Abstract
A chimpanzee (C-499) infected for more than 9 years with two subtype B isolates of human immunodeficiency virus type 1 (HIV-1), one (HIV-1SF2) that replicates poorly and one (HIV-1LAV-1b) that replicates efficiently in chimpanzees, died of AIDS 11 years after initial infection (F. J. Novembre et al., J. Virol. 71:4086–4091, 1997). Nucleotide sequence and phylogenetic analyses of the C2 to V5 region of env (C2-V5env) in proviral DNA from peripheral blood lymphocytes obtained 22 months before death revealed two distinct virus populations. One of these populations appeared to be a recombinant in env, having the V3 loop from HIV-1SF2 and the V4-V5 region from HIV-1LAV-1b; the other population had evolved from HIV-1LAV-1b. In addition to C2-V5env, the entire p17gag and nef genes were sequenced; however, based on nucleotide sequences and phlyogeny, whether the progenitor of the p17gag and nef genes was SF2 or LAV-1b could not be determined. Compared to the two original viruses, the divergence of all clones of C2-V5env ranged from 9.37 to 20.2%, that of p17gag ranged from 3.11 to 9.29%, and that of nef ranged from 4.02 to 7.9%. In contrast, compared to the maximum variation of 20.2% in C2-V5env for C-499, the maximum diversities in C2-V5env in proviruses from two chimpanzees infected with HIV-1LAV-1b for 9 and 10 years were 9.65 and 2.48%, respectively. These results demonstrate that (i) two distinct HIV-1 populations can coexist and undergo extensive diversification in chimpanzees with progressive HIV-1-induced disease and (ii) recombination between two subtype B strains occurred even though the second strain was inoculated 15 months after the first one. Furthermore, evaluation of env genes from three chimpanzees infected with the same strain suggests that the magnitude of HIV-1 diversification could be related to higher viral burdens, manifestations of disease, and/or dual infection.
The only nonhuman primate species that can be reliably infected with multiple isolates of human immunodeficiency virus type 1 (HIV-1) is the chimpanzee (Pan troglodytes). Compared to primary HIV-1 infection in humans, the virus can be transmitted to chimpanzees by the same routes, viral burdens in peripheral blood and lymph nodes peak during the first 4 to 8 weeks, and detection of HIV-1-specific immune responses coincides with partial clearance of free virus from plasma and decreased numbers of infected cells in blood and lymph nodes (1, 18, 21). Although chimpanzees have become infected after exposure to multiple clade (subtype) B isolates as well as isolates from other clades (3, 16, 21, 52, 64), in general, long-term persistent infections can be established only with HIV-1 strains classified as dualtropic and syncytium inducing (SI) (3, 17, 64). This limitation to productive infection with HIV-1 SI strains in vivo is also true in vitro (17, 63, 64). The only exception appears to be a non-SI strain that was isolated from a naturally infected chimpanzee (57). Despite the association in humans of HIV-1 strains having the SI phenotype with loss of CD4+ cells and progression to AIDS (62), reports of HIV-1-related disease in infected chimpanzees have been rare (1, 23, 39, 51). Because of this failure to develop AIDS by a subset of more than 150 chimpanzees that have been infected with HIV-1 for up to 13 years, many investigators feel that HIV-1 infection of chimpanzees is not a good model to use for evaluating candidate vaccines. However, immunization with HIV-1 antigens and challenge of chimpanzees, with either cell-free or cell-associated, homologous or heterologous virus, have provided valuable information about the feasibility of certain vaccine approaches and their protective efficacy (4–6, 22, 28–31).
Some possible reasons for lack of HIV-1-induced disease in chimpanzees include innate resistance to the pathogenic effects of HIV-1; lack of cytopathic effects of HIV-1 on lymphocytes; high levels of CD8+ lymphocytes (normal CD4/CD8 ratio of approximately 1.0) that mediate suppression of HIV-1 replication (8, 38); no virus-mediated aberrant effects on lymphocytes, such as induction of anergy and apoptosis (15, 32–34, 63); limited exposure to other pathogens due to isolation in restricted-access housing; and infection with HIV-1 strains having limited pathogenicity for chimpanzees. However, there is now evidence that most of these reasons are not valid, and only the last two possibilities—limited exposure to other pathogens and inherent properties of the HIV-1 strains with which chimpanzees have been inoculated—are likely to be relevant. The recent report of a chimpanzee (C-499) euthanized because of HIV-1-related severe immunodeficiency and AIDS, as defined by the Centers for Disease Control and Prevention, demonstrated that this species can develop AIDS (54). Furthermore, virus transfused in blood from C-499 to a naive chimpanzee induced a rapid decline in numbers of CD4+ lymphocytes, indicating that this strain, called HIV-1JC, was pathogenic for chimpanzees (54).
Chimpanzee C-499, which succumbed to AIDS, had been infected with the HIV-1SF2 strain for 15 months when it was exposed intravenously to a high dose of and became infected with HIV-1LAV-1b (Fig. 1) (24). This dually infected chimpanzee was inoculated 6 months later with a third HIV-1 strain, NDK, now known to belong to subtype D (53); however, no evidence that the third strain actually established infection was obtained. (This early study was performed before PCR was used routinely for genetic analysis.) Approximately 1 year after exposure to HIV-1NDK, chimpanzee C-499 was immune stimulated by intramuscular inoculation of purified p53gag formulated in adjuvant (20). This inoculation coincided with development of severe lymphopenia, characterized by a significant decline in CD4+ peripheral blood lymphocytes (to a nadir of 134 cells per μl of blood), loss of lymphocyte proliferative responses to mitogens, and thrombocytopenia; all of these conditions persisted for more than 1 year and then gradually returned to normal levels (23). When C-499 was euthanized, it had been infected with HIV-1SF2 for almost 11 years and with HIV-1LAV-1b for about 9.5 years. We report here the genetic characterization of the HIV-1 quasispecies in peripheral blood mononuclear cells (PBMC) obtained from this dually infected chimpanzee 22 months before its death, provide evidence for recombinant genomes, and compare its quasispecies with those present in two chimpanzees infected only with HIV-1LAV-1b for a comparable time.
FIG. 1.
Inoculation history of chimpanzee C-499 (24). The asterisk indicates the time at which blood was obtained from C-499 for the studies reported here. The virus identified as HIV-1JC by Novembre et al. (54) was obtained, as indicated, when C-499 had clinical AIDS.
MATERIALS AND METHODS
Chimpanzees and virus strains.
Three chimpanzees that had been infected with HIV-1 for either 9 or 10 years were evaluated; two of these animals (C-459 and C-499) were housed at the Yerkes Regional Primate Research Center, and the other one (C-487) was housed at the Laboratory for Experimental Medicine and Surgery in Primates (LEMSIP), New York University. C-459 had been inoculated with the first HIV-1 isolate, originally designated LAV-1 and later LAI (68), before it was passaged in T-cell lines (21). Approximately 5 months after inoculation, whole blood from C-459 was transfused into chimpanzee C-463, and 2 weeks later, when virus was isolated from C-463’s PBMC by coculture with normal human PBMC, a large stock of virus was cryopreserved and designated LAV-1b; this virus stock, which was passaged only in primary PBMC, was used to inoculate both C-499 and C-487 in separate studies (19, 21). At the time C-499 was inoculated with HIV-1LAV-1b, it had been infected with HIV-1SF2 for 15 months (Fig. 1); however, the secondary infection was not prevented, and C-499 became persistently coinfected with two HIV-1 strains (24). Housing and care of chimpanzees were provided according to institutional guidelines and standard practices for the humane care and use of chimpanzees in biomedical research. Animals were anesthetized with ketamine hydrochoride for all blood collections. (Note that chimpanzee C-487, housed at LEMSIP, was identified previously in reference 19; this animal is not the same C-487 housed at Yerkes and described in reference 21.)
Virus cultures.
To isolate virus from chimpanzee PBMC, approximately 107 cells were cocultured with normal human PBMC that had been stimulated for 3 days with phytohemagglutinin (PHA)-P as described previously (21). Virus replication was monitored by the production of reverse transcriptase activity in cell-free culture supernatants. To determine the least number of CD4+ lymphocytes required to isolate virus from PBMC, CD8+ cells were removed by using Dynabeads coated with antibodies to CD8. Aliquots of 10-fold serial dilutions of the CD4+-enriched cells were added in replicates of six to phytohemagglutinin-stimulated human PBMC in 24-well plates and monitored as described above. The genetic analysis of proviruses in cultured PBMC was done with DNA extracted after 4 weeks of culture.
Genetic analysis.
