Abstract
Biomolecule immobilization on nanomaterials is attractive for biosensors since it enables the capture of a higher concentration of bioreceptor units while also serving as a transduction element. The technique could enhance the accuracy, specificity, and sensitivity of the analytical measurements of biomolecules. However, it was found that the limitation in chemically binding biomolecules on nanoparticle surfaces could only cross-link between the C-terminal and N-terminal. Here, we report the facile one-step synthesis of amine-functionalized silica nanoparticles (AFSNPs). (3-Aminopropyl)triethoxysilane was used as a precursor to modify the functional surface of nanoparticles via the Stöber process. The biomolecules were immobilized to the AFSNPs through itaconic acid, a novel cross-linker that binds between the N-terminal and N-terminal and potentially improves proteins and nucleic acid immobilization onto the nanoparticle surface. The newly developed immobilization approach on AFSNPs for biomolecular detection enhanced the efficiency of ELISA, resulting in increased sensitivity. It might also be easily used to identify different pathogens for clinical diagnostics.
Introduction
A rapid growth of nanomaterial technology has been contributing significantly to a recent advancement in biosensor development.1 Nanoparticles (NPs) are defined as small particles, and they usually exhibit distinct properties due to their high surface-to-volume ratio, which are useful for biosensing. In principle, a biosensor relies on a specific interaction between an analyte and a biorecognition element on the biosensor, such as substrate–enzyme or antigen–antibody.2,3 Therefore, immobilization of biomolecules on nanoparticles is a promising strategy to enhance the sensitivity and selectivity of the biosensor.4,5 Various nanoparticles functionalized with biomolecules have been established and applied on biosensing applications, such as carbon nanotubes, gold nanoparticles, and other metallic nanoparticles.6−8
Silica nanoparticles (SNPs) are one of the inorganic nanomaterials that have been of interest for biomolecule immobilization. SNPs possess desirable characteristics, such as biocompatibility, mechanical stability, and tunable size and shape.9 SNPs could be formed in various shapes and structures resulting in different properties, for example, conventional nonporous SNPs,10 mesoporous silica nanoparticles,11,12 hollow mesoporous silica nanoparticles,13 and core–shell silica.14 Due to their versatile properties, SNPs have been exploited in a wide range of biomedical applications, such as drug delivery, therapeutics, and diagnostics.9,15,16 It has been previously reported that amine-functionalized SNPs are biocompatible and suitable for use as a drug delivery system.17 (3-Aminopropyl)triethoxysilane (APTES) has been used to provide an amine group for generating amine-functionalized silica nanoparticles (AFSNPs).18 In the conventional method, APTES was coated on the surface of the SNP via condensation reaction.18−21 However, there were some limitations, such as the uncontrollable number of functional groups, limited shelf-life stability, and random orientation of the molecules, leading to reduced effectiveness of biomolecule immobilization. It could ultimately decrease the sensitivity in biosensing applications.
There are several strategies employed for immobilization;22,23 however, they can be classified into two main categories: noncovalent and covalent attachment. Noncovalent bonding relies on electrostatic, hydrophobic, polar, and affinity interactions, such as biotin–avidin bonding and histidine–metal chelation. Meanwhile, covalent bonding involves the formation of bonds between the functional groups of biomolecules and the surface of materials, such as amine, carboxyl, thiol, aldehyde, or alkyne.24 To immobilize a biomolecule on the SNPs, surface functionalization of the SNPs is required before the addition of a cross-link between the SNPs and biomolecules. An amine functionalization is commonly utilized for the surface modification of various silicon-based materials, including silicon oxide and silicon nitride, which could be applied for immobilization of biomolecules. The presence of an amine functional group on the surface provides a versatile platform for many chemical cross-linkers. The most commonly used cross-linkers for biomolecules immobilization are glutaraldehyde and 1-ethyl-3-(3-dimethyl aminopropyl)-carbodiimide/N-hydroxysuccinimide (EDC/NHS).25 However, the limitation of these cross-linkers is that they can only cross-link between the N-terminal and C-terminal. Therefore, the challenge of this work is to use itaconic acid as a novel cross-linker for the immobilization of biomolecules through the N-terminal and N-terminal. The two carboxylic groups at two ends of itaconic acid could react with EDC/NHS and form two amide bonds, one with an amine group of biomolecules, such as proteins and amine-tagged nucleotides, and another with any amine-functionalized substrates, including the SNPs. The ability to bind between the N-terminal and N-terminal is beneficial for biosensor applications, especially amine-functionalized silicon-based platforms,25 for example, silicon photonic chips, silicon nanowires, and fiber optics.
In the present work, a one-step synthesis of AFSNPs was proposed. The AFSNPs were successfully synthesized via the Stöber process under relatively mild conditions in a relatively short time compared with the conventional method. The morphologies and chemical bonding of AFSNPs were confirmed for validation and determination of detection efficiency. Furthermore, the efficiency of biomolecule immobilization on the proposed AFSNPs and the feasibility of biosensing applications were investigated via the enzyme-linked immunosorbent assay (ELISA) method. The performance of the AFSNP-conjugated ELISA was evaluated and compared with that of the conventional ELISA.
