOW software v22.10.7 for sequencing.
2.5. Genomic Data Analysis
Raw FASTQ files were processed with the Connecticut Public Health Laboratory (CT-PHL) pipeline, also known as C-BIRD v0.9 “https://github.com/Kincekara/C-BIRD (accessed on 9 May 2023)”. The assembled contigs were further checked for contamination using CheckM [36]. Isolates with a bracken taxon ratio <0.7, genome estimated ratio >1.1, estimated sequencing depth <40×, and genome completeness <90% were excluded from subsequent analyses, as those isolates were either deemed contaminated with other bacterial species, had an unusual larger genome size, had low genome coverage, or had a low sequencing depth. Further analyses, including genome annotation, plasmid detection, antimicrobial resistance, virulence prediction, and MLST typing, were performed with the Public Health Bacterial Genomics (PHBG) v1.3.0 “https://github.com/theiagen/public_health_bacterial_genomics (accessed on 9 May 2023)”. Genome assemblies of isolates were refined with a unicycler v0.4.9 [37] hybrid assembler using Illumina and Oxford nanopore sequence reads. The plasmids were reconstructed and typed with MOB-suite v3.1.0 [38] and annotated with pLannotate v1.2.0 [39] and Prokka v1.14.6 [40].
3. Result
3.1. Isolate of Bacteria Strains
The selected 13 multidrug-resistant isolates were Enterobacter hormaechei (n = 1), Citrobacter sp. FDAARGOS_156 (n = 1), Enterobacter cloacae (n = 1), Klebsiella quasivariicola (n = 1), Klebsiella pneumoniae (n = 3), and Escherichia coli (n = 6). All were isolated from fecal samples obtained from healthy children.
3.2. Antibiotic Susceptibility Test and AMR Prediction
The highest resistance (85%) was observed in ciprofloxacin and cefotaxime, and the least resistance (46%) was seen in tetracycline. All 13 isolates subjected to antibiotic susceptibility tests were multidrug-resistant, defined as resistance to two or more classes of antibiotics (Table 1).
Table 1.
Multiple antibiotic resistance (MAR) pattern of enteric isolates from healthy children in Osun State.
| Sample ID | Species | Antibiotics Profile (Resistant) |
|---|---|---|
| OL31NF | Enterobacter hormaechei | ATM; CTX; CAZ; SXT; CIP. |
| OL13NFPC1 | Escherichia coli | ATM; CRO; CTX; SXT; C. |
| OL19NF | Citrobacter sp. FDAARGOS_156 | ATM; CRO; CTX; CAZ; SXT; C. |
| OL44NF | Escherichia coli | TE; SXT; CIP; CN. |
| OE28NF | Escherichia coli | ATM; CRO; CTX; TE; SXT; CIP; CN; C |
| OE36NF | Escherichia coli | CTX; TE; SXT; CIP; CN. |
| OE41NF | Escherichia coli | CTX; CIP; C. |
| OE43NF | Enterobacter cloacae | ATM; CRO; CTX; CAZ; TE; SXT; CIP; CN. |
| OE54NF | Klebsiella pneumoniae | ATM; CRO; CTX; CAZ; CIP; CN. |
| OE71NF | Klebsiella quasivariicola | CAZ; CIP; C; CN. |
| OE73NF | Klebsiella pneumoniae | ATM; CRO; CTX; CAZ; TET; SXT; CIP; C; CN. |
| OE75NF | Klebsiella pneumoniae | CTX; SXT; CIP; CN |
| J21 | Escherichia coli | ATM; CRO; CTX; CAZ; TE; SXT; CIP; CN; C. |
ATM: Aztreonam, CRO: Ceftriaxone, CTX: Cefotaxime, CAZ: Ceftazidime, TE: Tetracycline, SXT: Trimethoprim/sulfamethoxazole, CIP: Ciprofloxacin, CN: Gentamicin, C: Chloramphenicol.
3.3. Genome Sequence Analysis of the Isolates
Genome sequencing data based on the taxon ratio analysis showed all isolates harbored a wide range of AMR genes, plasmid replicons, and virulent genes. All of the isolates harbored beta-lactam-resistant genes, with 23% of the isolates seen to harbor tetracycline-resistant genes tetA, and macrolides pmrB_R256G and pmrB_Y358N resistance genes, which are responsible for colistin resistance via the efflux pump mechanism. Also, 92% of the isolates had different efflux pump genes responsible for at least two classes of antibiotic resistance (Table 2). Virulence genes were seen in 62% of the isolates, while plasmids were detected in 92% of the isolates detailed in Table 2.
Table 2.
Summary of genomic characterization of whole-genome-sequenced Enteric isolates.
