Abstract
The UL15 gene of herpes simplex virus (HSV) is one of several genes required for the packaging of viral DNA into intranuclear B capsids to produce C capsids that become enveloped at the inner nuclear membrane. A rabbit antiserum directed against UL15-encoded protein recognized three proteins with apparent Mrs of 79,000, 80,000, and 83,000 in highly purified B capsids. The 83,000-Mr protein was detected in type C capsids and comigrated with the product of a UL15 cDNA transcribed and translated in vitro. The 83,000- and 80,000-Mr proteins were readily detected in purified virions. Inasmuch as (i) none of these proteins were detectable in capsids purified from cells infected with HSV-1(ΔUL15), a virus lacking an intact UL15 gene, and (ii) corresponding proteins in capsids purified from cells infected with a recombinant virus [HSV-1(R7244), containing a 20-codon tag at the 3′ end of UL15] were decreased in electrophoretic mobility relative to the wild-type proteins, we conclude that the proteins with apparent Mrs of 83,000, 80,000, and 79,000 are products of UL15 with identical C termini. The 79,000-, 80,000-, and 83,000-Mr proteins remained associated with B capsids in the presence of 0.5 M guanidine HCl and remained detectable in capsids treated with 2.0 M guanidine HCl and lacking proteins associated with the capsid core. These data, therefore, indicate that UL15-encoded proteins are integral components of B capsids. Only the 83,000-Mr protein was detected in B capsids purified from cells infected with viruses lacking the UL6, UL17, or UL28 genes, which are required for DNA cleavage and packaging, suggesting that capsid association of the 80,000- and 79,000-Mr proteins requires intact cleavage and packaging machinery. These data, therefore, indicate that capsid association of the 80,000- and 79,000-Mr UL15-encoded proteins reflects a previously unrecognized step in the DNA cleavage and packaging reaction.
At least three types of capsids, designated A, B, and C, that differ in density and electron microscopic appearance accumulate in the nuclei of cells infected with wild-type herpes simplex virus type 1 (HSV-1). All three capsid types have an outer protein shell approximately 120 nm in diameter made from hexons and pentons of VP5, the major capsid protein. The hexons and pentons are linked by triplexes composed of VP19c and VP23 (22). The culmination of HSV capsid assembly is the insertion of viral DNA into B capsids to produce C capsids that become enveloped at the inner nuclear membrane. During DNA packaging, internal proteins of B capsids (including a scaffold composed of VP22a [or ICP35] and a viral protease, VP24) are lost as DNA is inserted (12, 14). A capsids are believed to be the products of an aborted packaging reaction in which the internal proteins are lost but DNA is not inserted.
Two types of B capsids, distinguished by the appearance of the internal scaffold, which can be of large diameter (large-cored B capsids, or procapsids [21]) or small diameter (small-cored B capsids), can be seen in electron micrographs of infected cell nuclei. Cleavage of ICP35 in procapsids by the viral protease likely releases the scaffold from the inner side of the outer capsid shell, allowing the scaffold to collapse inward to produce small-cored B capsids (33, 34, 36). Such capsids are readily purified from HSV-infected cells (14). Whether it is the large-cored or small-cored B capsid that receives viral DNA is currently controversial (15, 25, 34).
Mutations in at least seven genes (UL6, UL15, UL17, UL25, UL28, UL32, and UL33) prevent production of C capsids but are dispensable for assembly of B-like capsids (1, 2, 18, 24, 27, 29, 30, 35, 37). Although the functions of the proteins encoded by these genes are not known, at least the UL6 and UL25 proteins can associate with capsids (18, 23). Such capsid-associated proteins could play roles analogous to those of bacteriophages that comprise portal vertices into which DNA is inserted or could act as terminases which link the portal vertices to DNA and help mediate DNA packaging (9). Given the observation that UL15 and its homologs in other herpesviruses display homology to the T4 bacteriophage terminase gp17 (11), the primary goal of the present study was to determine if the UL15-encoded protein is a capsid component.
MATERIALS AND METHODS
Viruses and cells.
G5 transformed cells were derived from Vero cells and contain HSV-1 DNA from UL16 to UL21 (13). Clone 17 cells were derived from rabbit skin cells and contain a cDNA copy of the UL15 gene (3). The C1 cell line was derived from Vero cells and contains the entire UL28 gene and the UL27 gene minus a 969-bp BstEII fragment at the 5′ end (32). The G33 cell line was derived from Vero cells and contains HSV-1 DNA from UL6 to UL8 (24). Vero, rabbit skin, HEp-2, G5, C1, G33, and clone 17 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% newborn calf serum, penicillin, and streptomycin as previously described (3, 4, 7, 13, 24, 32).
