Abstract
Feline calicivirus (FCV), a member of the Caliciviridae, produces its major structural protein as a precursor polyprotein from a subgenomic-sized mRNA. In this study, we show that the proteinase responsible for processing this precursor into the mature capsid protein is encoded by the viral genome at the 3′-terminal portion of open reading frame 1 (ORF1). Protein expression studies of either the entire or partial ORF1 indicate that the proteinase is active when expressed either in in vitro translation or in bacterial cells. Site-directed mutagenesis was used to characterize the proteinase Glu-Ala cleavage site in the capsid precursor, utilizing an in vitro cleavage assay in which mutant precursor proteins translated from cDNA clones were used as substrates for trans cleavage by the proteinase. In general, amino acid substitutions in the P1 position (Glu) of the cleavage site were less well tolerated by the proteinase than those in the P1′ position (Ala). The precursor cleavage site mutations were introduced into an infectious cDNA clone of the FCV genome, and transfection of RNA derived from these clones into feline kidney cells showed that efficient cleavage of the capsid precursor by the virus-encoded proteinase is a critical determinant in the growth of the virus.
Feline calicivirus (FCV) causes an upper respiratory tract disease in cats and is an important veterinary pathogen (10). Virions are nonenveloped capsids of icosahedral symmetry that carry a VPg-linked 7.7-kb single-strand positive-sense RNA genome that is polyadenylated at the 3′ end. The genome is organized into three open reading frames (ORFs). The first (ORF1) encodes a large polyprotein that apparently undergoes proteolytic processing during infection to produce the mature nonstructural proteins. The second ORF (ORF2) of the FCV genome encodes the major structural protein that is translated as a 73-kDa precursor protein that is rapidly cleaved to yield the mature 62-kDa capsid protein by the removal of the first 125 amino acids (aa) from the N terminus (6, 26). The rate of this processing is relatively high, and the capsid precursor accumulates in infected cells only under conditions that inhibit this cleavage process, such as the addition of protease inhibitors to the growth medium or propagation of the virus at elevated temperature (4). It has been suggested that this cleavage is mediated by a virus-encoded enzyme because studies of the FCV capsid precursor in an eukaryotic expression system found no evidence for autocatalytic or host cell-mediated cleavage (30). The third and smallest ORF (ORF3) in FCV encodes a predicted basic polyprotein of 106 aa. Recently, a protein corresponding to this small ORF was detected with specific antisera in FCV-infected cells, but the function of this protein has not yet been determined (14).
Analysis of the proteins synthesized in feline kidney cells following infection with FCV indicates that the mature 62-kDa capsid protein is the predominant viral protein produced. However, additional viral proteins of 96, 75, 39, 36, and 27 kDa have been described (4). It has been proposed that these latter proteins correspond to cleavage products from a larger polyprotein and that they may represent nonstructural proteins. However, the boundaries of these cleavage products have not yet been mapped for FCV, and the role of proteolytic processing in the replication of this virus has not been fully defined.
The regions of homologous sequence between viruses in the Caliciviridae and those in the Picornaviridae in both the structural and nonstructural proteins suggest similarities in their replication strategies (3, 17, 18, 25, 32). Picornavirus structural and nonstructural proteins are synthesized as part of a single large polyprotein that matures in a cascade of proteolytic events. However, a major difference between the two families is that the calicivirus structural capsid protein is translated from an abundant subgenomic RNA coterminal with the 3′ end of the genome (2, 5, 7, 8, 23, 24, 26). For many members of the Caliciviridae, the mature capsid protein is thought to be translated directly from the subgenomic RNA. Direct sequence analysis of the N terminus of the rabbit hemorrhagic disease virus (RHDV) capsid protein in purified virus particles mapped it to the precise 5′ end of the subgenomic RNA (28). However, in a subset of viruses in the Caliciviridae, represented by FCV and San Miguel sea lion virus (SMSV), a larger capsid precursor protein undergoes proteolytic processing (4, 9). The purpose of the present study was to characterize the proteolytic activity involved in FCV capsid protein maturation from this larger precursor protein and examine the role of this cleavage event in the FCV replication strategy.
MATERIALS AND METHODS
Cells and virus.
Crandell-Rees feline kidney (CRFK) cell monolayers were grown in Eagle’s minimum essential medium supplemented with 10% heat-inactivated fetal bovine serum, amphotericin B (2.5 μg/ml), chlortetracycline (25 μg/ml), penicillin (250 U/ml), and streptomycin (250 μg/ml). The growth, purification, and molecular characterization of the Urbana (URB) strain of FCV have been described elsewhere (31).
Preparation of cell lysates.
CRFK monolayers (7 × 105 cells) were mock infected or infected with FCV at a multiplicity of infection of 10 and incubated at 37°C. At 6 h postinfection, the cells were washed once with phosphate-buffered saline (PBS), removed by scraping into fresh PBS, and pelleted by centrifugation at 8,000 × g for 5 min. The pellet was suspended in 300 μl of PBS and subjected to three cycles of freezing-thawing. The resulting lysate was clarified by centrifugation at 15,000 × g for 20 min and stored at −70°C.
For radiolabeling of virus-specific proteins, FCV-infected CRFK cells were washed at 4 h postinfection with methionine-free growth medium and incubated in the same medium for 30 min. [35S]methionine (>1,000 Ci/mmol; Amersham) was added to cells at a concentration of 150 μCi/ml, and the mixture was incubated for 3 h. The cells were washed with PBS before lysis in 300 μl of radioimmunoprecipitation assay (RIPA) buffer (29).
Plasmid construction.
Standard recombinant DNA methods were used for plasmid constructions (29). The numbering of the FCV nucleotide sequence in this study was derived from the complete genome sequence of FCV strain URB deposited in GenBank under accession no. L40021.
Plasmids pfI-21, pfI-20, pfI-45, pfI-9, pfI-19, pfI-28, and pfI-34 were selected by hybridization and restriction analysis from a cDNA library of the FCV RNA genome cloned into the pSPORT1 vector (31). These clones all contained ORF2, ORF3, a poly(A) tract, and various lengths of the C-terminal part of ORF1 upstream of ORF2 (Fig. 1A). Sequence analysis mapped the 5′ ends of these clones to nucleotides (nt) 5316, 5194, 4820, 4704, 4370, 3094, and 2989, respectively, of the FCV genome.
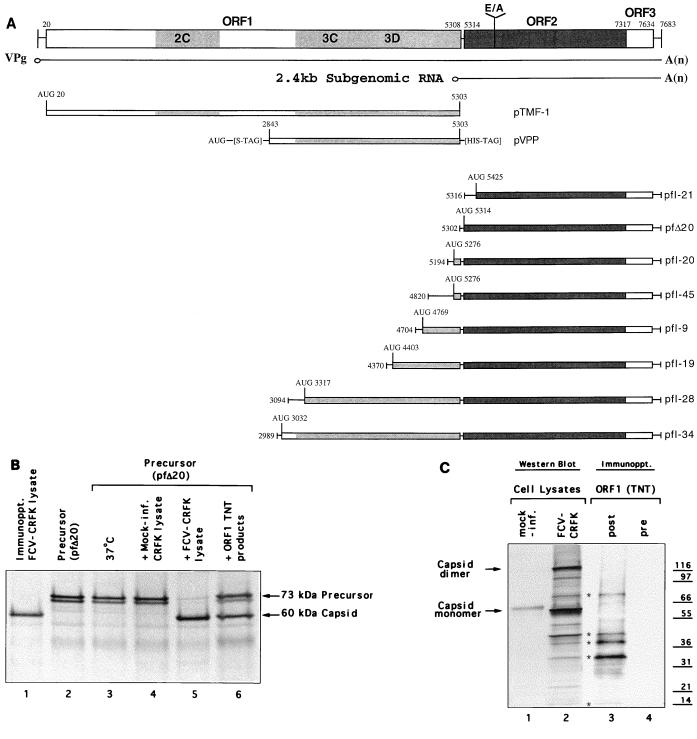
FIG. 1.
