Abstract
Extracellular vesicles (EVs) contain a plethora of biomolecules, including nucleic acids, with diverse diagnostic and therapeutic application potential. Although reverse transcription‐quantitative PCR (RT‐qPCR) is the most widely applied laboratory technique to evaluate gene expression, its applicability in EV research is challenged by the lack of universal and stably present reference genes (RGs). In this study, we identify, validate and establish SNRPG, OST4, TOMM7 and NOP10 as RGs for the normalization of EV‐associated genes by RT‐qPCR. We show the stable presence of SNRPG, OST4, TOMM7 and NOP10 in multiple cell lines and their secreted EVs (n = 12) under different (patho)physiological conditions as well as in human‐derived biofluids (n = 3). Enzymatic treatments confirm the presence of SNRPG, OST4, TOMM7 and NOP10 inside EVs. In addition, the four EV‐associated RGs are stably detected in a size‐range of EV subpopulations. RefFinder analysis reveals that SNRPG, OST4, TOMM7 and NOP10 are more stable compared to RGs established specifically for cultured cells or tissues such as HMBS, YWHAZ, SDHA and GAPDH. In summary, we present four universal and stably present EV‐associated RGs to enable normalization and thus steer the implementation of RT‐qPCR for the analysis of EV‐associated RNA cargo for research or clinical applications.
Keywords: blood, conditioned medium, exosomes, extracellular vesicles, microvesicles, mRNA, protease, reference genes, RNase, tissue, urine
1. INTRODUCTION
Extracellular vesicles (EVs) are spherical, nanometre‐sized, phospholipid bilayer‐enclosed particles, shed by all cells into their environment (Hendrix, 2021; Hendrix et al., 2023; van Niel et al., 2022). Amongst a variety of other biomolecules, cells produce EVs carrying RNA cargo that can alter the host's cell gene expression and function at the paracrine and systemic levels (Tkach & Théry, 2016; Valadi et al., 2007; Vergauwen et al., 2021). The past decade has delivered increased understanding on EVs and their RNA cargo resulting in the exploration of EV‐based biomarker and therapeutic applications. Despite these advances, many challenges remain related to RNA cargo validation and batch‐to‐batch quality control which are indispensable to translate EV‐based applications into clinical use (De Wever & Hendrix, 2019; Geeurickx & Hendrix, 2020).
Reverse transcription‐quantitative PCR (RT‐qPCR) is one of the most widely used laboratory techniques to evaluate gene expression. Its concept and simplicity, along with a combination of sensitivity, specificity and speed have made it the standard for nucleic acid research (Bustin et al., 2009). A prerequisite for the reliable interpretation of RT‐qPCR data is accurate normalization. Hereto, the selection of proper reference genes (RGs) with stable expression across different cell types and experimental conditions is fundamental (Eisenberg & Levanon, 2013; Zhu et al., 2008).
RGs are well established for cell lines and tissues, but their identification and validation in the EV research field remains challenging (Vandesompele et al., 2002). Although RGs show stable expression in the source cell or tissue, their stable presence in EVs and EV subpopulations has not been addressed (van Niel et al., 2022). Also, disease state, cell type, or treatment condition affects EV cargo, challenging the identification of RGs that are widely applicable across different experimental conditions (Gouin et al., 2017). The use of different separation methods, each with different recovery efficiency and specificity that can vary depending on biofluid, resulting in variable co‐separation of EVs and other RNA containing extracellular particles (e.g., protein aggregates and lipoproteins), further adds complexity to the search for suitable EV RGs (Louka et al., 2020; Van Deun et al., 2017, 2014; Vickers et al., 2011). To date, only a few efforts, mainly focused on microRNAs (miRNAs), have been made to validate RGs for EVs derived from cell lines and tissues or to identify cell type specific RGs (Chiba et al., 2012; Crossland et al., 2016; Gorji‐Bahri et al., 2021; Gouin et al., 2017; Ragni et al., 2019; Singh et al., 2022) (Figure S1).
In this study, we (1) identify SNRPG, OST4, TOMM7 and NOP10 as candidate RGs, (2) demonstrate their universal presence in EVs prepared from a wide variety of cellular and biofluid sources under diverse experimental and disease conditions, (3) confirm their detection inside EVs using enzymatic treatments and (4) analyze and compare their stability using four different algorithms to RGs established for cell lines and tissues, namely HMBS, YWHAZ, SDHA,GAPDH and U6. We provide guidance for the use of SNRPG, OST4, TOMM7 and NOP10 to normalize RT‐qPCR data from EV preparations (Box 1). Overall, the availability of these universal and stably present RGs will increase the applicability of RT‐qPCR for the analysis of EV‐associated RNA cargo for different research and clinical applications.
BOX 1: How to use SNRPG, OST4, TOMM7 and NOP10 to normalize RT‐qPCR data from EV preparations
Separate EVs from the desired source and perform quality control in compliance with current state‐of‐the‐art (e.g., MISEV guidelines and the EV‐TRACK knowledgebase) (Théry et al., 2018; Van Deun et al., 2017).
-
Isolate RNA from bulk or fractionated EV preparations.
*Normalization*: Normalization to the initial number of particles or protein concentration prior to RNA isolation is possible. However, this will not address variability introduced during RNA isolation or cDNA conversion. Normalization to RNA concentration is challenging due to the minute amounts of nucleic acids in highly specific EV preparations, depending on the amount of starting material (Gunasekaran et al., 2019). Alternatively equal volumes of RNA eluates or external RNA spikes can be used.
-
Assess the presence of SNRPG, OST4, TOMM7 and NOP10 in the EV preparations via RT‐qPCR (minimum of 3 technical replicates).
*Optional*: If validation of SNRPG, OST4, TOMM7 and NOP10 in the EV preparations is proven to be challenging, consider using the source as a positive control.
Perform stability analysis of results (using, e.g., RefFinder) obtained in step 3 and select most stable reference genes.
Implement the selected reference genes in compliance with the current state‐of‐the‐art (e.g., MIQE guidelines) to normalize relative quantification of RT‐qPCR data from EV preparations (Bustin et al., 2009).
Perform necessary quality controls (e.g., protease + RNase) to verify association between EVs and intended nucleic acids.
2. MATERIALS AND METHODS
2.1. Cell culture
The human brain cancer cell line U‐87 MG, lung cancer cell line A549, pancreatic cancer cell line PANC‐1, colon cancer cell lines HCT 116 and HT‐29, ovarian cancer cell lines SK‐OV‐3 and OVCAR‐3, breast cancer cell lines ZR‐75‐1, BT‐474, SK‐BR‐3, MDA‐MB‐231, MDA‐MB‐453, T‐47D and MCF7‐Rab27b (Hendrix et al., 2010), kidney cell line HEK293T, prostate cancer cell lines PC‐3 and LNCaP and bronchial epithelium cell line BEAS‐2B were obtained from ATCC (Manassas, VA, USA) and maintained in T175 flasks in either Dulbecco's Modified Eagle Medium (DMEM; cat. no. 41965039, ThermoFisher) (HCT 116, HT‐29, SK‐OV‐3, BT‐474, SK‐BR‐3, MDA‐MB‐231, MDA‐MB‐453 and PC‐3), Minimum Essential Media (MEM; cat. no. 10370‐047, ThermoFisher) (BEAS‐2B) or Roswell Park Memorial Institute 1640 medium (RPMI 1640; cat. no. 21875091, ThermoFisher) (ZR‐75‐1, T‐47D, LNCaP and OVACAR‐3). Additionally, previously immortalized cancer associated fibroblasts (CAFs) CT5.3 hTeRT were cultured in DMEM (De Vlieghere et al., 2015). All media were supplemented with 10% heat‐inactivated fetal bovine serum (cat. no. ATCC‐30‐2030, LGC Standards), 100 IU/mL penicillin and 100 mg/mL streptomycin (cat. no. 15070063, ThermoFisher). All cell lines were incubated at 37°C in 5% or 10% CO2 (Binder, Tuttlingen) according to provider's recommendations. The presence of Mycoplasma contamination was regularly tested using the MycoAlert Mycoplasma Detection Kit (cat. no. LT07‐318, Lonza).
2.2. Biofluid collection
Collection of patient samples was according to relevant guidelines and has been approved by the ethical committee (EC/2014/0655) of Ghent University Hospital.
Urine sample preparation was performed as previously described (Dhondt, et al., 2020; Dhondt, et al., 2020). Briefly, 50 mL of second morning urine was collected from non‐fasting healthy volunteers (n = 3) using sterile urine containers. Collected urine was immediately followed by centrifugation for 10 min at 1000 × g and 4°C. Cell‐free urine supernatants were collected by leaving approximately 0.5 cm urine above the cell pellet. The supernatant was concentrated using a Centricon plus‐70 centrifugal filter with 10 kDa molecular weight cut‐off (MWCO) (cat. no. UFC701008, Merck Life Science) and adjusted to 500 µL with phosphate‐buffered saline (PBS, cat. no. 20012019, ThermoFisher) (pH 7.2) Urine samples were immediately processed for EV separation (cfr. EV separation from urine section).
Blood plasma collection and platelet‐depleted plasma (PDP) preparation was performed as previously described (Dhondt et al., 2023; Tulkens et al., 2020; Vergauwen et al., 2021). Briefly, venous blood from cancer patients (breast cancer n = 2 and ovarian cancer n = 2) was collected using sterile 9 mL Vacuatte 9 NC Coagulation sodium citrate 3.2% tubes (cat. no. 455322, Greiner Bio‐One). PDP was then obtained within 120 min after collection by two consecutive centrifugation steps at 2500 × g for 15 min at room temperature. After each centrifugation step, the supernatant was transferred to a new sterile plastic tube and the pellet was discarded. PDP was stored at −80°C until further use.
Resected tumour tissue specimens (sarcoma, n = 3) were transported in PBS buffer and processed within 2 h after surgery. Tumour specimens were manual sliced into pieces of 2–3 mm3 and washed by placing them on a 70 µm cell strainer and gentle flushing three times with 5 mL of PBS. These pieces were weighed and then added to a 6‐well plate, each well containing 2 mL of PBS with 40% DMEM containing 0.5% EV‐depleted serum (EDS). EDS was obtained by centrifuging fetal bovine serum at 100,000 × g for 18 h at 4°C followed by a 0.2 µm filtration. The plate was incubated for 3 h on a rotating device at 37°C, 5% CO2 (Dhondt, et al., 2020). After incubation, tissue pieces were recovered and kept for histology and protein extraction. The supernatant, that is, tumour conditioned medium (TCM), was centrifuged for 10 min at 200 × g and 20 min at 2000 × g to remove cells and cell debris. After the final centrifugation step, supernatant was collected and stored at −80°C until EV separation.