Proviral DNA was amplified by nested PCR in three regions of the HIV-1 genome: a fragment spanning the C2 to V5 regions of the env gene (C2-V5env), and the entire p17gag and nef genes. Genomic DNA from cultured and uncultured chimpanzee PBMC was extracted with a QIAamp blood kit (Qiagen, Chatsworth, Calif.) and used as templates for nested PCR in a PTC-100 Programmable Thermal Cycler (MJ Research). First-round amplifications were done with 0.25 to 1 μg of genomic DNA for 25 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. A 5-μl (or 1/10 volume) aliquot of the first-round PCR products was subjected to a second round of PCR for 35 cycles. The positions of all primers are given relative to bases in the HIV-1HXB2 molecular clone in the HIV-1 database (53). The outer (p17/1 and p17/2) and inner (p17/3 and p17/4) primer pairs used to amplify 443 bp encoding p17gag were as follows: p17/1 (bases 681 to 704), 5′-TCTCGACGCAGGACTCGGCTTGCT-3′); p17/2 (bases 1242 to 1219), 5′-AAGTTCTAGGTGATATGGCCTGAT-3′; p17/3 (bases 757 to 780), 5′-AATTTTGACTAGCGGATCCTAGAA-3′); and p17/4 (bases 1199 to 1177), 5′-GTTCTGCAGTATAGGGTAATTTT-3′). The inner (D and F) and outer (C and H) primer pairs used to amplify a 681-bp fragment spanning C2-V5env were reported previously (25). For the entire nef gene, a 769-bp fragment was amplified with the outer (nef1 and nef2) and inner (nef3 and nef4) primer pairs: nef1 (bases 8682 to 8707), 5′-TAGCAGTAGCTGAGGGGACAGATAGG-3′; nef2 (bases 9557 to 9532), 5′-TGGTCTAACCAGAGAGACCCAGTACA-3′; nef3 (bases 8749 to 8775), 5′-ATACCTAGAAGAATAAGACAGGGCTT-3′; and nef4 (bases 9518 to 9496), 5′-TGCTTATATGCAGGATCTGAGGG-3′. The resultant PCR products were separated on 1% agarose gels stained with ethidium bromide, and the bands were excised; the DNA fragments were purified by using a Qiagen gel extraction kit and then cloned into the pCRII vector (75), using conditions specified by the manufacturer (Invitrogen, San Diego, Calif.). After transformation into competent Escherichia coli cells, plasmid DNA from independent bacterial colonies was prepared by using a Qiaprep 8 miniprep kit (Qiagen) and then was digested with EcoRI to check the inserts.
To estimate the minimum number of proviruses in 1 μg of genomic DNA (equivalent to DNA from approximately 1.5 × 105 cells), 1 μg of purified DNA was serially diluted 1:2 and added to DNA from a normal chimpanzee to give a total of 1 μg of DNA. At least two independent, nested PCRs were performed with each DNA mixture, and the last dilution to yield at least one positive reaction was considered to contain a minimum of one provirus. The products of each dilution were analyzed for heterogeneity by heteroduplex mobility assay (HMA), and we also considered the number of unique banding patterns present for each dilution of DNA when estimating the minimum number of proviruses present in a sample. For example, if a 1:32, but not a 1:64, dilution of 1 μg of DNA was PCR positive, and two distinct HMA patterns were seen, then the 1 μg or 1.5 × 105 cellular genomic equivalents was considered to contain a minimum of 64 proviruses, or approximately one provirus per 2,344 cells.
To minimize the possibility of contamination during PCR, positive-displacement pipettors with filtered tips were used, and reactions were performed in a sterile biosafety hood in a separate room away from the main laboratory. That no sequences similar to those reported here have been amplified from DNA samples from other HIV-1-infected chimpanzees during hundreds of PCRs in our laboratory indicates that the sequences are unique to these animals.
DNA heteroduplex assay.
To assess the diversity of clones generated by PCR, 60 clones containing the p17gag gene and 83 clones containing C2-V5env were selected for analysis by HMA, performed essentially as described by Delwart et al. (12, 13) except that we included 2.7 M urea to stabilize mismatched heteroduplexes. The HMA was used primarily as a prescreen to ensure that divergent clones were sequenced and that the extent of diversity was as accurate as possible. One clone each from env and gag (clone 6 for C2-V5env and clone 1C for p17gag) was selected at random as a probe to evaluate the other clones for heterogeneity. Plasmid DNA (5 ng) from each cloned fragment or 2.5 μl of the lysed (heated) bacterial culture was amplified again by PCR to provide sufficient product, and then equal volumes of the PCR products of the probe and other TA clones were used for the HMA. After denaturation at 95°C and slow cooling of the mixtures to room temperature to allow reannealing, aliquots of the reactions were electrophoresed on 5% polyacrylamide gels containing 2.7 M urea. Gels were stained with ethidium bromide, and heteroduplexes were visualized under UV light.
DNA sequencing and phylogenetic analysis.
Nucleotide sequences of fragments cloned from plasmid DNA were determined by the standard dideoxynucleotide chain termination method, using Sequenase version 2 (U.S. Biochemicals, Cleveland, Ohio) according to the manufacturer’s protocol. For sequencing p17gag and C2-V5env, the two inner PCR primers were used to sequence in both forward and reverse directions. In the gag gene, an overlap of approximately 50 bp was read. One internal primer was required for env; this primer was B3A (5′-GCACAGTTTTAATTGTGGAG-3′ [bases 7342 to 7361 in the HXB2 molecular clone]), and 10- to 20-base overlaps were generated. For the nef gene, three forward primers, which allowed overlaps of approximately 20 bases to be read, were used. These primers, also based on the HXB2 molecular clone, were nef0 (5′-CTTGGAAAGGATTTTGCTATA-3′ [bases 8773 to 8793]), nef5 (5′-TGGCTAGAAGCACAAGAGGA-3′ [bases 8964 to 8983]), and nef7 (5′-AGCTAGTACCAGTTGAGCCA-3′ [bases 9226 to 9245]).
The parental HIV-1LAI sequences were used for all analyses because DNA sequences of 10 clones of the region surrounding the V3 loop of the LAV-1b virus stock were identical to those of the original isolate (data not shown), demonstrating limited diversification during the 5 months of passage in C-459 before this stock was prepared. Translation to amino acids and amino acid sequence alignments were done with MacVector version 5.0 (Eastman Kodak Co., New Haven, Conn.), with minor manual adjustments. Genetic distances were determined by pairwise comparisons using the two-parameter method of Kimura, excluding gaps caused by insertions or deletions (PHYLIP package, version 3.572). The neighbor-joining method was used to analyze sequence relationships and to construct phylogenetic trees, which were evaluated statistically by 100 bootstrap replicates (PHYLIP).
Nucleotide sequence accession numbers.
GenBank accession numbers for the env sequences from C-499 are U56866 to U56887 and AF027771 to AF027785, those for p17gag are U56888 to U56899, and those for nef are AF027786 to AF027806.
RESULTS
Status of chimpanzees.
Approximately 1 year before diagnosis of AIDS and 9 years after initial infection with HIV-1SF2 (Fig. 1), peripheral blood was obtained from C-499. At the same time, blood was collected from another chimpanzee, C-459, that had been infected for more than 10 years with HIV-1LAI(LAV); C-459 was the animal through which the LAI(LAV) strain had been passaged for 5 months before the LAV-1b virus stock that was used to inoculate C-499 was generated (21). Also included in the study was chimpanzee C-487, which had been infected for 9 years with an aliquot of the same HIV-1LAV-1b stock with which C-499 was inoculated. At the time blood was collected from each animal, anti-HIV-1 antibody titers, viral burdens, and percentages of CD4+ and CD8+ T cells were determined (Table 1). C-487 had the highest levels of both antibody titers and copies of virion RNA in plasma. C-499 also had high antibody titers and a CD4/CD8 ratio of 0.24, which was comparable to that of C-487. The CD4/CD8 ratio for the third animal, C-459, was normal. At the time this analysis was done, HIV-1 was isolated from PBMC from C-499 and C-487 but not from C-459. Serial dilutions of purified CD4+ lymphocytes from C-499 indicated that 104 CD4+ T lymphocytes contained a minimum of one infectious cell.
TABLE 1.
Status of chimpanzees at time of genetic analysis
| Chimp | No. of yr infected | Anti-HIV antibody titera | No. of copies of RNA/mlb | % CD4 | % CD8 | CD4/CD8 ratio |
|---|---|---|---|---|---|---|
| C-499 | 9 | 204,800 | 724c | 15.7 | 66.2 | 0.24 |
| C-459 | 10 | 25,600 | 740c | 31.0 | 34.0 | 0.91 |
| C-487 | 9 | 819,200 | 12,900 | 16.5 | 70.9 | 0.23 |
HIV-1-specific antibody titers were defined as the reciprocal of the last serum dilution giving an optical density reading above the cutoff value and were determined with Genetic Systems LAV EIA kit (Sanofi Diagnostics Pasteur, Seattle, Wash.) according to the manufacturer’s instructions.
The number of copies of HIV-1 genomic RNA in plasma was determined with the Amplicor HIV Monitor kit, a quantitative reverse transcription-PCR assay, according to the manufacturer’s instructions (Roche Diagnostic Systems, Branchburg, N.J.).
Values are likely to be below the actual number of copies/milliliter because the plasma samples contained heparin which can interfere with the assay.
Genetic analysis of p17gag.