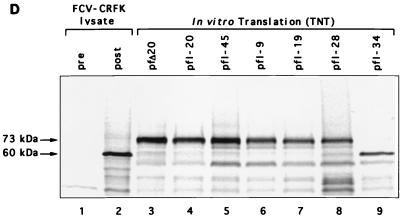
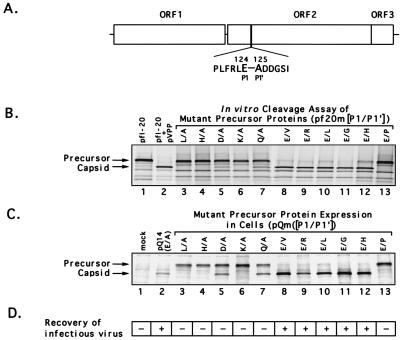
(A) Organization of the FCV genome (URB strain) and depiction of cDNA clones analyzed in this study. The corresponding genomic locations of the two major positive-sense RNA species (7.6 and 2.4 kb) found in infected cells and the location of the cleavage site (E124/A125) of the capsid precursor protein encoded in ORF2 are indicated. Clones pfI-21, pfI-20, pfI-45, pfI-9, pfI-19, pfI-28, and pfI-34 were selected from a cDNA library of the URB strain that was constructed by using the pSPORT plasmid (31) and contained nt 5316, 5194, 4820, 4704, 4370, 3094, and 2989, respectively, through a poly(A) tract from the URB genome, each under control of the T7 promoter. The location of the first in-frame AUG following the T7 promoter is indicated for each clone. Plasmids pTMF1, pVPP, and pfΔ20 were engineered as described in Materials and Methods. (B) Conditions under which in vitro cleavage of the FCV capsid precursor protein derived from plasmid pfΔ20 were observed. Lane 1, 60-kDa protein corresponding to the mature viral capsid protein immunoprecipitated (Immunoppt.) from an FCV-infected CRFK lysate with gp α-FCV; lane 2, in vitro TNT translation products synthesized from pfΔ20 without treatment. The translation products from pfΔ20 were treated as follows prior to analysis by SDS-PAGE: lane 3, incubation at 37°C for 3 h; lane 4, incubation (3 h, 37°C) with a nonradiolabeled CRFK lysate prepared from mock-infected cells; lane 5, incubation (3 h, 37°C) with a nonradiolabeled CRFK lysate prepared from FCV-infected cells; lane 6, incubation (3 h, 37°C) with nonradiolabeled translation products synthesized from pTMF-1. (C) Comparison of radiolabeled translation products derived from pTMF-1 (encoding the URB ORF1) with proteins produced in FCV-infected CRFK cells. Proteins produced in mock-infected (lane 1) or FCV-infected (lane 2) CRFK cells were analyzed in a Western blot reacted with cat FCV postinfection serum. The same infection serum was used to immunoprecipitate (Immunoppt.) radiolabeled TNT products derived from pTMF-1 (lane 3). Lane 4, immunoprecipitation analysis of radiolabeled TNT products derived from pTMF-1 with cat preinfection serum. The location of the mature capsid protein and its dimeric form in the FCV-infected CRFK cell lysate is shown. Asterisks denote proteins similar in size (indicated in kilodaltons) between ORF1 TNT products and the FCV-infected CRFK cell lysate. (D) Comparison of products produced by in vitro translation of cDNA clones encoding the ORF2 (with various lengths of upstream ORF1 sequence) with the mature 60-kDa capsid protein produced in virus-infected cells. Radiolabeled products (in all lanes) underwent immunoprecipitation with either preimmunization (lane 1) or postimmunization (lanes 2 to 9) gp α-FCV, as follows: lanes 1 and 2, FCV-infected CRFK lysate; lane 3, pfΔ20; lane 4, pfI-20; lane 5, pfI-45; lane 6, pfI-9; lane 7, pfI-19; lane 8, pfI-28; lane 9, pfI-34.
Plasmid pfΔ20 was created by excision of the NspV-SmaI fragment from plasmid pfI-20, followed by treatment with the Klenow fragment of DNA polymerase I and recircularization of the plasmid, resulting in a clone that contained the 3′-terminal part of the FCV genome starting at nt 5302.
Plasmid pTMF-1 was constructed by subcloning the 5,282-bp AspI-NspV fragment of plasmid pQ14 (31) into NcoI-digested pTM-1 vector (provided by B. Moss, National Institute of Allergy and Infectious Diseases) after filling in of the protruding restriction ends of the plasmid and the fragment. The resulting plasmid contained the T7 RNA polymerase promoter, the encephalomyocarditis virus internal ribosome entry site, and the entire ORF1 of the FCV genome with the exception of the two C-terminal codons.
Plasmid pVPP was constructed as follows. pTMF-1 was cleaved with BsaI, followed by incubation with Klenow enzyme first in the presence of dGTP and then in the presence of dATP. The fragments with partially filled-in protruding ends were further treated with XhoI, and a 2,503-bp fragment was isolated. This fragment was ligated into the Acc65.1 (modified by treatment with Klenow enzyme in the presence of dGTP and dTTP) and XhoI sites of the pET-29c plasmid (Novagen) downstream from the T7 promoter, and the selected clone was designated pVPP.
Site-directed mutagenesis of the capsid precursor cleavage site.
An intermediate plasmid, designated pfKBS11, was prepared by ligation of a 1,012-bp KpnI/SpeI fragment obtained from pfI-20 into the same sites of the pLITMUS28 vector (New England Biolabs). DNA fragments were amplified from plasmid pfKBS11 by PCR using a sense primer corresponding to nt 5297 to 5319 of the genome and an antisense primer corresponding to nt 5678 to 5703. The antisense primers were used to introduce mutations into either the P1 or P1′ position of the wild-type cleavage site sequence (Glu124/Ala125:5683GAAGCT5688). The P1 position mutations were GAA to CTC for Leu, CAC for His, GAT for Asp, AAG for Lys, and CAG for Gln. The P1′ position mutations were GCT to GTT for Val, CGT for Arg, CTT for Leu, GGT for Gly, CAT for His, and CCT for Pro. Purified PCR-generated DNA fragments were treated with NspV and BamHI and ligated back into the compatible sites of pfKBS11. Clones were screened by sequence analysis, and plasmids containing desired mutations in the cleavage site were selected and designated pm(P1/P1′), with the amino acid sequence of P1 and P1′ for each mutant designated by the single-letter code. The FCV-specific fragments were then excised from the pfKBS11 derivatives by digestion with NspV and SpeI and used to replace the wild-type sequences in pfI-20. The resulting clones were designated pf20m(P1/P1′). The same NspV-SpeI fragments were used to introduce capsid precursor cleavage site mutations into the full-length genomic clone pQ14, and the resulting clones were designated pQm(P1/P1′). Mutations in the selected clones were confirmed by sequence analysis.