2.3. Recombinant EVs
Generation and preparation of recombinant EVs (rEVs) was performed as previously described (Geeurickx et al., 2021, 2019). rEVs can also be commercially procured from Merck (cat. no. SAE0193, Merck Life Science).
2.4. Preparation of conditioned medium for EV separation
Preparation and collection of conditioned medium (CM) was performed as previously described (Van Deun et al., 2014). Briefly, CM was prepared from 3 × 108 cells grown at 60%–80% confluency in four Falcon Cell Culture Multi‐Flasks (cat. no. 353144, Corning). Cells were washed once with serum‐free (SF) medium, followed by two washing steps with 0.5% EDS supplemented medium. Flasks were incubated at 37°C in 5 or 10% CO2 for 24 h. After 24 h CM was collected and centrifuged for 10 min at 300 × g at 4°C. Cell counting was performed with trypan blue staining to assess cell viability (Countess II Automated Cell Counter; ThermoFisher). The supernatant was filtered using a 0.45 µm cellulose acetate filter (cat. no. 734‐5064, VWR International) and CM was concentrated up to 1 mL using a 10 kDa Centricon Plus‐70 centrifugal unit at 4°C (cat. no. UFC701008, Merck Life Science). Concentrated conditioned medium (CCM) was stored at −80°C until further use.
2.5. EV separation from blood plasma
EV separation from blood plasma was performed via size exclusion chromatography (SEC) as previously described (Tulkens et al., 2020; Vergauwen et al., 2021).
For SEC separation, Sepharose CL‐2B, was stored in ethanol and washed three times overnight with PBS (ThermoFisher) (pH 7.2) before use. A 10 mL syringe (cat. no. A18645, Romed Holland) was used where a nylon net with 20 µm pore size (cat. no. NY2002500, Merck Life Science) was placed at the bottom. The syringe was then filled up to 10 mL with Sepharose CL‐2B using a circular motion. To avoid drying of the Sepharose CL‐2B beads, PBS containing 0.32% trisodiumcitrate dihydrate (cat. no. S4641‐25G, Merck Life Science) was added. On top of the SEC column, PDP was loaded and eluates of 1 mL were collected. EV‐enriched eluates 4–6 (3 mL) were collected followed by a 3 h ultracentrifugation at 100,000 × g and 4°C using a SW32.1 Ti rotor. Resulting pellets were resuspended in 200 µL Qiazol and stored at −80°C.
2.6. EV separation from conditioned medium
EVs were separated from CCM via top‐down OptiPrep density gradient ultracentrifugation (DGUC) as previously described (Van Deun et al., 2014; Vergauwen et al., 2017). Briefly, a discontinuous iodixanol gradient was prepared by layering 4 mL of 40%, 4 mL of 20%, 4 mL of 10% and 3.5 mL of 5% iodixanol on top of each other in a 16.8 mL open top polyallomer tube (cat. no. 337986, Beckman Coulter). Iodixanol solutions were made by mixing appropriate amounts of a homogenization buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris‐HCL, [pH 7.4]) and a 50% iodixanol working solution. The working solution was prepared by combining a working solution buffer (0.25 M sucrose, 6 mM EDTA, 60 Mm Tris‐HCL, [pH 7.4]) and a stock solution of OptiPrep (60% (w/v) aqueous iodixanol solution). One milliliter of CCM sample was overlaid on top of the gradient, followed by 18 h ultracentrifugation at 100,000 × g and 4°C using a SW32.1 Ti rotor (Beckman Coulter). Afterwards, 16 DGUC fractions of 1 mL were collected from the top of the gradient (EV‐enriched fractions 9 and 10 ‐ corresponding to 1.09–1.11 g/mL densities) and kept. DGUC fractions were collected and diluted to 15 mL with PBS, followed by a 3 h ultracentrifugation at 100,000 × g and 4°C using a SW32.1 Ti rotor. Resulting pellets were resuspended in 100 µL PBS and stored at −80°C until further use. When indicated, 3 h ultracentrifugation was replaced by SEC‐based separation using Sepharose CL‐2B (cat. no. GE17‐0140‐01, GE Healthcare) where eluates 4–7 were collected and concentrated to 100 µL using an Amicon Ultra‐2 mL centrifugal filter with a 10 kDa cut‐off (cat. no. UFC201024, Merck Life Science). EV preparations were stored at −80°C until further use.
2.7. EV separation from urine
EV separation from urine was performed via a bottom‐up OptiPrep DGUC as previously described (Dhondt, et al., 2020). Shortly, 500 µL of concentrated urine was diluted in Tris buffer containing 1 mM Tris‐HCL (pH 7.4), 1 mM EDTA and 0.25 M sucrose to obtain a sample volume of 800 µL. Solutions of 5%, 10% and 20% iodixanol were made by mixing appropriate amounts of homogenization buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris‐HCL (pH 7.4)) and iodixanol working solution. The working solution was prepared by combining a working solution buffer (0.25 M sucrose, 6 mM EDTA, 60 mM Tris‐HCl, pH 7.4). The 800 µL urine sample was resuspended in 3.2 mL working solution, obtaining a 40% iodixanol suspension, and layered on the bottom of a 16.8 mL open top polyallomer tube. A discontinuous bottom‐up DGUC was prepared by overlaying the urine suspension with 4 mL 20%, 4 mL 10% and 3.5 mL 5% iodixanol solutions, and 1 mL PBS (pH 7.2), respectively. The DGUC was centrifuged for 18 h at 100,000 × g and 4°C (SW32.1 Ti rotor, Beckman Coulter). Afterwards, DGUC fractions of 1 mL were collected from the top of the gradient and EV‐enriched fractions 9 and 10 (corresponding to 1.09–1.11 g/mL densities) were further processed by SEC (cfr. EV separation from blood plasma section) (Van Dorpe et al., 2023). Eluates 4–7 were collected and concentrated to 100 µL using an Amicon Ultra‐2 mL centrifugal filter with a 10 kDa cut‐off. EV preparations were stored at −80°C until further use.
2.8. Dual size‐exclusion chromatography
For the preparation of size‐based EV subpopulations, a dual SEC approach was applied. In brief, SEC columns were prepared using Econo‐Pac chromatography columns (cat. no. 732‐1010, Bio‐Rad) with a bed volume of 20 mL and a reservoir of 10 mL containing a porous 30 µm polyethylene bed support. Sepharose CL‐2B was washed three times by sedimentation in PBS. A total of 20 mL washed Sepharose CL‐2B was added to the column. DGUC EV‐enriched fractions (corresponding to fractions 9 and 10) were loaded onto the SEC column followed by elution by PBS. Individual fractions of 0.5 mL were collected. Fractions 16 to 19 of the first SEC column were pooled and loaded on a second SEC column. Fractions of 0.5 mL were collected, pooled and concentrated to 65 µL using Amicon Ultra‐2 10K filters. Fractions 16 to 19 collected from the second SEC column were referred to as large EVs (lEVs ‐ >120 and <250 nm). Fractions 20–22 and 23–28 of the first SEC column were respectively referred to as medium‐sized EVs (mEVs ‐ >90 and <120 nm) and small‐sized EVs (sEVs ‐ >50 and <90 nm).
2.9. Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was performed using a NanoSight LM10‐HS microscope (Malvern Instruments, Malvern, UK) equipped with a 488 nm laser. During the measurements, an infusion speed of 20 was applied with an automatic syringe pump system. Scattered light was captured and three videos of 30 s were recorded of each sample with camera level set at 13 and detection threshold set at 3. Throughout the measurements, the temperature was monitored and a medium viscosity of 0.929 cP was assumed. The recorded videos were analysed with NTA Software version 3.4. The samples were diluted in PBS until the particle concentration was within the optimal concentration range of the NTA software (3 × 108–1 × 109 particles/mL).
2.10. Transmission electron microscopy
EV preparations were deposited on Formvar carbon‐coated, glow discharged grids, which were stained with uranylacetate and embedded in methylcellulose/uranylacetate. Prepared grids were then examined using a Tecnai Spirit transmission electron microscope (FEI) and images were captured with Quemasa charge‐coupled device camera (Olympus Soft Imaging Solutions).
2.11. Antibodies
The following primary antibodies (and dilutions) were used for immunostaining: anti‐Syntenin‐1 (1:1000) (cat. no. ab133267, Abcam), anti‐Flotillin‐1 (1:1000) (cat. no. 610820, Becton Dickinson), anti‐Alix (1:1000) (cat. no. 3A9, Bioké), anti‐TSG101 (1:1000) (cat. no. SC‐7964, Bio‐connect), anti‐Ago2 (1:1000) (cat. no. ab32381, Abcam) and anti‐CD9 (1:1000) (D3H4P, Bioké). As secondary antibodies, sheep anti‐mouse horseradish peroxidase‐linked antibody (1:3000) (cat. no. NA931V, VWR International) and donkey anti‐rabbit horseradish peroxidase‐linked antibody (1:4000) (cat. no. NA934V, VWR International) were used.
2.12. Protein analysis
Protein concentration and protein analysis were performed as previously described (Vergauwen et al., 2017). Briefly, protein concentrations were measured using the fluorometric Qubit protein assay kit (cat. no. 10543343, ThermoFisher) with the Qubit Fluorometer 3.0 (cat. no. Q33216, ThermoFisher) following manufacturer's instructions. Samples were prepared by a 1:1 dilution with 0.4% sodium dodecyl sulfate (SDS). Protein concentration of cell lysates obtained in Laemmli lysis buffer (0.125 M Tris‐HCl [pH 6.8], 10% glycerol, 2.3% SDS), were determined using the Bio‐Rad DC Protein Assay (cat. no. 500‐0113; 500‐0114; 500‐0115, Bio‐Rad).
For protein analysis, samples were dissolved in reducing sample buffer (0.5 M Tris‐HCl [pH 6.8], 40% glycerol, 9.2% SDS, 3% 2‐mercaptoethanol, 0.005% bromophenol blue) and boiled at 95°C for 5 min. Proteins were separated by SDS‐polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (cat. no. 162‐0115, Bio‐Rad). After blocking the membranes for 30 min with 5% non‐fat milk in PBS with 0.5% Tween 20, the blots were incubated overnight at 4°C with primary antibodies. Incubation with secondary antibodies was performed for 60 min at room temperature after extensive washing of the membranes with blocking buffer and PBS with 20. After final washing, chemiluminescence substrate (WesternBright Sirius, cat. no K‐12043‐D20, IsoGen) was added and imaging was performed using Proxima 2850 Imager (IsoGen Life Sciences). The data analysis was performed with the ProXima AQ‐4 software.