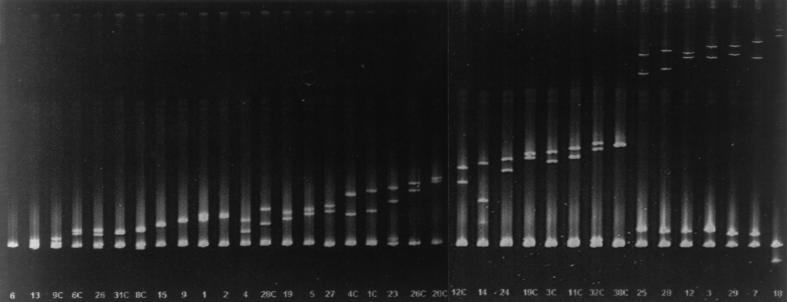
For p17gag, one of the most variable proteins encoded in HIV-1 gag (44), HMA of 17 clones PCR-amplified from approximately 1.5 × 105 uncultured PBMC from C-499 revealed nine different heteroduplex banding patterns (Fig. 2), with six patterns represented by one clone each. PCR amplification of p17gag from cocultured PBMC followed by HMA resulted in seven HMA patterns, three of which were not represented by clones from uncultured PBMC (Fig. 2, lanes 1C, 38C, and 36C). (Note that clones from cultured PBMC are indicated by a C after the clone number.) That the 12 HMA patterns identified in the 53 p17gag clones analyzed reflected the true extent of diversity is supported by data obtained from the analysis of the C2-V5env region, which indicated that at least 36 distinct proviruses were present (see below). Interestingly, the HMA pattern represented by the most clones (6 of 17 clones, or 35%) from uncultured PBMC also was most prevalent (17 of 36 clones, or 47%) among those from cultured PBMC (represented by clone 39C). Since the most diverse HMA pattern (Fig. 2, lane 8), relative to the probe, was represented by clones from both cultured and uncultured PBMC, the extents of diversity in the two PBMC populations were similar. Nucleotide sequencing of two clones with the same HMA pattern revealed that the difference between these two clones was less than the nucleotide differences between clones with different patterns; for example, clone 39C differed from the probe, clone 1C, by only 2 bp (Fig. 2). For this reason, only one clone representing each pattern was selected for nucleotide sequence analysis.
FIG. 2.
HMA of PCR amplicons of the p17gag gene from PBMC of C-499. Clone 1C was chosen at random and used as the probe; the first lane is the 1C homoduplex control. Each lane represents heteroduplexes of individual clones with 1C, ordered by increasing diversity. Clone numbers followed by a “C” indicate amplification from cultured PBMC, whereas numbers alone identify clones from uncultured PBMC. The top of the figure is coincident with the top of the gel.
Pairwise comparisons of each DNA sequence with all others, including those of the original infecting viruses, revealed that the percent nucleotide distances of the 12 p17gag sequences relative to SF2 and LAI were equivalent (Table 2); the intraclone distance ranged from 0.45 to 6.3%. (The nucleotide difference between p17gag encoded in the parental HIV-1 strains SF2 and LAI is 4.17%.) Although these comparisons did not allow the parentage of the clones, that is, whether they were derived from the SF2 or LAV-1b strain, to be determined, all of the clones contained a 6-bp (two-amino-acid) insertion near the 3′ end of the p17gag gene in the SF2 strain (Fig. 3). These six bases are not in LAI and are rarely found in other subtype B strains (53). In an attempt to clarify the relationships, a phylogenetic tree of the p17gag clones was constructed (Fig. 4). Included in the analysis were sequences from HIV-1 strains LAI and SF2, seven clade B strains chosen at random from the database (53), and the clade D strain NDK, to which C-499 also was exposed. Although all of the p17gag sequences branched with the HIV-1LAI strain, the bootstrap value for 100 trees was only 26; therefore, the parental virus could not be identified with certainty.
TABLE 2.
Genetic distances between HIV-1JC499 and the parental strains, LAI(LAV) and SF2
| Gene | Groupa | Genetic distance (%)
|
||
|---|---|---|---|---|
| LAI | SF2 | Intraclone | ||
| p17gag | All | 3.11–8.3 | 4.63–9.29 | 0.45–6.3 |
| C2-V5env | All | 9.54–15.2 | 9.37–20.2 | 0.78–19.28 |
| Minor | 9.54–11.47 | 9.37–11.4 | 1.4–4.48 | |
| Major | 11.98–15.2 | 16.13–20.2 | 0.78–8.47 | |
| 5′-C2-V3 | Minor | 12.6–15.33 | 6.1–9.42 | |
| Major | 13.25–15.56 | 18.52–21.91 | ||
| V4-V5-3′ | Minor | 7.4–9.42 | 11.6–14.92 | |
| Major | 9.74–15.55 | 12.93–20.25 | ||
| nef | All | 6.33–7.9 | 4.02–5.55 | 0.16–3.81 |
With respect to whether all clones or, in the case of C2-V5env, only a subset of clones were analyzed. Minor, the C2-V5env group represented by the smaller number of clones; major, the larger group of clones (Fig. 6).
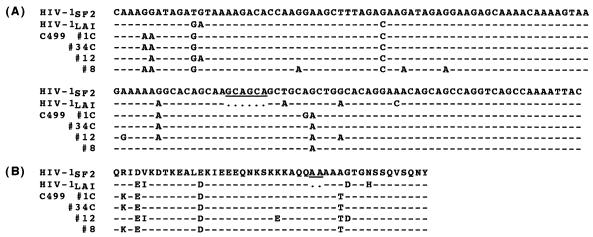
FIG. 3.
Nucleotide (A) and amino acid (B) sequences of the 3′ termini of the p17gag genes of HIV-1SF2, HIV-1LAI, and representative clones from C-499. Dashes indicate identity with HIV-1SF2, and dots indicate deletions. The six base pairs (two amino acids) in HIV-1SF2 that are not present in HIV-1LAI are underlined.
FIG. 4.
Phylogenetic relationship of the p17gag nucleotide sequences of HIV-1LAI and HIV-1SF2 to clones derived from proviral DNA in cultured (denoted by a “C”) and uncultured PBMC from chimpanzee C-499. The tree was rooted to the clade D strain NDK and includes seven HIV-1 clade B strains chosen at random from the database (53). Only bootstrap values greater than 50 are shown at nodes, unless the text refers to specific nodes with lower bootstrap values. Horizontal branch lengths reflect the genetic distance between sequences.
Genetic analysis of C2-V5env.
Because the diversity in env genes is generally greater than that in gag, 30 TA clones, generated by PCR from proviral DNA isolated from uncultured PBMC, were analyzed by HMA to estimate the extent of genetic diversity in C2-V5env. HMA of these 30 clones revealed 21 distinct patterns; 16 patterns were represented by only one clone, four patterns were represented by two clones, and the most prevalent group was represented by five clones (24%), indicating that the majority of PCR products were amplified from different proviruses. Furthermore, these HIV-1 proviruses in C-499’s uncultured PBMC formed two distinct populations: the larger population was represented by 21 clones (70%), and the smaller population was represented by 9 clones (30%). Proviral DNA was also isolated from C-499’s PBMC that had been cocultured with normal human PBMC for 15 days; all 53 clones from two independent PCRs were related to the major population of clones identified among proviruses in the uncultured PBMC. In contrast to the clones from uncultured PBMC, 19 (36%) of the 53 clones from cultured PBMC had essentially the same HMA pattern, indicating that one variant (represented by one clone among the uncultured PBMC PCR products) predominated during culture (Fig. 5, clone 3c). However, 14 additional HMA patterns not identified among the uncultured PBMC were observed after culture; 12 of these 14 patterns were represented by a single clone. That most HMA patterns from both uncultured and cultured PBMC were represented by only one or two clones illustrates the extreme diversity of C-499’s quasispecies and that most clones were derived independently.
FIG. 5.
Heteroduplex mobility assay of PCR amplicons of C2-V5env from cultured and uncultured PBMC of C-499. Clone 6 was chosen at random and used as the probe; the first lane shows the clone 6 homoduplex control. Each lane represents heteroduplexes of clone 6 with individual clones, ordered by increasing diversity. Two distinct populations were identified; the major and minor groups are seen in the first 29 lanes and the last 7 lanes, respectively. The heteroduplex banding pattern for clone 18 reflects a deletion in this region.
