Abstract
The vpr gene of human immunodeficiency virus type 1 (HIV-1) encodes a 96-amino-acid 14-kDa protein (viral protein R [Vpr]), which is produced late in the viral life cycle and is incorporated into the virion. Although Vpr is not required for viral replication in transformed cell lines and primary T lymphocytes, it is essential for productive infection of macrophages and monocytes and appears to be important for pathogenesis in vivo. To establish the role of Vpr in HIV-1 replication and pathogenesis, we have isolated cellular proteins with which Vpr interacts. By using the yeast two-hybrid system, Lys-tRNA synthetase (LysRS) was identified as a Vpr-interacting protein. The interaction between Vpr and LysRS was characterized both in vitro and in vivo, and the domains of Vpr required for the interaction were defined. In the presence of Vpr, LysRS-mediated aminoacylation of tRNALys is inhibited. Since tRNALys is the primer for reverse transcription of the HIV-1 genome, this suggests that the interaction between Vpr and LysRS may influence the initiation of HIV-1 reverse transcription.
Human immunodeficiency virus (HIV), the causative agent of AIDS, is a member of the Retroviridae family of viruses (reviewed in references 5 and 8). Primate immunodeficiency viruses are unique members of this family in that, in addition to the obligatory Gag, Pol, and Env proteins, their mRNA encodes six regulatory proteins, namely, Tat, Rev, Vif, Vpr, Vpu, and Nef (11, 62, 64). While Tat and Rev are essential for viral replication in all cell types, Vif, Vpr, Vpu, and Nef are dispensable for productive infection of transformed T lymphocytes and have therefore been termed accessory proteins. However, functional analysis of these so-called accessory proteins has shown their involvement in almost every stage of the viral life cycle including the infection process, nuclear migration of the preintegration complex, transcription of the provirus, and exit of the mature virion (51). In fact, the ability of HIV-1 to establish latent and chronic infections and to induce disease in vivo has been attributed to these proteins. To further understand the role of HIV-1 regulatory proteins during viral replication and pathogenesis, we focused on the Vpr gene product.
The vpr gene of HIV-1 encodes a 96-amino-acid 14-kDa protein (viral protein R [Vpr]), which is produced late in the viral life cycle and is incorporated into the virion through an interaction with the p6 region of Gag (36, 38, 46, 53). Vpr is the only accessory protein found within the virion in substantial amounts and is present at molar amounts equivalent to those of Gag. In vitro, Vpr is not required for viral replication in transformed cell lines and primary T lymphocytes; however, it is essential for productive infection of cells such as macrophages and monocytes (3, 22, 70). This is extremely important for viral pathogenesis in vivo because terminally differentiated macrophages are a natural cell target for HIV and provide a reservoir of viral production during the asymptomatic stages of disease (2). The most convincing evidence that Vpr plays an important role in vivo comes from experiments showing that rhesus monkeys infected with simian immunodeficiency virus (SIV) with Vpr deleted have a low viral burden and slow disease progression compared to those infected with the wild-type virus (25, 29, 37). Throughout the evolution of the lentiviruses, the vpr gene has been highly conserved, suggesting that it has an important function in pathogenesis (63).
The precise mechanisms by which Vpr influences viral replication are still unclear, but there is evidence to suggest that it is involved in nuclear transport of the preintegration complex in nondividing cells (24), a role that is in keeping with its virion association and accumulation in the nuclei of infected cells (45, 71). Vpr has also been shown to act as a weak transcriptional activator of the HIV long terminal repeat and other heterologous promoters (1, 67, 68), which may explain the fact that exogenous Vpr reactivates viral gene expression in latently infected T-cell lines (40, 41). Several groups have demonstrated that Vpr causes primary CD4+ lymphocytes and other cell types to accumulate in the G2 phase of the cell cycle (4, 17, 23, 34, 39, 54, 55, 73), possibly through its ability to associate with the HIV-1-encoded nucleocapsid protein p7 (NCp7), and activate protein phosphatase 2A (15, 42, 65). It has been postulated that, by blocking cell division prior to mitosis, Vpr prevents chronic viral infection and drives cells into apoptosis, thus contributing to the immunopathogenic effect of HIV (54, 57). Both the small size and simple genomic organization of HIV suggest that individual gene products have several distinct functions. Indeed, HIV virions which have packaged a wild-type Vpr but contain a mutation in the vpr gene replicate poorly in primary macrophages, suggesting that Vpr plays unique functional roles at different times in the viral life cycle (10). To further establish the role of Vpr in HIV-1 replication and pathogenesis, the yeast two-hybrid system was used to identify cellular proteins which interact with Vpr. We isolated a cDNA clone that coded for Lys-tRNA synthetase (LysRS) and characterized the interaction between Vpr and LysRS both in vitro and ex vivo. A functional consequence of the interaction is that in the presence of Vpr, LysRS-mediated aminoacylation of tRNALys is inhibited. The fact that tRNALys is the primer for reverse transcription of the HIV-1 genome suggests that the interaction between Vpr and LysRS may influence the initiation of HIV-1 reverse transcription.
MATERIALS AND METHODS
Plasmid construction.
pV44ER.LexA and pACT/pACT-cDNA were received from Colin Goding (Marie Curie Research Institute, Oxted, United Kingdom) and Stephen Elledge (Baylor College of Medicine, Houston, Tex.), respectively, and both have been described previously (16, 32). pLexA-vprwt, the bait plasmid used in this study, was obtained by subcloning the HIV-1 LAI vpr gene from the BamHI site of pGEX-vprwt (a generous gift from F. Bachelerie, Institut Pasteur, Paris, France) into BglII-digested pV44ER.LexA. pLexA-vprwt expressed a chimeric protein containing the LexA bacterial repressor (amino acids 1 to 211) at its N terminus and Vpr (amino acids 2 to 96) at its C terminus. pLexA-VprΔC contained the 186-bp BamHI-EcoRI fragment of pGEX-vprwt cloned into BamHI-EcoRI-digested pV44ER.LexA. pLexA-vprN was constructed by subcloning the 118-bp BamHI-NcoI fragment of pGEX-vprwt into pV44ER.LexA cut with the same enzymes. To obtain pLexA-vprC, the vpr open reading frame was amplified with oligonucleotides vpr1 (5′) and vpr2 (3′), which contained BamHI and ClaI restriction sites, respectively. The resultant product was digested with the appropriate enzymes and cloned into BglII-EcoRI-cut pV44ER.LexA. pLexA-vprΔC, pLexA-vprN, and pLexA-vprC expressed LexA in fusion with amino acids 2 to 62, 2 to 39, and 35 to 96 of Vpr respectively. The control bait plasmids used in the yeast two-hybrid system, pLexA-Da and pLexA-Rb, expressed the daughterless gene of Drosophila and the retinoblastoma protein, respectively, as LexA hybrids. These plasmids were provided by C. Goding. pACT-cDNA plasmids contained the Gal4 activation domain (Gal4AD) fused to a cDNA expression library generated from an Epstein-Barr virus-transformed B-cell line.
