Abstract
Nine glycoproteins (gB, gC, gD, gE, gG, gH, gI, gK, and gL) have been identified in bovine herpesvirus 1 (BHV-1). gM has been identified in many other alpha-, beta-, and gammaherpesviruses, in which it appears to play a role in membrane penetration and cell-to-cell fusion. We sought to express BHV-1 open reading frame UL10, which encodes gM, and specifically identify the glycoprotein. We corrected a frameshift error in the published sequence and used the corrected sequence to design coterminal peptides from the C terminus. These were expressed as glutathione S-transferase fusion proteins in Escherichia coli. The fusion protein containing the 63 C-terminal amino acids from the corrected gM sequence engendered antibodies that immunoprecipitated a 30-kDa protein from in vitro translation reactions programmed with the UL10 gene. Proteins immunoprecipitated by this antibody from virus-infected cells ran at 36 and 43 kDa in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and 43 and 48 kDa in nonreducing SDS-PAGE. Only the larger of the pair was present in virions. A 7-kDa protein was released from gM by reducing agents. The 7-kDa protein was not recognized in Western blots probed with the anti-gM antibody but reacted specifically with antibodies prepared against BHV-1 UL49.5, previously reported to be a 9-kDa protein associated with an unidentified 39-kDa protein (X. Liang, B. Chow, C. Raggo, and L. A. Babiuk, J. Virol. 70:1448–1454, 1996). This is the first report of a small protein covalently bound to any herpesvirus gM. Similar patterns of hydrophobic domains and cysteines in all known gM and UL49.5 homologs suggest that these two proteins may be linked by disulfide bonds in all herpesviruses.
Bovine herpesvirus 1 (BHV-1) is a common pathogen causing respiratory, ocular, and reproductive disease in cattle worldwide. Nine BHV-1 glycoproteins (gB, gC, gD, gE, gG, gH, gI, gK, and gL) have been identified and partially characterized. The BHV-1 sequence (27, 28) also contains open reading frames (ORFs) UL10 and UL49.5, encoding homologs of gM (2) and gN (10), respectively.
The BHV-1 UL10 gene sequence (28) predicts a 411-amino-acid protein with all of the features of the gMs found in every other herpesvirus sequenced to date. Herpesvirus gM gene sequences predict proteins about 350 to 475 amino acids long with little amino acid identity but nearly identical patterns of large hydrophobic domains separated by 20- to 40-amino-acid hydrophilic domains, a proline-cysteine pair 40 to 70 amino acids from the N terminus, and an N-linked glycosylation site exactly 12 amino acids to the right of the conserved proline-cysteine (13). The major physical difference between gMs appears to be the length of the C-terminal hydrophilic region, which varies from 20 to 110 amino acids. The gM genes that have been studied are expressed late (3, 13, 26). Transcripts (3) and gM protein (23) are described as abundant. The herpes simplex virus (HSV), human cytomegalovirus, pseudorabies virus (PRV), and equine herpesvirus 1 (EHV-1) glycoproteins have been detected by antipeptide or monoclonal antibodies as 46- to 63-kDa glycoproteins in infected cells and virions (2, 7, 11, 16, 21, 23). Consistent with the presence of many hydrophobic domains, gM is associated with cellular and virion membranes. Deletion of the majority of the gene or insertional mutagenesis shows that the gM genes of HSV, EHV-1, and PRV are not essential for replication in vitro (1, 6, 7, 17, 21) or in vivo (17), although the deletants grow to lower titers, penetrate cells more slowly than wild type virus (7, 21), and are defective for cell-to-cell fusion (6), suggesting that gM may play a role in membrane penetration.
The BHV-1 UL49.5 gene codes for a nonglycosylated virion surface protein (14, 15). It appears to have a homolog in every herpesvirus (5). Typical examples include HSV types 1 and 2 (HSV-1 and -2) UL49a or UL49.5, PRV UL49.5, EHV-1 gene 10, human and murine cytomegalovirus UL73, human herpesvirus 6 and 7 protein U46, Epstein-Barr virus BLRF1, and human herpesvirus 8 ORF 53. Sequences vary widely, but all code for proteins 84 to 138 amino acids in length, having a putative signal sequence approximately 20 amino acids long, a 15- to 81-amino-acid hydrophilic, presumably extracellular, domain containing a single cysteine 4 to 13 amino acids N terminal to a 19- to 35-amino-acid putative membrane anchor, and a 5- to 17-amino-acid cytoplasmic tail containing one or two cysteines immediately C terminal to the membrane anchor. The UL49.5 proteins are nonessential in BHV-1 (15), HSV-1 (24), and varicella-zoster virus (25).
In this study, we sought to identify the BHV-1 gM protein. We attempted to make antibodies against the predicted C-terminal domain and generated antibodies that failed to react with any viral protein. Therefore, we resequenced the 3′ end of UL10 and discovered a single nucleotide that was omitted from the original sequence. Using the corrected sequence, we expressed three C-terminal predicted peptides as glutathione S-transferase (GST) fusion proteins in Escherichia coli, made antibodies against the fusion proteins, and used antibodies against one fusion protein to immunoprecipitate a 30-kDa protein from in vitro translation reactions programmed with the UL10 gene and glycoproteins from virus-infected cells that ran at 36 and 43 kDa in reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and 43 and 48 kDa in nonreducing SDS-PAGE. The larger of the pair was precipitated from virion envelopes. Reducing conditions released a 7-kDa protein from gM immunoprecipitated from virus-infected cells and virion envelopes, suggesting that it was linked to gM by a disulfide bond. The 7-kDa protein was identified by specific antibodies as the BHV-1 UL49.5 protein.
MATERIALS AND METHODS
Virus, cells, and media.
BHV-1 (Cooper strain; ATCC VR-864) was replicated in Madin-Darby bovine kidney (MDBK; ATCC CCL22) cells in minimum essential medium (Gibco Laboratories, Life Technologies, Inc.) supplemented with 5% fetal bovine serum (HyClone Laboratories, Inc.) at 35°C in a 5% CO2 humidified atmosphere. The virus was semipurified by centrifugation through a 30% sucrose cushion and stored at −80°C. Virus for one experiment was further purified by isopycnic centrifugation on a potassium tartrate gradient (20).
E. coli JM109 (Promega) was used for plasmid maintenance and transformation, and E. coli BL-21 (Novagen) was used for GST fusion protein expression.
Production of antibodies against GST-gM C-terminal fusion proteins.