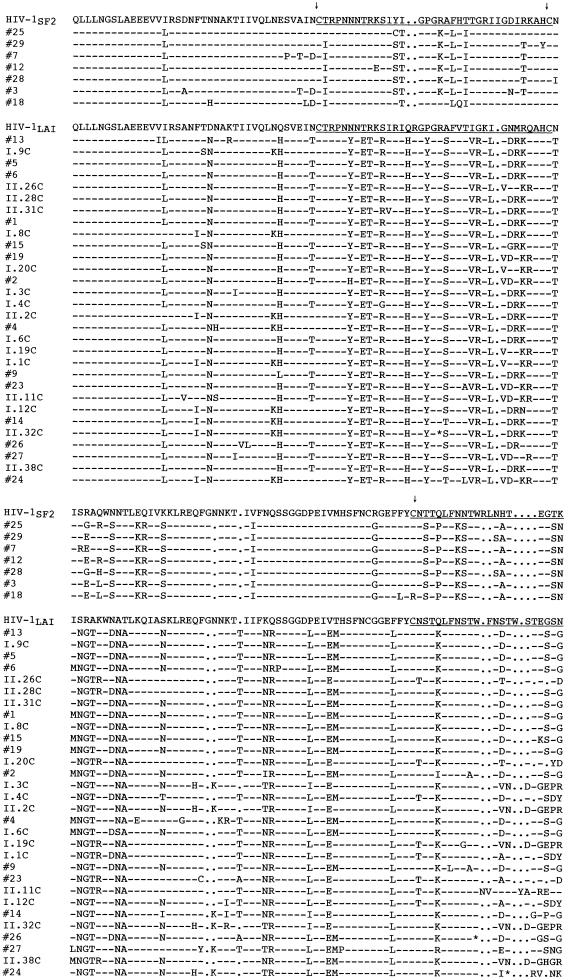
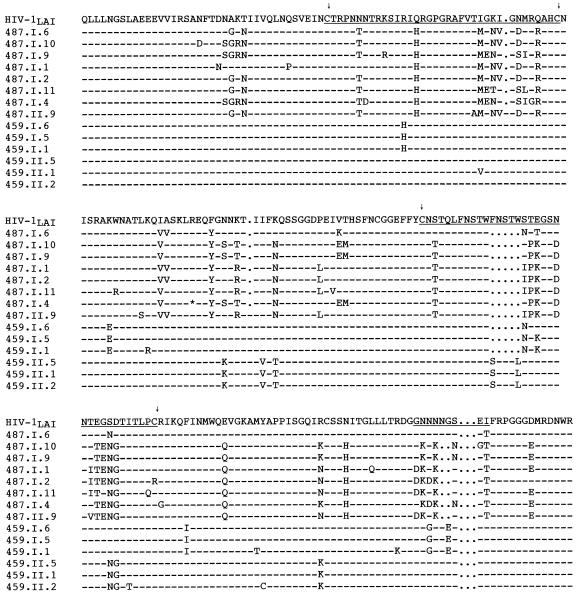
When all 83 clones from uncultured and cultured PBMC were compared by HMA, the major group was represented by 29 different banding patterns (Fig. 5), while seven patterns were identified for the minor group. Analyses of nucleotide and predicted amino acid sequences of the C2-V5env fragments confirmed that clones in the most prevalent group were derived from HIV-1LAV-1b, whereas the more divergent minor population of seven clones appeared to contain regions similar to both HIV-1LAV-1b and HIV-1SF2 (Fig. 6; see below). The extent of env divergence of all clones from either parental strain was significantly greater than that observed for the p17gag gene (Table 2). This extreme diversity, which was comparable to that documented by Novembre et al. (54) in the V1 and V2 env regions of HIV-1JC, resulted not only from nucleotide substitutions but also from insertions and deletions, many of which were localized within the V4 and V5 regions of gp120env. Only four clones appeared to be defective because of premature stop codons in V3 (clone II.32C) or V4 (clones 26, 24, and 18).
FIG. 6.
Amino acid sequence alignment of C2-V5env clones from C-499’s cultured and uncultured PBMC with HIV-1SF2 and HIV-1LAI. Dashes indicate identity with the parental strains, and dots signify gaps. Clone numbers preceded by “I” or “II” identify clones from two independent PCRs. The positions of V3, V4, and V5 are underlined and delineated by the arrows above cysteine residues. ∗, stop codon.
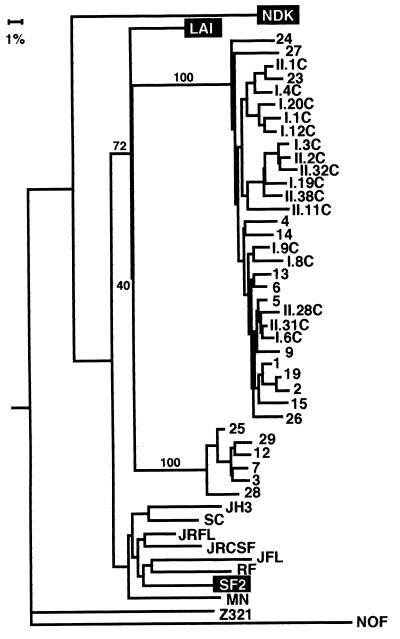
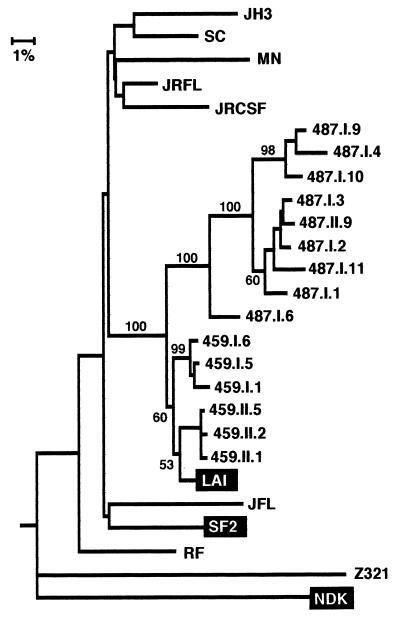
A phylogenetic tree was constructed with the sequences generated in this study, the three HIV-1 strains to which C-499 was exposed (LAI, SF2, and NDK), seven clade B strains chosen at random from the database (53), a clade A strain, and a clade C strain, to which the tree was rooted (Fig. 7). This phylogenetic analysis not only confirmed that two distinct env populations, the minor group of which formed a monophyletic cluster, were present in C-499’s PBMC but also suggested that both groups were derived from HIV-1LAV-1b; however, the bootstrap value for this node was 40, indicating a weak association. That clones from the cultured PBMC were interspersed among those in the major population from the uncultured PBMC indicated that multiple diverse genotypes may have been replication competent. When the sequences of the minor population were compared pairwise to LAI and SF2, the genetic distances from the two progenitor strains were essentially the same (Table 2), which did not allow a distinction of parentage. Similar pairwise comparisons of only the major group of C2-V5env clones, however, indicated that they were probably derived from LAV-1b.
FIG. 7.
Phylogenetic relationship of C2-V5env clones from C-499 to the parental strains and randomly chosen strains from clade B and clades A (Z321), D (NDK), and C (NOF), to which the tree is rooted. Clones designated by numbers only are from uncultured PBMC; the remaining clones are from two independent PCRs (I and II) of cultured (C) PBMC. See the legend to Fig. 4 for other explanations.
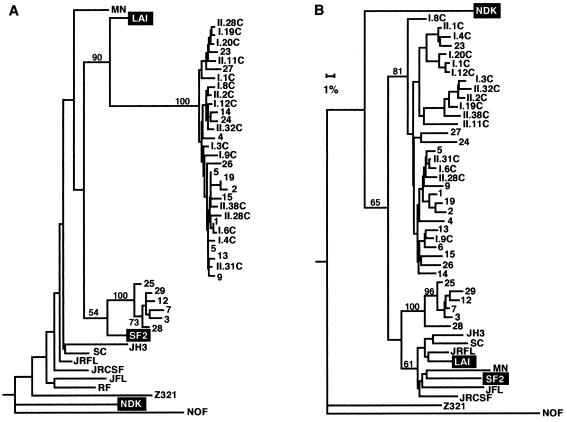
To determine whether the extreme degree of diversification in C2-V5env of HIV-1SF2 and HIV-1LAV-1b during 9 years of infection of C-499 was representative of that in other chimpanzees infected with HIV-1LAI-derived strains for comparable times, this region of proviral DNA in PBMC from chimpanzees C-459 (10 years) and C-487 (9 years) was analyzed. For each animal, clones from two independent PCR amplifications of proviral DNA were sequenced (Fig. 8). Among six clones from C-459’s proviral DNA, the intraisolate nucleotide distances ranged from 0.45 to 3.76%, whereas pairwise differences with HIV-1LAI ranged from 1.82 to 2.48%. For C-487, the differences among eight clones ranged from 0.93 to 7.51%; pairwise distances from HIV-1LAI ranged from 4.1 to 9.65%. Similar to the low frequency of apparently defective clones from C-499, none of six and only one of eight clones from C-459 and C-487, respectively, appeared to encode a defective Env protein. That there was greater diversity among clones from C-487 than those from C-459 is consistent with the histories of these two chimpanzees and the higher viral burden in C-487, which is reflected, in part, by this animal’s high HIV-1-specific antibody titer (Table 1). In addition, virus was isolated from C-487’s PBMC on 100% of attempts throughout its 9 years of infection, which was not true for similar virus isolation attempts from C-459. Furthermore, DNA sequence and phylogenetic analyses of the clones obtained from C-459 revealed two distinct populations of closely related sequences (Fig. 9). However, the intraisolate pairwise distances are well above the error frequency for Taq polymerase (<0.05%), suggesting that most clones were derived from different proviruses. Although not measured directly because of insufficient sample, it is likely that the proviral copy number in C-459’s PBMC was probably substantially lower than that in C-487’s PBMC. Serial dilution of C-487’s genomic DNA and subsequent PCR resulted in amplification of HIV-1 sequences at a 1:32 dilution, and the products of this reaction had four distinct patterns on HMA gels (data not shown). These results indicated that the minimum number of proviruses in 1 μg of DNA from C-487’s PBMC (∼1.5 × 105 cells) was between 32 and 128 (within the limitations of the assay), supporting the conclusion that the clones more than likely did not arise from amplification of one or only a few proviruses.