In vitro coupled transcription-translation experiments.
One microgram of plasmid DNA was used as the template in a coupled transcription-translation reaction (TNT T7 coupled reticulocyte lysate system; Promega). For radiolabeling of synthesized protein, [35S]methionine (>1,000 Ci/mmol) from ICN or Amersham was used at a concentration of 1.5 mCi/ml.
Precursor cleavage assay and analysis of protease inhibitors.
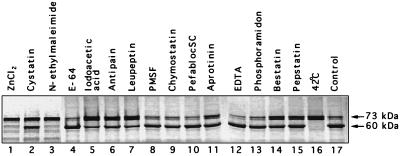
Nonradiolabeled lysates of FCV-infected CRFK cells were incubated for 1 to 3 h at 37°C with the radiolabeled capsid precursor protein produced by transcription and translation of pfΔ20. The integrity of the capsid precursor was then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a minigel system (Novex). Protease inhibitors were added to the FCV-infected CRFK cell lysates and incubated for 15 min at 37°C prior to mixing with radiolabeled capsid precursor protein. Inhibitors tested were antipain dihydrochloride (50 μg/ml), aprotinin (2 μg/ml), bestatin (40 μg/ml), chymostatin (60 μg/ml), EDTA (2 mM), E-64 (10 μg/ml), leupeptin (0.5 μg/ml), phenylmethylsulfonyl fluoride (PMSF; 170 μg/ml), PefablocSC (1 μg/ml), pepstatin (0.7 μg/ml), and phosphoramidon (200 μg/ml), purchased from Boehringer Mannheim; N-ethylmaleimide (1 mM), iodoacetic acid (2 mM), cystatin (100 μg/ml), N-tosyl-l-phenylalanine chloromethyl ketone (TPCK; 100 μg/ml), and Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK; 50 μg/ml), from Sigma Chemical Co.; and ZnCl2 (1 mM), from Fluka AG.
Immunoprecipitation and Western blot analysis.
Fifty microliters of the radiolabeled FCV-infected CRFK cell lysate was diluted with 50 μl of RIPA buffer and incubated at 4°C for 1 h with 5 μl of guinea pig antiserum raised against purified viral particles (gp α-FCV) or guinea pig preimmunization serum. After incubation with serum, 50 μl of a 50% slurry of protein G beads (Pharmacia) prewashed with RIPA buffer was added to the lysates. The mixture was gently rotated overnight at 4°C. The beads were pelleted by centrifugation at 12,000 × g and then washed twice with 1 ml of RIPA buffer, twice with 1 ml of washing buffer (1 M NaCl, 0.01 M Tris-HCl [pH 7.2], 0.1% Nonidet P-40) and twice with 1 ml of 0.01 M Tris-HCl (pH 7.2). The precipitated proteins were extracted from the beads by boiling in SDS-PAGE sample buffer with 2% mercaptoethanol for 5 min prior to analysis by SDS-PAGE.
For the immunoprecipitation of FCV nonstructural proteins synthesized in vitro, 15 μl of the TNT translation reaction was diluted with 80 μl of RIPA buffer and heated for 10 min at 60°C. The proteins were then incubated for 1 h at room temperature with 5 μl of cat FCV infection or preinfection sera (gifts from W. Mengeling, U.S. Department of Agriculture), and the immune complexes were precipitated with protein A beads (Sigma). Binding and washing conditions were the same as those described above.
Western blot analysis was performed by using standard techniques (29). Following electrophoretic transfer of proteins to nitrocellulose, the membrane was incubated with cat FCV infection serum (1:1,000 dilution). The binding of cat antibodies was detected with anti-cat immunoglobulins conjugated with phosphatase (Kirkegaard & Perry Laboratories) followed by development with alkaline phosphatase (AP) substrate reagents (Gibco BRL). In the Western blot analysis to detect histidine-tagged proteins, a nickel-nitrilotriacetic acid (Ni-NTA)–AP conjugate (Qiagen) was used according to the manufacturer’s protocol.
Recovery of recombinant virus.
In vitro transcription of synthetic, capped RNA from full-length genomic cDNA clones and RNA transfection experiments were conducted as described previously (31). Following incubation for 16 or 26 h at 37°C, transfected CRFK cell monolayers were analyzed by immunofluorescence with gp α-FCV and α-gp immunoglobulins conjugated with fluorescein (31). In parallel experiments, monolayers were incubated for 30 min with methionine-free medium, and proteins were radiolabeled as described above. Recovery of infectious virus was confirmed by transferring an aliquot of culture medium from transfected cells onto fresh CRFK monolayers.
Amino acid substitutions in the capsid protein from recovered viruses purified by CsCl gradient centrifugation (24) were confirmed by direct N-terminal sequence analysis. Viral proteins were separated by SDS-PAGE using tricine running buffer (Novex), transferred to a ProBlott membrane (Applied Biosystems), and visualized on the membrane by staining with 0.1% Coomassie blue R-250–40% methanol–1% acetic acid and destaining in 50% methanol. The band of interest was excised and subjected to N-terminal sequence analysis with a model 477A protein sequencer coupled to a model 120A PTH Analyzer (Applied Biosystems) according to the manufacturer’s program, NORMAL-1.
The presence of engineered mutations in the genomes of recovered viruses was also confirmed by direct sequence analysis of reverse transcriptase (RT)-mediated PCR (RT)-PCR products derived from viral RNA. Primers used for amplification were a sense primer corresponding to nt 5297 to 5319 of the genome, described above, and an antisense primer corresponding to nt 6196 to 6216. In addition, as a control for the presence of DNA from the original plasmid used to synthesize the RNA for transfection, the PCR was performed without an initial RT reaction. The agarose gel-purified DNA fragments were sequenced by using a cycle sequencing kit from Gibco BRL and a sense primer corresponding to nt 5530 to 5550 of the genome.
Expression of the protease in Escherichia coli.
E. coli BL21(DE3) cells were transformed with either plasmid pVPP or plasmid pET-29c, and transformed cells were grown in the presence of carbenicillin (50 μg/ml) in LB medium at 37°C. When the A600 of the culture reached 0.8, expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h, cells were collected by centrifugation at 4,000 × g for 15 min and the pellets were suspended in buffer containing 300 mM NaCl and 50 mM Na2HPO4 (pH 7.8) in 1/10 of the original culture volume. After freezing-thawing, the bacteria were sonicated (crude lysate) and subjected to centrifugation at 12,000 × g for 20 min. The supernatant (soluble fraction) was collected, and the pellet (insoluble fraction) was suspended in the same buffer, sonicated, and collected again by centrifugation.
RESULTS
Expression of FCV capsid precursor protein in a coupled in vitro transcription-translation system.