| S/N | Sample ID | Species | Sequence Type | AMR Genes | Virulence Genes | Plasmid Replicons |
|---|---|---|---|---|---|---|
| 1 | OL31NF * | Enterobacter hormaechei | No ST predicted | blaACT-16, qnrS1, fosA, oqxB, oqxA | No VIRULENCE genes detected by NCBI-AMRFinderPlus | Col(pHAD28), IncFIB(K), IncFII(pECLA) |
| 2 | OL13NFPC1 | Escherichia coli | ST409 | acrF, mdtM, glpT_E448K, blaEC | espX1 | No plasmids detected in database |
| 3 | OL19NF | Citrobacter sp. FDAARGOS_156 | ST187 | blaCMY | No VIRULENCE genes detected by NCBI-AMRFinderPlus | Col(pHAD28), ColRNAI |
| 4 | OL44NF | Escherichia coli | No ST predicted | blaEC, aadA1, dfrA7, sul1, tet(A), aph(3′)-Ia, mdtM, pmrB_Y358N, acrF, qnrS, blaTEM-1, aph(6)-Id, aph(3″)-Ib, sul2, qnrB19, mdtM | nfaE, afaC, lpfA, senB, iss, espX1, papA, sat, iutA, iucD, iucC, iucB, iucA, iha, f17a, f17g, ybtQ, ybtP, lpfA | Col(MG828), Col(MP18), Col(pHAD28), Col(pHAD28), Col(pHAD28), Col(pHAD28), Col156, IncFIB(AP001918), IncFII(pRSB107), IncI(Gamma), IncQ1 |
| 5 | OE28NF * | Escherichia coli | No ST predicted | mdtM, blaEC, blaEC, blaTEM, acrF, tet(A) | espX1, iutA, iucD, iucC, iucB, iucA, fdeC, lpfA, iss, ireA, sigA, iha, lpfA, ybtP, ybtQ | Col(pHAD28), Col(pHAD28), IncB/O/K/Z, IncFIA(HI1), IncR |
| 6 | OE36NF * | Escherichia coli | ST 219 | blaTEM-1, pmrB_Y358N, mdtM, acrF, emrD, gyrA_S83L, glpT_E448K, blaEC, sul2, aph(3″)-Ib, aph(6)-Id, sul1, dfrA7, tet(A) | eilA, lpfA1, iucA, iucB, iucC, iucD, iutA, iha, sigA, espX1, fdeC | IncB/O/K/Z, IncI2, IncQ1 |
| 7 | OE41NF | Escherichia coli | No ST predicted | qnrB, oqxA, emrD, fosA7, glpT_E448K, mdtM, blaCMY, blaEC | iss, espX1, fdeC | Col156, IncFIA(HI1), IncFIB(AP001918), IncFIB(K), IncFIB(pHCM2), IncY |
| 8 | OE43NF | Enterobacter cloacae | ST1236 | emrD, satA, mdtM, blaCMH, vmlR, aadK, mphK, blaEC, blaCTX-M | lpfA1, espX1, eilA | IncFII(K), IncR |
| 9 | OE54NF | Klebsiella pneumoniae | No ST predicted | fosA, oqxB, oqxA, emrD, blaSHV | No VIRULENCE genes detected by NCBI-AMRFinderPlus | Col440II, IncFIA(HI1), IncFIB(K) |
| 10 | OE71NF * | Klebsiella quasivariicola | ST3897 | emrD, fosA, kdeA, oqxB, oqxA, blaOKP-D | No VIRULENCE genes detected by NCBI-AMRFinderPlus | Col440I, Col440II, FII(pBK30683), IncFIB(K), IncFII(K) |
| 11 | OE73NF | Klebsiella pneumoniae | ST185 | blaTEM, fosA, blaSHV-11, dfrA15, aadA1, sul1, oqxB, oqxA, emrD | alo, plcR | Col(pHAD28), Col440I, IncFIB(K), IncFII(pKP91), IncR |
| 12 | OE75NF | Klebsiella pneumoniae | No ST predicted | fosA, emrD, pmrB_R256G, blaOKP-B, blaSHV-11, oqxB, oqxA, fosA | No VIRULENCE genes detected by NCBI-AMRFinderPlus | IncFIA(HI1), IncFIB(K), IncFIB(pNDM-Mar), IncFII(K), IncHI1B(pNDM-MAR) |
| 13 | J21 * | Escherichia coli | ST450 | blaTEM-1, parC_S80I, parE_S458A, catA1, gyrA_D87N, gyrA_S83L, dfrA17, acrF, mdtM, blaEC, aadA1, blaOXA-1, qepA4, tet(A), aph(6)-Id, aph(3″)-Ib, sul2 | sigA, sat, astA, ybtQ, ybtP, espX1, fdeC, iha, papH, papC, papF, papG-II, iucA, iucB, iucC, iucD, iutA, senB | Col(MG828), Col156, IncFIA, IncFIB(AP001918), IncFII(pRSB107), IncFII(pRSB107) |
* Samples which passed the QC step.
We then considered sequence data based on the genome estimated ratio > 1.1, estimated sequencing depth <40×, and genome completeness (<90%). Only five out of the thirteen sequenced isolates passed this quality control step. These isolates asterisked in Table 2 (E. coli = 3, K. quasiveriicola = 1, and E. hormaechei = 1) were selected for further downstream sequence analysis and characterization. The sequence data have been submitted to NCBI under the BioProject accession number PRJNA838568.
Genome lengths of 4.8 Mbp to 6 Mbp, 5.6 Mbp, and 5.8 Mbp for E. coli, K. quasiveriicola, and E. hormaechei were observed. The assembled contigs of the bacteria genome antimicrobial resistance (AMR) detection software (AMRFinder+ v3.10.42 and Kleborate v2.0.4) embedded in the PHBG pipeline showed the presence of beta-lactam-resistance genes such as blaTEM-1, blaEC, blaACT, and blaOKP-D, (Table 1). In addition, all five isolates contained other AMR genes associated with either fluoroquinolone (gyrAS83L, qnrS1), aminoglycoside (aph(6)-Id, aph(3″)-Ib), tetracycline (tetA), or sulfonamide (sul1, sul2) resistance. Multilocus sequence typing (MLST) predicted the ST type for three isolates, E. coli ST 219, ST450, and K. quasiveriicola ST 3897, while the remaining two isolates, E. hormaechei, and E. coli, had no ST prediction. This study revealed the presence of fluoroquinolone-resistant genes with more than triple mutations (gyrA_S83L, gyrA_D87N, parC_S80I, and parE_S458A) in E. coli ST219 and the Colistin-resistant gene (pmrB_Y358N) in E. coli ST450 reported in XDR E. coli.