FIG. 8.
Amino acid sequence alignment of C2-V5env clones from C-459’s and C-487’s uncultured PBMC with HIV-1LAI. See the legend to Fig. 6.
FIG. 9.
Phylogenetic relationships of C2-V5env clones from C-459 and C-487 to the parental HIV-1LAI. See legends to Fig. 4 and 7 for other explanations.
Evidence for recombination.
As discussed above, the minor population of C2-V5env sequences from C-499 appeared to branch with HIV-1LAV-1b (Fig. 7); however, visual comparison of the amino acid sequences of the V3 loops strongly indicated that this minor population was derived from HIV-1SF2 (Fig. 6). The ambiguity in the parentage of the minor population suggested that this group was comprised of recombinant viruses. To test this possibility, we evaluated separately the 5′ half of the C2-V5env fragment, which included C2, V3, and part of C3 (5′-C2-V3), and the 3′ half, which included part of C3, V4, C4, and V5 (V4-V5-3′). Phylogenetic trees of these two subfragments confirmed that the region encompassing the V3 loop of the minor and major populations were derived from SF2 and LAV-1b, respectively (Fig. 10A). In the V4-V5-3′ region, the two populations, while still distinct, did not branch with either parental strain (Fig. 10B); however, a comparison of the pairwise distances from SF2 and LAV-1b revealed that most clones in both populations were more closely related to LAV-1b than SF2 and that, more importantly, the smaller population was clearly more closely related to LAV-1b than to SF2 (Table 2).
FIG. 10.
Phylogenetic trees of the 5′-C2-V3env (A) and V4-V5env-3′ (B) fragments of clones from C-499. See legends to Fig. 4 and 7.
Genetic analysis of nef.
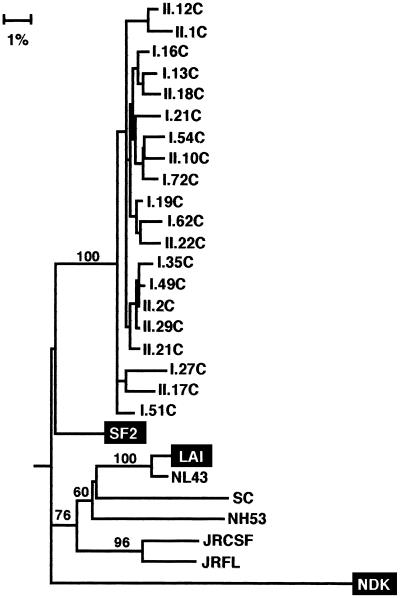
Because Nef appears to be important for pathogenicity in the simian immunodeficiency virus-macaque model (40), the entire nef gene in proviral DNA from C-499’s cultured PBMC was PCR amplified, and multiple clones from two reactions were sequenced. Of the 21 clones sequenced, only 3 (14.3%) were defective, and all of these encoded premature stop codons. The charged acidic motif at amino acids 62 to 65 (EEEE), the SH3-binding motif [(PXX)4] at amino acids 69 to 80, and the β-turn motif (GPGI) at amino acids 130 to 133 were conserved in all full-length clones (47, 66). Pairwise differences for all nef clones revealed a closer sequence relationship to the SF2 strain, and all of the nef clones branched with the SF2 strain in the phylogenetic tree (Fig. 11). However, none of these clones contained a 12-bp (four-amino-acid) insertion near the 5′ end of HIV-1SF2 that is not found in HIV-1LAI (53).
FIG. 11.
Phylogenetic relationship of nucleotide sequences of nef genes from C-499’s cultured PBMC to those of HIV-1SF2 and HIV-1LAI. The tree was rooted to NDK. See legends to Fig. 4 and 7.
DISCUSSION
The results of this study show that during 9 years of infection of a chimpanzee with two HIV-1 subtype B strains, extensive diversification occurred in C2-V5env and, to a lesser extent, in p17gag and nef. The divergence from the parental strains in env was greater than that for p17gag and nef, suggesting that Env had evolved independently from Gag and Nef during the 9 years of infection, which is consistent with observations during HIV-1 infection of humans (11, 46). Furthermore, in the C2-V5env clones generated from proviral DNA in uncultured PBMC, there were two distinct populations of related genomes, with one population more prevalent than the other (approximate ratio, 3:1). The 5′-C2-V3 region of the minor population clearly was derived from HIV-1SF2, and the major population was derived from HIV-1LAV-1b. Since both the genetic distances and the phylogenetic tree indicated that in the V4-V5-3′ region of Env, the minor population was more closely related to HIV-1LAV-1b, these results support the conclusion that the quasispecies in the minor population are recombinant genomes in C2-V5env. No evidence for sequences related to HIV-1NDK was obtained. Identifying the progenitors of p17gag and nef was more problematic. If one considers only the phylogenetic trees, then p17gag was derived from LAV-1b and nef was derived from SF2; however, the nucleotide and amino acid sequences of p17gag have features unique to SF2, and those of nef have features unique to LAV-1b. If either p17gag or nef was derived from SF2, then additional recombinational events may have occurred. To verify this, however, will require sequencing a molecularly cloned virus (or viruses) from C-499’s PBMC and performing a breakpoint analysis throughout the entire genome (60).
Analysis of the Env V3 loop.
In the V3 loop there were substantially more amino acid changes in proviruses derived from HIV-1LAV-1b than in those that evolved from HIV-1SF2. In the majority of V3 loop amino acid sequences in the LAV-1b-derived group, more than one-third (36%) of the amino acids had changed, compared with only 20% for the SF2-derived group. This latter percentage was comparable to the 19% amino acid differences in the V3 loop for C-487. Similarly, proviral DNA from a laboratory worker infected with the closely related HIV-1LAI(IIIB) strain for 5 years had accumulated only 11% diversity (59). If one compares these percent changes with those in unrelated strains, the amino acid difference in the V3 loop between viruses present in an HIV-infected hemophiliac at seroconversion and 7 years later was 11.4% (36). These observations indicate that greater diversification had occurred in C-499’s quasispecies with the LAV-1b-related V3 loop; furthermore, one of these changes was unusual. Although the original GPGR motifs in the tip of the loops of both parental strains had accumulated mutations, they were intrinsically different. All HIV-1SF2-derived viruses had acquired a GPGK motif, which is not uncommon among clade B isolates. In contrast, all HIV-1LAV-1b-derived clones from C-499 encoded GYGR, a tetrad not found in any of the 707 HIV-1 V3 sequences in the database, irrespective of clade designation (53).
Other genetic factors that might influence properties of HIV-1, such as cell tropism, include the net charge of the V3 loop and the number and placement of N-linked glycosylation sites. In C-499’s quasispecies, the number of positively charged amino acids in the V3 loop of the HIV-1LAV-1b-related viruses had increased from 9 to 12. Such increases in positive charge tend to increase macrophage tropism of HIV-1 isolates (9, 16, 41); however, since the HIV-1LAV-1b stock replicates efficiently in chimpanzee macrophages (27, 70), the effects of increased charge, if any, on this property cannot be predicted.
Analysis of the Env V4-V5 region.
In the V4 loop, most of the minor population of clones had 15 changes among the 29 amino acid residues (52%), which included an insertion of three and a deletion of two amino acids, whereas most of the clones in the major population had 12 amino acid changes, which included a deletion of 6 of the original 34 amino acids (35%). As observed in the V4 loop consensus sequence for C-487 and one group of clones from C-459, one copy of the FNSTW repeat was deleted; with the exception of three of the six clones from C-459, this deletion has been found in all isolates from chimpanzees infected with HIV-1LAI-related strains that we have sequenced (17). However, two amino acids of the second FNSTW in the consensus sequence for C-459’s quasispecies are different, suggesting that this repeat motif may be specifically selected against in chimpanzees. The more limited evolutionary changes in the quasispecies from the two asymptomatic chimpanzees were confirmed in the phylogenetic analysis (Fig. 9) and are consistent with the results of Reitz et al. (59). These investigators reported only 1.2% nucleotide divergence in 246 nucleotides encompassing the V4 loop in proviral DNA tested 5 years after a laboratory worker was infected with the closely related HIV-1LAI(IIIB) strain.
The most extensive changes were found in the V5 region of the clones from C-499. In a majority of the minor population of clones, 6 of 10 amino acids were altered, and 50% of the clones had a two-amino-acid insertion and/or a one-amino-acid deletion. In the major population of clones, 7 of 9 (78%) of the amino acids were altered in combination with various insertions and deletions. Consistent with diversification of other regions of C2-V5env, the quasispecies from C-459 had no or only two altered amino acids, while the frequency of mutation in V5 of proviral genomes from C-487 were intermediate to that of C-459 and C-499, resulting in changes in 4 of 9 (44%) of the V5 amino acids, with two others deleted in all but one clone. Thus, the concentration of mutations in the V3, V4, and V5 regions of all clones from C-499 and C-487 indicates that these variable regions were subject to considerable selective pressure in these two long-term HIV-1-infected chimpanzees.