Clones containing ORF2 from a cDNA library of the URB strain of FCV were selected for analysis of capsid precursor expression in the TNT system (Fig. 1A). To optimize protein synthesis in this system, plasmid pfI-20 was engineered to remove ORF1 sequences upstream of the beginning of ORF2 that contained additional AUG codons. The resulting plasmid, designated pfΔ20, contained the first AUG of ORF2 directly under control of the T7 RNA polymerase promoter. Analysis of the translation products from pfΔ20 by SDS-PAGE revealed the efficient synthesis of two major bands corresponding to proteins with molecular sizes of 73 and 70 kDa (Fig. 1B, lane 2). The 73-kDa protein was consistent in size with the predicted full-length product of synthesis from ORF2, while the 70-kDa protein was consistent with the predicted product (69.2 kDa) resulting from efficient initiation at the second AUG of ORF2. In some translation experiments of clones containing ORF2, we observed additional minor bands corresponding to proteins of 64 and 56 kDa, sizes consistent with those calculated for proteins produced by internal initiation at other AUGs in the template. All proteins described above were recognized by gp α-FCV in immunoprecipitation experiments and were not recognized by gp preimmunization serum (data not shown). Translation of the capsid proteins in the presence of canine microsomal membranes did not lead to changes in the observed mobility of the proteins (data not shown).
Evidence for cleavage of the FCV capsid precursor by a virus-encoded protease.
The conditions under which the capsid precursor could be processed into the mature capsid protein were examined. Reticulocyte lysates containing the translated capsid protein from clone pfΔ20 were incubated at 37°C for various lengths of time up to 12 h, but no evidence for autocatalytic cleavage of the precursor in the reticulocyte lysates was found (Fig. 1B, lane 3). Similarly, no evidence for cleavage was observed following incubation of the precursor with lysates prepared from noninfected CRFK cells (Fig. 1B, lane 4). However, incubation of the precursor with a lysate prepared from FCV-infected CRFK cells led to the appearance of a band (Fig. 1B, lane 5) corresponding in size to the mature radiolabeled URB FCV capsid protein (60 kDa) immunoprecipitated from an FCV-infected CRFK cell lysate (Fig. 1B, lane 1). Taken together, these data suggested the presence of a proteinase in FCV-infected cells that could cleave the capsid precursor substrate in trans.
To address whether the ORF1 of the FCV genome encoded a proteinase responsible for the cleavage of the capsid precursor, we cloned the URB ORF1 into the pTM-1 plasmid (designated pTMF-1). Analysis of the in vitro translation products synthesized from the pTMF-1 translation mixture showed evidence for autocatalytic cleavage of the encoded ORF1 polyprotein. Instead of the predicted polyprotein with an estimated size of 195 kDa, we observed at least four major protein bands with sizes ranging from approximately 30 to 80 kDa and several minor bands including a protein of approximately 14 kDa that were immunoprecipitated with cat FCV infection serum (Fig. 1C, lane 3). The proteins were not recognized by cat preinfection serum (Fig. 1C, lane 4). These four major proteins corresponded in size to four bands detected in FCV-infected CRFK cells in a Western blot reacted with the cat FCV infection serum (Fig. 1C, lane 2). These proteins were not recognized by cat preinfection serum (Fig. 1C, lane 1), although the cat preinfection serum did show reactivity with an apparently nonviral 62-kDa protein present in both FCV-infected and mock-infected cell lysates. The similarity between these proteins produced from in vitro translation of ORF1 and those in FCV-infected cells suggested that the protease sequences encoded in ORF1 were sufficient to mediate cleavage of the ORF1 polyprotein in the absence of cellular factors and that the majority of proteins synthesized in vitro were identical in size to mature forms of viral nonstructural proteins in infected cells. The nonradiolabeled ORF1 translational products derived from pTMF-1 were incubated with radiolabeled capsid precursor protein derived from in vitro translation of pfΔ20. Analysis of the products of this incubation by SDS-PAGE revealed the presence of mature, cleaved capsid protein, consistent with trans cleavage of the capsid precursor by a proteinase encoded in ORF1 (Fig. 1B, lane 6). Polypeptides consistent in size with the cleaved N-terminal part of the capsid precursor could be detected as faint bands in a high-percentage polyacrylamide gel (data not shown).
Mapping the FCV protease gene responsible for cleavage of the capsid precursor.
Further localization of the region of the FCV genome encoding the protease was facilitated by the observation that the capsid precursor protein could be efficiently translated from clones that contained various lengths of the viral genome upstream of the first AUG of ORF2. The 73-kDa precursor protein was detected following transcription and translation of plasmids pfΔ20 and pfI-20 (Fig. 1D, lanes 3 and 4), which contained either 12 or 120 nt of upstream ORF1 sequence, respectively. The 73-kDa protein was also produced efficiently from plasmids pfI-45, pfI-9, and pfI-19, containing the upstream 489, 605, and 939 nt, respectively, of ORF1 (Fig. 1D, lanes 5 to 7). Efficient synthesis of the capsid precursor was observed for noncapped RNA in the TNT system as well as for capped RNA in a noncoupled translation reaction (data not shown). We next examined the synthesis of capsid protein from two plasmids, pfI-28 and pfI-34, that contained upstream ORF1 sequences (a total of 2215 and 2319 nt, respectively) analogous to the 3C protease region of the picornaviruses that had been defined by Neill (25) as encompassing nt 3284 to 3694 (aa 1089 to 1225). In contrast to the plasmids that did not contain 3C protease region sequences, a 60-kDa band was observed among the products of translation of pfI-28 and pfI-34 that was immunoprecipitated with gp α-FCV (Fig. 1D, lanes 8 and 9). This 60-kDa band was consistent with the mature form of the capsid protein immunoprecipitated from FCV-infected CRFK cells (Fig. 1D, lane 2). The efficient cotranslational cleavage of the precursor encoded in pfI-34 was indirect evidence for the synthesis of an active proteinase. The first predicted methionine in the upstream ORF1 sequence of pfI-34 was located at nt 3032, and initiation at this methionine (aa 1005) would result in the translation of a protein that included the entire 3C region defined by Neill. In contrast, initiation at the first methionine (aa 1100) of pfI-28 would result in the synthesis of a truncated 3C region product.
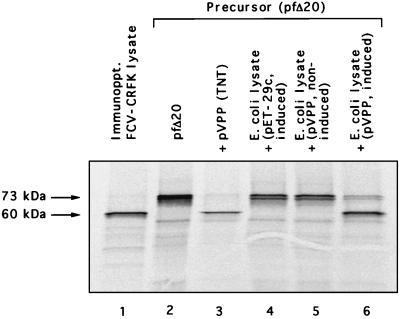
Subcloning of the C-terminal part of the ORF1 was carried out to further characterize the proteins encoded in this region of the FCV genome. pVPP contained nt 2843 to 5303 of the URB genome in the pET-29c vector so that the sequence encoding the 820 amino acid residues of the C-terminal part of the ORF1 could be expressed in either the TNT system or in bacteria as a protein fused to a His6tag at its C terminus. Incubation of nonradiolabeled TNT products derived from pVPP with the radiolabeled capsid precursor synthesized from pfΔ20 led to the efficient processing of the precursor into the mature capsid form (Fig. 2, lane 3), indicating that the proteinase encoded in pVPP was active in the in vitro trans cleavage assay. Cleavage of the radiolabeled capsid precursor was observed also after incubation with crude lysates of IPTG-induced E. coli cells harboring pVPP (Fig. 2, lane 6) but not after incubation with lysates of noninduced pVPP-transformed cells (Fig. 2, lane 5) or IPTG-induced cells harboring vector plasmid pET-29c (Fig. 2, lane 4). Virus-specific protease activity was detected in soluble as well as insoluble fractions of bacterial cell lysates (data not shown).
FIG. 2.