Results and Discussion
Fabrication Methods of Functional Nanoparticles
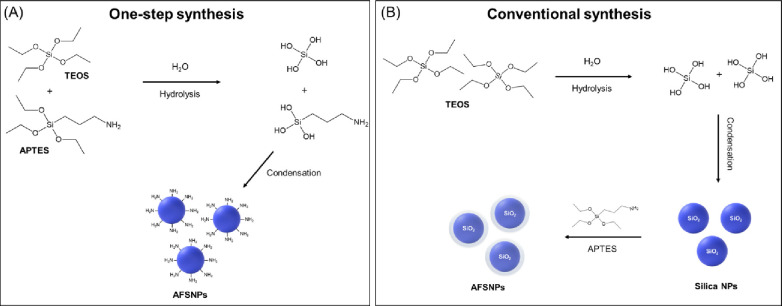
Amine-functionalized silica nanoparticles (AFSNPs) are fabricated through a one-step and conventional synthesis as illustrated in Scheme 1. In a one-step synthesis, the silicon-based monomers of TEOS and APTES were used to produce and modify surface-functionalized nanoparticles (Scheme 1A). The hydrolysis and condensation reactions were induced by adding base catalyst to the mixed solution, whereas the conventional synthesis of AFSNPs began with the fabrication of bare SNPs via the modified Stöber method using TEOS as the silica source (Scheme 1B). Amine functionalization was obtained by adding APTES later to cover the surface of SNPs.21
Scheme 1. Schematic of the Synthesis of Amine-Functionalized Silica Nanoparticles (AFSNPs) through One-Step Synthesis (A) and Conventional Synthesis (B).
Characterization of SNPs and AFSNPs
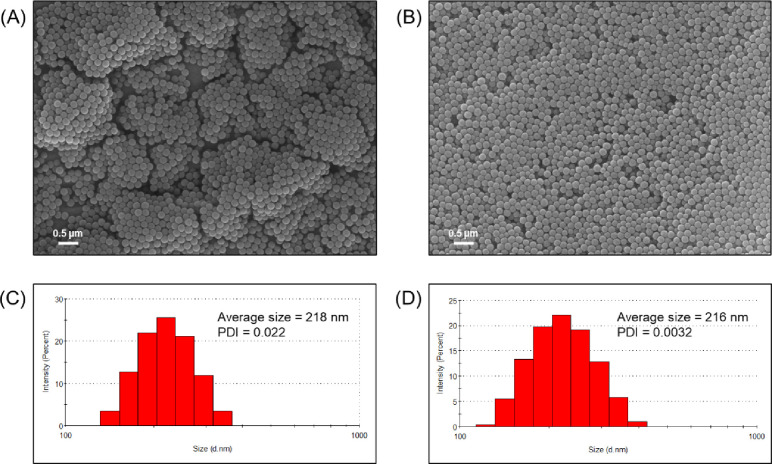
The scanning electron microscopy (SEM) images of nanoparticles are shown in Figure 1. SNPs (Figure 1A) and AFSNPs (Figure 1B) displayed the most prominent morphologies in a spherical shape with a smooth surface and homogeneous diameter. The AFSNPs are slightly smaller in size (diameter around 161 ± 5.9 nm) compared to the bare SNPs (diameter around 164 ± 17.4 nm). Particle size distribution was estimated by the dynamic light scattering (DLS). The histograms of the particle size distribution for the SNPs and AFSNPs are presented in Figure 1C,D, respectively. The histograms were fitted to the log-normal distribution function, and the peak values were taken as the average particle size of around 218 ± 2.5 nm (SNPs) and 216 ± 1.2 nm (AFSNPs) with a low polydispersity index (PDI), which represents a perfectly uniform particle size and is consistent with the SEM results. However, the different measured particle sizes between SEM and DLS are caused by the heterogeneous populations of the weighting procedure, which are inevitable.26
Figure 1.
Morphologies and size distribution of silica NPs (A and C) and AFSNPs (B and D), determined by SEM and DLS, respectively.
Figure S1 displays energy-dispersive spectroscopy (EDS) analysis of the elemental composition and mapping images of the SNPs (Figure S1A) and AFSNPs (Figure S1B). The red, green, and pink color images are associated with the Si, O, and N elements, respectively. The mapping images were acquired using the K-line spectra of the elements. In addition, the overlap image is shown. It can be observed that the elements have an inhomogeneous distribution in the nanoparticle, where there exist zones with high content of O and Si for both samples, whereas the zones with low content of N are shown in Figure 2B. The results are consistent with the elemental composition of the O, Si, and N (Figure S2), which shows an atomic ratio (inset table) close to 4.5:1:0 for SNPs and 3.1:1:0.3 for AFSNPs, respectively. This indicates that the elements are localized in the nanoparticle randomly. Therefore, this confirms the silica-based nanoparticle structure.