2.13. Protease and RNase treatment
Sixteen pelleted DGUC fractions were resuspended in 500 µL of PBS (pH 7.2). Next, the samples were treated with 2 µg/mL proteinase K from Tritirachium album (PK; cat. no P6556‐5MG, Merck Life Science) for 30 min at 37°C, followed by 5 mM protease inhibitor phenylmethylsulfonyl fluoride (PMSF; cat. no. 36978, ThermoFisher) for 10 min at room temperature. After PK treatment, the sample was treated with 8 µg/mL RNase A (cat. no. 10109142001, Merck Life Science) for 15 min at 37°C under gentle agitation and then 10× concentrated RNase inhibitor (cat. no 10430725, ThermoFisher) was added to the sample for 5 min at room temperature. Residual PK, PMSF, RNase and RNase inhibitor were removed by diluting the treated EV samples with PBS and performing a SEC as described above (crf. EV separation from blood plasma). For detergent treatment of samples, 1% (v/v) triton X‐100 cat. no. 1610407, Bio‐Rad) was added to a final concentration of 0.1% and incubated for 30 seconds at room temperature. When no treatment (i.e., negative control) was applied an equivalent volume of PBS was added to the samples. Treatment of samples was immediately followed by RNA isolation, cDNA conversion and RT‐qPCR analysis.
2.14. RNA measurement
Immediately following RNA isolation, cell pellet RNA was measured using the nanodrop technology. Samples were evaluated for RNA yield (ng/µL) and purity (A260/A280) (Table S1).
2.15. RNA isolation and RT‐qPCR
Total RNA was isolated using miRNeasy Micro kit (cat. no 217084, Qiagen) according to the manufacturer's instructions and eluted in 14 µL DNase/RNase‐free water. Twelve microlitres of RNA was reverse transcribed in a 20 µL reaction using iScript cDNA Synthesis kit (unique blend of oligo(dT) and random hexamer primers – cat. no. 170‐8891, Bio‐Rad) according to manufacturer instructions. Cell pellets were normalized for 1 µg of RNA prior to cDNA conversion. mRNA expression analysis was performed via RT‐qPCR using the SsoAdvanced Universal SYBR Green Supermix (cat. no. 1725272, Bio‐Rad). The 5 µL PCR reaction mix contained reverse and forward primers (0.5 µL of a 5 mM stock solution), SsoAdvanced Universal SYBR Green Supermix (2x) (2.5 µL) and cDNA (2 µL of a 1:4 dilution for EVs and 2 µL of a 1:10 dilution for cell pellets, corresponding to the cDNA reversed transcribed from 12 µL RNA). The 384‐well plate was then run on the LC480 (Roche Applied Sciences) at 95°C for 5 s, then 60°C for 30 s and 72°C for 1 s (for 44 cycles). Primers used for amplification of SNRPG, OST4, TOMM7 and NOP10 were designed using standard setting with Primer Express software (ThermoFisher) whereas primers used for the amplification of GAPDH, HMBS, SDHA and YWHAZ were retrieved based on literature findings (Vandesompele et al., 2002). Sequence of primers used throughout the study can be found in Table 1. Additionally, to better control RT‐qPCR runs, two negative controls were used: (1) a no template control (NTC) to control for extraneous nucleic acid contamination and primer dimer formation and (2) a no reverse transcriptase control (‐RT) to assess DNA contamination present in the final RNA preparation. Data were analysed using the LightCycler 480 SW 1.5.1 software. Efficiency was calculated from the slope of the standard curve using the equation: efficiency = −1 + 10(−1/slope).
TABLE 1.
oligonucleotides primers used for RT‐qPCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| SNRPG | AGCGGGAGCGTGAGGAA | TCGGGAGGGTGAGCTTTG |
| OST4 | ACGTGCAGCTCGCCATCT | TGTTGACGGCCACGTAGTGA |
| TOMM7 | GATTCCCGACGCTGTGGTT | TCCCCTTGAAGAGCTGCTGTA |
| NOP10 | GAAACGCTTCAAGGTGCTCAT | GGTGGCAGAAAAGACATCAGTTTA |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| HMBS | GGCAATGCGGCTGCAA | GGGTACCCACGCGAATCAC |
| SDHA | TGGGAACAAGAGGGCATCTG | CCACCACTGCATCAAATTCATG |
| YWHAZ | ACTTTTGGTACATTGTGGCTTCAA | CCGCCAGGACAAACCAGTAT |
| U6 | UGGAACGATACAGAGAAGAUUAGCAUGGCCC | CUGCGCAAGGAUGACACGCAAAUUCGUGAAG |
2.16. RNA dataset search
Investigation of selected RG's presence in publicly available datasets was performed by selecting five high quality EV RNA datasets. In order to qualify, datasets were queried by (1) main focus being on investigation of EV‐associated RNA cargo; (2) EV characterization compiling with MISEV guidelines; (3) Total RNA sequencing applied to EV preparations and (4) raw data publicly available in the gene expression omnibus database (GEO) (Clough & Barrett, 2016).
2.17. Irradiation
Cancer associated fibroblasts CT5.3 hTeRT were irradiated with 6MV photons from a linear accelerator (Synergy, Elekta) at a dose rate of 5 Gy/min. One single beam was used with a 1 cm Perspex plate to compensate for build‐up effects.
2.18. Data analysis, visualization
Statistical analysis and data visualization were performed using GraphPad Prism version 8 (GraphPad Software, San Diego, California, USA). Data are expressed as mean with standard deviation. Unpaired comparisons were conducted using a Kruskal–Wallis test with Dunn's multiple comparison test. Comparison of protease and RNase samples was conducted using a Mann–Whitney test. Stability evaluation was performed using the online tool RefFinder (https://www.heartcure.com.au/reffinder/) (miRDeepFinder). Unsupervised hierarchical clustering was generated using the Morpheus (https://software.broadinstitute.org/morpheus) analysis software.
2.19. EV‐TRACK
We have submitted all relevant experimental parameters of the described experiments to the EV‐TRACK knowledgebase (EV‐TRACK ID: EV220300) (Van Deun et al., 2017).
3. RESULTS
3.1. Identification of EV reference genes SNRPG, OST4, TOMM7 and NOP10
We searched published high quality RNA data to identify EV RGs based upon their differential enrichment in EV preparations obtained with high specificity from CCM by density gradient ultracentrifugation (DGUC) compared to other separation methods obtaining EV preparations with lower specificity (differential ultracentrifugation or polymer‐based precipitation) (Van Deun et al., 2014; Vergauwen et al., 2017). In contrast to commonly used and established RGs for cells and tissues such as SDHA and YWHAZ, we identified four protein coding genes SNRPG, OST4, TOMM7 and NOP10 that were consistently differentially enriched in DGUC EV preparations (Figure S2). The SNRPG (small nuclear ribonucleoprotein polypeptide G) gene translates to a core component of the small nuclear ribonucleoprotein (snRNP) complex (SNRPG). Oligosaccharyltransferase complex subunit 4 (OST4) codes for a protein that is a subunit of the eukaryotic membrane‐embedded oligosaccharyltransferase (OST) protein complex (Dumax‐Vorzet et al., 2013, [Link]). TOMM7, or translocase of outer mitochondrial membrane 7, encodes a subunit of the outer mitochondrial membrane that regulates both assembly and stability of the translocase from the outer mitochondrial membrane (TOM) complex (TOMM7). The NOP10 ribonucleoprotein coding gene, is a member of the H/ACA small nucleolar ribonucleoproteins (snoRNPs) gene family (NOP10). The presence of SNRPG, OST4, TOMM7 and NOP10 as well as four established RGs for cell lines and tissues was confirmed in five independent RNA datasets (Chen et al., 2016; Hinger et al., 2018; Morhayim et al., 2017; Rodosthenous et al., 2020; Yu et al., 2020) obtained from EV preparations of diverse cell types and biofluids using different EV preparation methods (Figure 1a).
FIGURE 1.
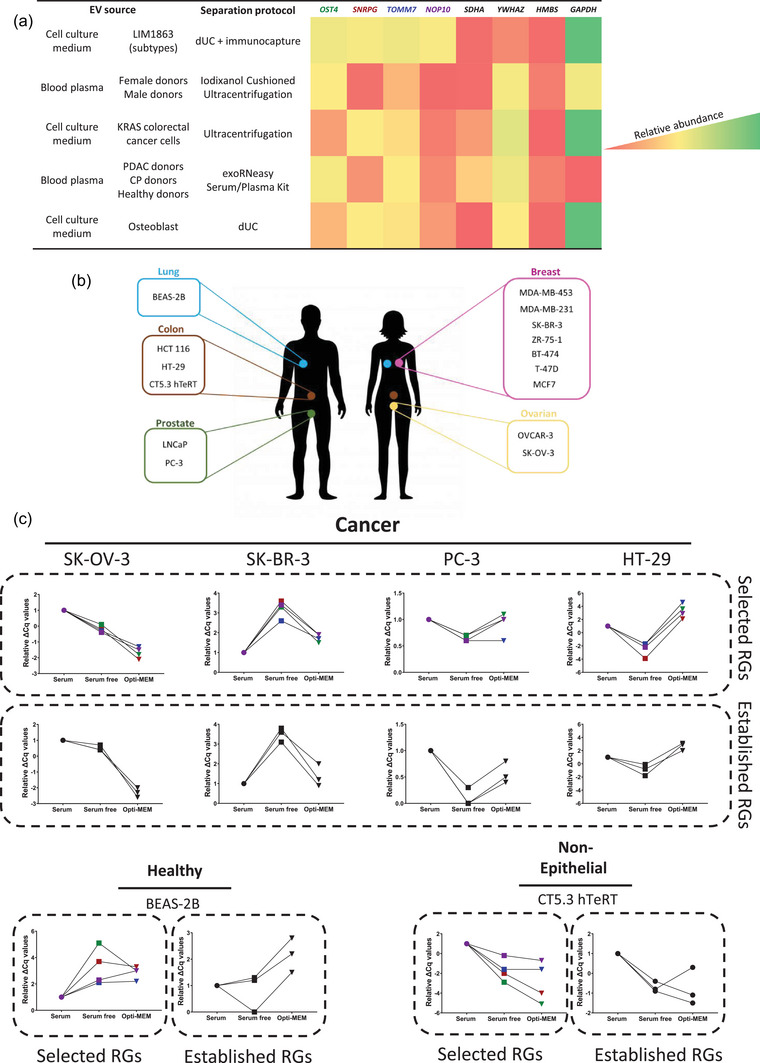
Identification of SNRPG, OST4, TOMM7, NOP10 and their evaluation versus established cell line and tissue reference genes (RGs) in cell lines of diverse origin. (a) Evaluation of the presence of identified (EV) and established (cell lines and tissue) RGs in five publicly available EV RNA datasets. Heatmap is based on relative abundance within each respective study. (b) Selected panel of 15 cell lines for the evaluation of RGs. (c) Expression profiles obtained by plotting the Cq value deviations relative to the serum condition (ΔCq serum free = avg Cq (RG) in serum – avg Cq (gene) in serum free; ΔCq opti‐MEM = avg Cq (gene) in serum free – avg Cq (gene) in opti‐MEM; data was normalized to average Cq value per RG in serum). Cell lines were cultured for 24 h under three different conditions: 10% serum (circles); serum‐ free (squares) and in Opti‐MEM medium (inverted triangle). Results are represented as expression profiles of tested genes in average delta Cq values after analysis of three technical replicates. CP, chronic pancreatitis; PDAC, pancreatic ductal adenocarcinoma.