3.4. Detection and Characterization of Virulence Genes and Mobile Genetic Elements in the Five Isolates
Of the five whole genome sequences that passed the quality control step, virulence genes were detected in all three E. coli isolates. In contrast, none were detected in K. quasivariicola and E. hormaechei (Table 2). Virulence genes responsible for bacteria iron uptake were detected in all the E. coli isolates. Aerobactin virulence genes iucABCD and iutA were also present in all the E. coli isolates, but only two E. coli isolates harbored the Yersiniabactin virulence genes ybtP and ybtQ (Table 2). Other virulence genes detected include the gene-encoding enterotoxin senB, the increased serum survival iss gene sigA, the secreted autotransporter toxin -sat, the east-1 heat-stable toxin -astA, the long polar fimbria -ipfA, the LEE encoded type III secretion system effector- espX1, for adherence- fdeC, the P-fimbria operon- papHCFGII, the Salmonella HilA homolog -eilA, and the iron-regulated outer membrane virulence protein–ireA gene.
A total of 28 plasmids were also detected in these five isolates, with each harboring between three and nine plasmid replicons known to be associated with antibiotic-resistant genes (ARGs). The Inc plasmid type is the most common plasmid type across all isolates. IncF and Col plasmids were detected in 90% (4/5) of the isolates. The IncF replicons detected included IncFII(pBK30683) (1/5), InFIB(K) (2/5), IncFII(K) (1/5), IncFII(pECLA) (1/5), IncFIA(HI1) (1/5), IncFIA (1/5), IncFIB(AP001918) (1/5), and IncFII(pRSB107) (1/5). The Col plasmid detected had replicons which included Col440I (2/5), Col440II (1/5), ColpHAD28 (3/5), ColMG828 (1/5), and Col156 (1/5). Other plasmid replicons detected were IncR (1/5), IncI2 (1/5), IncQ1 (1/5), and IncB/O/K/Z (2/5).
3.5. Plasmid Reconstruction Using Oxford Nanopore Sequencing Platform
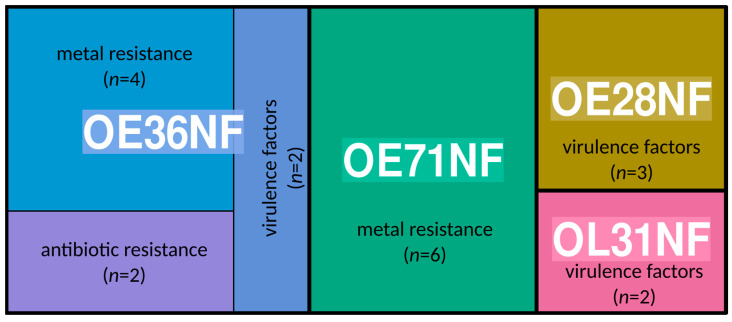
We reconstructed 13 plasmids from 3536 to 163,036 bp detected in E. hormaechei, K. quasivariicola, and two E. coli isolates (Supplementary Table S2). Plasmid reconstruction for the other E. coli was not conducted due to low sequencing library concentration. The K. quasivariicola plasmids were either conjugative, non-mobilizable, or mobilizable. The largest plasmid contained 11 contigs with IncFIB, IncFII, rep_cluster_2183, rep_cluster_2327, and rep_cluster_2358 replicon types. None of the plasmids in K. quasivariicola harbored any antibiotic-resistance genes; however, several metal-resistance genes and efflux proteins such as copA, copB, silP, cusA, cusB, and cusC were integrated into the plasmid (Figure 1). Interestingly, we detected the tetracycline resistance gene tetA and some mercury resistance genes such as merA and merC integrated into the plasmids of the two E. coli isolates that code for colicin polypeptide toxins, while the E. hormaechei isolate had plasmids carrying ylpA virulence genes (Figure 1 and Supplementary Table S2).
Figure 1.
Treemap showing the proportion of virulence factors, metal resistance, and antibiotic resistance genes curated from plasmids (n = 13) of the isolates that were successfully sequenced on the Oxford Nanopore (OE36NF-E. coli, OE71NF-K. quasipneumoniae, OE28NF-E. coli, and OL31NF-E. hormaechei).
4. Discussion
Antimicrobial resistance, especially in enteric bacteria that are of public health interest, continues to be of global concern, especially in low and middle-income countries [35]. In Nigeria, there is a paucity of information on the genomic characterization of circulating multidrug-resistant enteric bacteria, especially in healthy individuals that serve as reservoirs. Therefore, we recovered enteric pathogens that are of public health interest in this study from presumptively healthy children such as Enterobacter hormaechei, Citrobacter sp. FDAARGOS_156, Enterobacter cloacae, Klebsiella quasivariicola, Klebsiella pneumoniae, and Escherichia coli. Most importantly, all the isolates harbored antibiotic-resistant genes for at least one class of antibiotics; some harbored virulence genes and plasmid replicons carrying resistant and virulence genes. The presence of these pathogens and, importantly, the carriage of AMR genes, plasmids, and virulence genes conferring pathogenicity and resistant phenotypes in these healthy children are of great concern to public health. The occurrence of these pathogenic enteric bacteria in febrile individuals is well documented across the country [22,35,41,42]; however, there is limited information on healthy individuals in Sub-Saharan Africa as far as the authors know, which makes this study one of the very few studies with a genomic surveillance report on healthy children in Nigeria.