Analysis of the proteolytic activity of the proteins encoded in plasmid pVPP in the capsid precursor trans cleavage assay. Lane 1, immunoprecipitation (Immunoppt.) of radiolabeled FCV-infected CRFK cell lysate with gp α-FCV. Lane 2, pfΔ20 translation products, without treatment. The radiolabeled capsid precursor protein was incubated with the following nonradiolabeled preparations prior to analysis by SDS-PAGE: lane 3, in vitro TNT translation products synthesized from pVPP; lane 4, E. coli crude cell lysate prepared from IPTG-induced bacteria containing the pET-29c vector plasmid; lane 5, E. coli crude cell lysate from noninduced bacteria carrying plasmid pVPP; lane 6, E. coli crude cell lysate from IPTG-induced bacteria carrying plasmid pVPP.
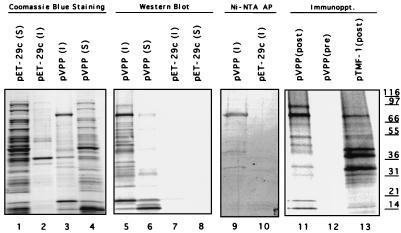
Analysis of radiolabeled TNT products derived from pVPP showed the presence of several proteins with sizes ranging from approximately 14 to 95 kDa that could be immunoprecipitated with cat FCV infection serum (Fig. 3, lane 11). A faint 95-kDa band corresponded in size to the predicted full-length product of translation from pVPP, but the major protein observed was approximately 78 kDa. Of interest, an approximately 78-kDa protein was present in a FCV-infected cell lysate (Fig. 1C, lane 2) and in the in vitro translation products derived from pTMF-1 (encoding the entire ORF1) (Fig. 3, lane 13) that was recognized by antibodies in the cat FCV infection serum.
FIG. 3.
Analysis of proteins encoded in plasmid pVPP. Bacteria carrying either plasmid pVPP or the vector plasmid pET-29c were induced with IPTG, and the soluble (S) or insoluble (I) bacterial products were prepared as described in the text. The bacterial products were subjected to SDS-PAGE and visualized with Coomassie blue stain: lane 1, pET-29c, soluble fraction; lane 2, pET-29c, insoluble fraction; lane 3, pVPP, insoluble fraction; lane 4, pVPP, soluble fraction. The same products were analyzed in a Western blot developed with cat postinfection serum: lane 5, pVPP, insoluble fraction; lane 6, pVPP, soluble fraction; lane 7, pET-29C, insoluble fraction; lane 8, pET-29c, soluble fraction. The insoluble fractions of pVPP or pET-29c were transferred to nitrocellose and probed with Ni-NTA–AP: lane 9, pVPP; lane 10, pET-29c. The radiolabeled TNT in vitro translation products derived from pVPP were immunoprecipitated (Immunoppt.) with either postinfection (lane 11) or preinfection (lane 12) cat serum. Lane 13, radiolabeled TNT products derived from pTMF-1 immunoprecipitated with cat postinfection serum.
The proteins expressed by bacteria carrying plasmid pVPP were analyzed by SDS-PAGE and Western blotting. Coomassie blue staining showed two major proteins of 78 and 18 kDa in the insoluble fraction (Fig. 3, lane 3) that did not correspond to the major bands of the pET-29c soluble and insoluble fraction controls (Fig. 3, lanes 1 and 2). A major 14-kDa protein was observed in the soluble fraction of bacterial cells harboring pVPP (Fig. 3, lane 4) that comigrated with a protein of similar size in the soluble fraction of bacterial cells harboring pET-29c (Fig. 3, lane 1). A Western blot of these fractions reacted with cat infection serum indicating that the 78-, 18-, and 14-kDa proteins were derived from FCV (Fig. 3, lanes 5 and 6). The proteins on nitrocellulose were probed also with a Ni-NTA–AP conjugate in order to localize the C-terminal His tag. The 78-kDa protein was recognized efficiently, mapping it to the 3′ end of pVPP and, therefore, the carboxy-terminal end of the FCV ORF1 (Fig. 3, lane 9). Cleavage of the 78-kDa protein from a 95-kDa precursor would result in an N-terminal cleavage product of approximately 18 kDa. The in vitro translation reaction of pVPP contained both 18- and 14-kDa proteins (Fig. 3, lane 11), and a time course analysis showed an accumulation of the 14-kDa protein with a relative decrease in the amount of 18-kDa protein (data not shown). Thus, it appeared that the 14-kDa protein expressed from pVPP that was observed in the TNT reactions and in the bacteria resulted from further proteolytic processing of the 18-kDa N-terminal sequence. The absence of intermediate-sized proteins between 95 and 78 kDa suggested that this cleavage event occurred in trans following the release of the 18-kDa protein from the 95-kDa precursor.
A panel of commercially available protease inhibitors was examined for its effect on the proteinase and tested at either the working concentrations recommended by the manufacturer or at concentrations 10 times higher. The effect of the inhibitors on the ability of the proteinase present in FCV-infected CRFK cell lysates to cleave the precursor in trans is shown in Fig. 4. A strong inhibitory effect was observed for the cysteine protease inhibitors ZnCl2 and N-ethylmaleimide at their recommended working concentrations (Fig. 4, lanes 1 and 3) but was not observed for iodoacetic acid (lane 5). Strong inhibition was not observed with papain-like cysteine protease inhibitors (cystatin and E-64; lanes 2 and 4), a papain protease inhibitor (antipain; lane 6), serine protease inhibitors (leupeptin, PMSF, chymostatin, PefablocSC, and aprotinin; lanes 7 to 11), metalloprotease inhibitors (EDTA and phosphoramidon; lanes 12 and 13), an amino peptidase inhibitor (bestatin; lane 14), or an aspartic acid protease inhibitor (pepstatin; lane 15) at their recommended concentrations (Fig. 4). However, the protease activity appeared to be sensitive to TPCK, TLCK, and concentrated quantities of iodoacetic acid (data not shown). Cleavage of the precursor was not observed when the FCV-infected cell lysate was incubated at 42°C for 15 min prior to incubation with the precursor (Fig. 4, lane 16).
FIG. 4.
Effects of selected protease inhibitors or temperature on the ability of the protease present in FCV-infected CRFK lysates to cleave in trans the capsid precursor protein translated from pfΔ20. In lanes 1 to 15, the indicated protease inhibitors were incubated with the FCV-infected CRFK lysate prior to incubation with the translated precursor protein under conditions described in the text. Lane 16, incubation of FCV-infected cell lysate at 42°C for 20 min prior to incubation with the capsid precursor; lane 17, precursor protein incubated with nontreated FCV-infected CRFK lysate.
Mutagenesis of the FCV proteinase cleavage site in the capsid protein precursor.