3.2. SNRPG, OST4, TOMM7 and NOP10 are expressed across multiple cell lines and culture conditions
Evaluation of reported gene expression levels in the Cancer Cell Line Encyclopedia (CCLE) mRNA database revealed a wide range of expression levels, from low to high, for SNRPG, OST4, TOMM7 and NOP10 in the available cell lines. To validate the expression of SNRPG, OST4, TOMM7 and NOP10, we selected 15 cell lines spanning low, medium or high relative expression for the four EV RGs according to CCLE, including 13 cancer cell lines and 2 non‐cancer cell lines (De Vlieghere et al., 2015) (Figure 1b) (Table S2). Considering the potential impact of culture medium on EV analysis (Urzì et al., 2022), we also evaluated, for all selected cell lines, the expression of SNRPG, OST4, TOMM7 and NOP10 and established cell line RGs (SDHA, YWHAZ and HMBS) using three different culture media (Vandesompele et al., 2002): (1) ATCC cell line recommended medium with serum addition, (2) ATCC cell line recommended medium without serum addition and (3) Opti‐MEM. Results of average quantification cycle (Cq) values after a 24 h incubation in the selected culture media, confirmed that SNRPG, OST4, TOMM7 and NOP10 are expressed across the different cell lines and this expression is not inhibited by the selected culture condition (Figures S3 and S4). Additionally, analysis of Cq value deviations relative to ATCC cell line recommended medium (calculated as follows: ΔCq serum free = avg Cq (RG) in serum – avg Cq (gene) in serum free; ΔCq opti‐MEM = avg Cq (gene) in serum free – avg Cq (gene) in opti‐MEM; data was normalized to average Cq value per RG in serum) revealed that expression of SNRPG, OST4, TOMM7 and NOP10 and established RGs is cell line dependent, but highly consistent (Figure 1c) (Vandesompele et al., 2002).
3.3. SNRPG, OST4, TOMM7 and NOP10 are present in EV preparations derived from multiple cell lines and biofluids
Next, we evaluated the presence of SNRPG, OST4, TOMM7 and NOP10 in bulk EV preparations from conditioned medium of cell lines, body fluids and EV reference material (recombinant extracellular vesicles (rEV)) in three technical replicates. In total, EVs were separated from 10 cell lines by DGUC (A549, BEAS‐2B, HCT 116, CT5.3 hTeRT, LNCaP, OVCAR‐3, SK‐OV‐3, PANC‐1, T‐47D, U‐87 MG) and two body fluids by SEC and/or DGUC (blood plasma and urine) as described in the material and methods section. In addition, we assessed the expression of SNRPG, OST4, TOMM7 and NOP10 in size‐based EV subpopulations: sEVs (>50 and <90 nm), mEVs (>90 and <120 nm) and lEVs (>120 and <250 nm) as characterised by transmission electron microscopy (TEM) and western blot (WB) analysis (Figures 2a and S5). A selection of EV preparations from conditioned media, blood plasma and urine were evaluated by NTA, TEM and WB analysis (analysing EV‐enriched proteins Alix, Flotillin‐1, TSG101, Syntenin‐1 and/or CD9 and/or non‐EV enriched protein Argonaute‐2) to confirm to preparation of EV‐enriched fractions in compliance with the current state‐of‐the‐art and as previously extensively reported by our research group (Figure S6) (Dhondt, et al., 2020; Théry et al., 2018; Tulkens et al., 2020; Vergauwen et al., 2021).
FIGURE 2.
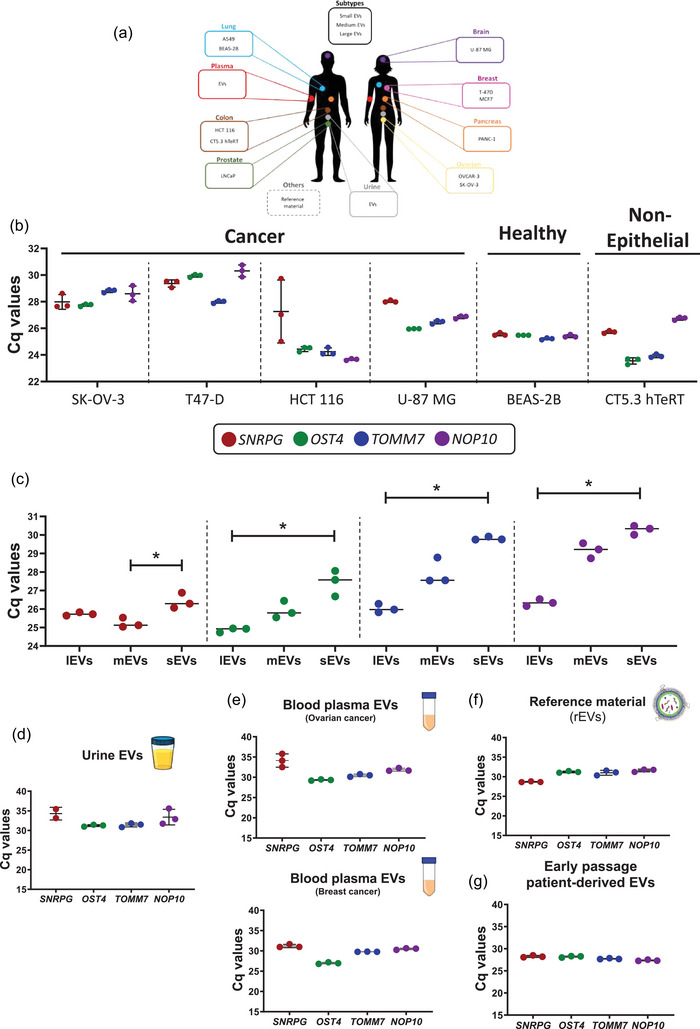
Identification of SNRPG, OST4, TOMM7 and NOP10 in EV preparations from diverse origins. (a) Selected panel of different origins from which EVs were prepared. (b) RT‐qPCR analysis of EVs prepared from media conditioned by cancer, healthy or non‐epithelial cell lines using DGUC. (c) RT‐qPCR analysis of a size range of EV subpopulations fractionated media conditioned by MCF7 cells using dual SEC (Kruskal–Wallis test, adjusted p‐value for Dunns's multiple comparison test = 0.0219). (d–f) RT‐qPCR analysis of EVs prepared from urine by DGUC (pool of healthy donors (n = 3)) (d), (e) EVs prepared from blood plasma using SEC (ovarian (n = 2) and breast (n = 2) cancer patients), (f) EV reference material (rEVs) and (g) early passage patient‐derived sarcoma cells. Results were obtained by analysis of three technical replicates. * Adjusted p‐value >0.05.
Quantification of SNRPG, OST4, TOMM7 and NOP10 by RT‐qPCR confirmed their presence in bulk EV preparations obtained from multiple cell lines (Figures 2b, S7 and Table S3). SNRPG, OST4, TOMM7 and NOP10 were detectable across size‐based EV subpopulations with increasing Cq values from lEVs to sEVs (Kruskal–Wallis test, adjusted p‐value for Dunns's multiple comparison test = 0.0219) (Figure 2c and Table S4).
To demonstrate the presence of SNRPG, OST4, TOMM7 and NOP10 across different biofluids and diverse donors we evaluated their presence in urine from healthy donors and blood plasma from patients with distinct cancer types. SNRPG, OST4, TOMM7 and NOP10 were identified in bulk EV preparations from pooled urine of three healthy donors (SNRPG: 34.29 ± 1.61; OST4: 31.24 ± 0.243; TOMM7: 31.39 ± 0.487; NOP10: 33.40 ± 1.98) in three technical replicates (Figure 2d and Table S5). Moreover, as a proof of concept, and as to obtain sufficient EVs for RT‐qPCR analysis, the four EV RGs were detected in bulk EV preparations from pooled blood plasma of two ovarian cancer patients (SNRPG: 34.13 ± 1.63; OST4: 29.32 ± 0.145; TOMM7: 30.47 ± 0.331; NOP10: 31.86 ± 0.376) and two breast cancer patients (SNRPG: 31.20 ± 0.3098; OST4: 26.98 ± 0.207; TOMM7: 29.78 ± 0.020; NOP10: 30.48 ± 0.206) in three technical replicates (Figure 2e).
Lastly, the presence of SNRPG, OST4, TOMM7 and NOP10 was further confirmed in rEVs (28.69 ± 0.102; 31.22 ± 0.216; 31.01 ± 0.621; 31.60 ± 0.339 for SNRPG, OST4, TOMM7 and NOP10, respectively) and early passage patient‐derived sarcoma cells (28.26 ± 0.227; 258.21 ± 0.165; 27.73 ± 0.118; 27.37 ± 0.144 for SNRPG, OST4, TOMM7 and NOP10 respectively) (Figure 2f,g). A serial dilution of rEVs (1 × 106–1 × 1011 particles as measured by fluorescent NTA) revealed a linear regression between 108 and 1011 particles (R 2 of 0.96, 0.99, 0.97 and 0.98 for SNRPG, OST4, TOMM7 and NOP10, respectively) (Figure S8).
3.4. SNRPG, OST4, TOMM7 and NOP10 are present inside EVs
To investigate the presence of SNRPG, OST4, TOMM7 and NOP10 inside EVs, we implemented two complementary approaches.
First, we analysed the distribution of the four EV RGs over sixteen 1 mL fractions collected from the top (density 1.040 g/mL) to the bottom (density 1.255 g/mL) of density gradients from CCM of two representative cell lines (HCT 116 and LNCaP ‐ Figure S2) (Figures 3a and S9). While SNRPG, OST4, TOMM7 and NOP10 were detectable in all fractions, the highest Cq values were identified in the top (1.040 g/mL – soluble proteins (SPs)) and bottom (1.255 g/mL – protein complexes) fractions of the density gradients. In contrast, the lowest Cq values were identified in the EV‐enriched density fractions (1.09–1.11 g/mL) (Figures 3a, S9 and Table S6).
FIGURE 3.