For in vitro protein-protein interactions, Vpr deletions were expressed as glutathione S-transferase (GST) fusion proteins. pGEX-vprwt contains the vpr gene of the LAI HIV-1 isolate cloned into the BamHI site of pGex2T. This vector expressed amino acids 2 to 96 of Vpr as a fusion protein with GST at the N terminus. pGEX-vprΔC was obtained by cloning the 186-bp BamHI-EcoRI fragment of pGEX-vprwt into pGex2T (Pharmacia) digested with the same enzymes. Oligonucleotides vpr3 (5′, BamHI site) and vpr4 (3′, EcoRI site) were used to amplify bp 4 to 114 of the vpr gene, which was then cloned into BamHI-EcoRI-digested pGex2T to generate pGex-vprN. pcDNA-lysRS was constructed by amplification of pACT-c2.10 with oligonucleotides c2.10(3) (5′, upstream ATG; BamHI site) and c2.10(4) (3′, EcoRI site). The PCR product was digested with BamHI and EcoRI and cloned into pCDNA3 (Invitrogen) cleaved with the same enzymes. This plasmid expressed LysRS in fusion with the 9-amino-acid hemagglutinin (HA) tag (YPYDVPDYA). To generate pGEX-lysRS, oligonucleotides c2.10(1) and c2.10(2) were used to amplify pACT-c2.10 DNA. The BamHI-EcoRI-digested product was then cloned into pGEX-2T digested with BamHI and EcoRI. Plasmid constructions were confirmed by automated DNA sequencing (ABI 377 sequencer).
Yeast two-hybrid system.
Growth and manipulation of yeast strains were carried out by standard procedures (21). The Saccharomyces cerevisiae reporter strain L40 was a kind gift from S. Hollenberg (Fred Hutchinson Cancer Research Center, Seattle, Wash.) and contains HIS3 and lacZ reporter genes under the control of LexA DNA binding sites. The procedure of Gietz et al. (19) was used to transform the L40 reporter strain with pLexA-vprwt. Yeast cells carrying the bait plasmid were then transformed with the activation domain-tagged cDNA library and grown on synthetic medium deficient in tryptophan, leucine, and histidine (SC Trp−, Leu−, His−). Aliquots were taken from each transformation mix before plating and grown on SC Tryp−, Leu− to determine the transformation efficiency. After 3 to 5 days of growth, His+ colonies were screened for β-galactosidase activity by a filter lift assay (7). Colonies that were positive in this screen were then “cured” of the bait vector by several rounds of growth in synthetic medium lacking leucine but containing tryptophan (SC Leu−, Trp+). Cured yeast strains were retransformed with the bait plasmid and control plasmids expressing nonspecific LexA fusion proteins. DNA was extracted from true-positive clones by the method of Hoffman and Winston (26), electroporated into Escherichia coli DH5α, and subjected to miniprep DNA analysis by automated sequencing.
To identify domains of Vpr that interact with c2.10, L40 was cotransformed with pLexA-vprwt and deletion mutants plus pACT-c2.10 and then grown on selective medium containing 0 to 50 mM 3-aminotriazole (3-AT). For quantification of β-galactosidase activity, single yeast colonies were picked from SC Trp−, Leu−, His+ plates into 50 μl of 100 mM potassium phosphate containing 0.2% Triton X-100. Then 2.5 μl of 0.1% sodium dodecyl sulfate (SDS) and 7.5 μl of chloroform were added, and the yeast cells were lysed by vigorous vortexing for 10 s. The Galacto-Light Plus (Tropix) chemiluminescent reporter assay was used to measure the relative light units (RLU) produced by interacting proteins. Values reported are the means from duplicate assays of four independent transformants.
Purification of GST fusion proteins.
GST fusion proteins were purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced E. coli essentially as described previously (61). After elution of GST-LysRS, peak fractions were pooled and a Centricon 30 (Amincon) was used to concentrate and exchange the protein into 100 mM Tris.HCl (pH 7.8). When fusion proteins bound to glutathione-agarose were required, the bacterial supernatant was incubated for 1 h at 4°C with 1 ml of a 50% (vol/vol) suspension of glutathione-agarose. The protein-bound agarose beads were washed four times with lysis buffer and resuspended in lysis buffer containing 0.02% sodium azide.
Aminoacylation assays.
Aminoacylation reactions were carried out by the method of Senger et al. (60). Assay mixtures, in a final volume of 20 μl, contained 0.5 ng (300 nM) of GST-LysRS, 144 mM Tris.HCl (pH 7.8), 5 mM dithiothreitol, 2 mM ATP, 10 mM MgCl2, 0.1 mg of bovine serum albumin (BSA) per ml, 5 kBq of l-[14C]lysine(1.85 MBq/ml; Amersham), and 20 μM bacterial tRNALys (Sigma). Following incubation for various times at 25°C, the reactions were stopped by the addition of ice-cold 10% trichloroacetic acid (TCA). Precipitates were collected by filtration through fiberglass disks, which were washed three times in 10% TCA, three times in 5% TCA, and once in 70% ethanol. Bound radioactivity was measured in a scintillation counter. To determine the effect of Vpr on LysRS activity 0.5 ng of GST-LysRS was incubated with either 0.5 ng of wild-type GST-Vpr or 0.5 ng of GST (in 1 M Tris HCl [pH 7.8]) before use in aminoacylation assays. The concentrations of all other assay components remained as above.
In vitro binding studies.