We resequenced nucleotides 17942 to 17708 from plasmid pSD58 (19) and confirmed the published sequence (28) except that we found a single extra G between positions 17836 and 17835, causing a reading frameshift and resulting in a predicted protein of 438 amino acids, 27 amino acids longer than the published gM, with an unglycosylated size of 43 kDa. The N-terminal primer TGGATCCCCGCTCGAAGGCGACGCA and C-terminal primer GGAGAATTCTTTATTTGACGTGCGCGG were used to amplify the corrected gM C-terminal 63 codons from plasmid pSD58. The gene fragment was amplified with Pfu polymerase and cloned into pGEX-KG (8) in frame with the GST gene to create the construct gMC-63. The recombinant plasmid was transformed into E. coli BL-21 and induced by isopropylthiogalactopyranoside at a final concentration of 0.2 mM overnight with gentle shaking at room temperature to restrict the formation of inclusion bodies. The cells were suspended in phosphate-buffered saline (PBS) and lysed by sonication. Triton X-100 was added at a final concentration of 1% to aid in solubilization of the fusion proteins. A 50% slurry of glutathione-Sepharose 4B equilibrated with 1× PBS was added and incubated with gentle agitation at room temperature for 30 min. The glutathione-Sepharose pellet was washed twice with 10 bed volumes of PBS. The fusion protein was eluted in buffer (10 mM glutathione, 50 mM Tris-HCl [pH 8.0]) and analyzed by SDS-PAGE. A preparation of GST lacking a fusion partner was similarly prepared. The proteins were emulsified in Freund’s complete adjuvant and injected subcutaneously into BALB/c mice. Mice were boosted twice at 3-week intervals with fusion protein emulsified with Freund’s incomplete adjuvant. Sera were sampled 2 weeks following the final dose.
Production of antibodies against GST-UL49.5 full-length and truncated fusion proteins.
Primers TGAGGATCCATGCCGCGGTCGCCGCTCATC and TCATCTAGATCAGCCCCGCCCCCGCGACT were used to amplify the entire 96-codon UL49.5 ORF from plasmid pSD57 (19). Primers ACTGGATCCATGGCCATCGTGCGCGGCCGCGA and TCATCTAGATCAGCCCCGCCCCCGCGACT were used to amplify codons 17 to 96. Both the full-length and truncated (UL49.5T) products were digested with BamHI and XbaI and cloned into similarly digested pGEX and pCDNA3 (Invitrogen). The fusion proteins were expressed as inclusion bodies which were solubilized in 7 M urea or 0.3% sarcosyl before injection into mice as described above.
In vitro transcription and translation of the gM and UL49.5 genes.
Based on the corrected sequence, we amplified the entire gM gene, bases 17628 to 18948, from plasmid pSD58, using primers CAGAATTCATGGCGGGCTCCGCGCAGCCT and GGAGAATTCTTTATTTGACGTGCGCGG and Pfu polymerase. The amplified fragment was ligated to itself, cut with EcoRI, and cloned into the EcoRI site in pcDNA3 downstream of the T7 promoter. The resulting plasmid was confirmed by restriction enzyme analysis.
The in vitro transcription was carried out according to the AmpliScribe protocol (Epicentre Technologies). Briefly, the following components are combined in a 20-μl total volume: 1 μg of EcoRI-cleaved plasmid, 2 μl of 10× T7 reaction buffer, 1.5 μl of 100 mM each ATP, CTP, GTP, and UTP, 2 μl of 100 mM dithiothreitol (DTT), and 2 μl of AmpliScribe T7 enzyme. The reaction mixture was incubated at 37°C for 2 h.
Transcripts were translated in an in vitro translation reaction mixture that included 8 μl of 12.5× translation mixture, 2 μl of 25 mM magnesium acetate, 2 μl of 2.5 M potassium acetate, 40 μl of reticulocyte lysate (Promega), 10 μg of gM RNA transcript, 10 μl of [35S]methionine, and water to 100 μl. The mixture was incubated at 30°C for 60 min. A sample was treated at 56°C for 10 min with sample preparation buffer containing 40 mM DTT as a reducing agent, subjected to SDS-PAGE in a 12 or 18% acrylamide gel, and autoradiographed at −70°C.
Radioimmunoprecipitation.
Radiolabeled uninfected and BHV-1-infected MDBK cells and virions were prepared as described by Marshall et al. (18). MDBK cells were infected at a multiplicity of infection (MOI) of 10 and labeled with [35S]methionine and -cysteine (ICN Pharmaceuticals Inc.) from 6 to 22 h after infection. The 35S-labeled cells and virions were lysed in NET buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 8]) containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 0.5% Nonidet P-40 (NP-40), and 0.5% deoxycholate. Radiolabeled virion envelopes were prepared by lysing purified virions with 0.5% NP-40 and 0.5% deoxycholate and centrifuging the lysate over a 30% sucrose cushion to remove nucleocapsids. Radiolabeled BHV-1-infected MDBK cell membranes were prepared by Dounce homogenizing cells in the presence of homogenizing buffer (20 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 1 mM PMSF), centrifuging the cells for 1 min at 2,000 rpm to remove cell debris, and centrifuging the supernatant for 20 min at 12,000 rpm to pellet membranes. The membrane pellet was washed once in homogenizing buffer and centrifuged at 40,000 rpm, producing the 40K supernatant. To test the strength of gM binding to membranes, some membrane pellets were washed once in 1 M NaCl. Washed membranes were resuspended in lysing buffer (NET, 0.5% NP-40, 0.5% deoxycholate). Immunoprecipitations were done with 10 μl of antiserum for each 106 cells.
Proteins were immunoprecipitated from in vitro translation reactions or from lysates of BHV-1-infected and uninfected MDBK cells or cell membranes, BHV-1 virions, or BHV-1 envelopes on Staphylococcus aureus (Gibco Laboratories, Life Technologies, Inc.) coated successively with rabbit anti-mouse antibodies (Cappel) and murine polyclonal antibodies. Precipitates were treated at 56°C with SDS-PAGE sample buffer with or without reducing agents, analyzed by reducing or nonreducing SDS-PAGE, and autoradiographed at −70°C.
Analysis of N-linked glycosylation.
N-linked glycosylation was analyzed as described previously (30). Briefly, radiolabeled gM immunoprecipitated from infected cell membranes was eluted from S. aureus with 0.8% SDS at 56 or 100°C and digested with various amounts of endo-β-N-acetylglucosaminidase H (endo H; Boehringer Mannheim) in 100 mM sodium acetate (pH 5)–150 mM NaCl–1% Triton X-100–1% 2-mercaptoethanol–0.2% SDS–0.5 mM PMSF for 18 h at 37° or digested with various amounts of peptide:N-glycosidase F (PNGase F; New England BioLabs) in 50 mM sodium phosphate (pH 7.5)–1% NP-40 for 5 h at 37°C. Proteins were analyzed by reducing SDS-PAGE and autoradiography.
To monitor the glycosylation of gM, cells were pulse-labeled with [35S]methionine and -cysteine from 7.5 to 8 h after infection in methionine- and cysteine-deficient medium and chased for 0 to 60 min with unlabeled methionine and cysteine. Uninfected cells were labeled at the same time. The glycosylation inhibitor tunicamycin was added to some infected cultures 30 min prior to labeling and was not removed for the 60-min chase. The cells were lysed, centrifuged to remove debris and nuclei, and immunoprecipitated as described above. The immunoprecipitated products were incubated at 56° in SDS-PAGE sample buffer with and without 100 mM DTT and analyzed by SDS-PAGE in 10 to 20% gradient gels with or without reducing agents.