The titers of the wild-type viruses HSV-1(F) and HSV-1(17) and the mutant virus R7244 were determined on Vero cell monolayers. Virus R7244 contains a 20-amino-acid epitope of the human cytomegalovirus glycoprotein B gene incorporated into the C terminus of UL15 protein encoded by a cDNA inserted into the native position of UL15 exon I (4). Stocks of R7244 were grown in clone 17 cells. HSV-1(ΔUL17) contains a lacZ expression cassette from a NotI site 105 bp from the 5′ end of UL17 to a XhoI site 516 bp from the 3′ end of UL17; it was grown and its titers were determined on G5 cells (29). HSV-1(ΔUL15) contains a lacZ expression cassette in place of 226 codons of exon II of UL15; it was grown and its titers were determined on clone 17 cells (3). The mutant gCB contains a 1,881-bp deletion in the UL28 gene; it was grown and its titers were determined on the C1 cell line (32). Cos-UL6− was derived from the HSV-1(17) strain and contains a 4-bp insertion at a site corresponding to amino acid residue 381; it was grown and its titers were determined on G33 cells (24).
In vitro expression of UL15 protein.
PRB4503 contains a cDNA of the UL15 gene inserted into the pGEM3Z vector (Promega) and delimited at the 5′ end by a PstI site at position 28840 and a Bsu36I site at the 3′ end at position 35093 of the published HSV-1(17) sequence (4, 17). PRB4503 was transcribed and translated for 1 h at 30°C with the TNTR T7/SP6 coupled reticulocyte system (Promega) according to the manufacturer’s protocol.
Capsid purification and analysis.
In a typical purification, Vero cell monolayers from three or four 850-cm2 roller bottles were infected at a multiplicity of infection of 5.0 PFU per cell and incubated at 34°C for 18 h. Nuclear lysates were prepared as described previously (26) and were separated on a 20 to 50% continuous sucrose gradient at 23,000 rpm for 1 h in a Beckman SW41 rotor. Light-scattering bands near the middle of the tube were collected with either a Pasteur pipette or a fractionating device (Haake Buchler) starting at the top of the tube. The collected material was diluted in TNE (0.5 M NaCl, 20 mM Tris-HCl [pH 7.4], and 1 mM EDTA) and pelleted at 20,000 rpm in a SW41 rotor for 2 h. The capsid-containing pellets were resuspended in TNE by sonication and were separated by centrifugation on a second continuous sucrose gradient. For fractionation experiments, 0.5-ml fractions were collected and pelleted by centrifugation. Fractionated material was either (i) dialyzed against TNE, negatively stained, and viewed by electron microscopy or (ii) resuspended in a buffer containing sodium dodecyl sulfate followed by separation on a denaturing 10% polyacrylamide gel. Electrophoretically separated proteins were either stained with Coomassie brilliant blue or transferred to nitrocellulose and probed with specific antibodies. The intensities of protein bands on Coomassie blue-stained gels or immunoblots were determined from digitized video images produced on a Stratagene Eagle Eye II digital camera with Eagle Sight software (version 3.1). Capsid purity was verified by (i) examination by electron microscopy, (ii) Coomassie blue and silver staining of electrophoretically separated capsid proteins, and (iii) immunoblotting with antibodies against HSV tegument proteins encoded by UL17 and UL16 that do not associate with highly purified capsids (19, 29).
Electron microscopy.
Purified capsids were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and adsorbed to standard carbon-Formvar-coated copper (300-mesh) electron microscopic grids. Samples were stained for 30 s in a 1:1 solution of potassium phosphotungstic acid and bacitracin and viewed through a Zeiss 902 transmission electron microscope at an accelerated voltage of 80 kV.
GuHCl treatment of capsids.
Purified capsids to be extracted with guanidine hydrochloride (GuHCl) were resuspended in TNE at 4°C at 0.5 to 1.5 mg of protein per ml, essentially as described previously (20) except that five 4.5-ml GuHCl solutions were prepared such that addition of 50 μl of purified capsids produced final concentrations of 0.05, 0.1, 0.5, 1.0, and 2.0 M GuHCl, respectively. The extracted capsids were pelleted through a 300-μl cushion of 25% (wt/wt) sucrose by centrifugation in a Beckman SW50.1 rotor for 1 h at 23,000 rpm and were analyzed on denaturing 10% polyacrylamide gels.
Virion purification.
Virions were purified essentially as described previously (31). Briefly, Vero cell monolayers in roller bottles were infected at a multiplicity of infection of 3.0 PFU per cell and were held at 34°C. Forty hours later, the cells were resuspended in 1.0 mM NaPO4 (pH 7.4) and homogenized in a Dounce homogenizer; the nuclei were immediately stabilized with 1.25 M sucrose in 1.0 mM NaPO4, and the virions were pelleted. The supernatant was then loaded onto a linear dextran T10 gradient and centrifuged in a Beckman SW28 rotor at 20,000 rpm for 1 h. The virion band, visible as a hazy region just above the middle of the tube, was collected and pelleted at 25,000 rpm for 2 h. For the second purification, virions were resuspended by sonication and trituration, solid sucrose was added to produce a solution of 50% sucrose (wt/wt), and the material was placed in a SW28 tube and overlaid with a discontinuous gradient formed by successive layers of 40, 30, and 20% sucrose (wt/wt). The gradients were then centrifuged at 25,000 rpm for 18 h in a SW28 rotor. The bands containing virions were collected with a Pasteur pipette. The virions were pelleted and suspended in a small volume of 0.01 M Tris buffer, and associated proteins were denatured in buffer-containing sodium dodecyl sulfate and separated on a denaturing polyacrylamide gel.