Glutathione-agarose beads (20 μl), bound to wild-type and mutant GST-Vpr fusion proteins (ca. 2 μg), were incubated for 30 min at 4°C with 1 ml of 3% BSA. After one wash in interaction buffer (750 mM potassium acetate, 0.1% Tween 20, 2 mg of BSA per ml), the beads were resuspended in 200 μl of interaction buffer containing either 10 μl of in vitro-translated LysRS or 100 μg of U937 cytoplasmic extract (made by standard detergent lysis techniques) and incubated for 2 h at 4°C. Samples were washed six times with 1 ml of interaction buffer without BSA, denatured (by boiling in the presence of 1.25% SDS and 0.35 M 2-mercaptoethenol), and subjected to polyacrylamide gel electrophoresis (PAGE).
In vitro transcription and translation.
To generate 35S-labelled LysRS, pcDNA-LysRS was used as template in the TNT-coupled wheat germ extract system (Promega) as specified by the manufacturer. Proteins were translated in a final volume of 50 μl in the presence of [35S]methionine (>1,000 Ci/mmol; Amersham), and 10 μl was assayed for binding to wild-type and mutant GST-Vpr proteins bound to the beads. Bound proteins were analyzed with a phosphorimager (Fuji BAS1000).
Western blotting analysis and antibodies.
To detect cellular LysRS, proteins were transferred to polyvinylidene difluoride (Sigma) membranes and subjected to Western blotting by standard procedures (69). Human antiserum derived from a patient with the anti-synthetase syndrome (a kind gift from C. Gelpi, Hospital de la Santa Cruz and San Pablo, Barcelona, Spain) (18) was used at a 1:1,000 dilution as the primary antibody in these studies. The serum was tested against 1,000 to 50 ng of recombinant GST-LysRS and 100 to 10 μg of cellular extract before use in interaction experiments.
Oligonucleotide primers.
Oligonucleotide sequences (shown in the sense orientation with restriction sites underlined) are as follows: vpr1 (upC120-310), GCACGGATCCCCTAGGATTTGGCTCCATAACTTA; vpr2, GGCAATCGATCTAGGATCTACTGGCTCCATT; vpr3, AATCAGGATCCGAACAAGCCCCAGGAGACC; vpr4, GTTCAGAATTCCCATGGAGCCAAATCCTAGG; c2.10(1), CGAATAGATCTCAGCTGAAGGTCAATGG; c2.10(2), GACGCAGATCTTCCTTGTCTCTCTTCTG; c2.10(3), CGAATAGATCTATGCAGCTGAAGGTCAATGG; c2.10(4), TAATCGAATTCAGCGTAATCAGGTACATCATATGGATACCTTGCAGACCTTGA.
RESULTS
Isolation of cDNA clones encoding Vpr-interacting proteins.
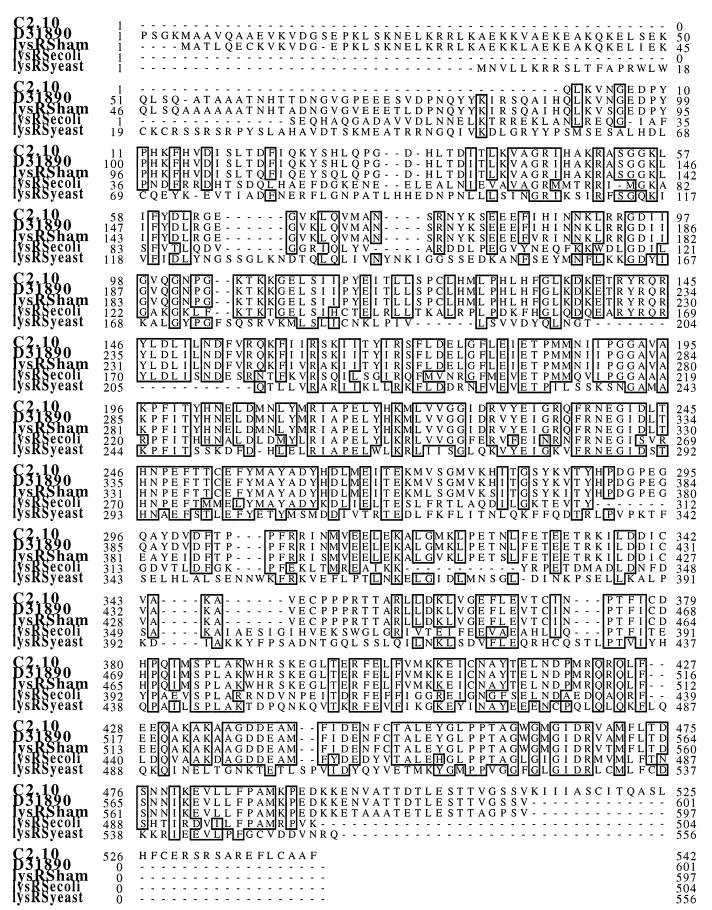
One of the key approaches to understanding HIV-1 pathogenesis is the elucidation of interactions between specific HIV gene products and the host cell. Therefore, we used the yeast two-hybrid system to identify cellular proteins that interact with the HIV accessory protein Vpr. The bait plasmid was constructed by fusing DNA encoding gene wild-type Vpr to the 3′ end of the LexA (encoding amino acids 1 to 211) in the yeast expression vector pV44ER.Lex (32). This was transformed into the S. cerevisiae reporter strain L40, which contains the yeast HIS3 and the bacterial lacZ genes under the control of synthetic promoters bearing LexA-binding sites (66). The resulting yeast strain was cotransformed with an activation domain-tagged human B-cell library (16), and clones containing interacting proteins were selected for by growth on histidine-deficient medium. From approximately 106 transformants screened, 18 that grew in the absence of histidine were identified; of these, 10 also expressed β-galactosidase activity. Double-positive clones were “cured” of the bait plasmid by repeated growth in the presence of tryptophan, leaving yeast strains that contained only the library plasmids. To test the specificity of the interactions between the candidate interacting partners and Vpr, cured clones were retransformed with control heterologous baits. DNA was extracted from the nine clones isolated that specifically interacted with Vpr. Sequence analysis (Fig. 1) revealed that clone 2.10 was 100% identical to a cDNA sequence (52) in the GenBank database (accession no. D31890) which coded for a putative human LysRS.