Western blotting.
Western blotting was carried out as described previously (30). Lysates were incubated at 56°C for 10 min in SDS-PAGE sample buffer with or without 100 mM DTT. Samples were analyzed by SDS-PAGE in 10 to 20% gradient gels and transferred to nitrocellulose paper (Bio-Rad) at 500 mA for 30 to 60 min. The nitrocellulose was blocked for 30 min with 20 mM Tris-HCl (pH 7.5)–150 mM NaCl–0.05% Tween 20–5% powdered skim milk, incubated for 60 min with antiviral antibody diluted 1:500 in blocking buffer, and then reacted with peroxidase-labeled anti-mouse antibody (1:5,000; Amersham). The final result was visualized with an enhanced chemiluminescence (ECL) reaction (Amersham). For reprobing, blots were incubated at 55°C for 30 min in stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl [pH 6.7]) and washed fully to remove residual reducing agent before repetition of the Western blotting with a second antibody.
RESULTS
In vitro translation of BHV-1 gM.
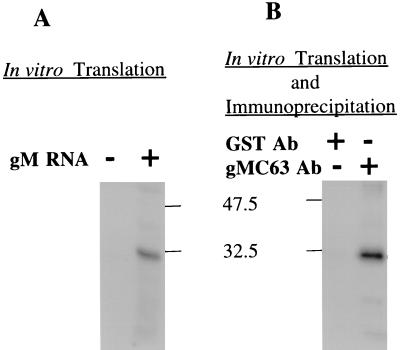
The entire gM gene was amplified by PCR using Pfu polymerase and inserted into pcDNA3 downstream of the T7 promoter. The gM mRNA transcript from this construct was translated in a rabbit reticulocyte lysate in the absence of membranes. A protein of 30 kDa was detected in reactions programmed with gM mRNA but not in control reactions (Fig. 1A). Antibody from mice immunized with gMC-63 but not GST precipitated the 30-kDa gM from in vitro translation reactions (Fig. 1B). Purified gMC-63, but not GST, blocked the immunoprecipitation (data not shown). The gMC-63 antibody was designated gMC antibody and was used for all subsequent experiments.
FIG. 1.
Antibodies (Ab) against the 3′ end of BHV-1 UL10 immunoprecipitate the UL10 in vitro translation product. (A) A 30-kDa protein was synthesized in a reticulocyte lysate in the presence but not the absence of the UL10 RNA transcript. The sample was treated at 56°C for 10 min in the presence of reducing agent, subjected SDS-PAGE in a 12% gel, and autoradiographed. (B) Sera from mice immunized with gMC-63 precipitated the 30-kDa protein synthesized in an in vitro translation reaction programmed with UL10 mRNA. Sera from mice immunized with GST did not precipitate the 30-kDa protein.
Immunoprecipitation of gM from BHV-1-infected cells and virions.
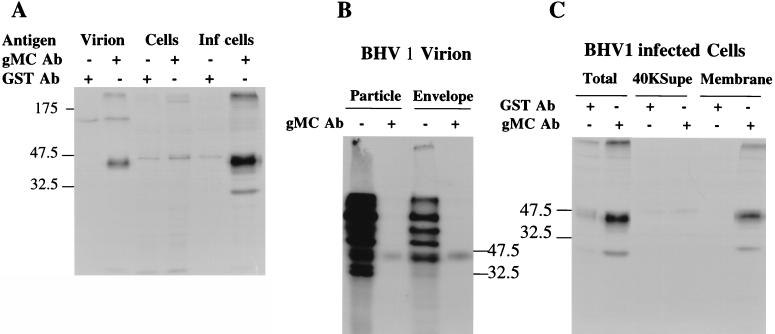
To identify gM in viral materials, detergent-solubilized virions and lysates of uninfected and BHV-1-infected cells were immunoprecipitated with gMC or GST antibody. A 43-kDa protein was precipitated from virions by gMC but not GST antibody (Fig. 2A). A 100-kDa protein was precipitated from virions by both gMC and GST antibodies, suggesting that it was not precipitated specifically. A major 43-kDa protein and lesser amounts of 36- and 30-kDa proteins were precipitated from infected but not uninfected cells by gMC antibody. Antibody against GST did not precipitate these proteins. Samples having a 43-kDa protein precipitated by gMC antibody also had radiolabeled proteins that failed to enter the gel. It is probable that these were aggregates containing the 43-kDa protein because all of the 43-kDa protein appeared at the top of the gel if the samples were boiled prior to electrophoresis (data not shown). However, this aggregate also contained cellular protein because the GST and gMC antibodies precipitated from uninfected cells small amounts of protein that also failed to enter the gel. These data suggest that the 43-kDa protein found in both virions and infected cells is mature gM and that the 36- and 30-kDa proteins are partially glycosylated, unglycosylated, or proteolytically processed gM.
FIG. 2.
BHV-1 gM can be immunoprecipitated from virions, infected cells, virion envelopes, and the membranes but not the cytosol of infected cells. (A) MDBK cells were infected (Inf) at an MOI of 10 or mock infected and labeled with [35S]methionine and -cysteine for 16 h beginning 6 h after infection. Labeled virions were semipurified by centrifugation through a 30% sucrose cushion. The cells and virions were lysed with NP-40 and sodium deoxycholate. Samples were immunoprecipitated with the gMC or GST antibody (Ab), treated at 56°C with SDS-PAGE sample buffer in the presence of the reducing agent DTT, analyzed by SDS-PAGE on a 12% gel, and autoradiographed. (B) Metabolically radiolabeled virions were lysed in NP-40 and sodium deoxycholate and either loaded directly on the gel (−) or immunoprecipitated with gMC antibody (+). Another sample of virus was treated with detergents, and the envelope fraction was cleared of nucleocapsids by centrifugation over a 30% sucrose cushion and either loaded directly on the gel (−) or precipitated with gMC antibody (+). Samples were treated with DTT at 56°C, analyzed by SDS-PAGE on a 12% gel, and autoradiographed. (C) Cells were infected and labeled as for panel A. Cell membranes were obtained from cells disrupted in a Dounce homogenizer, centrifuged at low speed to remove cell debris, pelleted at 12,000 rpm, washed with homogenizing buffer, and repelleted at 40,000 rpm. Lysates of total cells, the 40K supernatant from washed membranes (40KSupe), and the washed membranes themselves were immunoprecipitated with antibody against GST or gM. Precipitates were treated with DTT at 56°C, analyzed by SDS-PAGE on a 12% gel, and autoradiographed.
BHV-1 gM is associated with virion and cell membranes.