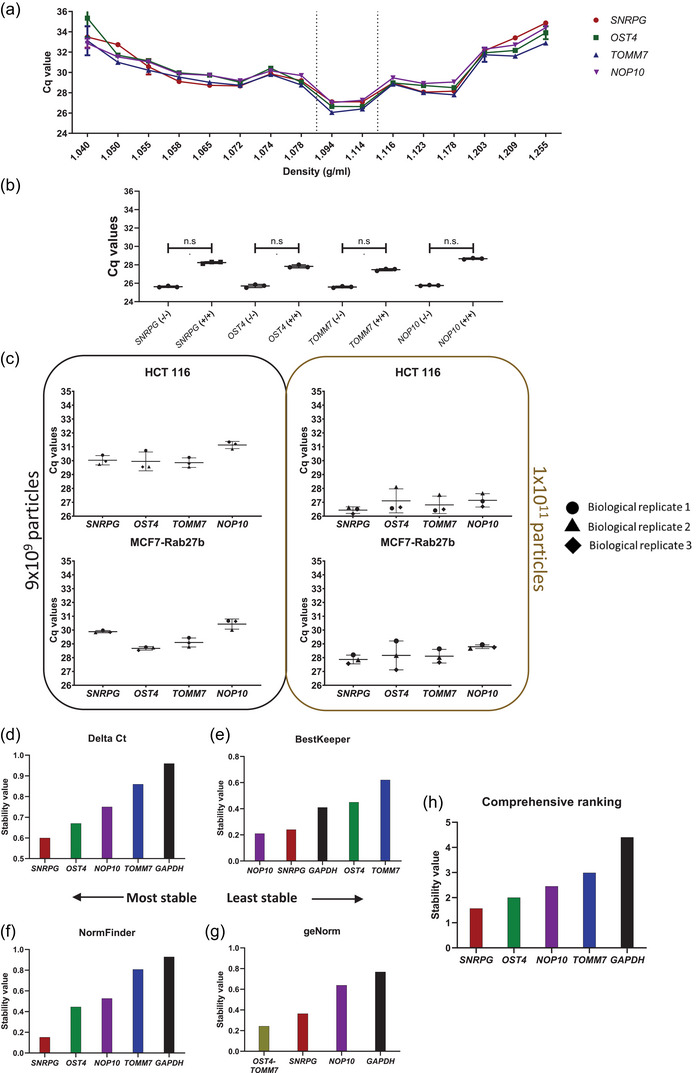
Validation of SNRPG, OST4, TOMM7 and NOP10 as suitable EV reference genes (RGs) using enzymatic treatments, biological replicates and stability algorithms. (a) RT‐qPCR analysis of all density fractions prepared from media conditioned by HCT 116 cells using DGUC. (b) RT‐qPCR analysis of SNRPG, OST4, TOMM7 and NOP10 in all density fractions prepared from media conditioned by HCT 116 cells using DGUC prior (blue) and after (red) proteinase K and RNase A treatment. Dotted lines represent average Cq values of selected RGs in soluble protein fractions (1.06–1.07 g/mL). (c) Intrinsic biological variability of EVs prepared from media conditioned by HCT 116 or MCF7 cells using DGUC in three biological replicates. Normalization to particle number was performed via nanoparticle tracking analysis (NTA). Biological results are shown as average Cq values of three technical replicates. (d–h) Stability evaluation using the RefFinder tool of EV RGs versus the established tissue and cell lines RG GAPDH in EVs prepared from media conditioned by LnCap via DGUC. Results of four algorithms are shown, (d) Delta Ct, (e) BestKeeper, (f) NormFinder and (g) geNorm alongside their respective (h) geometric mean.
Second, we performed a protease K (PK) and RNase A treatment of sixteen 1 mL density fractions from CCM prepared of a representative cell line (HCT 116 – Figures 3a, S3 and S7) (Figure 3b). The efficacy of the PK treatment was confirmed by WB analysis, demonstrating absence of the outer membrane loop of tetraspanins CD9 and CD81 but retained presence of intraluminal proteins Alix, flotillin‐1 and syntenin in PK‐treated EV‐enriched density fractions (Figure S10A). PK treatment was followed by RNase A treatment prior to RNA isolation and RT‐qPCR analysis as described in the materials and methods section. The functionality of RNase was confirmed by RT‐qPCR analysis, with significantly increased Cq values of selected RGs when applying detergent treatment prior to enzymatic digestion (Figure S10B; Kruskal–Wallis test, adjusted p‐value for Dunns's multiple comparison test = 0.0146). Extravesicular depletion of RNA by this enzymatic treatment revealed that SNRPG, OST4, TOMM7 and NOP10 remained detectable in the EV‐enriched density fractions (SNRPG Cq: EV 26.95 ± 0.191/SPs 29.85 ± 0.226, OST4 Cq: EV 27.71 ± 0.342/SPs 31.84 ± 0.211, TOMM7 Cq: EV 26.89 ± 0.445/SPs 31.03 ± 0.249, NOP10 Cq: EV 28.53 ± 0.449/SPs 29.95 ± 0.147) (Figure 3b).
3.5. SNRPG, OST4, TOMM7 and NOP10 are stably present across biological replicates
To study the impact of biological variability on SNRPG, OST4, TOMM7 and NOP10, EVs were prepared from conditioned medium of seven cell lines (SK‐OV‐3, HCT 116, PANC‐1, MCF7, U‐87 MG, A549 and BEAS‐2B with >91% viability) as well as two biofluids (urine and tumour conditioned medium (TCM)) in three biological replicates (i.e., three different cell passages for the indicated cell lines and three different donors for the indicated biofluids) (Table S7). For all EV preparations, RNA was isolated from 9 × 1010 particles as quantified by NTA and analysed by RT‐qPCR.
SNRPG, OST4, TOMM7 and NOP10 were detected across all biological replicates (Figures 3c and S12). Despite normalization to equal particle number (9 × 1010 particles), Cq values between different biological replicates showed high variability (Figures 3c and S11). To investigate if this Cq value discrepancy was due to biological differences between the randomized sets, we compared the variability within each individual replicate by analysing the technical replication (Figure S12). This revealed low variability between technical replicates for all biological samples.
To evaluate if Cq values associate with particle concentrations, we next normalized to equal particle number (1 × 1011 particles) (Figure 3c right panel, S13 and Table S8). Overall, the average Cq values of the three biological replicates decreased with increasing particle number (as compared to 9 × 1010 particles) (Figures 3c and S13).
3.6. Stability evaluation of SNRPG, OST4, TOMM7 and NOP10 using RefFinder
Stability evaluation of the four EV RGs was performed using the web‐based tool RefFinder (miRDeepFinder). We evaluated the four top algorithms used in RG identification: NormFinder, geNorm, BestKeeper and Delta Ct. NormFinder is based on a mathematical model of gene expression to estimate intergroup and intragroup sample variability (Andersen et al., 2004). geNorm determines pairwise standard deviation values of all genes and eliminates the least stable genes until only two remain, which are considered the most stable (Vandesompele et al., 2002). BestKeeper creates an index of stability based on amplification efficiencies and Cq values followed by pairwise correlation analysis of each selected index until it reaches its final ranking (Pfaffl et al., 2004). The Delta Ct approach designates stability by comparing relative expression of genes within each sample and ranks genes based on their Cq standard deviation (Silver et al., 2006).
We evaluated the stability of SNRPG, OST4, TOMM7 and NOP10 in EVs versus the most commonly used established cell line RG, GAPDH, using EV‐enriched fractions from one representative cell line (LNCaP – within the same biological replicate). Delta Ct and NormFinder identified SNRPG as the most stable gene (Figure 3d,f), while BestKeeper identified NOP10 as the most stable to normalize EV RT‐qPCR data. The geNorm algorithm identified OST4 and TOMM7 as a stable gene pair. Overall, 3 out of 4 algorithms identified GAPDH as the least stable RG to normalize EV RT‐qPCR data (Figure 3d,f,g). Comprehensive ranking of the four algorithms identified SNRPG as the most stable gene and GAPDH as the least stable gene (Figure 3h). A similar stability analysis of SNRPG, OST4, TOMM7 and NOP10 in EVs was performed versus five additional established cell line RGs (HMBS, YHWAZ, SDHA, U6 and B2M) for EV‐enriched fractions of three additional cell lines (HCT 116, MCF7, PANC1 – within the same biological replicate) and two biofluids (TCM and urine ‐ within the same biological replicate) (Figure S14). Delta Ct and NormFinder algorithms identified SNRPG, OST4 and TOMM7 while BestKeeper identified OST4 as the most stable gene(s) to normalize EV RT‐qPCR data. The geNorm algorithm identified OST4 and TOMM7 as the most stable combination. Overall, comprehensive ranking of the four algorithms confirmed that SNRPG, OST4, TOMM7 and NOP10 are more stable compared to established cell line RGs even when considering diverse EV sources. Extended gene stability analysis by RefFinder for all cell lines and biofluids evaluated in this study, revealed that the comprehensive ranking of SNRPG, OST4, TOMM7 and NOP10 depends on the source (Figure S15).
3.7. SNRPG, OST4, TOMM7 and NOP10 remain stable under cellular perturbation
To assess their stability upon cellular perturbation which may alter their presence in EVs, we evaluated SNRPG, OST4, TOMM7 and NOP10 in bulk EV preparations obtained by DGUC from conditioned medium of cancer‐associated fibroblasts (CAFs) irradiated (10 Gy) or not. For both conditions, RNA was isolated from 9 × 1010 particles as quantified by NTA and analysed by RT‐qPCR (Table S8).
SNRPG, OST4, TOMM7 and NOP10 are and remain present in EV preparations from CAFs upon irradiation (Figure 4a). A stability analysis with Delta Ct and NormFinder algorithms identified OST4 as the most stable gene (Figure 4b,d). BestKeeper identified NOP10 as the most stable gene whilst geNorm identified OST4 and TOMM7 as the most stable gene pair for the normalization of EV RT‐qPCR data. Notably, 3 out of 4 algorithms identified SNRPG as the least stable gene (Delta Ct, NormFinder and geNorm). In accordance, comprehensive ranking of the four algorithms identified OST4 as most stable RG and SNRPG as the least stable RG for the normalization of this particular dataset.
FIGURE 4.
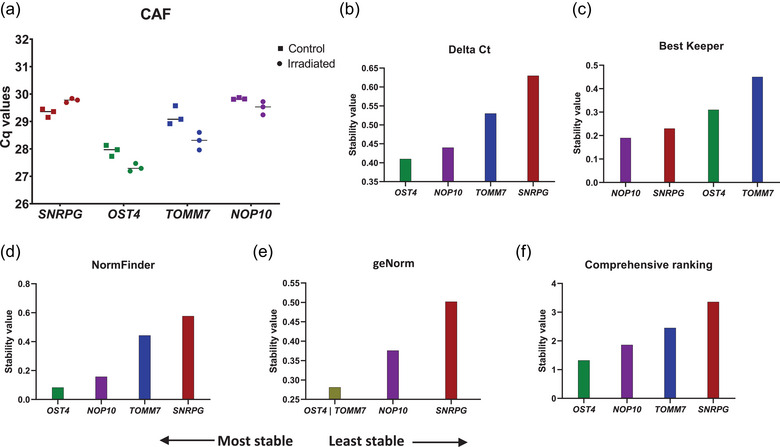
Validation of SNRPG, OST4, TOMM7 and NOP10 as suitable reference genes using destabilizing conditions. (a) RT‐qPCR analysis of EVs prepared from media conditioned by CAFs (irradiated (10 Gy) or not) by DGUC in three technical replicates. Stability analysis of SNRPG, OST4, TOMM7 and NOP10 was performed using the RefFinder tool. Results of four algorithms are shown, (b) Delta Ct, (c) BestKeeper, (d) NormFinder and (e) geNorm alongside their respective (f) geometric mean.