Similar resistant genes (gyrA_S83L, gyrA_D87N, parC_S80I, and parE_S458A) identified from this study in presumptively healthy children have been reported in disease cases of children admitted to hospitals in Bangladesh and Benin [43,44]. The presence of these enteric pathogens isolated from presumptive healthy children is of great concern as these children serve as potential reservoirs for transmission of these pathogens to other susceptible individuals. In addition to transmission, the presence of resistance genes indicates the possibility of treatment failure, further promoting the spread of AMR in society and its longer existence in the population. It is possible that these pathogens developed resistance due to consistent drug pressure, as most drugs are readily available to citizens in the country, encouraging indiscriminate use of these drugs [18].
Our study provides a glimpse into the genetic variation of enteric pathogens in healthy children in Osun State in the Southwestern part of Nigeria. Virulence genes identified amongst the E. coli isolates are known to significantly influence the degree of pathogenicity of bacterial infection in confirmed disease patients in some reported cases [45,46]. It is unclear how these genes in the bacteria are not causing any symptoms in these children. However, reports have shown that colonization with AMR enteric pathogens in healthy individuals could lead to pulmonary infection, urinary infection, and bacteremia [47,48]. K. quasivariicola and E. hormaechei harbored no virulence genes. The presence of more than three plasmid replicons associated with AMR is of significant concern as reports of colonizing enteric bacteria resulting in infections are facilitated by MDR and plasmid carriage [49].
The pathogenic enteric bacteria in this study are known as commensals of the gut microbiota and have been reported in other studies [22,50]. Once seeded with AMR, commensal organisms may significantly contribute to the dissemination of resistance due to the connectivity in microbial communities [51]. The enteric bacteria (E. coli, K. quasiveriicola, and E. hormaechei) detected in this study have been reported in many cases to cause diarrhea and sepsis in children and also serve as colonizers in healthy children [4,52,53]. In addition, the pathogens were observed to be resistant to almost all of the classes of antibiotics. The presence of AMR genes and plasmids harbored by these bacteria in healthy children further confirms the in vitro resistance observed, and it is of concern as children in this community will enable the persistent transmission of diseases caused by these enteric bacteria, leading to increased community-acquired infections [54]. This could increase morbidity, mortality, and healthcare costs, especially in immunocompromised individuals. This, therefore, necessitates active surveillance of these AMR bacteria in both asymptomatic and clinical cases to give a total overview of the transmission dynamics and evolution.
Although not surprising, the detection of AMR genes in this population of healthy children is alarming, as antibiotic abuse has been established in this part of the world [26,55]. This is seen in the AST result and genomic data, as most isolates were resistant to folate-pathway drugs, quinolones, and beta-lactam drugs, typically first and second-line antibiotics. The presence of efflux pump genes responsible for MDR resistance increases the expression of resistance in these isolates. Resistance to these drugs, some of the most common antibiotics used in treating diarrhea diseases in children, could pose a challenge in future infections with these resistant strains. Also, reports have shown that most cases of infection with MDR enteric bacteria are not just from a person-to-person transmission or from contaminated water/food, but could also be transmitted from the individual colonized by these MDR bacteria through horizontal gene transfer [49]. This is worrisome as almost all isolates sequenced in this study, as seen in Table 1, harbored AMR genes for beta-lactams, aminoglycosides, quinolones, colistin, and fosfomycin drugs.
A significant driver of the spread of antibiotic resistance in bacteria is the presence of mobile genetic elements [56]. Plasmids spread ARG to other bacteria through horizontal gene transfer, increasing antibiotic resistance prevalence. They are known to contain genes responsible for antibiotic resistance, colonization, and virulence, which provides an advantage for bacteria survival [56]. As seen in this study, the plasmid recovered from the E. coli ST219 strains harbored a tetA gene responsible for tetracycline resistance and a caa gene that codes for colicin polypeptide toxins known to destroy cells of other organisms by depolarizing the cytoplasmic membrane of the cells [57,58]. Also, the plasmid in the K. quasivariicola ST3897 harbored the cusC gene associated with copper and silver resistance, which, when found in pathogenic K. pneumoniae, facilitates the invasion of the brain microvascular endothelial cells, thereby causing neonatal meningitis [59,60].
Apart from the presence of virulence genes and mobile genetic elements as a driver of MDR, the indiscriminate use of antibiotics in this part of the world is also a significant contributing factor to the high prevalence of MDR bacteria [27]. Although the healthy children in this study were not on any antibiotics drugs at the time of sample collection, it has been reported that children are exposed to antibiotics early on, mainly without a physician’s prescription [61,62,63]. To mitigate the continuous spread of MDR enteric bacteria and the possible emergence of XDR enteric bacteria, there is a need for proper surveillance in healthy individuals to achieve proper monitoring and control of AMR in the country. Although some control measures such as infection prevention control and antimicrobial stewardship have been implemented to monitor AMR in the population [55,64,65], the non-inclusion of healthy individuals in these control measures will truncate all efforts. Another critical factor in proper surveillance is the use of sensitive techniques. In addition to confirming the AST analysis, the whole genome sequencing employed in this study revealed genetic elements that drive pathogenicity and the spread of MDR enteric bacteria from healthy children. This might not have been possible by culture or a PCR alone. This proves the importance of the WGS technique in pathogen enteric bacteria surveillance and its implementation in healthy individuals to achieve disease elimination and eradication.
Limitation
The limitation of this study is the small number of the isolates (13) considered for the whole genome sequence from the overall sample size or pure isolates obtained from the fecal samples. The molecular characterization detected provided an insight into the existence of AMR genes and other mobile elements that may confer pathogenicity in these organisms and can further be transmitted to other organisms. Therefore, this study serves as an indicator for the general and adequate national surveillance of enteric bacteria in healthy individuals, especially children, for the total control of AMR. We therefore recommend further studies on the AMR of healthy individuals, especially children who are the major population at risk of AMR infection, to ensure proper monitoring and management policies.