To determine if gM was associated with the virion envelope, lysates of partially purified radiolabeled virions and virion envelopes were immunoprecipitated with gMC antibody (Fig. 2B). The envelope lysate had the major glycoproteins identified previously (18, 20). The whole-particle lysate had many additional proteins, most clearly visible below 43 kDa, probably representing nucleocapsid proteins. The gMC antibody precipitated a 43-kDa protein from both. Comigration of the protein precipitated with gMC antibody and the major 43-kDa envelope glycoprotein suggests that the previously recognized 45-kDa protein (18) might have been gM, although it is not clear that all of the 43-kDa band is gM.
To determine if gM was associated with the membranes of infected cells, cell membranes were prepared from radiolabeled BHV-1-infected MDBK cells and immunoprecipitated with gMC antibody (Fig. 2C). Proteins of 30, 36, and 43 kDa were specifically immunoprecipitated from whole cells (Fig. 2C, Total), but only the 30- and 43-kDa proteins were seen in cell membranes (Fig. 2C, Membrane). No gM was found in the soluble fraction of washed cell membranes (Fig. 2C, 40KSupe). To determine the strength of the association, membranes were washed with 1 M NaCl, but even this did not dislodge gM (data not shown). These data imply that both the 30- and 43-kDa proteins are integrated into the membrane, but only the 43-kDa protein is incorporated into virions.
BHV-1 gM has N-linked but not O-linked glycosylation.
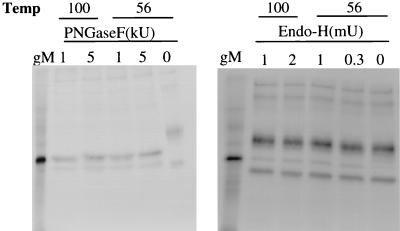
To determine if the 43-kDa protein was the glycosylated form of the 30-kDa protein and determine the type of glycosylation, we immunoprecipitated the 43-kDa protein from infected cells with the gMC antibody and digested it with glycolytic enzymes. Both endo H, which cleaves high-mannose structures, and PNGase F, which removes all N-linked oligosaccharides, reduced the size of the 43-kDa gM to 30 kDa, where it comigrated with gM synthesized in the in vitro translation system (Fig. 3), although endo H digestion was incomplete. Further evidence was obtained by treating infected cells with tunicamycin, an inhibitor of N-linked glycosylation, and immunoprecipitating gM with the gMC antibody. Only a molecule of 30 kDa was precipitated (see Fig. 5, lane 5), suggesting either that no O-linked carbohydrate is normally added or that tunicamycin inhibited O-linked glycosylation by blocking transport out of the endoplasmic reticulum. Thus, all unglycosylated gM, whether synthesized in a reticulocyte lysate in the absence of membranes, made in MDBK cells in the presence of tunicamycin, or deglycosylated by endo H or PNGase F, had an apparent molecular mass of 30 kDa. These data and the presence of only one N-linked glycosylation site strongly suggest that BHV-1 gM is synthesized as a protein with an apparent molecular mass of 30 kDa that is decorated with a single complex N-linked oligosaccharide to an apparent molecular mass of 43 kDa.
FIG. 3.
Deglycosylation of gM by PNGase F and endo H. Lysates of 35S-labeled infected cell membranes were immunoprecipitated with gMC antibody. Precipitates were treated with 0.8% SDS at 100 or 56°C and digested with 0 to 5 kU of PNGase F or 0 to 2 mU of endo H. The gM in the left lane of each panel was synthesized in an in vitro translation system programmed with the BHV-1 UL10 RNA transcript.
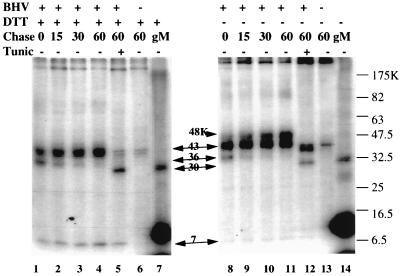
FIG. 5.
Pulse-chase and immunoprecipitation to show the maturation of BHV-1 gM. Proteins from 35S-labeled BHV-1-infected cells without (lanes 1 to 4 and 8 to 11) or with (lanes 5 and 12) tunicamycin treatment (Tunic), and similarly labeled uninfected cells (lanes 6 and 13), were chased into mature proteins for 0 to 60 min with unlabeled amino acids. gM was immunoprecipitated with gMC antibody and analyzed by SDS-PAGE on 10 to 20% gradient gels in the presence (lanes 1 to 7) or absence (lanes 8 to 14) of the reducing agent DTT. Unprecipitated UL10 in vitro translation products were analyzed in lanes 7 and 14. The strong signal at 8 to 10 kDa in the UL10 in vitro translation reaction was not present in reactions done in other reticulocyte lysates.
A 7-kDa protein coprecipitates with BHV-1 gM.
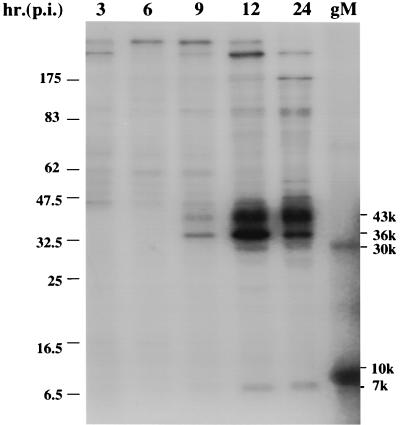
To define the kinetics of gM synthesis, infected cells were pulse-labeled for 1 h at 2, 5, 8, 11, and 23 h after infection. Labeled cells were washed, lysed, and immunoprecipitated with gMC antibody. Precipitated proteins were analyzed by SDS-PAGE in 10 to 20% gradient gels under reducing conditions (Fig. 4). Small amounts of the 43-kDa gM first appeared at 9 h, suggesting that BHV-1 gM is a late protein like other herpesvirus gMs (3, 12, 13, 26). Three other proteins with apparent molecular masses of 36, 30, and 7 kDa appeared at the same time. The density of the 43-kDa protein band increased with time. The 36- and 30-kDa proteins peaked at 12 h and then decreased dramatically, suggesting that the 36-kDa protein was a partially glycosylated intermediate of the 30-kDa unglycosylated gM. The 7-kDa protein appeared with the same kinetics as gM, suggesting that it either coprecipitated with gM or shared an epitope with the C-terminal 63 amino acids of gM. A 10-kDa protein in the UL10 in vitro translation reaction is probably the product of a contaminating RNA because it does not immunoprecipitate with the gMC antibody (data not shown).
FIG. 4.
Coprecipitation of gM and a 7-kDa protein by gMC antibody at various times postinfection (p.i.). MDBK cells were infected at an MOI of 10 and labeled with [35S]methionine and -cysteine for 1 h at the times indicated. Cells were lysed and immunoprecipitated with gMC antibody. The samples were analyzed by SDS-PAGE in a 10 to 20% gradient gel alongside in vitro-translated BHV-1 gM (gM). The 43-, 36-, 30-, 10-, and 7-kDa protein bands are indicated.
Evidence that the association of the 43-kDa gM with the 7-kDa protein requires disulfide bonding.