Immunoblotting.
Electrophoretically separated capsid or virion proteins were electrically transferred to nitrocellulose and probed with either a previously characterized polyclonal antiserum directed against UL15-encoded proteins at a dilution of 1:750 or with ICP5-specific polyclonal antiserum (NC1) at a dilution of 1:5,000 (10). The UL15-specific rabbit polyclonal antibody was generated by immunization with an affinity-purified bacterial protein (UL15-MBP) containing the malE gene product fused to the protein encoded by the majority of UL15 exon II (4). Bound antibody was visualized by the addition of alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin followed by the addition of chromogenic substrate (Bio-Rad) as previously described (6). The apparent Mrs of proteins were calculated based upon the migration of prestained standards (Bio-Rad). The migration of each standard in a given batch was verified by comparison to electrophoretically separated, unlabeled proteins.
RESULTS
Multiple UL15 proteins are components of B capsids.
To test the possibility that the UL15 protein is capsid associated, type B capsids were purified from HSV-1(F)-infected Vero cells on two successive continuous sucrose gradients (see Materials and Methods). The capsids were then separated on a third continuous sucrose gradient, fractions were collected, and the capsids were pelleted by centrifugation. Capsid-associated proteins were electrophoretically separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and reacted with a previously described antiserum directed against UL15 exon II-encoded protein sequences and maltose-binding protein (UL15-MBP) (4). Bound antibody was visualized by the addition of alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin followed by the addition of chromogenic substrate. The results are shown in Fig. 1.
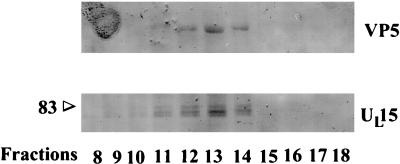
FIG. 1.
Scanned digital images of immunoblots probed with anti-UL15-MBP antibody. Fractions (0.5 ml) of a 14-ml continuous sucrose gradient containing purified B capsids were collected starting at the top of the tube, and pelleted material was electrophoretically separated, transferred to nitrocellulose, and reacted with the UL15-MBP antiserum. Bound immunoglobulin was visualized by addition of alkaline phosphatase-conjugated anti-rabbit antibody followed by fixation of colored substrate. The two panels show regions of the immunoblot containing VP5 (upper panel), and UL15-encoded proteins (lower panel). The positions of bands corresponding to the 83,000-Mr (83) UL15 proteins are indicated. Only fractions 8 to 18 are shown; fractions 1 to 7 and 19 to 25 did not contain detectable levels of UL15 or VP5.
Three proteins with Mrs of approximately 83,000, 80,000, and 79,000 were readily detected by the anti-UL15-MBP serum in fractions 11 to 14 and were faintly detectable in fractions 9, 10, and 15 of the sucrose gradient, whereas other fractions did not contain these proteins at detectable levels. Electron microscopic analysis revealed that fractions 11 to 14 contained capsids without other electron-dense material greater than 2.0 nm in diameter (data not shown). In some immunoblots of capsid proteins probed with the anti-UL15-MBP serum (such as that shown in Fig. 1), it was noted that the anti-UL15-MBP antibody reacted weakly with large amounts of VP5. Thus, lanes of the immunoblot that contained fractions 11 to 14 also contained a band corresponding to the major capsid protein, VP5. Both the VP5 and UL15 proteins were most readily detected in fraction 13. Gradient fractions 1 to 7 and 20 to 25 did not contain proteins that reacted with the anti-UL15-MBP antibody, nor was VP5 detectable (data not shown). Taken together, these data indicate that UL15 proteins with apparent Mrs of 83,000, 80,000, and 79,000 are readily detectable in purified capsids.
To insure that the proteins with apparent Mrs of 83,000, 80,000, and 79,000 recognized by the anti-UL15-MBP antiserum were encoded by UL15, B capsids were purified from Vero cells infected with HSV-1(F) or a virus lacking most of the second exon of UL15, designated HSV-1(ΔUL15) (3). Proteins associated with the purified capsids were electrophoretically separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and reacted with the anti-UL15 antiserum. As shown in Fig. 2A, the proteins with apparent Mrs of 83,000, 80,000, and 79,000 present in HSV-1(F) capsids were not detectable in capsids purified from HSV-1(ΔUL15)-infected cells. Cross-reactivity with VP5 in the same lanes of the immunoblot showed that similar amounts of capsid proteins were loaded in each lane of the polyacrylamide gel. We conclude that, inasmuch as they are not detectable in capsids purified from cells infected with the UL15 deletion mutant, the 83,000-, 80,000-, and 79,000-Mr proteins recognized by the UL15-MBP antiserum are products of the UL15 gene.