5. Conclusions
Our findings in this study showed the presence of MDR enteric bacteria harboring resistant genes, virulence genes, and plasmids in healthy children. This may contribute to the continuous, global, and widespread increase in AMR observed. Therefore, close inspection and surveillance of the healthy population in addition to clinical cases is recommended to control the spread and ultimately achieve eradication of disease. Applying WGS in epidemiological surveillance will improve the detection of MDR pathogens by overcoming the limitation of analyzing only a small part of the genome and providing more rapid management, thus controlling the emergence of new antibiotic-resistant strains and their evolution.
Acknowledgments
We thank the Ladoke Akintola Teaching Hospital for their collaborative support in conducting the study. We are also grateful for the African Center of Excellence for Genomics of Infectious Diseases (ACEGID) facility, Redeemer’s University, Nigeria, that enabled us to carry out this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12030505/s1, Table S1: Whole Genome Sequencing Parameters of Sequenced Isolates; Table S2: Detailed Plasmid Reconstruction Data of Four Enteric Pathogens.
Author Contributions
Study concept and design: J.N.U. and O.A.F.; sequencing and bioinformatics: J.N.U. and I.B.O.; analysis and interpretation of data: J.N.U. and I.B.O. authors; funding acquisition: C.T.H.; drafting of the manuscript: J.N.U. and I.B.O. critical revision of the manuscript for important intellectual content: J.N.U., I.B.O., C.T.H. and O.A.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was carried out according to the principles of the Declaration of Helsinki. Ethical clearance for this study was obtained from the research ethics committee of the Ladoke Akintola University of Technology Teaching Hospital (LAUTECH) (LTH/EC/2019/09/431) and Ministry of Health (OSHREC/PRS/569T/164) Oshogbo, Osun State, Nigeria.
Informed Consent Statement
Informed consent was obtained from the individuals who participated in this study. Also, written informed consent was obtained from the parents.
Data Availability Statement
Sequence data of isolates are deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA838568.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This work was funded by grants from the NIH-H3Africa (https://h3africa.org) (U01HG007480 and U54HG007480 to C.T.H); and the World Bank ACE impact grant (worldbank.org) (ACE – 019 project to C.T.H).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Rock C., Donnenberg M.S. Reference Module in Biomedical Sciences. Elsevier; Amsterdam, The Netherlands: 2014. Human Pathogenic Enterobacteriaceae. [Google Scholar]
- 2.Lee G.O., Eisenberg J.N.S., Uruchima J., Vasco G., Smith S.M., Van Engen A., Victor C., Reynolds E., MacKay R., Jesser K.J., et al. Gut Microbiome, Enteric Infections and Child Growth across a Rural–urban Gradient: Protocol for the ECoMiD Prospective Cohort Study. BMJ Open. 2021;11:e046241. doi: 10.1136/bmjopen-2020-046241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher S.M., Stark D., Ellis J. Prevalence of Gastrointestinal Pathogens in Sub-Saharan Africa: Systematic Review and Meta-Analysis. J. Public Health Afr. 2011;2:e30. doi: 10.4081/jphia.2011.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oppong T.B., Yang H., Amponsem-Boateng C., Kyere E.K.D., Abdulai T., Duan G., Opolot G. Enteric Pathogens Associated with Gastroenteritis among Children under 5 Years in Sub-Saharan Africa: A Systematic Review and Meta-Analysis. Epidemiol. Infect. 2020;148:e64. doi: 10.1017/S0950268820000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milton R., Gillespie D., Dyer C., Taiyari K., Carvalho M.J., Thomson K., Sands K., Portal E.A.R., Hood K., Ferreira A., et al. Neonatal Sepsis and Mortality in Low-Income and Middle-Income Countries from a Facility-Based Birth Cohort: An International Multisite Prospective Observational Study. Lancet Glob. Health. 2022;10:e661–e672. doi: 10.1016/S2214-109X(22)00043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim C.L., Cruz Espinoza L.M., Vannice K.S., Tadesse B.T., Owusu-Dabo E., Rakotozandrindrainy R., Jani I.V., Teferi M., Bassiahi Soura A., Lunguya O., et al. The Burden of Typhoid Fever in Sub-Saharan Africa: A Perspective. Res. Rep. Trop. Med. 2022;13:1–9. doi: 10.2147/RRTM.S282461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elimian K.O., Musah A., Mezue S., Oyebanji O., Yennan S., Jinadu A., Williams N., Ogunleye A., Fall I.S., Yao M., et al. Descriptive Epidemiology of Cholera Outbreak in Nigeria, January–November, 2018: Implications for the Global Roadmap Strategy. BMC Public Health. 2019;19:1264. doi: 10.1186/s12889-019-7559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogawski McQuade E.T., Shaheen F., Kabir F., Rizvi A., Platts-Mills J.A., Aziz F., Kalam A., Qureshi S., Elwood S., Liu J., et al. Epidemiology of Shigella Infections and Diarrhea in the First Two Years of Life Using Culture-Independent Diagnostics in 8 Low-Resource Settings. PLoS Negl. Trop. Dis. 2020;14:e0008536. doi: 10.1371/journal.pntd.0008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mero S., Timonen S., Lääveri T., Løfberg S., Kirveskari J., Ursing J., Rombo L., Kofoed P.