To clearly characterize the relationship between the various proteins immunoprecipitated by the gMC antibody, we compared immunoprecipitated proteins from pulse-chase-labeled BHV-1-infected MDBK cells, tunicamycin-treated BHV-1-infected cells, and in vitro-translated UL10 RNA by SDS-PAGE in the presence (Fig. 5, left panel) and absence (Fig. 5, right panel) of DTT. In the presence of DTT, a 36-kDa protein present at the beginning of the chase (lane 1) decreased as a 43-kDa protein increased, implying that the 36-kDa protein may be a precursor of the 43-kDa protein. The 36-kDa protein was obviously larger than the unglycosylated gM made in the presence of tunicamycin (lane 5) or by in vitro translation (lane 7), which suggests it may be partially glycosylated gM. No unglycosylated gM was detected in infected cells even without the chase, implying that oligosaccharide was transferred to the sole glycosylation site at amino acids 57 to 59 before synthesis of the C terminus, the site of the gMC epitope(s) between amino acids 375 and 438. The 7-kDa protein was present with glycosylated (lanes 1 to 4) and unglycosylated (lane 5) gM, suggesting that gM glycosylation is not required for formation of the gM–7-kDa protein heterodimer. In the absence of DTT, a small amount of a 36-kDa protein present after the pulse disappeared during the chase, perhaps being chased into a larger protein, either the 43-kDa band that was present during the entire chase or the 48-kDa protein that increased steadily during the chase. Again, the 30-kDa protein detected in tunicamycin-treated infected cells (lane 12) and the in vitro-translated UL10 (lane 14) was not detected during the chase (lanes 8 to 11). The 7-kDa protein is nearly undetectable in all lanes. The majority of the gMC-precipitable protein in the tunicamycin-treated culture was 43 kDa, a protein absent from the mock-infected culture (lane 13) and the tunicamycin-treated sample analyzed in the presence of DTT (lane 5) and perhaps identical to the 43-kDa protein detected in reducing conditions (lanes 1 to 4). The probable interpretation of lanes 8 to 11, therefore, is that the 36-kDa protein is partially glycosylated gM, the 43-kDa band is monomeric fully glycosylated gM, and the 48-kDa band is a fully glycosylated, disulfide-linked heterodimer of 43-kDa gM and the 7-kDa protein. An alternative interpretation is that gM and the 7-kDa protein may require intrachain disulfide bonding of one or both proteins for association. Most gM in tunicamycin-treated cells was linked to the 7-kDa protein, as shown by the presence of a 40-kDa band in nonreducing conditions (lane 12) and 30- and 7-kDa bands in reducing conditions (lane 5), suggesting that glycosylation is not required for dimerization. Again, the band at about 10 kDa in the in vitro translation reaction was not seen when the same RNA was translated in other reticulocyte lysates and was not precipitated by the gMC antibody. We concluded it was the product of a contaminating RNA.
The 7-kDa protein linked to gM is UL49.5.
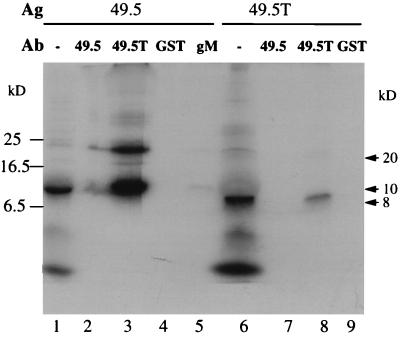
Liang et al. characterized BHV-1 UL49.5 as a nonglycosylated 9-kDa virion surface protein disulfide linked to an unidentified 39-kDa virion protein (14). We speculated that this 39-kDa protein might be gM. To determine if our 7-kDa protein was UL49.5, we produced antibodies against GST fusion proteins containing BHV-1 UL49.5. To confirm that the antibodies recognized the UL49.5 protein, we cloned full-length and N-terminally truncated UL49.5 ORFs downstream of the T7 promoter in pcDNA3, produced transcripts in vitro, and translated the transcripts in a rabbit reticulocyte lysate. A 10-kDa protein was synthesized from the UL49.5 transcript (Fig. 6, lane 1), and an 8-kDa protein was synthesized from the truncated UL49.5 transcript (lane 6). Bands in common between the in vitro translations may represent rabbit reticulocyte proteins or fragments of UL49.5. Antibodies directed against the truncated UL49.5 protein precipitated both UL49.5 and UL49.5T translation products (lanes 3 and 8). The 20-kDa band precipitated by the UL49.5T antibody only from full-length transcripts may be the UL49.5 dimer described by Liang et al. (14). Antibodies engendered by full-length UL49.5 precipitated UL49.5 very poorly (lane 2) and did not precipitate UL49.5T (lane 7). GST and gM antibodies did not precipitate either protein (lanes 4, 5, and 9). We interpret these results as evidence that the UL49.5T antibody recognizes either unprocessed or processed UL49.5 protein. This antibody was used in all subsequent experiments. It is not clear why the full-length UL49.5 protein did not engender a strong antibody response, but this question was not explored further.
FIG. 6.
Immunoprecipitation of proteins encoded by full-length (49.5) and N-terminally truncated UL49.5 (49.5T) synthesized in vitro with antibodies (Ab) directed against fusion proteins containing the full-length UL49.5 protein (lanes 2 and 7), the truncated protein (lanes 3 and 8), GST (lanes 4 and 9), or gM (lane 5). For comparison, 1 μl of the unprecipitated UL49.5 translation reaction (lane 1) and 5 μl of the UL49.5T translation reaction (lane 6) were loaded. Each immunoprecipitation was begun with 5 μl of translation reaction mixture. Samples were analyzed by SDS-PAGE in an 18% gel and autoradiographed. Ag, antigen.
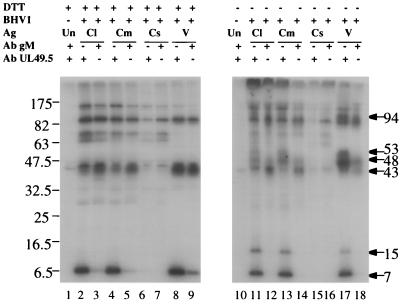
Coimmunoprecipitation of gM and UL49.5 from BHV-1 virions and infected cell lysates was used to show their association. Cells were labeled with [35S]methionine and -cysteine between 10 and 26 h after BHV-1 infection, and lysates were prepared. Virions were semipurified by centrifugation through a 30% sucrose cushion and lysed with nonionic detergents. Both gM and UL49.5 antibodies precipitated both 7- and 43-kDa proteins from infected cell lysate (Fig. 7, lanes 2 and 3), infected cell membranes (lanes 4 and 5), and virions (lanes 8 and 9). These proteins were not precipitated from a 40K supernatant of infected cell membranes (lanes 6 and 7). More 7-kDa protein than 43-kDa protein was precipitated by UL49.5 antibody (lanes 2, 4, and 8), and more 43-kDa protein than 7-kDa protein was precipitated by gM antibody (lanes 3, 5, and 9). Different ratios of 43- and 7-kDa proteins may reflect the presence of uncomplexed gM and UL49.5. Other bands appeared in all immunoprecipitations, including those from the 40K supernatant which contained very little gM or UL49.5, suggesting they may be unrelated to gM and UL49.5.