FIG. 1.
Clone 2.10 codes for an N-terminally deleted LysRS. Amino acid sequence comparison of the c2.10 open reading frame with the putative human LysRS (GenBank accession no. D31890), the putative Chinese hamster ovary LysRS (ham), the constitutive form of E. coli LysRS, and yeast LysRS. Amino acids 232 to 259 of c2.10 represent the consensus sequence for the active site of class II aminoacyl synthetases. Numbers indicate the amino acid positions relative to the N terminus of the corresponding protein. Dots represent gaps inserted to optimize the alignment. Identical residues are indicated by open boxes.
The product of clone 2.10 is a LysRS which interacts with the N terminus of Vpr.
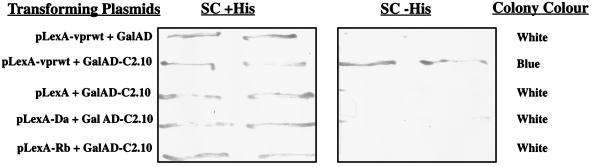
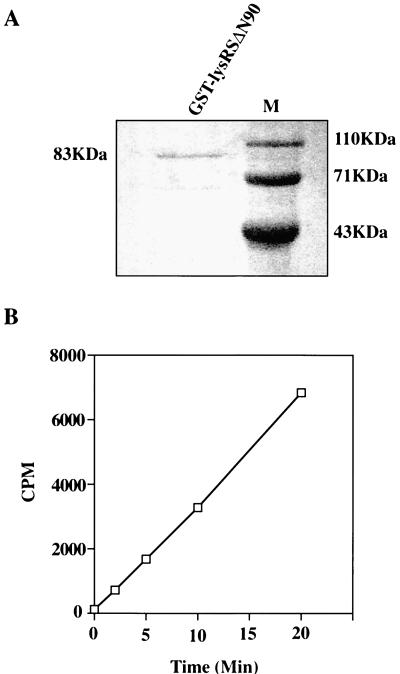
The cDNA present in clone 2.10 was incomplete, lacking sequences corresponding to the 5′ end of the mRNA. Thus, c2.10 encodes a protein of 512 amino acids (56.3 kDa), lacking 90 amino acids at its N terminus (LysRSΔN90). This clone conferred histidine prototrophy and β-galactosidase activity to yeast strain L40 when cotransformed with the LexA-Vpr hybrid but not with LexA alone or with the heterologous LexA-retinoblastoma and LexA-daughterless hybrids (pLexΔ-Rb and pLexA-Da, respectively) (Fig. 2). To confirm that c2.10 did indeed code for active human LysRS, the protein was expressed in bacteria as a GST fusion (as described in Materials and Methods) and purified protein was used in aminoacylation assays (Fig. 3A). These assays investigated the ability of the putative LysRS to catalyze the reaction between [14C]lysine and tRNALys by measuring [14C]Lys-tRNA conjugates as TCA-precipitable radioactivity. The kinetics of this reaction indicate that active, human LysRS was isolated from the two-hybrid screen (Fig. 3B) and that the N-terminal 90 amino acids of this enzyme are not required for its activity.
FIG. 2.
Vpr interacts specifically with the product of c2.10 in the yeast two-hybrid system. The L40 reporter strain, cotransformed with LexA and the Gal4 activation domain (GalAD) or GalAD-c2.10 fusion protein, was assayed for histidine prototrophy and β-galactosidase activity. Filter lift assays were used to detect β-galactosidase activity. LexA fusions are Vpr amino acids 12 to 96, Daughterless protein of Drosophila (Da), and retinoblastoma (Rb). Growth in the absence of histidine and a high level of β-galactosidase activity (represented by a strong blue color of the colony) are indications of an interaction between the hybrid proteins. The results obtained with two individual transformants are shown for each transformation.
FIG. 3.
Isolated LysRSΔN90, expressed by c2.10, is active in aminoacylation assays. (A) Coomassie blue-stained gel of purified GST-lysRSΔN90. (B) Aminoacylation assay in which the above protein was used to catalyze the reaction between [14C]lysine and tRNALys. TCA-precipitable radioactivity was measured with a scintillation counter. The background, determined by activity in the absence of tRNALys, was subtracted from the experimental values.
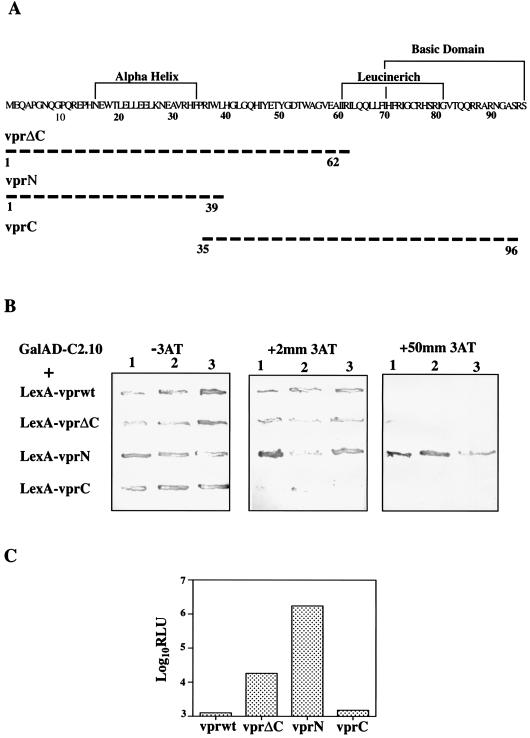
Previous studies (47, 50, 71) have identified a number of putative structural domains of Vpr (Fig. 4A). To localize the domain that mediates binding to LysRS, a series of N- and C-terminal deletions of Vpr were expressed in L40 as LexA fusions and assayed for their ability to interact with GalAD-c2.10. First, the level of transcriptional activation from the HIS3 reporter was determined by growth on SC His− containing increasing concentrations of the yeast HIS3 gene product inhibitor, 3-AT (35). As shown in Fig. 4B, yeast cells expressing LexA-VprN retained their ability to grow on His− medium in the presence of 50 mM 3-AT whereas those expressing LexA-VprC were unable to grow on His− medium when 3-AT was present. We subsequently quantified transcription from the lacZ reporter gene by using a chemiluminescence assay for β-galactosidase activity (Fig. 4C). On average, β-galactosidase activity was 1,000 times greater in extracts from yeasts expressing the Vpr N-terminal hybrid than from those expressing the C-terminal hybrid. These results indicate that LysRS binds to the N terminus of Vpr with a much higher affinity than to the C terminus. In both these assays, the behavior of the VprΔC hybrid was intermediate between those of Vpr N- and C-terminal isolated domains, suggesting that sequences within the central portion of Vpr (amino acids 39 to 62) prevent high-affinity binding of the N terminus to LysRS.