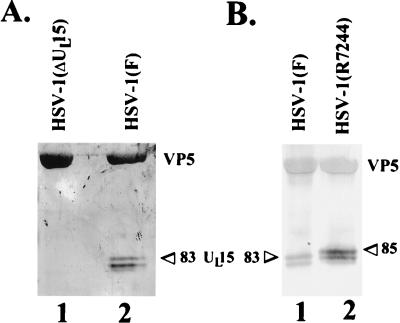
FIG. 2.
Scanned digital images of immunoblots probed with anti-UL15-MBP antibody. Electrophoretically separated proteins were transferred to nitrocellulose and probed with the antiserum directed against the UL15-MBP fusion protein. (A) Type B capsids were purified from cells infected with wild-type HSV-1(F) or with HSV-1(ΔUL15), lacking most of the UL15 second exon. (B) B capsids were purified from cells infected with HSV-1(F) or with HSV-1(R7244), containing a 20-codon epitopic tag at the 3′ end of the UL15 gene. The positions of the bands corresponding to the 83,000- and the 85,000-Mr UL15 proteins (83 and 85, respectively) are indicated.
To further demonstrate that the 83,000-, 80,000-, and 79,000-Mr proteins were derived from translation of the UL15 gene, we took advantage of an available recombinant virus, designated R7244, which contained DNA encoding a 20-amino-acid epitopic tag inserted within the 3′ end of a UL15 cDNA. Lysates of cells infected with R7244 contain UL15 protein with decreased electrophoretic mobility relative to the UL15 protein in HSV-1(F)-infected cell lysates (4). B capsids were purified from Vero cells infected with HSV-1(F) or HSV-1(R7244). Capsid-associated proteins were then electrophoretically separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with the anti-UL15-MBP serum. As shown in Fig. 2B, HSV-1(F) B capsids contained proteins with Mrs of 83,000, 80,000, and 79,000 that were recognized by the UL15-MBP-specific antiserum. In contrast, the anti-UL15-MBP serum recognized three proteins with apparent Mrs of 85,000, 82,000, and 81,000 in lysates of B capsids purified from R7244-infected cells. Thus, capsids purified from R7244-infected cells contain capsid-associated UL15-encoded proteins with decreased electrophoretic mobilities relative to those of the corresponding wild-type proteins. These data demonstrate that the 83,000-, 80,000-, and 79,000-Mr proteins are products of UL15, since they contain protein encoded by the 3′ end of the UL15 gene.
One noteworthy observation is that the band with an apparent Mr of 79,000 was not detected in all capsid preparations, possibly due to the inability to resolve the 80,000- and 79,000-Mr proteins in some experiments. We also cannot exclude the possibility that the protein with an apparent Mr of 79,000 may arise from degradation of either the 83,000- or the 80,000-Mr protein. However, at least the 83,000- and 80,000-Mr UL15-encoded proteins were always detected in wild-type B capsids.
Effects of GuHCl extraction on the association of the UL15 protein with B-type capsids.
Treatment of purified capsids with various concentrations of GuHCl has been shown to result in removal of selected proteins. Specifically, the majority of the capsid pentons, comprised mostly of VP5, VP26 (which binds the tips of the hexons), and components of the capsid core (VP22a), are removed upon treatment with 2.0 M GuHCl. In contrast, VP5, comprising the capsid hexons, and VP19c and VP23, which comprise the capsid triplexes, are more resistant to extraction under this treatment (20, 22, 23).