Analysis of Nef.
As previously noted in HIV-1 quasispecies in humans (11, 37, 58, 66), other than multiple amino acid changes in an area of sequence polymorphism near the 5′ terminus of C-499’s nef genes, other mutations resulting in amino acid changes were scattered at random throughout Nef. This finding and the overall lower diversity in Nef, compared to that in the p17 Gag and Env proteins, suggest there is less (or no) selective pressure on the nef gene, which is consistent with evaluations of nef gene evolution over time in HIV-1-infected humans (11, 58). In fact, Plikat et al. (58) concluded that genetic drift was the major factor in evolution of nef and predicted that after 10 years of infection, a nef quasispecies would diverge 7.1%. The divergence of C-499’s nef sequences from both HIV-1SF2 and HIV-1LAV-1b is in remarkable agreement with this estimate (Table 2).
Diversification and disease.
While some studies of virus evolution in HIV-1-infected humans have concluded that disease progression is associated with more extensive genetic diversity (55, 65, 67), others have shown either that diversity correlates with longer asymptomatic periods (7, 12, 26, 43, 45, 48, 69, 72) or that there is no correlation (2, 71, 74). In one study, pairwise comparisons of the V3 consensus nucleotide sequences of isolates from six children infected by blood products from a single donor indicated that the intraperson variation (range, 0.3 to 2.9%) was similar to that observed in C-459’s proviruses (71). The aforementioned study also revealed no correlation between genetic heterogeneity and disease, but relative to the donor inoculum, the progressors tended to harbor viral genomes with less divergence than the nonprogressors. In a similar evaluation of six hemophiliacs who received factor VIII from the same donor, after 5 years the interpatient variation in the V4-V5 region ranged from 5.6 to 11.1%, but a relationship between intrapatient HIV-1 diversity and viral burden in PBMC was not observed (2). More recently, an analysis of nonsynonymous mutations in the V3 region of HIV-1 in 44 persons evaluated at seroconversion and 5 years later indicated that intrahost evolution was directly related to the duration of the immunocompetent period (45). This conclusion was supported by Wolinsky et al. (72), who found an inverse correlation between rapid disease progression and both genetic diversity in the V3-V5 region and the frequency of cytotoxic T-lymphocyte precursors. Most studies relate diversity in various regions of env to disease progression; however, Yoshimura et al. (73) evaluated the full-length gag gene and found that the extent of variation appeared to correlate with duration of infection.
In contrast to many studies of HIV-1 infections in humans, the genetic diversity was much greater in the HIV-1 proviruses in PBMC from chimpanzee C-499, which developed AIDS, than in PBMC from the other two chimpanzees that were infected for a comparable time but did not develop clinical disease. It is interesting, however, that C-487, with a level of diversification intermediate between that of C-459 and C-499, had experienced repeated stimulation of the immune system during the initial 2 years of infection, each episode of which was accompanied by transient increases in HIV-1 viral burden in peripheral blood (19). These manipulations may have contributed to maintenance of high viral loads and evidence of immune dysfunction—16.5% CD4+ lymphocytes, a CD4/CD8 ratio of 0.23 (Table 1), and elevated levels of apoptotic lymphocytes compared to normal chimpanzees (10). However, whether the observed genetic diversification contributed to or was a consequence of disease progression cannot be determined. Thus, no consistent correlation between genetic diversification in env and disease progression has emerged in cohorts of humans (or chimpanzees) infected with the same strain. It is likely that major factors in the degree of diversification and disease progression are virus-host interactions specific to each individual and the HIV-1 strain that they harbor (74).
The observed difference in diversification in the three chimpanzees cannot be explained by length of infection because C-459 had been infected longer than C-499. Likewise, it cannot be explained by viral burdens over the course of infection or at the time these blood samples were obtained (Table 1), which is consistent with a study of humans by Balfe et al. (2). Despite C-487’s extremely high HIV-1-specific antibody titers, which are directly proportional to viral burden in chimpanzees (29, 39), during the past 9 years virus has been isolated from its PBMC on every attempt using standard culture conditions, and for the last 2 to 3 years, this animal has maintained a level of virus between 2 × 103 and 1.3 × 104 RNA copies/ml of plasma. These quantities of virus are comparable to those in a group of patients who developed AIDS between 6 and 9 years after infection, described by Henrard et al. (35). The 100% success at isolating HIV-1 from C-487’s PBMC was not observed with C-459 or C-499 before evidence of disease developed. The low HIV-1 RNA copy number (724 copies/ml) that we found in plasma from C-499 may be an underestimate because the only plasma sample available contained heparin and had been frozen for almost 2 years. Using a different assay, Novembre et al. (54) found approximately 105 RNA equivalents/ml of plasma during the last few months before C-499 was euthanized. The values obtained in that and the present study indicate that some HIV-1-infected chimpanzees have levels of virion RNA in plasma comparable to that in humans (35, 49, 50). In addition, it is possible that the extreme genetic diversity in C-499’s quasispecies resulted from, or was influenced by, the long-term coexistence of two distinct HIV-1 strains that together enhanced the degree of evolution driven by immune selection. Because there are now several chimpanzees coinfected with two different strains of HIV-1, it will be possible to evaluate the influence of coinfection on genetic diversity.
An outcome of the present study was the identification of recombinant proviruses between HIV-1SF2 and HIV-1LAV-1b in C2-V5env and indications that additional recombinatorial events may have occurred. In one aspect this was surprising because, in comparison to HIV-1LAV-1b, HIV-1SF2 not only replicates poorly in chimpanzee PBMC in vitro but also establishes significantly lower viral burdens in infected animals and can rarely be isolated from PBMC after the first 2 months of infection (24, 29, 52). Since HIV-1SF2 was inoculated 15 months before HIV-1LAV-1b, then HIV-1SF2 had to be actively replicating in some body compartment so that individual cells were infected with both strains. It appears that recombination between distinct HIV-1 strains in dually infected chimpanzees may be as prevalent as it appears to be in humans (14, 42, 60, 61). We recently reported the identification of chimeric subtype B and E env genes in lymph node tissue obtained 24 weeks after a chimpanzee had been coinfected with HIV-1LAI(IIIB) and HIV-190CR402 (25). Furthermore, we subsequently obtained evidence of interclade recombination in another dually infected chimpanzee (17). Thus, the HIV-chimpanzee model may provide a way to document the timing and extent of recombination that can occur between two (or more) HIV-1 strains during coinfections.
Although the inherent pathogenicity of HIV-1 strains may influence disease progression, since all three chimpanzees were infected with the same strain, this factor should not have been important. The original HIV-1LAV-1b inoculum that C-499 received has an SI phenotype, is cytopathic for chimpanzee PBMC, and is both macrophage and T-cell tropic; it not only replicates well in chimpanzee bone marrow- and blood monocyte-derived macrophages but also induces syncytia in both MT2 cells and chimpanzee and human PBMC (27, 70). Although these properties may have contributed to disease progression, they are unlikely to be the determining factors because C-487 was inoculated with the same virus stock. In preliminary in vitro studies, the quasispecies recovered from C-499 has retained its SI phenotype for both MT2 cells and human PBMC, which is consistent with the results of Novembre et al. for HIV-1JC (54). It should be noted, however, that Novembre et al. (54) were incorrect in their statement that C-499 was not inoculated with a strain of HIV-1 that forms syncytia in chimpanzee PBMC. The cytopathic effects of HIV-1LAV-1b for chimpanzee cells, including syncytium formation, have been well documented (21, 27, 70). Comparisons of biologic and molecular properties of HIV-1 recovered from these and other long-term-infected chimpanzees with and without evidence of disease may provide insight into determinants of HIV-1 pathogenesis.
A logical extension of the characterization of this heterogeneous quasispecies from C-499 is to determine whether cell-free HIV-1 recovered from this chimpanzee that developed AIDS is pathogenic for other chimpanzees. Although Novembre et al. (54) reported a rapid decline in a chimpanzee inoculated with C-499’s virus, that animal was the recipient of a transfusion of 40 ml of whole blood, and therefore it received an extremely high dose of virus. We have inoculated two chimpanzees, one intravenously and one by atraumatic exposure to the cervical mucosa, with cell-free supernatant from a coculture of C-499’s PBMC with normal human PBMC; this virus stock established persistent high viral burdens in both chimpanzees. In addition, during 21 months of infection, both animals have exhibited steady declines in percentages and numbers of CD4+ lymphocytes (10). This same combination of high levels of plasma RNA and decreasing numbers of CD4+ cells is predictive of disease progression in humans (49, 50, 56). Thus, that chimpanzees can exhibit pathogenic sequelae as a result of HIV-1 infection validates their continued use to evaluate the potential efficacy of HIV-1 vaccines. The use of viruses, such as the one described here (which we call HIV-1JC499), will allow investigators to assess vaccine-mediated protection not only against infection but also against disease.
ACKNOWLEDGMENTS
We thank Jackie Stallworth for technical assistance, Dawn Grill for secretarial assistance, Beatrice Hahn for suggestions on the genetic analysis, and Harold McClure, Yerkes Primate Research Center, for providing blood samples from C-459 and C-499.