FIG. 4.
Characterization of the Vpr-LysRS interaction in vivo. (A) Amino acid sequence of wild-type Vpr with previously defined functional domains shown (47, 50, 71). N- and C-terminal Vpr deletion mutants were cloned into PV44ER.Lex and expressed in yeast. Broken lines represent the region of protein expressed by the mutants, and numbers indicate the amino acid positions. (B) Vpr deletion mutants were transformed into L40 along with GalAd-c2.10 and grown on His− plates containing 0 to 50 mM 3-AT to measure the level of transcription from the HIS3 gene. Yeast strains expressing strongly interacting proteins remain prototrophic for histidine in the presence of high concentrations of 3-AT. The assay was carried out on three separate transformants for each Vpr mutant. (C) The β-galactosidase activity of yeast strains expressing Vpr hybrid proteins along with GalAD-c2.10 was measured in a chemiluminescence assay. The background was determined by L40 expressing LexA-Vprwt and was around 200 RLU. Values are means of triplicate assays performed on four independent transformants.
Vpr interacts with recombinant LysRS.
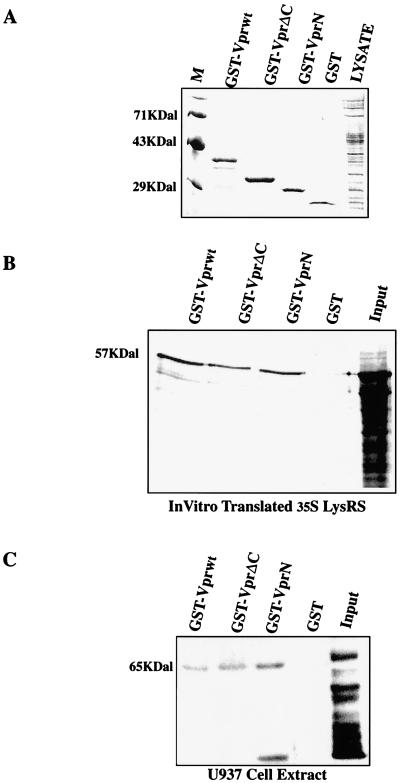
To confirm the binding data observed in yeast, we tested the ability of wild-type and mutant Vpr to bind to recombinant and cellular LysRS. Wild-type Vpr, VprΔC, and VprN were cloned into the pGex2T vector and expressed in bacteria as GST fusion proteins. Bacterial lysates were incubated with glutathione-agarose beads, and the bound proteins were observed by Coomassie blue staining of SDS-PAGE gels (Fig. 5A). To determine whether these proteins could interact with recombinant LysRS, LysRSΔN90 was cloned into the pcDNA3 vector with a C-terminal HA tag and 35S-labeled protein was made in an in vitro transcription-translation reaction. Radiolabelled LysRS was incubated with the glutathione-agarose beads described above, and after extensive washing, bound proteins were resolved by SDS-PAGE and revealed by phosphorimager analysis (Fig. 5B). As predicted, recombinant LysRS interacted with wild-type Vpr-GST but not with GST alone. Deletion of the C-terminal 61 amino acids (VprN-GST) did not impair this interaction, which is in agreement with the yeast two-hybrid studies and confirms that LysRS binds directly to the N terminus of Vpr.
FIG. 5.
Wild-type and C-terminally deleted Vpr bind to recombinant and cellular LysRS. (A) Representative Coomassie blue-stained gel showing concentrations of GST and GST-Vpr fusions used in in vitro interaction experiments. GST and GST-Vpr fusion proteins were affinity purified on glutathione-agarose beads, separated by SDS-PAGE, and analyzed by Coomassie blue staining. (B) In vitro-translated 35S-LysRS (10 μl), was incubated with equivalent amounts of GST and GST-Vpr (wild type and mutant) glutathione-agarose beads, as shown in panel A. Bound proteins were resolved by SDS-PAGE, the gel was dried, and the radioactive species were viewed by phosphorimager analysis. (C) GST and GST-Vpr (wild type and mutant) glutathione-agarose beads were incubated with 100 μg of U937 cell extract, and bound cellular proteins were resolved by PAGE. The gels were then immunoblotted with antiserum from a patient with anti-synthetase syndrome as the primary antibody.
Cellular LysRS binds to GST-Vpr.
To establish that Vpr could interact with full-length LysRS derived from human cells, we examined the interaction between Vpr and native LysRS derived from the HIV-permissive U937 monocytic cell line. GST and the above panel of GST-Vpr beads were incubated with detergent-lysed U937 cell extract. Bound proteins were eluted and subjected to SDS-PAGE and Western blotting. The primary antibody used in these experiments was serum derived from a patient with the anti-synthetase syndrome, which has previously been found to contain anti-LysRS antibodies (18). Before use in interaction experiments, the serum was shown to react with GST-LysRSΔN90 and 35S-LysRSΔN90 (data not shown). This reactivity was lost when the serum was preincubated with GST-LysRSΔN90 glutathione-agarose beads but not with GST beads alone (data not shown). As can be seen in Fig. 5C, wild-type and C-terminally deleted Vpr specifically bind to a 65-kDa protein which is immunoreactive with antiserum from the patient with anti-synthetase syndrome. The smaller species observed in Fig. 5C is a frequently detected degredation product of LysRS, and its presence in the GST-VprN lane indicates that the N terminus of Vpr binds to this domain of the protein. Similar to the results obtained with yeast, GST-VprN was found to bind to LysRS with a higher affinity than did the Vprwt and VprΔC fusions. This result confirms the previous data and demonstrates that the N terminus of Vpr interacts with full-length lysRS in its native state.