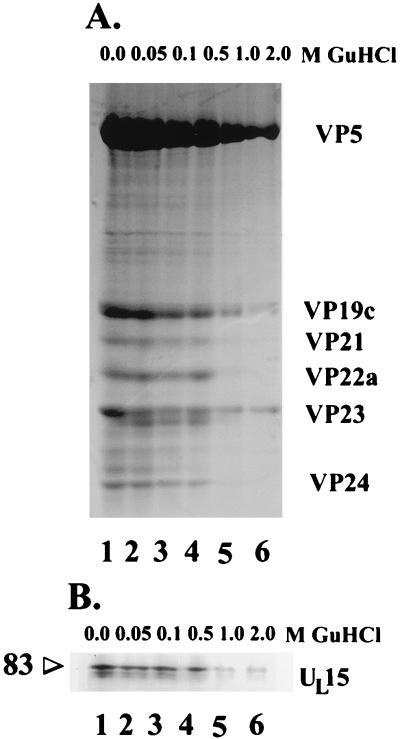
To determine whether UL15 proteins were tightly associated with capsids, B-type capsids were purified and equal amounts were extracted with 0.05, 0.1, 0.5, 1.0, or 2.0 M GuHCl, pelleted through a sucrose cushion to remove solubilized material, separated on a denaturing polyacrylamide gel, stained with Coomassie blue, and analyzed by densitometry (see Materials and Methods). Compared to the amounts in untreated capsids (Fig. 3A, lane 1), approximately 40% of VP5, 21% of VP19c, and 30% of VP23 were retained in pelletable material upon treatment with 2.0 M GuHCl (Fig. 3A, lane 6, and Table 1). The amounts of VP21, VP22a, and VP24 were diminished to undetectable levels upon extraction with 2.0 M GuHCl (Table 1 and Fig. 3A). These data suggest that in this experiment, treatment with 2.0 M GuHCl denatured some capsids and removed proteins associated with the cores of the remaining capsids.
FIG. 3.
(A) Scanned image of Coomassie blue-stained denaturing polyacrylamide gel. Equal amounts of purified B capsids were extracted with the indicated concentrations of GuHCl, and proteins which remained capsid associated were electrophoretically separated and stained with Coomassie blue. The identities of capsid proteins are indicated to the right. (B) Immunoblot of GuHCl-treated capsids. Samples of the preparations shown in panel A were electrophoretically separated, transferred to nitrocellulose, and reacted with the anti-UL15-MBP serum. The position of a band corresponding to the 83,000-Mr UL15 protein (83) is indicated.
TABLE 1.
Percentages of capsid proteins remaining after extraction with GuHCla
| Protein | % Protein after extraction with indicated concn (M) of GuHCl
|
||||
|---|---|---|---|---|---|
| 0.05 | 0.1 | 0.5 | 1.0 | 2.0 | |
| VP5 | 100.0 | 100.0 | 93.0 | 79.0 | 40.0 |
| VP19c | 95.0 | 81.0 | 67.0 | 40.0 | 21.0 |
| VP21 | 97.0 | 68.0 | 49.0 | 16.0 | NDb |
| VP22a | 96.0 | 68.0 | 74.0 | 12.0 | ND |
| VP23 | 89.0 | 60.0 | 54.0 | 35.0 | 30.0 |
| VP24 | 92.0 | 58.0 | 37.0 | 3.0 | ND |
| UL15 protein | 81.0 | 83.0 | 75.0 | 16.0 | 15.0 |
Percentages were calculated by dividing densitometric values of Coomassie blue-stained proteins in capsids extracted with GuHCl by values of corresponding proteins in untreated capsids. For the UL15 protein, densitometric values of the immunoblot reacted with the UL15-MBP antibody were used.
Nondetectable.
Electrophoretically separated capsid proteins from the above preparations were transferred to nitrocellulose and were reacted with the anti-UL15-MBP antibody. As shown in Fig. 3B, at least 75% of the reactivity with the proteins with apparent Mrs of 83,000, 80,000, and 79,000 was retained in capsids treated with 0.05, 0.1, and 0.5 M GuHCl whereas 15 and 16% of the reactivity was retained in capsids treated with 1.0 and 2.0 M GuHCl, respectively. The retention of UL15 protein immunoreactivity at 1.0 and 2.0 M GuHCl was less than that of VP23 (35 and 30% retained, respectively) and VP19c (40 and 21% retained, respectively) but more than that of core proteins such as VP24, which were reduced to undetectable levels. This suggested that UL15 proteins were more resistant to extraction than were core proteins such as VP24. A caveat of this interpretation is that a linear correlation between UL15 protein concentration and reactivity on immunoblots has not been formally established. We can conclude, however, that UL15 proteins remain associated with capsids extracted with 0.5 M GuHCl. This supports the conclusion that UL15 proteins are integral components of capsids rather than contaminants of the capsid preparations.
Capsids from cells infected with cleavage and packaging mutants contain only the UL15-encoded protein with an apparent Mr of 83,000.
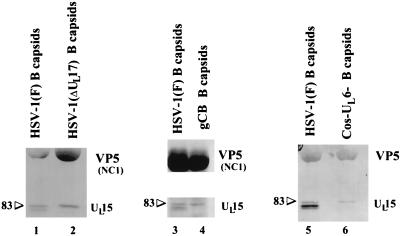
Because genes in addition to UL15 are required for DNA cleavage and packaging, it was of interest to determine if capsid association of UL15-encoded proteins was dependent on other gene products. To address this question, capsids were purified from Vero cell monolayers infected with the wild-type strain HSV-1(F) or with cleavage and packaging mutants lacking either the UL17, UL6, or UL28 gene [HSV-1(ΔUL17), Cos-UL6−, and gCB, respectively] (24, 29, 32). Proteins associated with the purified capsids were separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and probed with the anti-UL15-MBP serum. Surprisingly, while at least the 83,000- and 80,000-Mr UL15 proteins were present in capsids purified from cells infected with HSV-1(F), only the 83,000-Mr protein was detectable in capsids purified from cells infected with HSV-1(ΔUL17), Cos-UL6−, or gCB (Fig. 4). These data indicate that at least the UL6, UL17, and UL28 genes are individually required for association of the UL15 80,000-Mr protein with B capsids and suggest that the UL15 80,000-Mr protein associates with capsids only in the presence of intact cleavage and packaging machinery.