This work was supported in part by NIH grant AI28147 and Pasteur Merieux Serum and Vaccins.
REFERENCES
- 1.Alter H J, Eichberg J W, Masur H, Saxinger W C, Gallo R, Macher A M, Lane H C, Fauci A S. Transmission of HTLV-III infection from human plasma to chimpanzees: an animal model for AIDS. Science. 1984;226:549–552. doi: 10.1126/science.6093251. [DOI] [PubMed] [Google Scholar]
- 2.Balfe P, Simmonds P, Ludlum C A, Bishop J O, Brown A J L. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990;64:6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barre-Sinoussi F, Georges-Courbot M-C, Fultz P N, Tuyet H N T, Muchmore E, Saragosti S, Dubreuil G, Georges A, vander Ryst E, Girard M. Characterization and titration of an HIV type 1 subtype E chimpanzee challenge stock. AIDS Res Hum Retroviruses. 1997;13:583–591. doi: 10.1089/aid.1997.13.583. [DOI] [PubMed] [Google Scholar]
- 4.Berman P W, Gregory T J, Riddle L, Nakamura G R, Champe M A, Porter J P, Wurm F M, Hershberg R D, Cobb E K, Eichberg J W. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature. 1990;345:622–625. doi: 10.1038/345622a0. [DOI] [PubMed] [Google Scholar]
- 5.Berman P W, Murthy K K, Wrin T, Vennari J C, Cobb E K, Eastman D J, Champe M, Nakamura G R, Davison D, Powell M F, Bussiere J, Francis D P, Matthews T, Gregory T J, Obijeski J F. Protection of MN-rgp120-immunized chimpanzees from heterologous infection with a primary isolate of human immunodeficiency virus type 1. J Infect Dis. 1996;173:52–59. doi: 10.1093/infdis/173.1.52. [DOI] [PubMed] [Google Scholar]
- 6.Bruck C, Thiriart C, Fabry L, Francotte M, Pala P, Van Opstal O, Culp J, Rosenberg M, DeWilde M, Heidt P, Heeney J. HIV-1 envelope-elicited neutralizing antibody titres correlate with protection and virus load in chimpanzees. Vaccine. 1994;12:1141–1148. doi: 10.1016/0264-410x(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 7.Burger H, Weiser B, Flaherty K, Gulla J, Nguyen P-N, Gibbs R A. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc Natl Acad Sci USA. 1991;88:11236–11240. doi: 10.1073/pnas.88.24.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro B A, Walker C M, Eichberg J W, Levy J A. Suppression of human immunodeficiency virus replication by CD8+ cells from infected and uninfected chimpanzees. Cell Immunol. 1991;132:246–255. doi: 10.1016/0008-8749(91)90023-5. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, I. C., M. Girard, and P. N. Fultz. Loss of CD4+ T cells in HIV-1-infected chimpanzees is associated with increased lymphocyte apoptosis. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 11.Delassus S, Cheynier R, Wain-Hobson S. Evolution of human immunodeficiency virus type 1 nef and long terminal repeat sequences over 4 years in vivo and in vitro. J Virol. 1991;65:225–231. doi: 10.1128/jvi.65.1.225-231.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 14.Diaz R S, Sabino E C, Mayer A, Mosley J W, Busch M P the Transfusion Safety Study Group. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J Virol. 1995;69:3273–3281. doi: 10.1128/jvi.69.6.3273-3281.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estaquier J, Idziorek T, DeBels F, Barre-Sinoussi F, Hurtel B, Aubertin A-M, Venet A, Mehtali M, Muchmore E, Michel P, Mouton Y, Girard M, Ameisen J C. Programmed cell death and AIDS: significance of T-cell apoptosis in pathogenic and nonpathogenic primate lentiviral infections. Proc Natl Acad Sci USA. 1994;91:9431–9435. doi: 10.1073/pnas.91.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fultz, P. N. Unpublished data.
- 18.Fultz P N. Animal models for human immunodeficiency virus infection and disease. In: Wormser G P, editor. AIDS and other manifestations of HIV infection. 3rd ed. New York, N.Y: Lippincott-Raven; 1997. pp. 201–215. [Google Scholar]
- 19.Fultz P N, Gluckman J-C, Muchmore E, Girard M. Transient increases in numbers of infectious cells in an HIV-infected chimpanzee following immune stimulation. AIDS Res Hum Retroviruses. 1992;8:313–317. doi: 10.1089/aid.1992.8.313. [DOI] [PubMed] [Google Scholar]
- 20.Fultz P N, Horaist C, McClure H M, Steimer K S, Dina D, Mawle A C. Postinfection immunization of human immunodeficiency virus-infected chimpanzees with recombinant HIV-1 env and gag antigens. In: Chanock R, Brown F, Lerner R, Ginsberg H, editors. Vaccines 89. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 225–231. [Google Scholar]
- 21.Fultz P N, McClure H M, Swenson R B, McGrath C R, Brodie A, Getchell J P, Jensen F C, Anderson D C, Broderson J R, Francis D P. Persistent infection of chimpanzees with human T-cell leukemia virus type III/lymphadenopathy associated virus: a potential model for acquired immunodeficiency syndrome. J Virol. 1986;58:116–124. doi: 10.1128/jvi.58.1.116-124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fultz P N, Nara P, Barre-Sinoussi F, Chaput A, Greenberg M L, Muchmore E, Kieny M-P, Girard M. Vaccine protection of chimpanzees against challenge with HIV-1-infected peripheral blood mononuclear cells. Science. 1992;256:1687–1690. doi: 10.1126/science.256.5064.1687. [DOI] [PubMed] [Google Scholar]
- 23.Fultz P N, Siegel R L, Brodie A, Mawle A C, Stricker R B, Swenson R B, Anderson D C, McClure H M. Prolonged CD4+ lymphocytopenia and thrombocytopenia in a chimpanzee persistently infected with HIV-1. J Infect Dis. 1991;163:441–447. doi: 10.1093/infdis/163.3.441. [DOI] [PubMed] [Google Scholar]
- 24.Fultz P N, Srinivasan A, Greene C R, Butler D, Swenson R B, McClure H M. Superinfection of a chimpanzee with a second strain of human immunodeficiency virus. J Virol. 1987;61:4026–4029. doi: 10.1128/jvi.61.12.4026-4029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fultz P N, Yue L, Wei Q, Girard M. Human immunodeficiency virus type 1 intersubtype (B/E) recombination in a superinfected chimpanzee. J Virol. 1997;71:7990–7995. doi: 10.1128/jvi.71.10.7990-7995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganeshan S, Dickover R E, Korber B T M, Bryson Y J, Wolinsky S M. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gendelman H E, Ehrlich G D, Baca L M, Conley S, Ribas J, Kalter D C, Meltzer M S, Poiesz B J, Nara P. The inability of human immunodeficiency virus to infect chimpanzee monocytes can be overcome by serial viral passage in vivo. J Virol. 1991;65:3853–3863. doi: 10.1128/jvi.65.7.3853-3863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girard M, Kieny M-P, Pinter A, Barre-Sinoussi F, Nara P, Kolbe H, Kusumi K, Chaput A, Reinhart T, Muchmore E, Ronco J, Kaczorek M, Gomard E, Gluckman J-C, Fultz P N. Immunization of chimpanzees confers protection against challenge with human immunodeficiency virus. Proc Natl Acad Sci USA. 1991;88:542–546. doi: 10.1073/pnas.88.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard M, Meignier B, Barre-Sinoussi F, Kieny M-P, Matthews T, Muchmore E, Nara P L, Wei Q, Rimsky L, Weinhold K, Fultz P N. Vaccine-induced protection of chimpanzees against infection by a heterologous human immunodeficiency virus type 1. J Virol. 1995;69:6239–6248. doi: 10.1128/jvi.69.10.6239-6248.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girard M, van der Ryst E, Barre-Sinoussi F, Nara P, Tartaglia J, Paoletti E, Blondeau C, Jennings M, Verrier F, Meignier B, Fultz P N. Challenge of chimpanzees immunized with a recombinant canarypox-HIV-1 virus. Virology. 1997;232:98–104. doi: 10.1006/viro.1997.8560. [DOI] [PubMed] [Google Scholar]
- 31.Girard M, Yue L, Barre-Sinoussi F, van der Ryst E, Meignier B, Muchmore E, Fultz P N. Failure of a human immunodeficiency virus type 1 (HIV-1) subtype B-derived vaccine to prevent infection of chimpanzees by an HIV-1 subtype E strain. J Virol. 1996;70:8229–8233. doi: 10.1128/jvi.70.11.8229-8233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gougeon M-L, Garcia S, Heeney J, Tschopp R, Lecoeur H, Guetard D, Rame Y, Dauguet C, Montagnier L. Programmed cell death in AIDS-related HIV and SIV infections. AIDS Res Hum Retroviruses. 1993;9:553–563. doi: 10.1089/aid.1993.9.553. [DOI] [PubMed] [Google Scholar]
- 33.Gougeon M-L, Lecoeur H, Boudet F, Ledru E, Marzabal S, Boullier S, Roue R, Nagata S, Heeney J. Lack of chronic immune activation in HIV-infected chimpanzees correlates with the resistance of T cells to Fas/Apo-1 (CD95)-induced apoptosis and preservation of a T helper phenotype. J Immunol. 1997;158:2964–2976. [PubMed] [Google Scholar]
- 34.Heeney J, Jonker R, Koornstra W, Dubbes R, Niphuis H, DiRienzo A-M, Gougeon M-L, Montagnier L. The resistance of HIV-infected chimpanzees to progression to AIDS correlates with absence of HIV-related T-cell dysfunction. J Med Primatol. 1993;22:194–200. [PubMed] [Google Scholar]