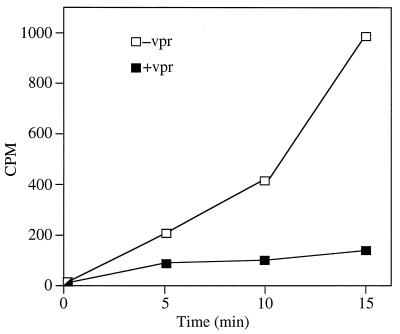
Vpr inhibits LysRS-catalyzed aminoacylation of tRNALys.
To test the functional significance of the Vpr-LysRS interaction, aminoacylation assays were carried out with GST-LysRSΔN90 preincubated with wild-type GST-Vpr or purified GST as a control. In the presence of wild-type Vpr, LysRS-catalyzed aminoacylation of tRNALys was strongly inhibited (Fig. 6).
FIG. 6.
Vpr inhibits aminoacylation of tRNALys. GST-LysRSΔN90, preincubated with either wild-type GST-Vpr (+vpr) or GST (−vpr), was tested for its ability to catalyze the reaction between tRNALys and [14C]lysine. [14C]lysine-tRNALys conjugates were TCA precipitated, and radioactivity was measured with a scintillation counter.
DISCUSSION
In this study, we used the yeast two-hybrid system to identify cellular proteins that interact with HIV-1 Vpr. One cDNA isolated (c2.10) was completely homologous to a sequence encoding a putative human LysRS (52). The function of this protein was confirmed, and the interaction between Vpr and LysRS was characterized in vivo and in vitro. LysRS belongs to the aminoacyl-tRNA synthetase group of enzymes, which catalyze the reaction between an amino acid and the 3′ end of its cognate tRNA (13, 14, 43, 59). Although the LysRS expressed by c2.10 lacked 90 amino acids at its N terminus (LysRSΔN90), it remained active in aminoacylation assays (Fig. 3). This result is in keeping with that of Martinez and Mirande (49), who found that the N-terminal extension (amino acids 1 to 69) of S. cerevisiae LysRS was dispensable in vivo for aminoacylation activities. Furthermore, structural data from the E. coli LysRS S protein indicated that deletion of the N-terminal 30 amino acids (corresponding to amino acids 64 to 93 of human LysRS by sequence comparison) did not affect the ability of the protein to specifically recognize the anticodon loop of tRNALys and catalyze aminoacylation (9, 58). Several highly conserved residues that are thought to be involved in binding to Mg2+ and ATP have been identified in class II aminoacyl synthetases (to which LysRS belongs) (20). None of these residues fall within the N-terminal 90 amino acids of human LysRS (12). These data indicate that the active portion of LysRS is the main target for interaction with Vpr.
There are many reports in the literature about the putative structure-function map of Vpr (45, 47, 50, 53, 71). Mutations in the alpha-helical region (Fig. 4) eliminate virion incorporation, as do deletions at the C terminus (53, 71). Amino acids in the core of the protein (amino acids 30 to 60) are important for nuclear localization of Vpr (24, 50, 71), and the C-terminal basic region is essential for protein stability and induction of cell cycle arrest (47, 50). Using a panel of deletion mutants, we demonstrated that LysRS interacts with both the N and C termini of Vpr (Fig. 4B and C and Fig. 5). However, the affinity of the interaction was much greater for the N-terminal domain of Vpr (amino acids 1 to 39) than for the C terminal domain (amino acids 35 to 96). In vivo and in vitro data from the wild-type Vpr and VprΔC constructs indicated that the presence of C-terminal sequences negatively influences binding of the N terminus to the enzyme. During HIV infection, cellular and/or viral proteins may interact with Vpr and induce conformational changes which expose the N terminus and thus regulate the affinity with which it binds to LysRS. Li et al. (42) previously demonstrated that Vpr forms a tight complex with the p7 nucleocapsid protein (NCp7) of HIV. During the early stages of HIV replication, binding to NCp7 may expose the N-terminal portion of Vpr, allowing high-affinity binding to LysRS. During the latter stages of infection, the N terminus of Vpr is complexed with the HIV-encoded p6 (36, 38, 53), which may also modulate the interaction with the synthetase. The uracil DNA glycosylase enzyme (6) and the glucocorticoid receptor type II complex (56, 72) are cellular proteins that were previously found to interact with Vpr. The finding in this study that the C-terminal domain of Vpr opposes the effects of the N-terminal region is not without precedent. Tung et al. (65) recently demonstrated that NCp7-VprN complexes increase the activity of protein phosphatase 2A while NCp7:VprC complexes inhibit this activity. Additional mutants, including those with point mutations within Vpr, will be needed to further characterize the interaction with LysRS and determine how it affects other functions of Vpr.
Reverse transcription is the first postentry stage in the retroviral replication cycle. In all retroviruses, this process is primed by a tRNA in which 18 nucleotides at the 3′ terminus is complementary to a region at the 5′ end of the viral RNA genome, described as the primer-binding site (reviewed in reference 44). In HIV-1, the primer for reverse transcription has been identified as tRNALys3 (27, 28, 30, 31, 48). During viral assembly, the tRNALys3 is selected for incorporation into the virus from over 100 host cell species (27, 28, 33, 48). At present, little is known about the signals on tRNALys3 that target it for virion incorporation, although it has been suggested that the aminoacylation state of the tRNA may be one such signal (27). By Western blotting of virion extracts with anti-LysRS antiserum (data not shown), we were unable to detect LysRS in HIV-1 virions; this suggested that the synthetase is not incorporated into the virion along with the primer. To function as a primer for reverse transcription, tRNALys3 must have a free 3′ end, which is not the case for aminoacylated tRNAs. It has previously been demonstrated that while all the tRNALys species in HIV-infected cells are acylated, all virion-associated tRNALys species are deacylated (27). Thus, the ability of Vpr to interact with LysRS and inhibit its enzymatic activity may be an important viral mechanism to prevent acylation of the primer and target it to the assembling virion. Since deacylation occurs as part of the translation process (14), this may be more important in resting cells, where low levels of translation are taking place and therefore there are lower levels of deacylated tRNA molecules. In the continuing search for improved anti-HIV agents, disruption of this specific Vpr-LysRS interaction might represent an alternative avenue for therapeutic intervention.