FIG. 4.
Scanned image of immunoblot of B capsids purified from cells infected with a wild-type strain, HSV-1(F), and mutants which do not cleave and package DNA. Capsids were purified from cells infected with HSV-1(F) or the DNA cleavage and packaging mutants Cos-UL6−, HSV-1(ΔUL17), or gCB, lacking functional UL6, UL17, and UL28, respectively. Proteins associated with purified capsids were electrophoretically separated, transferred to nitrocellulose, and reacted with the UL15-MBP-specific antibody (lanes 5 and 6) or anti-UL15-MBP serum and NC1, a polyclonal antibody directed against VP5 (lanes 1 to 4), to clearly demonstrate the relative amounts of capsid proteins loaded in the lanes. The positions of the bands corresponding to the 83,000-Mr UL15 protein (83) are indicated.
The 83,000-Mr UL15 protein is the predominant form in C capsids.
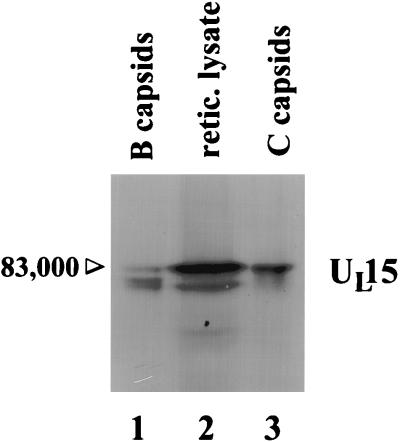
To determine what forms of UL15 proteins are associated with C capsids, Vero cell monolayers were infected with HSV-1(F) for 18 h and capsids were purified on a continuous 20 to 50% sucrose gradient. B and C capsids, distinguished by their sedimentations in the gradient, were individually collected, pelleted, and purified on separate sucrose gradients. Capsid-associated proteins were separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and reacted with the anti-UL15-MBP serum. As shown in Fig. 5, whereas B capsids contained the 83,000-, 80,000-, and 79,000-Mr UL15-encoded proteins (lane 1), the 83,000-Mr UL15 protein was the predominant form of UL15-encoded protein detected in C capsids (lane 3).
FIG. 5.
Scanned image of immunoblot probed with UL15-MBP-specific antiserum. B and C capsids were purified from HSV-1(F)-infected cells, and a cDNA of the known UL15 mRNA was transcribed and translated in a rabbit reticulocyte (retic. lysate). Proteins were electrophoretically separated and reacted with the UL15-MBP antiserum. The position of the band corresponding to the 83,000-Mr UL15 protein is indicated.
Origin of the UL15-encoded proteins.
To identify which of the 83,000-, 80,000-, and 79,000-Mr UL15 proteins constituted the full-length translation product of the UL15 cDNA detectable in HSV-1-infected cells (5), a UL15 cDNA was transcribed and translated in rabbit reticulocyte lysate. The proteins, and polypeptides associated with wild-type B and C capsids, were separated in separate lanes of a denaturing polyacrylamide gel, transferred to nitrocellulose, and reacted with the UL15-MBP-specific antiserum. As shown in Fig. 5, lane 2, the electrophoretic migration of the in vitro transcription-translation product was indistinguishable from that of the 83,000-Mr protein present in B and C capsids (lanes 1 and 3). Also present in the in vitro transcription-translation reaction was a UL15 product, possibly derived from initiation at a methionine codon (codon 35) in the UL15 cDNA (17), that comigrated with the protein with an apparent Mr of 79,000 but not the protein with an apparent Mr of 80,000. A protein that comigrated with the UL15-encoded protein with an apparent Mr of 80,000 in B capsids was not detected in the rabbit reticulocyte lysate programmed with the UL15 cDNA. We therefore conclude that the UL15-encoded protein with an apparent Mr of 83,000 comigrates with the translational product of the full-length UL15 cDNA whereas the 79,000-Mr protein comigrates with an additional protein produced in in vitro transcription-translation reactions. These data suggest that the 83,000-Mr protein is the product of the full-length UL15 gene.
At least two forms of the UL15 protein are associated with virions.