- 35.Henrard D R, Phillips J F, Muenz L R, Blattner W A, Wiesner D, Eyster M E, Goedert J J. Natural history of HIV-1 cell-free viremia. JAMA. 1995;274:554–558. [PubMed] [Google Scholar]
- 36.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Leigh-Brown A J. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Zhang L, Ho D D. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J Virol. 1995;69:93–100. doi: 10.1128/jvi.69.1.93-100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husch B, Eibl M M, Mannhalter J W. CD3, CD8 double positive cells from HIV-1-infected chimpanzees show group-specific inhibition of HIV-1 replication. AIDS Res Hum Retroviruses. 1993;9:405–413. doi: 10.1089/aid.1993.9.405. [DOI] [PubMed] [Google Scholar]
- 39.Johnson B K, Stone G A, Godec M S, Asher D M, Gajdusek D C, Gibbs C J. Long-term observations of human immunodeficiency virus-infected chimpanzees. AIDS Res Hum Retroviruses. 1993;9:375–378. doi: 10.1089/aid.1993.9.375. [DOI] [PubMed] [Google Scholar]
- 40.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 41.Korber B T M, MacInnes K, Smith R F, Myers G. Mutational trends in V3 loop protein sequences observed in different genetic lineages of human immunodeficiency virus type 1. J Virol. 1994;68:6730–6744. doi: 10.1128/jvi.68.10.6730-6744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leitner T, Escanilla D, Marquina S, Wahlberg J, Brostrom C, Hansson H B, Uhlen M, Albert J. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 43.Liu S-L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G-M, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 45.Lukashov V V, Kuiken C L, Goudsmit J. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J Virol. 1995;69:6911–6916. doi: 10.1128/jvi.69.11.6911-6916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martins L P, Chenciner N, Asjo B, Meyerhans A, Wain-Hobson S. Independent fluctuation of human immunodeficiency virus type 1 rev and gp41 quasispecies in vivo. J Virol. 1991;65:4502–4507. doi: 10.1128/jvi.65.8.4502-4507.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNearney T, Hornickova Z, Templeton A, Birdwell A, Arens M, Markham R, Saah A, Ratner L. Nef and LTR sequence variation from sequentially derived human immunodeficiency virus type 1 isolates. Virology. 1995;208:388–398. doi: 10.1006/viro.1995.1166. [DOI] [PubMed] [Google Scholar]
- 48.McNearney T, Westervelt P, Thielan B J, Trowbridge D B, Garcia J, Whittier R, Ratner L. Limited sequence heterogeneity among biologically distinct human immunodeficiency virus type 1 isolates from individuals involved in a clustered infectious outbreak. Proc Natl Acad Sci USA. 1990;87:1917–1921. doi: 10.1073/pnas.87.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellors J W, Kingsley L A, Rinaldo C R, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 50.Mellors J W, Rinaldo C R, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 51.Morrow W J W, Homsy J, Eichberg J W, Krowka J, Pan L-Z, Gaston I, Legg H, Lerche N, Thomas J, Levy J A. Long-term observation of baboons, rhesus monkeys, and chimpanzees inoculated with HIV and given periodic immunosuppressive treatment. AIDS Res Hum Retroviruses. 1989;5:233–245. doi: 10.1089/aid.1989.5.233. [DOI] [PubMed] [Google Scholar]
- 52.Murthy K K, Cobb E K, El-Amad Z, Ortega H, Hsueh F C, Satterfield W, Lee D R, Kalish M L, Haigwood N L, Kennedy R C, Steimer K S, Schultz A, Levy J A. Titration of a vaccine stock preparation of human immunodeficiency virus type 1SF2 in cultured lymphocytes and in chimpanzees. AIDS Res Hum Retroviruses. 1996;12:1341–1348. doi: 10.1089/aid.1996.12.1341. [DOI] [PubMed] [Google Scholar]
- 53.Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.Mex: Los Alamos National Laboratory; 1994. [Google Scholar]
- 54.Novembre F J, Saucier M, Anderson D C, Klumpp S A, O’Neil S P, Brown C R, Hart C E, Guenthner P C, Swenson R B, McClure H M. Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J Virol. 1997;71:4086–4091. doi: 10.1128/jvi.71.5.4086-4091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowak M A, Anderson R M, McLean A R, Wolfs T F W, Goudsmit J, May R M. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 56.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D the Veterans Affairs Cooperative Study Group on AIDS. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 57.Peeters M, Janssens W, vanden Haesevelde M, Fransen K, Willems B, Heyndrickx L, Kestens L, Piot P, Van Der Groen G, Heeney J. Virologic and serologic characteristics of a natural chimpanzee lentivirus infection. Virology. 1995;211:312–315. doi: 10.1006/viro.1995.1407. [DOI] [PubMed] [Google Scholar]
- 58.Plikat U, Nieselt-Struwe K, Meyerhans A. Genetic drift can dominate short-term human immunodeficiency virus type 1 nef quasispecies evolution in vivo. J Virol. 1997;71:4233–4240. doi: 10.1128/jvi.71.6.4233-4240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reitz M S, Hall L, Robert-Guroff M, Lautenberger J, Hahn B H, Shaw G M, Kong L I, Weiss S H, Waters D, Gallo R C, Blattner W. Viral variability and serum antibody response in a laboratory worker infected with HIV type 1 (HTLV type IIIB) AIDS Res Hum Retroviruses. 1994;10:1143–1155. doi: 10.1089/aid.1994.10.1143. [DOI] [PubMed] [Google Scholar]
- 60.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 61.Sabino E C, Shpaer E G, Morgado M G, Korber B T M, Diaz R S, Bongertz V, Cavalcante S, Galvao-Castro B, Mullins J I, Mayer A. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68:6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, deGoede R E Y, van Steenwijk R P, Lange J M A, Schattenkerk J K M E, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuitemaker H, Meyaard L, Kootstra N A, Dubbes R, Otto S A, Tersmette M, Heeney J L, Miedema F. Lack of T cell dysfunction and programmed cell death in human immunodeficiency virus type 1-infected chimpanzees correlates with absence of monocytotropic variants. J Infect Dis. 1993;168:1140–1147. doi: 10.1093/infdis/168.5.1140. [DOI] [PubMed] [Google Scholar]
- 64.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shpaer E G, Mullins J I. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J Mol Evol. 1993;37:57–65. doi: 10.1007/BF00170462. [DOI] [PubMed] [Google Scholar]
- 66.Shugars D C, Smith M S, Glueck D H, Nantermet P V, Seillier-Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67:4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strunnikova N, Ray S C, Livingston R A, Rubalcaba E, Viscidi R P. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J Virol. 1995;69:7548–7558. doi: 10.1128/jvi.69.12.7548-7558.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wain-Hobson S, Vartanian J-P, Henry M, Chenciner N, Cheynier R, Delassus S, Martins L P, Sala M, Nugeyre M-T, Guetard D, Klatzmann D, Gluckman J-C, Rozenbaum W, Barre-Sinoussi F, Montagnier L. LAV revisited: origins of the early HIV-1 isolates from Institut Pasteur. Science. 1991;252:961–965. doi: 10.1126/science.2035026. [DOI] [PubMed] [Google Scholar]
- 69.Wang B, Ge Y C, Jozwiak R, Bolton W, Palasanthiran P, Ziegler J, Chang J, Xiang S-H, Cunningham A L, Saksena N K. Molecular analyses of human immunodeficiency virus type 1 V3 region quasispecies derived from plasma and peripheral blood mononuclear cells of the first long-term nonprogressing mother and child pair. J Infect Dis. 1997;175:1510–1515. doi: 10.1086/516489. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe M, Ringler D J, Fultz P N, MacKey J J, Boyson J E, Levine C G, Letvin N L. A chimpanzee-passaged human immunodeficiency virus isolate is cytopathic for chimpanzee cells but does not induce disease. J Virol. 1991;65:3344–3348. doi: 10.1128/jvi.65.6.3344-3348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolfs T F W, de Jong J-J, vanden Berg H, Tunagel J M G H, Krone W J A, Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci USA. 1990;87:9938–9942. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 73.Yoshimura F K, Diem K, Learn G H, Riddell S, Corey L. Intrapatient sequence variation of the gag gene of human immunodeficiency virus type 1 plasma virions. J Virol. 1996;70:8879–8887. doi: 10.1128/jvi.70.12.8879-8887.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou M-Y, Clark S E, Gomez-Sanchez C E. Universal cloning method by TA strategy. BioTechniques. 1995;19:34–35. [PubMed] [Google Scholar]