-E., Kantele A. Prevalence of Diarrhoeal Pathogens among Children under Five Years of Age with and without Diarrhoea in Guinea-Bissau. PLoS Negl. Trop. Dis. 2021;15:e0009709. doi: 10.1371/journal.pntd.0009709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamel N.A., El-Tayeb W.N., El-Ansary M.R., Mansour M.T., Aboshanab K.M. XDR-Klebsiella Pneumoniae Isolates Harboring blaOXA-48: In Vitro and in Vivo Evaluation Using a Murine Thigh-Infection Model. Exp. Biol. Med. 2019;244:1658–1664. doi: 10.1177/1535370219886826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zakir M., Khan M., Umar M.I., Murtaza G., Ashraf M., Shamim S. Emerging Trends of Multidrug-Resistant (MDR) and Extensively Drug-Resistant (XDR) Salmonella Typhi in a Tertiary Care Hospital of Lahore, Pakistan. Microorganisms. 2021;9:2484. doi: 10.3390/microorganisms9122484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Duin D., Paterson D.L. Multidrug-Resistant Bacteria in the Community: An Update. Infect. Dis. Clin. N. Am. 2020;34:709–722. doi: 10.1016/j.idc.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D., Jeannot K., Xiao Y., Knapp C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019;10:1933. doi: 10.3389/fmicb.2019.01933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace M.J., Fishbein S.R.S., Dantas G. Antimicrobial Resistance in Enteric Bacteria: Current State and next-Generation Solutions. Gut Microbes. 2020;12:1799654. doi: 10.1080/19490976.2020.1799654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei S., Liljeros F., Shaman J. Identifying Asymptomatic Spreaders of Antimicrobial-Resistant Pathogens in Hospital Settings. Proc. Natl. Acad. Sci. USA. 2021;118:e2111190118. doi: 10.1073/pnas.2111190118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chisholm R.H., Campbell P.T., Wu Y., Tong S.Y.C., McVernon J., Geard N. Implications of Asymptomatic Carriers for Infectious Disease Transmission and Control. R. Soc. Open Sci. 2018;5:172341. doi: 10.1098/rsos.172341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar M., Sarma D.K., Shubham S., Kumawat M., Verma V., Nina P.B., Jp D., Kumar S., Singh B., Tiwari R.R. Futuristic Non-Antibiotic Therapies to Combat Antibiotic Resistance: A Review. Front. Microbiol. 2021;12:609459. doi: 10.3389/fmicb.2021.609459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aworh M.K., Kwaga J., Okolocha E., Mba N., Thakur S. Prevalence and Risk Factors for Multi-Drug Resistant Escherichia Coli among Poultry Workers in the Federal Capital Territory, Abuja, Nigeria. PLoS ONE. 2019;14:e0225379. doi: 10.1371/journal.pone.0225379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okeke I.N., Aboderin O.A., Byarugaba D.K., Ojo K.K., Opintan J.A. Growing Problem of Multidrug-Resistant Enteric Pathogens in Africa. Emerg. Infect. Dis. 2007;13:1640–1646. doi: 10.3201/eid1311.070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mokomane M., Kasvosve I., de Melo E., Pernica J.M., Goldfarb D.M. The Global Problem of Childhood Diarrhoeal Diseases: Emerging Strategies in Prevention and Management. Ther. Adv. Infect. Dis. 2018;5:29–43. doi: 10.1177/2049936117744429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ugboko H.U., Nwinyi O.C., Oranusi S.U., Oyewale J.O. Childhood Diarrhoeal Diseases in Developing Countries. Heliyon. 2020;6:e03690. doi: 10.1016/j.heliyon.2020.e03690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson K.M., Dyer C., Liu F., Sands K., Portal E., Carvalho M.J., Barrell M., Boostrom I., Dunachie S., Farzana R., et al. Effects of Antibiotic Resistance, Drug Target Attainment, Bacterial Pathogenicity and Virulence, and Antibiotic Access and Affordability on Outcomes in Neonatal Sepsis: An International Microbiology and Drug Evaluation Prospective Substudy (BARNARDS) Lancet Infect. Dis. 2021;21:1677–1688. doi: 10.1016/S1473-3099(21)00050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olorukooba A.A., Ifusemu W.R., Ibrahim M.S., Jibril M.B., Amadu L., Lawal B.B. Prevalence and Factors Associated with Neonatal Sepsis in a Tertiary Hospital, North West Nigeria. Niger. Med J. 2020;61:60–66. doi: 10.4103/nmj.NMJ_31_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shobowale E.O., Solarin A.U., Elikwu C.J., Onyedibe K.I., Akinola I.J., Faniran A.A. Neonatal Sepsis in a Nigerian Private Tertiary Hospital: Bacterial Isolates, Risk Factors, and Antibiotic Susceptibility Patterns. Ann. Afr. Med. 2017;16:52–58. doi: 10.4103/aam.aam_34_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uwe N.O., Ezenwa B.N., Fajolu I.B., Oshun P., Chukwuma S.T., Ezeaka V.C. Antimicrobial Susceptibility and Neonatal Sepsis in a Tertiary Care Facility in Nigeria: A Changing Trend? JAC Antimicrob. Resist. 2022;4:dlac100. doi: 10.1093/jacamr/dlac100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achi C.R., Ayobami O., Mark G., Egwuenu A., Ogbolu D., Kabir J. Operationalising One Health in Nigeria: Reflections from a High-Level Expert Panel Discussion Commemorating the 2020 World Antibiotics Awareness Week. Front. Public Health. 