To determine if UL15-encoded proteins were virion components, Vero cell monolayers were infected at 3.0 PFU per cell and were purified as described in Materials and Methods (31). Virions were separated on a denaturing polyacrylamide gel, transferred to nitrocellulose, and reacted with the anti-UL15-MBP antibody. As shown in Fig. 6, at least two different forms of UL15, corresponding to Mrs of approximately 83,000 and 80,000, were detectable in lanes containing both virions and B capsid polypeptides. To confirm these results, virions purified through a dextran gradient were further purified through a sucrose flotation gradient as described previously (31). Proteins associated with the purified virions were analyzed on immunoblots probed with the anti-UL15-MBP antibody as described above. The results reiterated those shown in Fig. 6 and indicated that both the 80,000- and 83,000-Mr proteins were detectable in virions (data not shown).
FIG. 6.
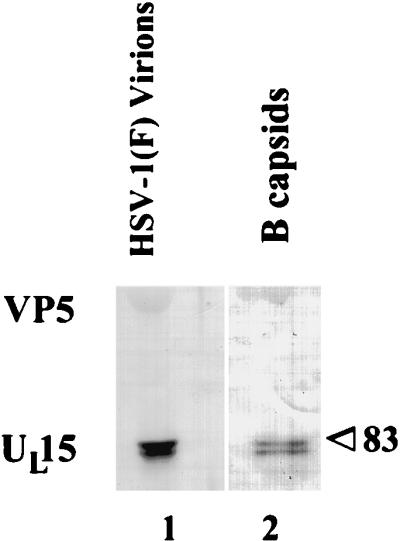
Scanned image of immunoblot probed with the UL15-MBP-specific antiserum. Virions and B capsids were purified from HSV-1(F)-infected cells, and associated proteins were electrophoretically separated and reacted with the UL15-MBP-specific antiserum. The position of the band corresponding to the 83,000-Mr UL15 protein (83) is indicated.
DISCUSSION
We have demonstrated that UL15 proteins are components of B and C capsids and virions. Evidence supporting the observation that B capsids contain UL15-encoded proteins includes the observations that UL15 proteins (i) cofractionate with viral capsids in a sucrose gradient, (ii) remain associated with capsids extracted with 0.5 M GuHCl, and (iii) are detectable in capsids extracted with 1.0 and 2.0 M GuHCl. The observation that UL15 proteins remain detectable in highly purified capsids treated with 2.0 M GuHCl argues that UL15 proteins are integral components of capsids. Parenthetically, the 35,000-Mr UL15 protein which has previously been reported (4) was not found to associate with capsids or virions (data not shown).
UL15-encoded proteins with apparent Mrs of 83,000, 80,000, and 79,000 are detectable in B capsids. Previous results indicated that the major UL15-encoded protein in electrophoretically separated lysates had an apparent Mr of 75,000 (4), but the use of more carefully calibrated standards in the present study indicated that the major protein in infected cell lysates, and the slowest-migrating UL15 protein associated with capsids, migrated with an apparent Mr of 83,000 (data not shown). The fact that the 83,000-, 80,000-, and 79,000-Mr capsid proteins are derived from UL15 was confirmed by the observations that (i) these proteins were not detectable in capsids purified from cells infected with HSV-1(ΔUL15), a virus lacking an intact UL15 gene, and (ii) corresponding proteins in capsids purified from cells infected with a recombinant virus [HSV-1(R7244)] containing a 20-codon tag at the 3′ end of UL15 had decreased electrophoretic mobilities relative to those of the wild-type proteins. The latter observation also indicated that the 80,000- and 79,000-Mr species did not result from carboxy-terminal cleavage of the 83,000-Mr protein, since the tag was retained in all three proteins.
The observations that the 80,000- and the 79,000-Mr proteins were not detectable in capsids from HSV-1(ΔUL17)-, Cos-UL6−-, or gCB-infected cells indicate that capsid association of these proteins is dependent on at least the UL6, UL17, and UL28 genes and argue that capsid association of the proteins with apparent Mrs of 80,000 and 79,000 requires intact cleavage and packaging machinery. Additionally, the 83,000-, 80,000-, and 79,000-Mr proteins were detectable in capsids purified from complementing cell lines infected with the UL17 and UL6 null viruses (data not shown). One possibility is that the 83,000-Mr UL15 protein associates with the capsid and is modified to produce the 80,000- and 79,000-Mr proteins, coincident with the conversion of large-cored B capsids to small-cored B capsids. Inasmuch as the 83,000-Mr protein comigrates with full-length UL15 protein produced in rabbit reticulocyte lysates, such modifications would have to account for the increased electrophoretic mobility of the other UL15-encoded proteins. We cannot rule out the possibility that the 80,000- and 79,000-Mr proteins are derived from (i) translation initiated at codon 35 (ATG) in the UL15 mRNA (17) or (ii) translation of an alternatively spliced RNA. We do not favor the latter possibility because the cononical splice donor site is destroyed in R7244 UL15 mRNA, suggesting that alternative splicing could not occur (4, 5). We also cannot rule out the possibility that the 79,000-Mr protein is a breakdown product of the 83,000- or 80,000-Mr proteins, which may account for its variability in different experiments.