ACKNOWLEDGMENTS
We thank S. Elledge (Baylor College of Medicine, Houston, Tex.), C. Goding (Marie Curie Research Institute, Oxted, England), and F. Bachelerie (Institut Pasteur, Paris, France) for providing plasmid constructs; S. Hollenberg (Fred Hutchinson Cancer Research Center, Seattle, Wash.) for providing the yeast strain L40; and C. Gelpi (Hospital Santa Cruz and San Pablo, Barcelona, Spain) for providing the anti-synthetase patient antisera. We are indebted to Alex Houston for DNA sequencing and to Margaret Wilson for secretarial assistance. Comments on the manuscript from Fernando Arenzana-Seisdedos are greatly appreciated.
This work was supported by the Medical Research Council AIDS Directed Programme and EC project ROCIO.
REFERENCES
- 1.Agostini I, Navarro J M, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator—cooperation with promoter bound activator domains and binding to TFIIB. J Mol Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 2.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizier J-L. HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature. 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 3.Balotta C, Lusso P, Crowley R, Gallo R C, Franchini G. Antisense phosphorothioate oligonucleotides targeted to the vpr gene inhibit human immunodefeciency virus type 1 replication in primary human macrophages. J Virol. 1993;67:4409–4414. doi: 10.1128/jvi.67.7.4409-4414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartz S R, Rogel M E, Emerman M. Human immunodefeciency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beiser C. Recent advances—HIV infection. 2. Br Med J. 1997;314:579–583. doi: 10.1136/bmj.314.7080.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouhamdan M, Benichou S, Rey F, Navarro J-M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodefeciency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 8.Cohn J A. Recent advances—HIV infection. 1. Br Med J. 1997;314:487–491. doi: 10.1136/bmj.314.7079.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Commans S, Plateau P, Blanquet S, Dardel F. Solution structure of the anticodon binding domain of Escherichia coli lysyl-tRNA synthetase and studies of its interaction with tRNAlys. J Mol Biol. 1995;253:100–113. doi: 10.1006/jmbi.1995.0539. [DOI] [PubMed] [Google Scholar]
- 10.Conner I B, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:936–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 11.Cullen B R, Green W C. Functions of the auxilary gene products of the human immunodeficiency virus type 1. Virology. 1990;178:1–5. doi: 10.1016/0042-6822(90)90373-y. [DOI] [PubMed] [Google Scholar]
- 12.Cusack S, Hartlein M, Leberman R. Sequence structural and evolutionary relationships between class II aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang C V, Glinski R L, Gainey P C, Hilderman R H. Physical and biochemical characterisation of a purified arginyl-tRNA synthetase-lysyl-tRNA synthetase complex from rat liver. Biochemistry. 1982;21:1959–1966. doi: 10.1021/bi00537a040. [DOI] [PubMed] [Google Scholar]
- 14.Delarue M. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1995;5:48–55. doi: 10.1016/0959-440x(95)80008-o. [DOI] [PubMed] [Google Scholar]
- 15.DeNoronha C M C, Takaorikondo A, McEntree M, Greene W C. The HIV vpr protein causes G2 cell cycle arrest by interacting with and enhancing protein phosphatase 2A function. Mol Biol Cell. 1996;7:2064. [Google Scholar]
- 16.Durfee T, Becherer K, Chen P-L, Yeh S H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 17.Emerman M. HIV-1, Vpr and the cell cycle. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 18.Gelpi C, Kanterewicz E, Gratacos J, Targoff I N, Rodriguez-Sanchez J L. Coexistence of two antisynthetases in a patient with the antisynthetase syndrome. Arthritis Rheum. 1996;39:692–697. doi: 10.1002/art.1780390424. [DOI] [PubMed] [Google Scholar]
- 19.Gietz D, St Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grigoreva A Y. Structure of aminoacyl transfer RNA synthetases in higher eukaryotes based on molecular cloning data. Mol Biol. 1994;28:630–638. [Google Scholar]
- 21.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–21. [PubMed] [Google Scholar]
- 22.Hattori N, Michaels F, Fargnoli K, Marcon L, Gallo R C, Franchini G. The human immunodeficiency type 2 gene is essential for productive infection of human macrophages. Proc Natl Acad Sci USA. 1990;87:8080–8084. doi: 10.1073/pnas.87.20.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in non dividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoch J, Lang S M, Weeger M, Stahlhennig C, Coulibaly C, Dittmer U, Hunsmann G, Fuchs D, Muller J, Sopper S, Fleckenstein B, Uberla K T. Vpr deletion mutant of simian immunodefeciency virus induces AIDS in rhesus monkeys. J Virol. 1995;69:4807–4813. doi: 10.1128/jvi.69.8.4807-4813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast effeciently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1985;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Mak J, Cao Q, Li Z, Wainberg M A, Kleiman L. Incorporation of excess wild-type and mutant tRNA3Lys into human immunodeficiency virus type 1. J Virol. 1994;68:7676–7683. doi: 10.1128/jvi.68.12.7676-7683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Shalom A, Li Z, Wang J, Mak J, Wainberg M A, Kleiman L. Effects of modifying the tRNA3Lys anticodon on the initiation of human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4700–4706. doi: 10.1128/jvi.70.7.4700-4706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Igarashi T, Ami Y, Yamamoto H, Shibata R, Kuwata T, Mukai R, Shinohara K, Komatsu T, Adachi A, Hayami M. Protection of monkeys vaccinated with vpr- and/or nef-defective simian immunodeficiency virus strain mac human immunodefeciency virus type 1 chimeric viruses: a potential candidate live-attenuated human AIDS vaccine. J Gen Virol. 1997;78:985–989. doi: 10.1099/0022-1317-78-5-985. [DOI] [PubMed] [Google Scholar]