2021;9:673504. doi: 10.3389/fpubh.2021.673504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chukwu E.E., Oladele D.A., Enwuru C.A., Gogwan P.L., Abuh D., Audu R.A., Ogunsola F.T. Antimicrobial Resistance Awareness and Antibiotic Prescribing Behavior among Healthcare Workers in Nigeria: A National Survey. BMC Infect. Dis. 2021;21:22. doi: 10.1186/s12879-020-05689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medugu N., Tickler I.A., Duru C., Egah R., James A.O., Odili V., Hanga F., Olateju E.K., Jibir B., Ebruke B.E., et al. Phenotypic and Molecular Characterization of Beta-Lactam Resistant Multidrug-Resistant Enterobacterales Isolated from Patients Attending Six Hospitals in Northern Nigeria. Sci. Rep. 2023;13:10306. doi: 10.1038/s41598-023-37621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afolayan A.O., Oaikhena A.O., Aboderin A.O., Olabisi O.F., Amupitan A.A., Abiri O.V., Ogunleye V.O., Odih E.E., Adeyemo A.T., Adeyemo A.T., et al. Clones and Clusters of Antimicrobial-Resistant Klebsiella from Southwestern Nigeria. Clin. Infect. Dis. 2021;73:S308–S315. doi: 10.1093/cid/ciab769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolourchi N., Giske C.G., Nematzadeh S., Mirzaie A., Abhari S.S., Solgi H., Badmasti F. Comparative Resistome and Virulome Analysis of Clinical NDM-1–producing Carbapenem-Resistant Enterobacter Cloacae Complex. J. Glob. Antimicrob. Resist. 2022;28:254–263. doi: 10.1016/j.jgar.2022.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Sim E.M., Wang Q., Howard P., Kim R., Lim L., Hope K., Sintchenko V. Persistent Salmonella Enterica Serovar Typhi Sub-Populations within Host Interrogated by Whole Genome Sequencing and Metagenomics. PLoS ONE. 2023;18:e0289070. doi: 10.1371/journal.pone.0289070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jesser K.J., Trueba G., Konstantinidis K.T., Levy K. Why Are so Many Enteric Pathogen Infections Asymptomatic? Pathogen and Gut Microbiome Characteristics Associated with Diarrhea Symptoms and Carriage of Diarrheagenic E. coli in Northern Ecuador. Gut Microbes. 2023;15:2281010. doi: 10.1080/19490976.2023.2281010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uwanibe J.N., Kayode T.A., Oluniyi P.E., Akano K., Olawoye I.B., Ugwu C.A., Happi C.T., Folarin O.A. The Prevalence of Undiagnosed Salmonella Enterica Serovar Typhi in Healthy School-Aged Children in Osun State, Nigeria. Pathogens. 2023;12:594. doi: 10.3390/pathogens12040594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kariuki S., Dyson Z.A., Mbae C., Ngetich R., Kavai S.M., Wairimu C., Anyona S., Gitau N., Onsare R.S., Ongandi B., et al. Multiple Introductions of Multidrug-Resistant Typhoid Associated with Acute Infection and Asymptomatic Carriage, Kenya. eLife. 2021;10:e67852. doi: 10.7554/eLife.67852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayode A., Okunrounmu P., Olagbende A., Adedokun O., Hassan A.-W., Atilola G. High Prevalence of Multiple Drug Resistant Enteric Bacteria: Evidence from a Teaching Hospital in Southwest Nigeria. J. Infect. Public Health. 2020;13:651–656. doi: 10.1016/j.jiph.2019.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson J., Nash J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018;4:e000206. doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuffie M.J., Barrick J.E. pLannotate: Engineered Plasmid Annotation. Nucleic Acids Res. 2021;49:W516–W522. doi: 10.1093/nar/gkab374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 41.Oyejobi G.K., Sule W.F., Akinde S.B., Khan F.M., Ogolla F. Multidrug-Resistant Enteric Bacteria in Nigeria and Potential Use of Bacteriophages as Biocontrol. Sci. Total Environ. 2022;824:153842. doi: 10.1016/j.scitotenv.2022.153842. [DOI] [PubMed] [Google Scholar]
- 42.Obasi A.I., Ugoji E.O., Nwachukwu S.U. Incidence and Molecular Characterization of Multidrug Resistance in Gram-negative Bacteria of Clinical Importance from Pharmaceutical Wastewaters in South-western Nigeria. Environ. DNA. 2019;1:268–280. doi: 10.1002/edn3.28. [DOI] [Google Scholar]
- 43.Manik R.K., Mahmud Z., Mishu I.D., Hossen M.S., Howlader Z.H., Nabi A.H.M.N. Multidrug Resistance Profiles and Resistance Mechanisms to β-Lactams and Fluoroquinolones in Bacterial Isolates from Hospital Wastewater in Bangladesh. Curr. Issues Mol. Biol. 2023;45:6485–6502. doi: 10.3390/cimb45080409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yehouenou C.L., Bogaerts B., De Keersmaecker S.C.J., Roosens N.H.C., Marchal K., Tchiakpe E., Affolabi D., Simon A., Dossou F.M., Vanneste K., et al. Whole-Genome Sequencing-Based Antimicrobial Resistance Characterization and Phylogenomic Investigation of 19 Multidrug-Resistant and Extended-Spectrum Beta-Lactamase-Positive Escherichia Coli Strains Collected from Hospital Patients in Benin in 2019. Front. Microbiol. 2021;12:752883. doi: 10.3389/fmicb.2021.752883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sora V.M., Meroni G., Martino P.A., Soggiu A., Bonizzi L., Zecconi A. Extraintestinal Pathogenic Escherichia Coli: Virulence Factors and Antibiotic Resistance. Pathogens. 2021;10:1355. doi: 10.3390/pathogens10111355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duan Y., Gao H., Zheng L., Liu S., Cao Y., Zhu S., Wu Z., Ren H., Mao D., Luo Y. Antibiotic Resistance and Virulence of Extraintestinal Pathogenic Escherichia Coli (ExPEC) Vary According to Molecular Types. Front. Microbiol. 2020;11:598305. doi: 10.3389/fmicb.2020.598305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonnet M., Eckert C., Tournebize R. Decolonization of Asymptomatic Carriage of Multi-Drug Resistant Bacteria by Bacteriophages? Front. Microbiol. 