We have noted that at least the 83,000- and 80,000-Mr UL15 proteins are associated with virions, whereas the 83,000-Mr protein is the predominant form associated with C capsids, the progenitors of virions. We favor the hypothesis that the 80,000- and the 79,000-Mr UL15 proteins are bound tightly to B capsids and become incorporated into the virion tegument by weak association with C capsids. Thus, the 79,000- and 80,000-Mr proteins are mostly lost during purification of C capsids. Alternatively, the 79,000- and 80,000-Mr proteins could associate with B capsids, disassociate from C capsids, and reassociate with C capsids as they mature into virions.
It is noteworthy that in the T4 bacteriophage system, a protein (gp23*) which has DNA-dependent ATPase and non-sequence-specific endonuclease activities remains enzymatically inactive until it is proteolytically cleaved during the cleavage and packaging reaction (8, 16, 28). This could provide a means to regulate enzymatic activity until all components required for DNA packaging are properly assembled. If such regulation were to occur during the HSV cleavage and packaging reaction, it could also serve to coordinate the onset of DNA cleavage with DNA packaging and scaffold cleavage and expulsion. In this scenario, the 83,000-Mr UL15-encoded protein could represent an inactive form of UL15-encoded protein, whereas the 80,000- and 79,000-Mr proteins might perform tightly regulated functions in the DNA-packaging reaction. Determining whether this is the case or whether the proteins perform entirely different functions will require additional studies.
ACKNOWLEDGMENTS
We thank Fred Homa, Arvind Patel, and Stan Person for recombinant viruses and the cell lines necessary for their propagation. The NC1 antibody was kindly provided by Roselyn Eisenberg and Gary Cohen. We also thank Jarek Okulicz-Kozaryn for excellent technical assistance.
These studies were supported by NIH grant R01 GM50740.
REFERENCES
- 1.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 2.Al-Kobashi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 3.Baines J D, Cunningham C, Nalwanga D, Davison A J. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines J D, Poon A P W, Rovnak J, Roizman B. The UL15 gene of herpes simplex virus encodes two proteins and is required for cleavage of viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines J D, Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992;66:5621–5626. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines J D, Roizman B. The UL21 gene of herpes simplex virus 1 is dispensable for replication in cell culture. J Virol. 1994;68:2929–2936. doi: 10.1128/jvi.68.5.2929-2936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black L W. In vitro packaging of bacteriophage T4 DNA. Virology. 1981;113:336–344. doi: 10.1016/0042-6822(81)90160-4. [DOI] [PubMed] [Google Scholar]
- 9.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 10.Cohen G, Ponce de Leon H M, Diggelmann H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 12.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 13.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J Y, Irmiere A, Gibson W. Primate cytomegalovirus assembly: evidence that DNA packaging occurs subsequently to B capsid assembly. Virology. 1988;167:87–96. doi: 10.1016/0042-6822(88)90057-8. [DOI] [PubMed] [Google Scholar]
- 16.Manne V. A bacteriophage T4 DNA packaging related DNA-dependent ATPase-endonuclease. J Biol Chem. 1982;257:13223–13232. [PubMed] [Google Scholar]
- 17.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 18.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalwanga D, Rempel S, Roizman B, Baines J D. The UL16 gene product of herpes simplex virus is a virion protein that colocalizes with intranuclear capsid proteins. Virology. 1996;226:236–242. doi: 10.1006/viro.1996.0651. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb W W, Homa F L, Thomsen D R, Booy F P, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown S C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 23.Patel A H, Maclean J B. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–478. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 24.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 25.Perdue M L, Cohen J C, Kemp M C, Randall C C, O’Callaghan D J. Characterization of three species of nucleocapsids of equine herpesvirus type-1 (EHV-1) Virology. 1975;64:187–204. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 26.Perdue M L, Kemp M C, Randall C C, O’Callaghan D J. Studies of the molecular anatomy of the L-M cell strain of equine herpesvirus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974;59:201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- 27.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao V B. Evidence that a phage T4 DNA packaging enzyme is a processed form of the major capsid gene product. Cell. 1985;42:967–977. doi: 10.1016/0092-8674(85)90293-4. [DOI] [PubMed] [Google Scholar]
- 29.Salmon, B., C. Cunningham, A. J. Davison, Wendy J. Harris, and J. D. Baines. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 30.Sherman G, Bachenheimer S L. DNA processing in temperature-sensitive morphogenetic mutants of HSV-1. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 31.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tengelsen L A, Pedersen N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and capsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen D R, Newcomb W W, Brown J C, Homa F L. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol. 1995;69:3690–3703. doi: 10.1128/jvi.69.6.3690-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 35.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 36.Wilson D W, Church G A. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]