PCR was used to introduce changes into either the P1 or P1′ positions of the capsid precursor protein wild-type cleavage site (E/A) (Fig. 5A), resulting in mutated precursor clones (p20m L/A, H/A, D/A, K/A, Q/A, E/V, E/R, E/L, E/G, E/H, and E/P) that were used as templates for in vitro synthesis of capsid precursor proteins in a TNT reaction. Synthesized precursors were incubated at 37°C with nonradiolabeled TNT mixtures derived from pVPP as the source of proteinase. Incubation of the proteinase with the radiolabeled TNT products from pfI-20 that contained the wild-type precursor cleavage site showed the cleavage of the precursor into the mature capsid protein (Fig. 5B, lane 2). No evidence for cleavage was observed when the proteinase was incubated with precursors that contained L, H, and K mutations in the P1 position (Fig. 5B, lanes 3, 4, and 6); however, weak specific cleavage was observed for precursors that possessed D and Q in the P1 position (Fig. 5B, lanes 5 and 7). Of the six amino acid substitutions introduced into the P1′ position of the cleavage site, five (V, R, L, G, and H) allowed cleavage of the precursor (Fig. 5B, lanes 8 to 12), while one (P) did not (Fig. 5B, lane 13). Differences in the rates of cleavage were detectable among the five mutants that showed complete cleavage early in the first hour of the reaction, but the final cleavage reaction appeared similar among the mutants at the end of the incubation period (data not shown).
FIG. 5.
Analysis of the effects of amino acid mutations introduced into the cleavage site of the FCV capsid precursor protein on the efficiency of cleavage by the viral proteinase as measured in the in vitro trans cleavage assay or by the ability of the virus to grow in cell culture. (A) Sequence of cleavage site between the precursor leader sequence and the mature capsid protein into which amino acid substitutions were introduced. (B) The radiolabeled capsid precursor proteins derived from in vitro translation of pfI-20 or engineered plasmids containing mutant cleavage sites [clones pf20m(P1/P1′)] were incubated with nonradiolabeled translation products derived from plasmid pVPP. The first lane contains the pfI-20 translation products without treatment. (C) Capped, synthetic RNA from individual full-length clones that contained the mutant cleavage sites [clones pQm(P1/P1′)] were transfected into CRFK cells, and proteins synthesized in cells were radiolabeled with [35S]methionine. The following cell lysates were analyzed by immunoprecipitation with gp α-FCV serum: lane 1, mock-infected CRFK cells; lane 2, CRFK cells transfected with RNA derived from pQ14; lanes 3 to 13, CRFK cells transfected with RNA derived from plasmids pQm(P1/P1′). (D) Effects of mutations in the P1 or P1′ position of the precursor cleavage site on the ability to recover viable virus from engineered full-length infectious clones. In a separate experiment, an aliquot of cell culture medium from cells transfected with RNA derived from each of the pQm(P1/P1′) clones was transferred to a fresh CRFK monolayer. Clones that did (+) and did not (−) yield viable progeny are indicated.
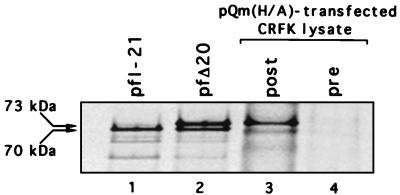
To analyze the effects of alterations in the capsid precursor cleavage site on virus replication, we constructed a series of full-length genomic clones by engineering DNA fragments with the corresponding precursor mutations into the infectious FCV clone, pQ14. The resulting clones (pQm L/A, H/A, D/A, K/A, Q/A, E/V, E/R, E/L, E/G, E/H, and E/P) were used to produce synthetic genomic RNAs for transfection experiments. Immunoprecipitation analysis of labeled capsid protein from transfected cells confirmed the expression of ORF2 for all the capsid precursor mutants (Fig. 5C). In addition, we observed a marked similarity between the processing of the capsid precursor in cells and that in the in vitro cleavage assay (Fig. 5B and C). The apparent accumulation of the 73-kDa protein in cells transfected with RNA from plasmids encoding mutant cleavage sites that were not cleaved by the proteinase suggested that the first AUG of the precursor may be preferentially utilized in cells. The immunoprecipitation products of the noncleaved capsid precursor from pQm H/A expressed in cells (Fig. 6, lane 3) were compared directly with those synthesized in the TNT system from pfI-21 (a cDNA clone in the library that lacked the first AUG of ORF2) (Fig. 6, lane 1) and pfΔ20 (a clone that contained the first AUG of ORF2) (Fig. 6, lane 2). The presence of only the 73-kDa protein in the cell lysates from pQm H/A indicated that the first AUG of ORF2 is utilized preferentially in cells.
FIG. 6.
Comparison of FCV capsid precursor polyprotein expressed in vitro and in transfected cells. Lanes 1 and 2, radiolabeled products of in vitro translation of pfI-21 and pfΔ20, respectively; lanes 3, and 4, immunoprecipitation of radiolabeled precursor with gp α-FCV and gp preimmunization serum, respectively, from CRFK cells transfected with capped genomic RNA that encoded the H/A mutant cleavage site.
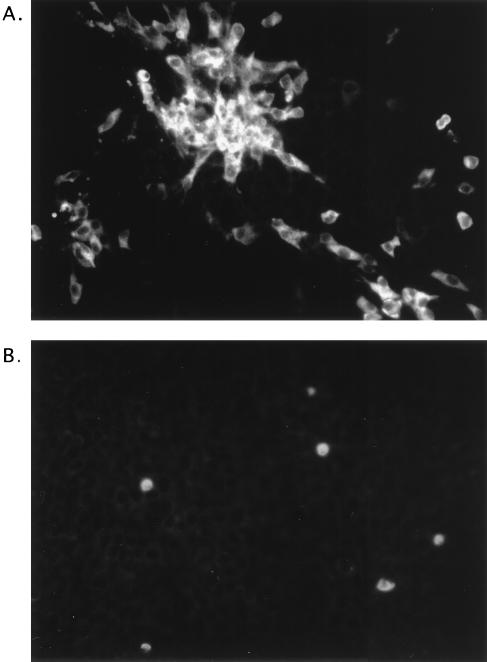
Transfer of medium from transfected cells to fresh CRFK monolayers allowed the recovery of FCV for mutants showing complete cleavage of the precursor in cells (pQm E/V, E/R, E/L, E/G, and E/H) (Fig. 5D). A time course titration for the recovered viruses from these mutants did not reveal significant differences in their growth characteristics in comparison with each other or with virus recovered from pQ14 (data not shown). Transfected CRFK cell monolayers were analyzed at 16 h posttransfection by immunofluorescence using gp α-FCV. All wells transfected with genomic RNAs contained intensely stained cells at 16 h posttransfection, including those mutants with capsid precursor cleavage sites that were not cleaved in the in vitro assay and from which virus could not be recovered (data not shown). However, the observation of transfected cells at 26 h posttransfection by immunofluorescence allowed the detection of marked differences in the distribution of positive cells between viable and nonviable mutants as illustrated in Fig. 7. Figure 7A shows the pattern of fluorescence from viable mutant pQm E/G that was similar in appearance to that of wild-type infectious FCV foci in CRFK monolayers. In contrast, Fig. 7B shows the pattern of fluorescence from nonviable mutant pQm Q/A in which single, positively stained cells remained distributed throughout the monolayer. Capsid expression was not observed by immunofluorescence in control transfection experiments in which full-length, capped transcript RNAs derived from cDNA clones of RNA replication-defective FCV mutants were used, indicating that the positive signal observed with the capsid precursor mutants was not due to translation of the newly-transfected RNA (data not shown). Recovered viruses were amplified in CRFK cells and purified by gradient centrifugation. Direct sequence analysis of the capsid protein from purified viruses confirmed the presence of corresponding mutations at the N terminus for the E/L, E/V and E/G mutant viruses. However, the identity of the N-terminal amino acid for the E/R and E/H mutants could not be determined by direct protein sequence analysis, and the mutations in these viruses were confirmed by sequence analysis of RT-PCR products derived from the genomic RNA of the purified viruses. Viruses could not be recovered from cells transfected with pQm Q/A and pQm D/A, although mature, processed capsid protein was observed in the radiolabeled cell lysates. To examine whether infectious virus was retained in an intracellular compartment. CRFK monolayers were transfected with RNA from these two plasmids in three separate experiments. After 24 h, the transfected cells were frozen, thawed, and passed onto fresh cells. No evidence for infectious virus was found.