2023;14:1266416. doi: 10.3389/fmicb.2023.1266416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arzilli G., Scardina G., Casigliani V., Petri D., Porretta A., Moi M., Lucenteforte E., Rello J., Lopalco P., Baggiani A., et al. Screening for Antimicrobial-Resistant Gram-Negative Bacteria in Hospitalised Patients, and Risk of Progression from Colonisation to Infection: Systematic Review. J. Infect. 2022;84:119–130. doi: 10.1016/j.jinf.2021.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Campos-Madueno E.I., Moradi M., Eddoubaji Y., Shahi F., Moradi S., Bernasconi O.J., Moser A.I., Endimiani A. Intestinal Colonization with Multidrug-Resistant Enterobacterales: Screening, Epidemiology, Clinical Impact, and Strategies to Decolonize Carriers. Eur. J. Clin. Microbiol. Infect. Dis. 2023;42:229–254. doi: 10.1007/s10096-023-04548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., Belzer C., Delgado P.S., Arboleya M., Mancabelli L., et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017;81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brinkac L., Voorhies A., Gomez A., Nelson K.E. The Threat of Antimicrobial Resistance on the Human Microbiome. Microb. Ecol. 2017;74:1001–1008. doi: 10.1007/s00248-017-0985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Silva C.L.P., Miranda L.E.V., Moreira B.M., Rebello D., Carson L.A., Kellum M.E., de Almeida M.C.L., Sampaio J.L.M., O’Hara C.M. Enterobacter Hormaechei Bloodstream Infection at Three Neonatal Intensive Care Units in Brazil. Pediatr. Infect. Dis. J. 2002;21:175–177. doi: 10.1097/00006454-200202000-00022. [DOI] [PubMed] [Google Scholar]
- 53.Foster-Nyarko E., Alikhan N.-F., Ikumapayi U.N., Sarwar G., Okoi C., Tientcheu P.-E.M., Defernez M., O’Grady J., Antonio M., Pallen M.J. Genomic Diversity of Escherichia Coli from Healthy Children in Rural Gambia. PeerJ. 2021;9:e10572. doi: 10.7717/peerj.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Duin D., Paterson D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016;30:377–390. doi: 10.1016/j.idc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chukwu E.E., Oladele D.A., Awoderu O.B., Afocha E.E., Lawal R.G., Abdus-Salam I., Ogunsola F.T., Audu R.A. A National Survey of Public Awareness of Antimicrobial Resistance in Nigeria. Antimicrob. Resist. Infect. Control. 2020;9:72. doi: 10.1186/s13756-020-00739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.San Millan A. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends Microbiol. 2018;26:978–985. doi: 10.1016/j.tim.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Baty D., Knibiehler M., Verheij H., Pattus F., Shire D., Bernadac A., Lazdunski C. Site-Directed Mutagenesis of the COOH-Terminal Region of Colicin A: Effect on Secretion and Voltage-Dependent Channel Activity. Proc. Natl. Acad. Sci. USA. 1987;84:1152–1156. doi: 10.1073/pnas.84.5.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marković K.G., Grujović M.Ž., Koraćević M.G., Nikodijević D.D., Milutinović M.G., Semedo-Lemsaddek T., Djilas M.D. Colicins and Microcins Produced by Enterobacteriaceae: Characterization, Mode of Action, and Putative Applications. Int. J. Environ. Res. Public Health. 2022;19:11825. doi: 10.3390/ijerph191811825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kucerova E., Clifton S.W., Xia X.-Q., Long F., Porwollik S., Fulton L., Fronick C., Minx P., Kyung K., Warren W., et al. Genome Sequence of Cronobacter Sakazakii BAA-894 and Comparative Genomic Hybridization Analysis with Other Cronobacter Species. PLoS ONE. 2010;5:e9556. doi: 10.1371/journal.pone.0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franke S., Grass G., Rensing C., Nies D.H. Molecular Analysis of the Copper-Transporting Efflux System CusCFBA of Escherichia Coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ecker L., Olarte L., Vilchez G., Ochoa T.J., Amemiya I., Gil A.I., Lanata C.F. Physicians’ Responsibility for Antibiotic Use in Infants from Periurban Lima, Peru. Rev. Panam. Salud Publica. 2011;30:574–579. [PubMed] [Google Scholar]
- 62.Adisa R., Orherhe O.M., Fakeye T.O. Evaluation of Antibiotic Prescriptions and Use in under-Five Children in Ibadan, SouthWestern Nigeria. Afr. Health Sci. 2018;18:1189–1201. doi: 10.4314/ahs.v18i4.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duong Q.A., Pittet L.F., Curtis N., Zimmermann P. Antibiotic Exposure and Adverse Long-Term Health Outcomes in Children: A Systematic Review and Meta-Analysis. J. Infect. 2022;85:213–300. doi: 10.1016/j.jinf.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Huang S., Eze U.A. Awareness and Knowledge of Antimicrobial Resistance, Antimicrobial Stewardship and Barriers to Implementing Antimicrobial Susceptibility Testing among Medical Laboratory Scientists in Nigeria: A Cross-Sectional Study. Antibiotics. 2023;12:815. doi: 10.3390/antibiotics12050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Babatola A.O., Fadare J.O., Olatunya O.S., Obiako R., Enwere O., Kalungia A., Ojo T.O., Sunmonu T.A., Desalu O., Godman B. Addressing Antimicrobial Resistance in Nigerian Hospitals: Exploring Physicians Prescribing Behavior, Knowledge, and Perception of Antimicrobial Resistance and Stewardship Programs. Expert. Rev. Anti-Infect. Ther. 2021;19:537–546. doi: 10.1080/14787210.2021.1829474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data of isolates are deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA838568.