4. DISCUSSION
Tools for precise quantification of biomolecular EV cargo are instrumental to advance our understanding of EV‐associated cargo and ensure its translation in diagnostic or therapeutic applications. RT‐qPCR holds the potential to analyze the RNA content of EVs with high sensitivity, specificity and reproducibility but requires RGs for accurate data interpretation. In our study, we identify and validate four EV mRNA RGs (SNRPG, OST4, TOMM7 and NOP10) for the normalization of RT‐qPCR datasets. We demonstrate universal presence and stable abundance, rendering SNRPG, OST4, TOMM7 and NOP10 suitable for wide‐spread use across different cell lines, cellular conditions and biofluids (Figure S1).
We show that SNRPG, OST4, TOMM7 and NOP10 are present in a broad range of cells regardless of their origin (epithelial, fibroblast, cancer associated and normal cell lines) (Figure S1). While beyond the scope of our study, this suggests that SNRPG, OST4, TOMM7 and NOP10 also hold potential for normalization of RT‐qPCR data from cells or tissues. The presence of these four RGs in cells maintained under diverse culture medium conditions is fundamental, considering that a variety of media are used to prepare EVs from cells for in vitro and in vivo experimentation (medium with or without serum addition) or biopharmaceutical production (chemically defined medium) (Urzì et al., 2022).
SNRPG, OST4, TOMM7 and NOP10 are universally present in EV preparations derived from various human biomaterials, including cell lines, blood plasma, urine and tumour conditioned media as well as reference materials (Geeurickx et al., 2019). Interestingly, SNRPG, OST4, TOMM7 and NOP10 associate with particle size (i.e., larger EVs have lower Cq values) and particle number (i.e., higher particle concentrations have lower Cq values). Although Cq values can be indicative for particle size and particle number within a particular experiment, we recommend careful consideration of (pre/post)‐analytical variables (such as EV source, EV preparation protocol and RNA extraction efficiency) that may impact Cq values (Hulstaert et al., 2021). In addition, Cq values vary between biological replicates (i.e., different passages of source cells, biofluids from different donors), which is in concordance with previous studies showing occurrence of disparities in transcription even between cells from the same passage (Battich et al., 2015; Moignard & Göttgens, 2014). Upon DGUC, SNRPG, OST4, TOMM7 and NOP10 enrich in fractions corresponding to EV density, namely 1.09–1.11 g/mL. In addition, the abundance of SNRPG, OST4, TOMM7 and NOP10 in these particular density fractions does not significantly alter upon proteinase K and RNase treatment, confirming their presence inside EVs.
Four well‐established pipelines for RG identification and validation (miRDeepFinder) were implemented and identified that SNRPG, OST4, TOMM7 and NOP10 possess the required properties of RGs: (1) they are present in EVs of all cell lines and biofluids tested, including small, medium and large EV subpopulations – so they are constitutively present and abundant; and (2) they display stable presence in EVs upon perturbation of cell source or using different EV preparation protocols (Van Deun et al., 2014) – so their presence is robust across different conditions (Bustin et al., 2009). Proper RG selection strategies prior to relative quantification analysis is fundamental in any given RT‐qPCR experiment, as is recommended by the MIQE guidelines (Bustin et al., 2009), considering the variability of SNRPG, OST4, TOMM7 and NOP10 within biological replicates as well as differences in stability values used by different RefFinder algorithms (e.g., geNorm M‐value thresholds) (Hellemans et al., 2007).
Established cell line and tissue RGs such as GAPDH, YWHAZ, HMBS and SDHA are often used to normalize mRNA RT‐qPCR data in EV research (Abumaghaid et al., 2022; Yang et al., 2022). Similarly to SNRPG, OST4, TOMM7 and NOP10, two out of three established RGs appear to be detectable across a size range of subpopulations (Kruskal–Wallis test, adjusted p‐value for Dunns's multiple comparison test = 0.0219) (Figure S16A). However, we did not observe differential enrichment of GAPDH, YWHAZ, HMBS and SDHA in DGUC EV preparations (Figure S2); PK and RNase A treatment of density gradient fractions from CCM prepared of a representative cell line (HCT 116 ‐ Figure S16B) depicted equal Cq values in EV‐enriched and soluble protein fractions; this in contrast to SNRPG, OST4, TOMM7 and NOP10 (Figure 3b).
Why (i.e., function), how (i.e., actively or passively) and how many (i.e., copy number) of SNRPG, OST4, TOMM7 and NOP10 are universally and continuously sorted into EVs remains to be explored. Considering the short amplicon sizes (81–112 bp) analyzed in this study, future studies implementing full‐length RNA sequencing should be considered to assess whether or not SNRPG, OST4, TOMM7 and NOP10 are fragmented (Vermeulen et al., 2011). Indeed, early studies have hypothesized that EV mRNA is likely fragmented and results from untranslated regions (Batagov & Kurochkin, 2013; Nolte‐’t Hoen et al., 2012). Furthermore, considering the bulk approach implemented, the percentage of EVs containing SNRPG, OST4, TOMM7 and NOP10 remains unclear (O'Brien et al., 2020). Interestingly, the use of selected RGs may not be bound to a singular species (e.g., homo sapiens) with confirmed expression of selected RGs homologs by a variety of different species (e.g., Mus musculus – Figure S17). Of note, primer modifications may be required due to the homologous nature of selected RGs in some species. Lastly, considering that small non‐coding RNAs (e.g., U6) have been described to normalize EV microRNA data, the use of SNRPG, OST4, TOMM7 and NOP10 as normalizers for EV microRNA data can be further explored (Mestdagh et al., 2009). Considering that our study focuses on the study of EV‐associated mRNAs, future studies employing more biological replicates and sample sizes should be considered to address these technical and biological questions.
In conclusion, we have identified and validated SNRPG, OST4, TOMM7 and NOP10 as suitable RGs for the normalization of EV RT‐qPCR data. The availability and implementation of these universal and stably present EV RGs will be instrumental to advance the use of RT‐qPCR for the analysis of EV‐associated RNA cargo for research or clinical applications.
AUTHOR CONTRIBUTIONS
Cláudio Pinheiro: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; validation; visualization; writing—original draft; writing—review and editing. Niké Guilbert: Data curation; formal analysis; investigation; resources; writing—original draft; writing—review and editing. Lien Lippens: Formal analysis; investigation; resources; visualization; writing—review and editing. Quentin ROUX: Data curation; investigation; resources; visualization; writing—review and editing. Robin Boiy: Data curation; investigation; resources; visualization; writing—review and editing. Suzanne Fischer: Investigation; resources; writing—review and editing. Sofie Van Dorpe: Investigation; resources; writing—review and editing. Bram De Craene: Investigation; resources; software; writing—review and editing. Geert Berx: Writing—review and editing. Tom Boterberg: Investigation; resources; writing—review and editing. Gwen Sys: Resources; writing—review and editing. Hannelore Denys: Resources; writing—review and editing. Ilkka Miinalainen: Investigation; writing—review and editing. Pieter Mestdagh: Investigation; methodology; resources; software; writing—review and editing. Jo Vandesompele: Investigation; methodology; resources; software; writing—review and editing. Olivier De Wever: Conceptualization; funding acquisition; methodology; project administration; resources; supervision; writing—original draft; writing—review and editing. An Hendrix: Conceptualization; funding acquisition; methodology; project administration; resources; supervision; writing—original draft; writing—review and editing.
CONFLICT OF INTEREST STATEMENT
A.H., O.D.W., J.V. and P.M. are inventors on the patent application covering the rEV technology (WO2019091964).
Supporting information
Table S1: RNA measurements obtained after RNA isolation of 15 cell pellets cultured under different conditions. RNA purity was assessed by interpretation of A260/A280 ratios.
Table S2: Transcripts Per Million (TPM) of SNRPG, OST4, TOMM7 and NOP10 in the 13 selected cancer cell lines. Data was obtained through the Cancer Cell Line Encyclopedia (CCLE) database.
Table S3: Cq values obtained after RT‐qPCR analysis of EV derived from different cell lines (n = 6). RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S4: Cq values obtained after RT‐qPCR analysis of different EV size subpopulations. RTqPCR results are presented as average Cq values of three technical replicates.
Table S5: Cq values obtained after RT‐qPCR analysis of urinary EVs. RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S6: RT‐qPCR analysis of top, EV‐enriched and bottom fractions prepared from media conditioned by HCT 116 using DGUC. RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S7: Assessment of cell viability, cell count and total particle number (measured by nanoparticle tracking analysis (NTA)) for diverse cell sources.
Table S8: Nanoparticle tracking analysis (NTA) characterization, including mean size and particle number, of EVs prepared from diverse sources in three biological replicates.
Fig. S1: Literature analysis of state‐of‐the‐art articles (including this study) investigating EV reference genes.
Fig. S2: Enrichment of SNRPG, OST4, TOMM7 and NOP10 with high specificity in EVs prepared from medium conditioned by breast cancer cells using DGUC versus ExoQuick (EQ) or ultracentrifugation (UC).
Fig. S3: Evaluation of SNRPG, OST4, TOMM7 and NOP10 cellular expression in 15 cell lines by RT‐qPCR.
Fig. S4: Evaluation of HMBS, YWHAZ and SDHA cellular expression in 15 cell lines by RTqPCR.
Fig. S5: Characterization of a size range of EV subpopulations by transmission electron microscopy and western blot analysis.
Fig. S6: Characterization of DGUC fractions obtained from media cultured by HCT 116, CT5.3 hTeRT or MCF7 cells.
Fig. S7: Evaluation of the presence of SNRPG, OST4, TOMM7 and NOP10 in of EVs prepared from media conditioned by different cell lines (n = 9) using DGUC.
Fig. S8: Analysis of SNRPG, OST4, TOMM7 and NOP10 in serial dilutions of rEV.
Fig. S9: RT‐qPCR analysis of all density fractions prepared from media conditioned by LNCaP cells using DGUC.
Fig. S10: Functionality evaluation of proteinase K (PK) and RNase treatment by western blot and RT‐qPCR analysis.