FIG. 7.
Immunofluorescence of CRFK cells transfected 26 h previously with full-length capped, synthetic RNA from pQm E/G (A) and pQm Q/A (B).
DISCUSSION
Synthesis of the FCV capsid precursor protein is detected in cells as early as 2 h following infection, but the protein is rapidly cleaved into the 62-kDa mature capsid (4). In the present work, we demonstrate that the proteinase responsible for this cleavage is encoded in the C-terminal part of ORF1 of the FCV genome. A virus-specific protein of approximately 78 kDa was consistently observed in FCV-infected cells and in clones containing the entire or C-terminal part of ORF1. We propose that this 78-kDa protein may be an active proteinase complex that corresponds to the 3CD protein complex of the picornaviruses. A similar 3CD-like complex has been observed in translational studies of the ORF1 of the Southampton human calicivirus (113 kDa) and RHDV (73 kDa) (20, 34). The picornavirus and RHDV proteases participate in the cleavage of the nonstructural as well as the structural capsid proteins from a large polyprotein (13, 19, 27). It is likely that the same FCV protease is responsible for the cleavage of the N terminus of the 3CD protein and perhaps other sites of the nonstructural polyprotein in addition to cleavage of the capsid precursor. However, as for RHDV and Southampton virus, the cleavage of the 3CD protein at the border between the protease and polymerase sequence appears to be inefficient in vitro. Further studies are in progress to identify the cleavage products observed in our expression studies and to determine whether they have counterparts in infected cells.
Among sequences found in the picornavirus-like 3C region of the FCV genome are domains containing H1110, E1131, C1193, and H1208 that are presumably involved in the formation of the catalytic site of the proteinase (3, 25). Picornavirus 3C cysteine proteinases are considered members of a family of chymotrypsin-like serine proteases that contain a cysteine instead of serine as the nucleophile in the active site (1, 11, 12). The picornavirus 3C proteinases are sensitive to inhibitors binding to thiol groups and are weakly inhibited by classical inhibitors of serine proteases (16, 33). In the present study, two classical cysteine protease inhibitors were the only chemicals that completely inhibited the FCV protease activity. Thus, the FCV proteinase apparently is similar to the picornavirus 3C proteinases in regard to inhibition with different classes of protease inhibitors. However, elucidation of the details of the FCV proteinase interaction with inhibitors will require further work with the purified enzyme. An interesting observation in this study was the sensitivity of the proteinase to increased temperature, which may, in part, explain why FCV is restricted for growth at higher temperatures. The inactivation of the proteinase by incubation at 42°C was irreversible, suggesting that the protease is highly sensitive to changes in conformation.
A feature of the picornavirus 3C proteinases that distinguishes them from cellular serine proteases is their high substrate specificity that is largely determined by the structure of the primary cleavage site sequence. Most wild-type cleavage sites in picornavirus polyproteins have the sequences Q (or E)/G, A, or S (13, 19, 27). Presumably, the structural element of the protease that determines selection of glutamine (Q) in the P1 position is the presence of a conserved histidine residue near the active site (1, 22). All calicivirus 3C-like protease sequences described thus far possess this conserved histidine residue, and there is similarity between the primary sequences of the picornavirus and calicivirus proteinase recognition sites, with the Southampton human calicivirus proteinase recognizing Q/G and the RHDV proteinase recognizing E/G and T (20, 21, 34). The FCV proteinase appears closer to that of RHDV in that the wild-type cleavage site of the capsid precursor is E/A. The FCV protease was less tolerant of changes in the P1 position, but it could recognize Q and D in the P1 position, similar to the RHDV protease (34). Certain mutations in the P1 (L, K, and H) and P1′ (P) positions completely inhibited the FCV precursor cleavage, confirming that the primary sequence of the cleavage site is important. However, it is of interest that several E/A sites exist near the primary cleavage site that are not utilized by the proteinase, raising the possibility that an additional factor for the preference of the known E/A cleavage site in the precursor could be its conformational presentation to the proteinase. In the present study, the conformation of the N-terminal leader sequence could be ruled out as an important aspect of cleavage because products of internal initiation from ORF2 were cleaved as efficiently as the full-length precursor. In regard to the cleavage site itself, the stretch of amino acids that comprise the precursor cleavage site is the most hydrophilic region of the protein predicted by computer analysis. However, the introduction of hydrophobic amino acids V and L into the P1′ position of the cleavage site did not significantly affect the cleavage efficiency. The cleavage site region exhibits also a local concentration of negatively charged amino acids, but the substitution of A with positively charged R and H did not affect the cleavage. We were also able to observe efficient cleavage of the precursor after the substitution of D in the P3′ position with amino acids that reduced local negative charge such as N, S, G, and R (data not shown). It is likely that the folding of the remainder of the capsid precursor molecule plays an important role in providing an accessible cleavage site to the proteinase. Processing of the precursor in the in vitro trans cleavage assay indicates that the cleavage site is accessible after translation of the entire protein.
The mutations analyzed in the in vitro cleavage assay were introduced into the full-length clone in order to examine their effects on the growth and replication of the virus. The accumulation of intact precursor molecules of a single size in cells transfected with genomic RNA carrying mutations in the precursor cleavage site confirmed that the E/A cleavage site at amino acid residue 125 is the unique site of the precursor recognized by the proteinase during viral replication. The appearance of numerous faint bands in studies of FCV grown at higher temperatures led to the suggestion by others (4, 26) that the capsid precursor may be processed into intermediate forms before final maturation. However, the data in the present study indicate that the removal of the leader sequence is a one-time event and not a process consisting of consecutive proteolytic steps. Furthermore, complete cleavage of the FCV capsid precursor is apparently critical in the production of infectious virions, consistent with previous studies by Carter (4) in which inhibition of the capsid precursor cleavage by p-fluorophenylalanine or elevated temperature prevented the development of FCV cytopathic effect and release of progeny virus. We could not recover virus following the transfection of RNA from genomic mutants carrying D/A and Q/A cleavage sites, in which incomplete cleavage occurred that resulted in the accumulation in cells of both noncleaved precursor and mature capsid protein with the correct N-terminal sequence. The relationship between these two forms of the capsid protein during virion maturation and assembly is not yet understood, but our data suggest that only the cleaved form of the capsid can assemble into infectious virions. The role of the leader polypeptide is not known, but deletion of this sequence from the FCV genome prevents the recovery of virus as well as the detection of capsid protein synthesis in transfected cells (unpublished data). It will be of interest to examine whether caliciviruses that translate a mature-sized capsid protein directly from the subgenomic RNA have a counterpart for the function of this sequence in their genomes.