- 30.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA tRNA3lys (template/primer) complex. J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 31.Isel C, Lanchy J-M, Le-Grice S F J, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type-1 reverse transcription require the post-transcriptional modifications of primer tRNA3lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 32.Jayaraman P-S, Hirst K, Goding C R. The activation domain of a basic-helix-loop-helix protein is masked by repressor interaction with domains distinct from that required for transcription regulation. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M, Mak J, Ladha A, Cohn E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into the wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodefeciency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishore G M, Sha D M. Amino acid biosynthesis inhibitors and herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- 36.Kondo E, Gottlinger H G. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J Virol. 1996;70:159–164. doi: 10.1128/jvi.70.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang S M, Weeger M, Stahl-Hennig C, Coulibaly C, Hunsmann G, Muller J, Muller-Hermelink H, Fuchs D, Wachter H, Daniel M M, Desrosiers R C, Fleckenstein B. Importance of Vpr for infection of rhesus monkeys with simian immunodeficiency virus. J Virol. 1993;67:902–912. doi: 10.1128/jvi.67.2.902-912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavallee C, Yao X J, Ladha A, Gottlinger H, Haseltine W A, Cohen E A. Requirement of the pr55gag precursor for incorporation of the vpr product into human immunodefiency virus type 1 viral particles. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy D N, Fernandes L S, Williams W V, Weiner D B. Induction of cell differentiation by human immunodeficiency virus 1 vpr. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-y. [DOI] [PubMed] [Google Scholar]
- 40.Levy D N, Refaeli Y, MacGregor R R, Weiner D B. Serum vpr regulates productive infection and latency of human immunodefeciency type 1. Proc Natl Acad Sci USA. 1994;91:10873–10877. doi: 10.1073/pnas.91.23.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levy D N, Refaeli Y, Weiner D B. Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J Virol. 1995;69:1243–1252. doi: 10.1128/jvi.69.2.1243-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M-S, Garcia-Asua G, Bhattacharyya U, Mascagni P, Austen B M, Roberts M M. The Vpr protein of human immunodefeciency virus type 1 binds to nucleocapsid protein p7 in vitro. Biochem Biophys Res Commun. 1996;218:352–355. doi: 10.1006/bbrc.1996.0061. [DOI] [PubMed] [Google Scholar]
- 43.Lin L, Hale S P, Schimmel P. Aminoacylation error correction. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 44.Litvak S, Sarih-Cottin L, Fournier M, Andreola M, Tarrago-Litvak L. Priming of HIV replication by tRNALys3. Role of reverse transcriptase. Trends Biochem Sci. 1994;3:114–118. doi: 10.1016/0968-0004(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y-L, Spearman P, Ratner L. Human immunodefeciency virus type 1 viral protein R localisation in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Y-L, Bennet R P, Wills J W, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macreadie I G, Castelli L A, Hewish D R, Kirkpatrick A, Ward A C, Azad A A. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc Natl Acad Sci USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mak J, Jiang M, Wainberg M A, Hammarskjold M-L, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNA-Lys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez R, Mirande M. The polyanion binding domain of cytoplasmic lys-transfer RNA synthetase from Saccharomyces cerevisiae is not essential for cell viability. Eur J Biochem. 1992;207:1–11. doi: 10.1111/j.1432-1033.1992.tb17012.x. [DOI] [PubMed] [Google Scholar]
- 50.Marzio P D, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller R H, Sarver N. HIV accessory proteins as therapeutic targets. Nat Med. 1997;3:389–394. doi: 10.1038/nm0497-389. [DOI] [PubMed] [Google Scholar]
- 52.Nomura, N., N. Miyajima, T. Sazuka, A. Tanaka, Y. Kawarabayashi, T. Nagase, K. Ishikawa, T. Seki, and S. Tabata. Unpublished data.
- 53.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Planelles V, Bachelerie F, Jowett J B M, Haislip A, Xie Y, Banooni P, Masuda T, Chen I S Y. Fate of human immunodeficiency virus type 1 provirus in infected cells—a role for Vpr. J Virol. 1995;69:5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Planelles V, Jowett J B M, Li Q-X, Xie Y, Hahn B, Chen I S Y. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Refaeli Y, Levy D N, Weiner D B. The glucocorticoid receptor type II complex is a target of the HIV-1 vpr gene product. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saluta M V, Hirshfield I N. The occurrence of duplicate lysyl-tRNA synthetase gene homologs in Escherichia coli and other procaryotes. J Bacteriol. 1995;177:1872–1878. doi: 10.1128/jb.177.7.1872-1878.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt E, Schimmel P. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science. 1995;264:265–267. doi: 10.1126/science.8146659. [DOI] [PubMed] [Google Scholar]
- 60.Senger B, Despons L, Walter P, Fasiolo F. The anticodon triplet is not sufficient to confer methionine acceptance to a transfer RNA. Proc Natl Acad Sci USA. 1992;89:10768–10771. doi: 10.1073/pnas.89.22.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 62.Subbramanian R A, Cohen E A. Molecular biology of the human immunodeficiency virus accessory proteins. J Virol. 1994;68:6831–6835. doi: 10.1128/jvi.68.11.6831-6835.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trono D. HIV accessory proteins—leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 65.Tung H Y L, De Rocquigny H, Zhao L-J, Cayla X, Roques B P, Ozon R. Direct activation of protein phosphatase-2A0 by HIV-1 encoded complex NCp7:Vpr. FEBS Lett. 1997;401:197–201. doi: 10.1016/s0014-5793(96)01470-6. [DOI] [PubMed] [Google Scholar]
- 66.Votjek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 67.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L-J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 68.Wang L L, Mukherjee S, Narayan O, Zhao L J. Characterization of a leucine zipper like domain in vpr protein of human immunodeficiency virus type 1. Gene. 1996;178:7–13. doi: 10.1016/0378-1119(96)00312-5. [DOI] [PubMed] [Google Scholar]
- 69.Webster A, Leith I R, Hay R T. Activation of adenovirus-coded protease and processing of the preterminal protein. J Virol. 1994;68:7292–7300. doi: 10.1128/jvi.68.11.7292-7300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westervelt P, Henkel T, Trowbridge D B, Orenstein J, Heuser J, Gendelman H E, Ratner L. Dual regulation of silent and productive infection in monocytes by distinct human immunodeficiency virus type 1 determinants. J Virol. 1992;66:3925–3931. doi: 10.1128/jvi.66.6.3925-3931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao X-J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao L-J, Mukherjee S, Narayan O. Biochemical mechanisms of HIV-1 vpr function. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 73.Zhao Y Q, Cao J A, Ogorman M R G, Yu M, Yogev R. Effect of human immunodeficiency virus type 1 protein R (Vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J Virol. 1996;70:5821–5826. doi: 10.1128/jvi.70.9.5821-5826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]