Fig. S11: Evaluation of biological variability using EVs prepared from diverse origins in three biological replicates.
Fig. S12: Evaluation of technical variability within individual biological replicates using EVs prepared from media conditioned by SK‐OV‐3 and U‐87‐MG by DGUC in three technical replicates.
Fig. S13: Evaluation of biological variability using EVs prepared from diverse origins in three biological replicates.
Fig. S14: Stability analysis using the RefFinder tool for SNRPG, OST4, TOMM7, NOP10, YWHAZ, HMBS, SDHA, U6 and B2M in EVs.
Fig. S15: Stability analysis using the RefFinder tool for SNRPG, OST4, TOMM7 and NOP10 in EVs.
Fig. S16: Evaluation of cell and tissue reference genes GAPDH, YHWAZ and SDHA in EVs.
Fig. S17: Identification of OST4 in EVs prepared from media conditioned by murine ID8 ovarian cancer cells and 4T1 mammary cancer cells.
ACKNOWLEDGEMENTS
We thank Sofie De Geyter (Laboratory of Experimental Cancer Research, Department of Human Structure and Repair, Ghent University, Ghent, Belgium) and Biocenter Oulu Electron Microscopy core facility (a member of Biocenter Finland) for experimental support. This work was supported by European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie grant agreement No [722148], Ghent University, the Fund for Scientific Research‐Flanders and Kom op tegen Kanker (Stand up to Cancer), the Flemish cancer society.
Pinheiro, C. , Guilbert, N. , Lippens, L. , Roux, Q. , Boiy, R. , Fischer, S. , Van Dorpe, S. , De Craene, B. , Berx, G. , Boterberg, T. , Sys, G. , Denys, H. , Miinalainen, I. , Mestdagh, P. , Vandesompele, J. , De Wever, O. , & Hendrix, A. (2024). Identification and validation of extracellular vesicle reference genes for the normalization of RT‐qPCR data. Journal of Extracellular Vesicles, 13, e12421. 10.1002/jev2.12421
DATA AVAILABILITY STATEMENT
All data and material related to the conclusions are presented in this manuscript and/or the supplementary materials.
REFERENCES
- Abumaghaid, M. M. , Abdelazim, A. M. , Belali, T. M. , Alhujaily, M. , & Saadeldin, I. M. (2022). Shuttle transfer of mRNA transcripts via extracellular vesicles from male reproductive tract cells to the cumulus‐oocyte complex in rabbits (Oryctolagus cuniculus). Frontiers in Veterinary Science, 9, 816080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, C. L. , Jensen, J. L. , & Ørntoft, T. F. (2004). Normalization of real‐time quantitative reverse transcription‐PCR data: A model‐based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250. [DOI] [PubMed] [Google Scholar]
- Batagov, A. O. , & Kurochkin, I. V. (2013). Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3'‐untranslated regions. Biology Direct, 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battich, N. , Stoeger, T. , & Pelkmans, L. (2015). Control of transcript variability in single mammalian cells. Cell, 163, 1596–1610. [DOI] [PubMed] [Google Scholar]
- Bustin, S. A. , Benes, V. , Garson, J. A. , Hellemans, J. , Huggett, J. , Kubista, M. , Mueller, R. , Nolan, T. , Pfaffl, M. W. , Shipley, G. L. , Vandesompele, J. , & Wittwer, C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real‐time PCR experiments. Clinical Chemistry, 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Chen, M. , Xu, R. , Ji, H. , Greening, D. W. , Rai, A. , Izumikawa, K. , Ishikawa, H. , Takahashi, N. , & Simpson, R. J. (2016). Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Scientific Reports, 6, 38397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, M. , Kimura, M. , & Asari, S. (2012). Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncology Reports, 28, 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, E. , & Barrett, T. (2016). The gene expression omnibus database. Methods in Molecular Biology (Clifton, N.J.), 1418, 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossland, R. E. , Norden, J. , Bibby, L. A. , Davis, J. , & Dickinson, A. M. (2016). Evaluation of optimal extracellular vesicle small RNA isolation and qRT‐PCR normalisation for serum and urine. Journal of Immunological Methods, 429, 39–49. [DOI] [PubMed] [Google Scholar]
- De Vlieghere, E. , Gremonprez, F. , Verset, L. , Mariën, L. , Jones, C. J. , De Craene, B. , Berx, G. , Descamps, B. , Vanhove, C. , Remon, J. P. , Ceelen, W. , Demetter, P. , Bracke, M. , De Geest, B. G. , & De Wever, O. (2015). Tumor‐environment biomimetics delay peritoneal metastasis formation by deceiving and redirecting disseminated cancer cells. Biomaterials, 54, 148–157. [DOI] [PubMed] [Google Scholar]
- De Wever, O. , & Hendrix, A. (2019). A supporting ecosystem to mature extracellular vesicles into clinical application. The EMBO Journal, 38, e101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt, B. , Geeurickx, E. , Tulkens, J. , Van Deun, J. , Vergauwen, G. , Lippens, L. , Miinalainen, I. , Rappu, P. , Heino, J. , Ost, P. , Lumen, N. , De Wever, O. , & Hendrix, A. (2020). Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density‐based fractionation of urine. Journal of Extracellular Vesicles, 9, 1736935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt, B. , Lumen, N. , De Wever, O. , & Hendrix, A. (2020). Preparation of multi‐omics grade extracellular vesicles by density‐based fractionation of urine. STAR Protocols, 1, 100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt, B. , Pinheiro, C. , Geeurickx, E. , Tulkens, J. , Vergauwen, G. , Van Der Pol, E. , Nieuwland, R. , Decock, A. , Miinalainen, I. , Rappu, P. , Schroth, G. , Kuersten, S. , Vandesompele, J. , Mestdagh, P. , Lumen, N. , De Wever, O. , & Hendrix, A. (2023). Benchmarking blood collection tubes and processing intervals for extracellular vesicle performance metrics. Journal of Extracellular Vesicles, 12, e12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumax‐Vorzet, A. , Roboti, P. , & High, S. (2013). OST4 is a subunit of the mammalian oligosaccharyltransferase required for efficient N‐glycosylation. Journal of Cell Science, 126, 2595–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, E. , & Levanon, E. Y. (2013). Human housekeeping genes, revisited. Trends in Genetics: TIG, 29, 569–574. [DOI] [PubMed] [Google Scholar]
- Geeurickx, E. , & Hendrix, A. (2020). Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Molecular Aspects of Medicine, 72, 100828. [DOI] [PubMed] [Google Scholar]
- Geeurickx, E. , Lippens, L. , Rappu, P. , De Geest, B. G. , De Wever, O. , & Hendrix, A. (2021). Recombinant extracellular vesicles as biological reference material for method development, data normalization and assessment of (pre‐)analytical variables. Nature Protocols, 16, 603–633. [DOI] [PubMed] [Google Scholar]
- Geeurickx, E. , Tulkens, J. , Dhondt, B. , Van Deun, J. , Lippens, L. , Vergauwen, G. , Heyrman, E. , De Sutter, D. , Gevaert, K. , Impens, F. , Miinalainen, I. , Van Bockstal, P. J. , De Beer, T. , Wauben, M. H. M. , Nolte‐’t‐Hoen, E. N. M. , Bloch, K. , Swinnen, J. V. , van der Pol, E. , Nieuwland, R. , … Hendrix, A. (2019). The generation and use of recombinant extracellular vesicles as biological reference material. Nature Communications, 10, 3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorji‐Bahri, G. , Moradtabrizi, N. , Vakhshiteh, F. , & Hashemi, A. (2021). Validation of common reference genes stability in exosomal mRNA‐isolated from liver and breast cancer cell lines. Cell Biology International, 45, 1098–1110. [DOI] [PubMed] [Google Scholar]
- Gouin, K. , Peck, K. , Antes, T. , Johnson, J. L. , Li, C. , Vaturi, S. D. , Middleton, R. , de Couto, G. , Walravens, A. S. , Rodriguez‐Borlado, L. , Smith, R. R. , Marbán, L. , Marbán, E. , & Ibrahim, A. G. (2017). A comprehensive method for identification of suitable reference genes in extracellular vesicles. Journal of Extracellular Vesicles, 6, 1347019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran, P. M. , Luther, J. M. , & Byrd, J. B. (2019). For what factors should we normalize urinary extracellular mRNA biomarkers? Biomolecular Detection and Quantification, 17, 100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans, J. , Mortier, G. , De Paepe, A. , Speleman, F. , & Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biology, 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix, A. (2021). The nature of blood(y) extracellular vesicles. Nature Reviews. Molecular Cell Biology, 22, 243. [DOI] [PubMed] [Google Scholar]
- Hendrix, A. , Lippens, L. , Pinheiro, C. , Théry, C. , Martin‐Jaular, L. , Lötvall, J. , Lässer, C. , Hill, A. F. , & Witwer, K. W. (2023). Extracellular vesicle analysis. Nature Reviews Methods Primers, 3, 56. [Google Scholar]
- Hendrix, A. , Maynard, D. , Pauwels, P. , Braems, G. , Denys, H. , Van den Broecke, R. , Lambert, J. , Van Belle, S. , Cocquyt, V. , Gespach, C. , Bracke, M. , Seabra, M. C. , Gahl, W. A. , De Wever, O. , & Westbroek, W. (2010). Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. Journal of the National Cancer Institute, 102, 866–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinger, S. A. , Cha, D. J. , Franklin, J. L. , Higginbotham, J. N. , Dou, Y. , Ping, J. , Shu, L. , Prasad, N. , Levy, S. , Zhang, B. , Liu, Q. , Weaver, A. M. , Coffey, R. J. , & Patton, J. G. (2018). Diverse long RNAs are differentially sorted into extracellular vesicles secreted by colorectal cancer cells. Cell Reports, 25, 715–725. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert, E. , Decock, A. , Morlion, A. , Everaert, C. , Verniers, K. , Nuytens, J. , Nijs, N. , Schroth, G. P. , Kuersten, S. , Gross, S. M. , Mestdagh, P. , & Vandesompele, J. (2021). Messenger RNA capture sequencing of extracellular RNA from human biofluids using a comprehensive set of spike‐in controls. STAR Protocols, 2, 100475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka, A. , Zacco, E. , Temussi, P. A. , Tartaglia, G. G. , & Pastore, A. (2020). RNA as the stone guest of protein aggregation. Nucleic Acids Research, 48, 11880–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh, P. , Van Vlierberghe, P. , De Weer, A. , Muth, D. , Westermann, F. , Speleman, F. , & Vandesompele, J. (2009). A novel and universal method for microRNA RT‐qPCR data normalization. Genome Biology, 10, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs—PubMed. https://pubmed.ncbi.nlm.nih.gov/22290409/ [DOI] [PubMed]