Using full-length genomic clones that possessed mutations that abolished precursor cleavage, we observed accumulation of a 73-kDa protein in transfected cells but found no evidence for additional synthesis of the 70-kDa protein that was detected in our in vitro translation experiments. This observation suggested that the first AUG of the subgenomic RNA is the optimal site for the positioning of the ribosome for translation of the capsid precursor protein in cells and that the initiation of protein synthesis takes place at the first AUG of this template. The relatively short noncoding regions at the 5′ end of the FCV genome (19 nt) and subgenomic RNA (18 nt) and lack of consensus motifs argue against a picornavirus-like internal ribosome entry site in the calicivirus genome but does not rule out a unique calicivirus translation-enhancing element. It was recently demonstrated that the removal of the VPg protein from the 5′ ends of both the genomic and subgenomic FCV RNAs with proteinase K dramatically reduced the translation efficiency of both viral RNAs in vitro (15), and it is possible that the VPg is involved in the positioning of the ribosome at the first AUG of the ORF2 in infected cells. Studies are in progress to examine the role of the VPg in FCV replication and to map critical regions of the FCV genome involved in translation of the viral proteins.
ACKNOWLEDGMENTS
We thank John Coligan and Mark Garfield, LMS, NIAID, NIH, for the protein sequence analysis and Stephen Leppla and Valerie Gordon, NIDR, NIH, for sharing protease inhibitors. We thank Jose Valdesuso for his dedicated technical support. We acknowledge Jerry M. Keith, NIDR, NIH, and Tina Schultheiss, LID, NIAID, NIH, for enthusiastic and constructive discussions. We extend our appreciation to Albert Z. Kapikian, and Robert M. Chanock, LID, NIAID, NIH, for continuing support and encouragement.
REFERENCES
- 1.Allaire M, Chernaia M M, Malcolm B A, James M N G. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 2.Black D N, Burroughs J N, Harris T J R, Brown F. The structure and replication of calicivirus RNA. Nature. 1978;274:614–615. doi: 10.1038/274614a0. [DOI] [PubMed] [Google Scholar]
- 3.Boniotti B M, Wirlblich C, Sibilia M, Meyers G, Thiel H-J, Rossi C. Identification and characterization of 3C-like protease from rabbit hemorrhagic disease virus, a calicivirus. J Virol. 1994;68:6487–6495. doi: 10.1128/jvi.68.10.6487-6495.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter M J. Feline calicivirus protein synthesis investigated by Western blotting. Arch Virol. 1989;108:69–79. doi: 10.1007/BF01313744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter M J. Transcription of feline calicivirus RNA. Arch Virol. 1990;114:143–152. doi: 10.1007/BF01310744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter M J, Milton I D, Turner P C, Meanger J, Bennett M, Gaskell R M. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch Virol. 1992;122:233–235. doi: 10.1007/BF01317185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehreshmann D W, Schaffer F L. RNA synthesized in calicivirus-infected cells is atypical of picornaviruses. J Virol. 1977;22:572–576. doi: 10.1128/jvi.22.2.572-576.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehreshmann D W, Schaffer F L. Calicivirus intracellular RNA: fractionation of 18-22 S RNA and lack of typical 5′-methylated cap on 36 S and 22 S San Miguel sea lion virus RNAs. Virology. 1979;95:251–255. doi: 10.1016/0042-6822(79)90426-4. [DOI] [PubMed] [Google Scholar]
- 9.Fretz M, Schaffer F L. Calicivirus proteins in infected cells: evidence for capsid polypeptide precursor. Virology. 1978;89:318–321. doi: 10.1016/0042-6822(78)90065-x. [DOI] [PubMed] [Google Scholar]
- 10.Gaskell R M. Feline medicine and therapeutics. In: Chandler E A, Gaskell C J, Hilbery A D R, editors. 1st ed. Oxford, England: Blackwell; 1985. pp. 257–270. [Google Scholar]
- 11.Gorbalenya A E, Donchenko A P, Blinov V M, Koonin E V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 12.Haemmerle T, Hellen C U T, Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem. 1991;266:5412–5416. [PubMed] [Google Scholar]
- 13.Harris K S, Hellen C U T, Wimmer E. Proteolytic processing in the replication of picornaviruses. Semin Virol. 1990;1:323–333. [Google Scholar]
- 14.Herbert T P, Brierley L, Brown T D K. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from single, functionally bicistronic, subgenomic mRNA. J Gen Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- 15.Herbert T P, Brierly I, Brown T D K. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78:1033–1040. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- 16.Jewell D A, Swietnicki W, Dunn B M, Malcolm B A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Wang M, Wang K, Estes M K. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- 18.Lambden P R, Caul E O, Asley C R, Clarke I N. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–519. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- 19.Lawson M A, Semler B L. Picornavirus protein processing - enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–88. [PubMed] [Google Scholar]
- 20.Liu B, Clarke I N, Lambden P R. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70:2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin Alonso J M, Casais R, Boga J A, Parra F. Processing of rabbit hemorrhagic disease virus polyprotein. J Virol. 1996;70:1261–1265. doi: 10.1128/jvi.70.2.1261-1265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews D A, Smith W W, Ferre R A, Condon B, Budahazi G, Sisson W, Villafranca J E, Janson C A, McElroy H E, Gribskov C L, Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 23.Meyers G, Wirblich C, Thiel H-J. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology. 1991;184:677–686. doi: 10.1016/0042-6822(91)90437-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neill J D, Mengeling W L. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 1988;11:59–72. doi: 10.1016/0168-1702(88)90067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neill J D. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA-polymerase, cysteine protease and 2C polypeptides. Nucleic Acids Res. 1990;17:145–160. doi: 10.1016/0168-1702(90)90061-f. [DOI] [PubMed] [Google Scholar]
- 26.Neill J D, Reardon I M, Heinrikson R L. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J Virol. 1991;65:5440–5447. doi: 10.1128/jvi.65.10.5440-5447.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 28.Parra F, Boga J A, Marin M A, Casais R. The amino terminal sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res. 1993;27:219–228. doi: 10.1016/0168-1702(93)90034-k. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Shin Y-S, Tohya Y, Oshikamo R, Kawaguchi Y, Tomonaga K, Miyazawa T, Kai C, Mikami T. Antigenic analysis of feline calicivirus capsid precursor protein and its deleted polypeptides produced in mammalian cDNA expression system. Virus Res. 1993;30:17–26. doi: 10.1016/0168-1702(93)90012-c. [DOI] [PubMed] [Google Scholar]
- 31.Sosnovtsev S V, Green K Y. RNA transcripts derived from cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology. 1995;210:383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 32.Tohya Y, Taniguchi Y, Takahashi E, Utagawa E, Takeda N, Miyamura K, Yamazaki S, Mikami T. Sequence analysis of the 3′-end of feline calicivirus genome. Virology. 1991;183:810–814. doi: 10.1016/0042-6822(91)91016-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidner J R, Dunn B M. Development of synthetic peptide substrates for the poliovirus 3C proteinase. Arch Biochem Biophys. 1991;286:402–408. doi: 10.1016/0003-9861(91)90058-q. [DOI] [PubMed] [Google Scholar]
- 34.Wirblich C, Sibilia M, Boniotti M B, Rossi C, Thiel H-J, Meyers G. 3C-like protease of rabbit hemorrhagic disease virus: identification of cleavage sites in the ORF1 polyprotein and analysis of cleavage specificity. J Virol. 1995;69:7159–7168. doi: 10.1128/jvi.69.11.7159-7168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]