FIG. 7.
Immunoprecipitation of the gM-UL49.5 complex with antibody against either gM or UL49.5. Lysates of radiolabeled uninfected cells (Un), BHV-1-infected cells (Cl), cell membranes (Cm), 40K cell membrane supernatants (Cs), and semipurified virions (V) were precipitated with antibody (Ab) against truncated UL49.5T or gM. Uninfected cells (lanes 1 and 10) were immunoprecipitated with antibodies against both gM and UL49.5. The immunoprecipitates were incubated at 56°C in the presence (lanes 1 to 9) or absence (lanes 10 to 18) of DTT, analyzed by SDS-PAGE in 10 to 20% gradient gels, and autoradiographed. Positions of molecular mass markers (left) and calculated molecular masses of antibody-precipitated bands (right), both in kilodaltons, are indicated. Ag, antigen.
To show that the 43-kDa gM and 7-kDa UL49.5 proteins were linked by disulfide bonds, the same immunoprecipitations were analyzed in the absence of DTT (Fig. 7, right panel). Two bands, 43 and 48 kDa, were precipitated from infected cells by gM antibody (lanes 12 and 14). This result was identical to that in Fig. 5. Five bands, 7, 15, 48, 53, and 94 kDa, were specifically precipitated from infected cell lysates, cell membranes, and virions by UL49.5 antibody (lanes 11, 13, and 17) but not from uninfected cells (lane 10) or the 40K supernatant (lane 15). We interpret these data as evidence that some UL49.5 (7 kDa) and its dimer (15 kDa) and some gM (43 kDa) are independent and that some UL49.5 and gM are coprecipitated with antibody to either. As reported by Liang et al. (14), the 15-kDa protein band is seen only in the absence of DTT, suggesting that it is a disulfide-linked UL 49.5 dimer. The 48-kDa protein may be a complex of gM and UL49.5 because it is the only protein precipitated by both gM and UL49.5T antibodies. The 53-kDa protein precipitated by UL49.5 antibody from all infected cell preparations and virions (lanes 11, 13, and 17) may be a complex of gM and the UL49.5 dimer. A possible explanation for it not being precipitated by gM antibody is that the UL49.5 dimer blocks the gM C-terminal epitope(s) or UL49.5 forms a heterodimer with an as yet unidentified partner slightly larger than gM. The linkage between gM and UL49.5 must depend on a disulfide bond because gM precipitated with UL49.5 antibody resolved as a 43-kDa protein in the presence of DTT (lanes 2, 4, and 8) and 48- and 53-kDa proteins in the absence of DTT (lanes 11, 13, and 17), and the UL49.5 precipitated with gM antibody appears as a 7-kDa protein in the presence (lanes 3, 5, and 9) but not in the absence (lanes 12, 14, and 18) of DTT.
The identities of other bands in the immunoprecipitates were less clear. A sharp band at 43 kDa was precipitated by all antibodies, including nonimmune sera (lanes 1 and 10), suggesting that it was a cellular protein precipitating nonspecifically. All bands above the 47.5-kDa marker in DTT-treated precipitates appeared in the absence of gM and UL49.5 (lanes 6 and 7), suggesting that they were unrelated to gM and UL49.5. Bands of similar size also were seen in nonreduced precipitates, and at least three of these bands were seen in the absence of gM and UL49.5 (lanes 15 and 16), suggesting that they also were unrelated to gM and UL49.5. An indistinct, DTT-sensitive band labeled 94 kDa is particularly prominent in precipitates from virions (lanes 17 and 18) and may represent a dimer of the gM-UL49.5 dimer because it was precipitated by both antibodies.
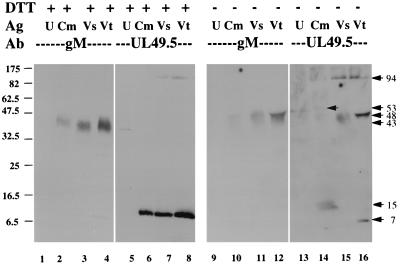
To further confirm the identification of proteins seen in immunoprecipitations, Western blots of uninfected cells, infected cell membranes, semipurified BHV-1, and highly purified BHV-1 separated by SDS-PAGE in the presence and absence of DTT were probed successively with antibodies against UL49.5 and gM (Fig. 8). Infected cells and virus analyzed in the presence of DTT contained a 7-kDa protein that reacted with antibody against UL49.5 (lanes 6 to 8). When the same blot was stripped and reprobed with antibody against gM, the 43-kDa protein reacted (lanes 2 to 4). This result showed that the antibodies reacted only with the intended target protein. When the same samples separated in nonreducing SDS-PAGE were probed with antibody against gM, a 48-kDa protein was identified by both gM (lanes 10 to 12) and UL49.5 (lanes 14 to 16) antibodies. Antibody against UL49.5 also identified 15- and 53-kDa proteins in infected cells (lane 14) and a 7-kDa protein in tartrate gradient-purified virions (lane 16). Antibody against UL49.5 also identified a larger protein labeled 94 kDa (lanes 15 and 16). It is not clear that this is the same protein labeled 94 kDa in Fig. 7. We interpret the reactivity of UL49.5 antiserum with a 7-kDa protein in the reducing blot and 48-kDa protein in the nonreducing blot as clear evidence that gM and UL49.5 are linked into a 48-kDa dimer by one or more disulfide bonds. Reactivity of UL49.5 antibody with the 94-kDa protein that does not react with antibody against gM suggests that UL49.5 may bind to some other protein as well, but the appearance of the 94-kDa protein in samples run with DTT suggests that this does not require a disulfide bond. Further, the reactivity of UL49.5 antibody with the 48- and 53-kDa proteins in the nonreducing blot of infected cell membranes (lane 14) but only the 48-kDa protein in the blot of purified virions (lane 16) suggests a maturation process in which UL49.5 dimers are linked to gM and then dissociated, leaving only monomers of UL49.5 attached to gM. The presence of free UL49.5 in purified virions suggests that either some UL49.5 is free in virions or the disulfide linkage to gM was occasionally cleaved during the preparation of virions.
FIG. 8.
Analysis of the gM-UL49.5 complex by Western blotting. Cells were infected at an MOI of 10 and collected 24 h later, and membranes were prepared (Cm). Mock-infected cells were collected at the same time, and membranes were similarly prepared (U). Virions were prepared from cells infected for 60 h at an MOI of 0.5 and either semipurified (Vs) or banded on a potassium tartrate gradient (Vt). Lysates were incubated at 56°C in the presence (lanes 1 to 8) or absence (lanes 9 to 16) of 40 mM DTT for 10 min, analyzed by SDS-PAGE in 10 to 20% gradient gels, and transferred to nitrocellulose paper. The blots were probed with UL49.5T antibody (Ab) first. Bound antibody was detected by ECL (Amersham). The blots were stripped, reprobed with gM antibody, and again detected by ECL. Positions of molecular mass markers on the (left) and calculated sizes of specific bands are indicated on the (right), both in kilodaltons, are indicated. Ag, antigen.