- Moignard, V. , & Göttgens, B. (2014). Transcriptional mechanisms of cell fate decisions revealed by single cell expression profiling. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 36, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morhayim, J. , van de Peppel, J. , Dudakovic, A. , Chiba, H. , van Wijnen, A. J. , & van Leeuwen, J. P. (2017). Molecular characterization of human osteoblast‐derived extracellular vesicle mRNA using next‐generation sequencing. Biochimica et biophysica acta. Molecular Cell Research, 1864, 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte‐’t Hoen, E. N. , Buermans, H. P. , Waasdorp, M. , Stoorvogel, W. , Wauben, M. H. , & ’t Hoen, P. A. (2012). Deep sequencing of RNA from immune cell‐derived vesicles uncovers the selective incorporation of small non‐coding RNA biotypes with potential regulatory functions. Nucleic acids research, 40, 9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOP10 . NOP10 ribonucleoprotein [Homo sapiens (human)] ‐ Gene ‐ NCBI. https://www.ncbi.nlm.nih.gov/gene/55505
- O'Brien, K. , Breyne, K. , Ughetto, S. , Laurent, L. C. , & Breakefield, X. O. (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nature Reviews. Molecular Cell Biology, 21, 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OST4 oligosaccharyltransferase complex subunit 4, non‐catalytic [Homo sapiens (human)] ‐ Gene ‐ NCBI. https://www.ncbi.nlm.nih.gov/gene/100128731
- Pfaffl, M. W. , Tichopad, A. , Prgomet, C. , & Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel‐based tool using pair‐wise correlations. Biotechnology Letters, 26, 509–515. [DOI] [PubMed] [Google Scholar]
- Ragni, E. , Perucca Orfei, C. , De Luca, P. , Colombini, A. , Viganò, M. , Lugano, G. , Bollati, V. , & de Girolamo, L. (2019). Identification of miRNA reference genes in extracellular vesicles from adipose derived mesenchymal stem cells for studying osteoarthritis. International Journal of Molecular Sciences, 20, 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodosthenous, R. S. , Hutchins, E. , Reiman, R. , Yeri, A. S. , Srinivasan, S. , Whitsett, T. G. , Ghiran, I. , Silverman, M. G. , Laurent, L. C. , Van Keuren‐Jensen, K. , & Das, S. (2020). Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. Iscience, 23, 101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, N. , Best, S. , Jiang, J. , & Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real‐time PCR. BMC Molecular Biology, 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. D. , Patnam, S. , Koyyada, R. , Samal, R. , Alvi, S. B. , Satyanaryana, G. , Andrews, R. , Panigrahi, A. K. , Rengan, A. K. , Mudigonda, S. S. , Maitra, S. , & Sasidhar, M. V. (2022). Identifying stable reference genes in polyethene glycol precipitated urinary extracellular vesicles for RT‐qPCR‐based gene expression studies in renal graft dysfunction patients. Transplant Immunology, 75, 101715. [DOI] [PubMed] [Google Scholar]
- SNRPG small nuclear ribonucleoprotein polypeptide G [Homo sapiens (human)] ‐ Gene ‐ NCBI. https://www.ncbi.nlm.nih.gov/gene/6637
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach, M. , & Théry, C. (2016). Communication by extracellular vesicles: Where we are and where we need to go. Cell, 164, 1226–1232. [DOI] [PubMed] [Google Scholar]
- TOMM7 translocase of outer mitochondrial membrane 7 [Homo sapiens (human)] ‐ Gene ‐ NCBI. https://www.ncbi.nlm.nih.gov/gene/54543
- Tulkens, J. , De Wever, O. , & Hendrix, A. (2020). Analyzing bacterial extracellular vesicles in human body fluids by orthogonal biophysical separation and biochemical characterization. Nature Protocols, 15, 40–67. [DOI] [PubMed] [Google Scholar]
- Urzì, O. , Olofsson Bagge, R. , & Crescitelli, R. (2022). The dark side of foetal bovine serum in extracellular vesicle studies. Journal of Extracellular Vesicles, 11, e12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. , & Speleman, F. (2002). Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , Berx, G. , Boere, J. , Boukouris, S. , Bremer, M. , Buschmann, D. , Byrd, J. B. , Casert, C. , Cheng, L. , Cmoch, A. , … Hendrix, A. (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14, 228–232. [DOI] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Sormunen, R. , Cocquyt, V. , Vermaelen, K. , Vandesompele, J. , Bracke, M. , De Wever, O. , & Hendrix, A. (2014). The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. Journal of Extracellular Vesicles, 3(1), 24858, 10.3402/jev.v3.24858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorpe, S. , Lippens, L. , Boiy, R. , Pinheiro, C. , Vergauwen, G. , Rappu, P. , Miinalainen, I. , Tummers, P. , Denys, H. , De Wever, O. , & Hendrix, A. (2023). Integrating automated liquid handling in the separation workflow of extracellular vesicles enhances specificity and reproducibility. Journal of Nanobiotechnology, 21, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel, G. , Carter, D. R. F. , Clayton, A. , Lambert, D. W. , Raposo, G. , & Vader, P. (2022). Challenges and directions in studying cell‐cell communication by extracellular vesicles. Nature Reviews. Molecular Cell Biology, 23, 369–382. [DOI] [PubMed] [Google Scholar]
- Vergauwen, G. , Dhondt, B. , Van Deun, J. , De Smedt, E. , Berx, G. , Timmerman, E. , Gevaert, K. , Miinalainen, I. , Cocquyt, V. , Braems, G. , Van den Broecke, R. , Denys, H. , De Wever, O. , & Hendrix, A. (2017). Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Scientific Reports, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergauwen, G. , Tulkens, J. , Pinheiro, C. , Avila Cobos, F. , Dedeyne, S. , De Scheerder, M. A. , Vandekerckhove, L. , Impens, F. , Miinalainen, I. , Braems, G. , Gevaert, K. , Mestdagh, P. , Vandesompele, J. , Denys, H. , De Wever, O. , & Hendrix, A. (2021). Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. Journal of Extracellular Vesicles, 10, e12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, J. , De Preter, K. , Lefever, S. , Nuytens, J. , De Vloed, F. , Derveaux, S. , Hellemans, J. , Speleman, F. , & Vandesompele, J. (2011). Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Research, 39, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, K. C. , Palmisano, B. T. , Shoucri, B. M. , Shamburek, R. D. , & Remaley, A. T. (2011). MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nature Cell Biology, 13, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Gao, J. , Gao, F. , Zhao, Y. , Deng, B. , Mu, X. , & Xu, L. (2022). Extracellular vesicles‐encapsulated microRNA‐29b‐3p from bone marrow‐derived mesenchymal stem cells promotes fracture healing via modulation of the PTEN/PI3K/AKT axis. Experimental Cell Research, 412, 113026. [DOI] [PubMed] [Google Scholar]
- Yu, S. , Li, Y. , Liao, Z. , Wang, Z. , Wang, Z. , Li, Y. , Qian, L. , Zhao, J. , Zong, H. , Kang, B. , Zou, W. B. , Chen, K. , He, X. , Meng, Z. , Chen, Z. , Huang, S. , & Wang, P. (2020). Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut, 69, 540–550. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , He, F. , Hu, S. , & Yu, J. (2008). On the nature of human housekeeping genes. Trends in Genetics: TIG, 24, 481–484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: RNA measurements obtained after RNA isolation of 15 cell pellets cultured under different conditions. RNA purity was assessed by interpretation of A260/A280 ratios.
Table S2: Transcripts Per Million (TPM) of SNRPG, OST4, TOMM7 and NOP10 in the 13 selected cancer cell lines. Data was obtained through the Cancer Cell Line Encyclopedia (CCLE) database.
Table S3: Cq values obtained after RT‐qPCR analysis of EV derived from different cell lines (n = 6). RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S4: Cq values obtained after RT‐qPCR analysis of different EV size subpopulations. RTqPCR results are presented as average Cq values of three technical replicates.
Table S5: Cq values obtained after RT‐qPCR analysis of urinary EVs. RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S6: RT‐qPCR analysis of top, EV‐enriched and bottom fractions prepared from media conditioned by HCT 116 using DGUC. RT‐qPCR results are presented as average Cq values of three technical replicates.
Table S7: Assessment of cell viability, cell count and total particle number (measured by nanoparticle tracking analysis (NTA)) for diverse cell sources.
Table S8: Nanoparticle tracking analysis (NTA) characterization, including mean size and particle number, of EVs prepared from diverse sources in three biological replicates.
Fig. S1: Literature analysis of state‐of‐the‐art articles (including this study) investigating EV reference genes.
Fig. S2: Enrichment of SNRPG, OST4, TOMM7 and NOP10 with high specificity in EVs prepared from medium conditioned by breast cancer cells using DGUC versus ExoQuick (EQ) or ultracentrifugation (UC).
Fig. S3: Evaluation of SNRPG, OST4, TOMM7 and NOP10 cellular expression in 15 cell lines by RT‐qPCR.
Fig. S4: Evaluation of HMBS, YWHAZ and SDHA cellular expression in 15 cell lines by RTqPCR.
Fig. S5: Characterization of a size range of EV subpopulations by transmission electron microscopy and western blot analysis.
Fig. S6: Characterization of DGUC fractions obtained from media cultured by HCT 116, CT5.3 hTeRT or MCF7 cells.
Fig. S7: Evaluation of the presence of SNRPG, OST4, TOMM7 and NOP10 in of EVs prepared from media conditioned by different cell lines (n = 9) using DGUC.
Fig. S8: Analysis of SNRPG, OST4, TOMM7 and NOP10 in serial dilutions of rEV.
Fig. S9: RT‐qPCR analysis of all density fractions prepared from media conditioned by LNCaP cells using DGUC.
Fig. S10: Functionality evaluation of proteinase K (PK) and RNase treatment by western blot and RT‐qPCR analysis.
Fig. S11: Evaluation of biological variability using EVs prepared from diverse origins in three biological replicates.
Fig. S12: Evaluation of technical variability within individual biological replicates using EVs prepared from media conditioned by SK‐OV‐3 and U‐87‐MG by DGUC in three technical replicates.
Fig. S13: Evaluation of biological variability using EVs prepared from diverse origins in three biological replicates.
Fig. S14: Stability analysis using the RefFinder tool for SNRPG, OST4, TOMM7, NOP10, YWHAZ, HMBS, SDHA, U6 and B2M in EVs.
Fig. S15: Stability analysis using the RefFinder tool for SNRPG, OST4, TOMM7 and NOP10 in EVs.
Fig. S16: Evaluation of cell and tissue reference genes GAPDH, YHWAZ and SDHA in EVs.
Fig. S17: Identification of OST4 in EVs prepared from media conditioned by murine ID8 ovarian cancer cells and 4T1 mammary cancer cells.
Data Availability Statement
All data and material related to the conclusions are presented in this manuscript and/or the supplementary materials.