DISCUSSION
We interpret our data as evidence that BHV-1 UL10 encodes gM, that nascent gM polypeptides obtain an immature oligosaccharide before the C-terminal epitope is synthesized, and that by the time gM becomes reactive with the gMC antibody, some is glycosylated to a 43-kDa protein lacking a UL49.5 partner and some has been both glycosylated and linked to UL49.5 to form the mature 48-kDa heterodimer. Further, our data suggest that all of the gM present in the cell membrane has been fully glycosylated but some lacks its UL49.5 partner, and that the gM incorporated into the virion has been fully glycosylated and most has been linked to UL49.5. Our data also suggest that UL49.5 is linked to the 43-kDa gM prior to incorporation into virions. The lack of a 53-kDa protein in complexes immunoprecipitated with the gMC antibody suggests that the UL49.5 dimer is cleaved before or shortly after linkage to gM.
The large disparity between the calculated molecular mass of 45 kDa for unglycosylated BHV-1 gM and the apparent size of 30 kDa measured for gM synthesized in vitro was not unexpected. There are three possible explanations. First, translation may have initiated after the first AUG or terminated prematurely. The first AUG is used to initiate HSV-1 (2) and EHV-1 (22) gM. Since the second AUG in BHV-1 gM is at codon 122 and would code for a protein of 33 kDa, our data might suggest initiation at the second AUG. However, the only N-linked glycosylation site in BHV-1 gM is at amino acids 71 to 73. Initiation at the second AUG would produce a protein with no N-linked glycosylation site, which is inconsistent with our data. Premature termination would result in a protein that would not react with our gMC antibody and also is inconsistent with our results. Second, full-length gM could be proteolytically cleaved near either the N or C terminus. It seems unlikely that it is cleaved at the N terminus because gM synthesized in the presence of tunicamycin (Fig. 5, lanes 5 and 12) has the same mobility as gM synthesized in an in vitro translation system without membranes (Fig. 5, lanes 7 and 14), strongly suggesting that no signal peptide is cleaved from gM. In addition, no signal peptide has been found in other herpesvirus gMs studied to date. Again, it is unlikely that the C terminus is cleaved from the gM because this is the site of the epitope recognized by the gMC antibody and only the C-terminal fragment would be precipitated. Third, anomalously rapid migration in SDS-PAGE has been noted for other herpesvirus proteins with multiple membrane-spanning domains such as PRV deglycosylated gM, which migrates with an apparent mass of slightly over 30 kDa despite its predicted size of 42 kDa (7), and HSV gK, which has an apparent mass of 29 kDa but has a predicted size of 36 kDa (9). Therefore, we concluded that BHV-1 gM was probably also migrating anomalously rapidly in SDS-PAGE and that the 30-kDa nonglycosylated protein made in in vitro translation reactions was probably the full-length 45-kDa BHV-1 gM. If this is true, the actual molecular mass of glycosylated gM may be around 60 kDa and the complex with the 7-kDa molecule may be 67 kDa. The anomalous migration of these proteins in SDS-PAGE casts some doubt on the conclusion that gM lacks O-linked carbohydrate because it is not certain that gM with and without O-linked glycosylation would migrate to different positions in a gel.
The 7-kDa protein associated with gM is the 96-amino-acid UL49.5. The BHV-1 UL49.5 sequence predicts a protein with a molecular mass of 10 kDa that would be reduced to 8 kDa if it were cleaved at amino acid 21 as predicted by the method of von Heijne (29). The first 16 amino acids constitute a potential signal sequence, and amino acids 35 to 51 of the putative mature protein constitute a probable membrane anchor, making it very similar to homologs in other herpesviruses (4, 5). A corresponding O-glycosylated protein, designated gN, has been identified in pseudorabies virion envelopes (10). Liang et al. (14) expressed BHV-1 UL49.5 as a fusion protein and made antibodies that identified a nonglycosylated 9-kDa virion surface protein. They showed it was disulfide linked to an unidentified 39-kDa virion protein and showed that it was disulfide linked to itself to make an 18-kDa dimer. This finding corresponds closely with our data.
How are gM and UL49.5 linked? All known gM homologs have a cysteine at the beginning of the second hydrophobic domain 40 to 70 amino acids from the N terminus (13), another 114 to 135 amino acids away from the first, near the end of the fifth hydrophobic domain, and others scattered throughout. Since each UL49.5 homolog has a cysteine about 14 amino acids proximal (15) and another immediately distal to the putative transmembrane anchor sequence, BHV-1 UL49.5 having only these cysteines, it is likely that one or both of the conserved cysteines in each molecule form disulfide bonds between the two molecules. Linkage of the two proteins by two disulfide bonds would likely constrain the conformation of both. An alternative explanation is that intrachain disulfide bonds are required to hold one or both of these molecules in a conformation that permits noncovalent interactions to bind the two molecules together. Our data suggest that this is unlikely because the dimer remained intact when it was treated with 1% SDS and heated to 56°C prior to Western blotting (Fig. 8).
Since other proteins immunoprecipitated with gM and UL49.5, it could be argued that these two proteins may be linked via an intermediary. However, when unreduced proteins were analyzed by Western blotting (Fig. 8), a band corresponding to the sum of the gM and UL49.5 molecular weights reacted with antibodies against both proteins, suggesting any additional member(s) of the complex would have to be very small.
Where and when are gM and UL49.5 linked? Since unglycosylated gM made in the presence of tunicamycin was linked to the UL49.5, the linkage must be possible prior to glycosylation and therefore may occur in the endoplasmic reticulum.
Why did it take so long to identify BHV-1 gM and its UL49.5 partner? The prominent 42-kDa protein seen in [3H]glucosamine-labeled virions (20) and 45-kDa glycoprotein seen in BHV-1 envelopes (18) were probably gM, but they have not been investigated. The fact that BHV-1 gM has only five methionines among its 438 amino acids suggests that if the 45-kDa protein is gM, either there must be a large amount of it in virion membranes or there is a second protein that comigrates with gM to account for the intensity of the 45-kDa band labeled with [35S]methionine. Perhaps this very hydrophobic protein escaped notice simply because it aggregates under the normal conditions for SDS-PAGE or because it is a relatively poor antigen.
Do the gMs of other herpesviruses have a smaller partner? gM has been particularly difficult to investigate because its many hydrophobic domains cause it to aggregate when boiled in SDS-PAGE sample buffer, and so it often fails to resolve on gels (2, 7, 21, 23). In addition, immunoprecipitates were often analyzed on low-percentage gels where a small protein would not have been resolved (2). In many studies, gM was reduced and analyzed by Western blotting, and so an associated protein could not have been observed (23). Nevertheless, Osterrieder et al. (22) noted that EHV-1 gM migrated more slowly in SDS-PAGE in the absence of 2-mercaptoethanol but reasoned that this was caused by intramolecular disulfide bonds. However, they observed a decrease in apparent mass from 59 to 61 kDa to 50 to 55 kDa, which is exactly what would be expected if the EHV-1 gene 10 protein was released by treatment with 2-mercaptoethanol.
What is the function of the gM-UL49.5 dimer? Deletion and insertion mutations in gM had minor effects on membrane penetration speed and replication efficiency (1, 7, 16, 17, 21). However, it is unclear if critical areas of gM were functional in these mutants because all changes were made distal to the conserved N-linked glycosylation site and the N-terminal proline-cysteine pair that may be the attachment site for UL49.5. Deletion of varicella-zoster virus ORF9A, the UL49.5 homolog, caused a slight decrease in viral replication and syncytium formation. Deletion of BHV-1 and HSV-1 UL49.5 had minor effects on viral replication (15, 24). Osterrieder et al. (22) speculated that gM might create an ion channel but did not observe functional ion channels when EHV-1 gM was expressed in Xenopus laevis oocytes (22). The absence of a covalently bound UL49.5 may help explain these negative data. Clearly, the gM-UL49.5 dimer requires further investigation.
ACKNOWLEDGMENTS
We thank Etienne Thiry and Eric Baranowski for their generous gift of the BH 44 monoclonal antibody, which was critical to preliminary experiments leading up to the present study. The assistance of Chad Johnson, Peter Evans, and Jing Wu is gratefully acknowledged.
This research was supported by USDA National Research Initiative grants 92-37204-7900 and 94-37204-0857 and a Shaw Scholarship to G.J.L. from the Milwaukee Foundation.
ADDENDUM IN PROOF
After this article was accepted for publication, Jöns et al. (A. Jöns, J. M. Dijkstra, and T. C. Mettenleiter, J. Virol. 72:550–557, 1998) published their finding that pseudorabies virus gM and gN form a disulfide-linked dimer.
REFERENCES
- 1.Baines J D, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baradaran K, Dabrowski C E, Schaffer P A. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC. J Virol. 1994;68:4251–4261. doi: 10.1128/jvi.68.7.4251-4261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker D E, Roizman B. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–566. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett B C, Dolan A, Telford E A R, Davison A J, McGeoch D J. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J Gen Virol. 1992;73:2167–2171. doi: 10.1099/0022-1317-73-8-2167. [DOI] [PubMed] [Google Scholar]
- 6.Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68:7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jons A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kari B, Li W, Cooper J, Goertz R, Radeke B. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J Gen Virol. 1994;75:3081–3086. doi: 10.1099/0022-1317-75-11-3081. [DOI] [PubMed] [Google Scholar]
- 12.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Eidman K, Gehrz R C, Kari B. Identification and molecular characterization of the murine cytomegalovirus homolog of the human cytomegalovirus UL100 gene. Virus Res. 1995;36:163–175. doi: 10.1016/0168-1702(94)00117-u. [DOI] [PubMed] [Google Scholar]
- 14.Liang X, Chow B, Raggo C, Babiuk L A. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J Virol. 1996;70:1448–1454. doi: 10.1128/jvi.70.3.1448-1454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang X, Tang M, Manns B, Babiuk L A, Zamb T J. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology. 1993;195:42–50. doi: 10.1006/viro.1993.1344. [DOI] [PubMed] [Google Scholar]
- 16.MacLean C A, Efstathiou S, Elliott M L, Jamieson F E, McGeoch D J. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol. 1991;72:897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- 17.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigation of its role in vivo. J Gen Virol. 1993;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 18.Marshall R L, Rodriguez L L, Letchworth G J. Characterization of envelope proteins of infectious bovine rhinotracheitis virus (bovine herpesvirus 1) by biochemical and immunological methods. J Virol. 1986;57:745–753. doi: 10.1128/jvi.57.3.745-753.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayfield J E, Good P J, VanOort H J, Campbell A R, Reed D E. Cloning and cleavage site mapping of DNA from bovine herpesvirus 1 (Cooper strain) J Virol. 1983;47:259–264. doi: 10.1128/jvi.47.1.259-264.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misra V, Blumenthal R M, Babiuk L A. Proteins specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus) J Virol. 1981;40:367–378. doi: 10.1128/jvi.40.2.367-378.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osterrieder N, Neubauer A, Brandmuller C, Braun B, Kaaden O R, Baines J D. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osterrieder N, Neubauer A, Fakler B, Brandmuller C, Seyboldt C, Kaaden O R, Baines J D. Synthesis and processing of the equine herpesvirus 1 glycoprotein M. Virology. 1997;232:230–239. doi: 10.1006/viro.1997.8561. [DOI] [PubMed] [Google Scholar]
- 23.Pilling A, Davison A J, Telford E A, Meredith D M. The equine herpesvirus type 1 glycoprotein homologous to herpes simplex virus type 1 glycoprotein M is a major constituent of the virus particle. J Gen Virol. 1994;75:439–442. doi: 10.1099/0022-1317-75-2-439. [DOI] [PubMed] [Google Scholar]
- 24.Pyles R B, Sawtell N M, Thompson R L. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol. 1992;66:6706–6713. doi: 10.1128/jvi.66.11.6706-6713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross J, Williams M, Cohen J I. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology. 1997;234:186–195. doi: 10.1006/viro.1997.8652. [DOI] [PubMed] [Google Scholar]
- 26.Scalzo A A, Forbes C A, Davis-Poynter N J, Farrell H E, Lyons P A. DNA sequence and transcriptional analysis of the glycoprotein M gene of murine cytomegalovirus. J Gen Virol. 1995;76:2895–2901. doi: 10.1099/0022-1317-76-11-2895. [DOI] [PubMed] [Google Scholar]
- 27.Schwyzer M, Styger D, Vogt B, Lowery D E, Simard C, LaBoissière S, Misra V, Vlcek C, Paces V. Gene contents in a 31-kb segment at the left genome end of bovine herpesvirus-1. Vet Microbiol. 1996;53:67–77. doi: 10.1016/s0378-1135(96)01235-7. [DOI] [PubMed] [Google Scholar]
- 28.Vlcek C, Benes V, Lu Z, Kutish G F, Paces V, Rock D, Letchworth G J, Schwyzer M. Nucleotide sequence analysis of a 30-kb region of the bovine herpesvirus 1 genome which exhibits a colinear gene arrangement with the UL21 to UL4 genes of herpes simplex virus. Virology. 1995;210:100–108. doi: 10.1006/viro.1995.1321. [DOI] [PubMed] [Google Scholar]
- 29.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X P, Wu S, Letchworth G J. Yeast-secreted bovine herpesvirus type 1 glycoprotein D has authentic conformational structure and immunogenicity. Vaccine. 1997;15:679–688. doi: 10.1016/s0264-410x(96)00234-4. [DOI] [PubMed] [Google Scholar]