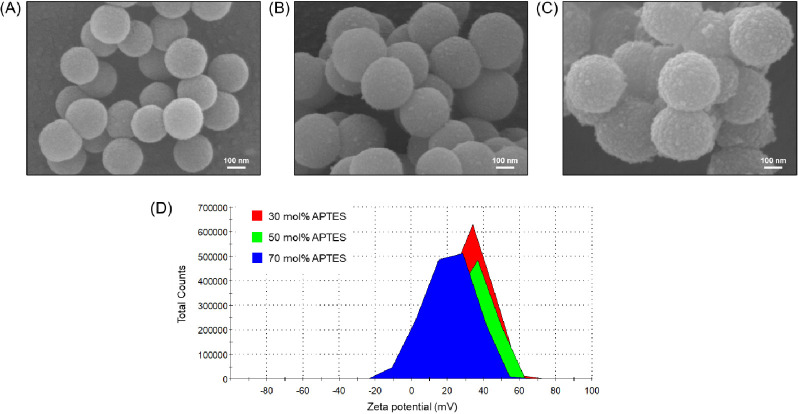
Figure 2.
SEM images of the AFSNPs with varying APTES concentrations (A) 30 mol %, (B) 50 mol %, (C) 70 mol %, and (D) ζ-potential measurement of AFSNPs.
Figure S3 shows the Fourier transform infrared (FTIR) spectroscopy of SNPs and AFSNPs (conventional and one-step). The spectrum of the samples shows peaks at 1049 to 1100 cm–1 and around 795 cm–1, which are attributed to stretching vibration of Si–O–Si bonds in the spine and free silanol group, respectively.10 The absorption peak at around 939 to 945 cm–1 is indicative of Si–OH vibration;27 when compared to SNPs and AFSNPs (conventional), the AFSNPs (one-step) showed weaker absorbance than the SNPs. The reason is that part of the Si–OH has been replaced by the bending vibrations of −NH (primary) in APTES as a new band appeared at 694 cm–1,28 while this peak did not appear in ASFNPs (conventional) because there was no cross-link between TEOS and APTES. Moreover, the AFSNPs (conventional) showed broad peaks at 3260 and 1628 cm–1, which could be attributed to −NH2 stretching and bending, respectively.29,30 It indicated that nanoparticle surface was successfully modified by comonomers of TEOS and APTES. It is noteworthy that the −NH2 stretching and bending bands of ASFNPs (one-step) are shifted to lower wavenumbers by the hydrogen bonding,31 which leads to high reactivity with molecules and specific interactions. In the comparison of the ASFNP (one-step) spectrum at different concentrations of added APTES (Figure S4), it was found that the intensity of 70 mol % APTES was higher than that of 50 mol % APTES at a peak of around 694 cm–1, which illustrated the amine concentration in the particle.
In addition, this research synthesized AFSNPs by mixing TEOS and APTES in different concentrations, as shown in Figure 2A–C. As the concentration of APTES was increased (30, 50, and 70 mol %), the morphological characteristics revealed that the AFSNP particle size increased to 224 ± 8.4, 279 ± 18.0, and 384 ± 44.6 nm, respectively, and the surface appearance of the particles became rougher due to the nonhydrolyzed and noncondensed amine function (−NH2).32Figure 2D shows the surface charge measured by the ζ-potential. It can be seen that the AFSNPs in all conditions showed positive charges compared to the bare SNPs that were negatively charged due to the oxygen atoms on their surface. This confirmed that the surface could be modified to have amine functional groups by adding APTES in the system.18,21 However, a similar charge in the range +32 to +39 mV could not confirm the presence of the number of amines because the number of particles in the solution was different.
Effect of Amine Concentrations to Immobilization of Biomolecules on AFSNPs
The synthesized AFSNPs were used to enhance the ELISA sensitivity. As illustrated in Scheme 2, a newly developed cross-linker was used to bind between amine-terminated biomolecules and amine-terminated ASFNPs through covalent bonding. The efficiency of the amine concentration, the stability of biomolecule immobilization, and the performance of AFSNPs were examined through the ELISA technique. Signals in antirabies virus detection by ELISA were compared between SNPs and AFSNPs synthesized in different concentrations of APTES. It was found that the concentration of APTES added to the particle synthesis system affected the particle morphology in terms of size and surface roughness. It also affects the amine content at the particle surface, which consequently affects the antigen conjugation since antigen is a protein that needs primary amines33 for binding on particles using the itaconic coupling principle. In Figure 3, the SNPs (0 mol % APTES) showed no ELISA signal, which indicated that the antigen could not be immobilized on the surface of the SNPs, whereas the AFSNPs showed a higher ELISA signal as the antigen concentration increased. At the start of the graph, despite having the lowest number of APTES, 30 mol % exhibited the highest slope due to its smallest particle size and largest surface area for antibody immobilization.34 However, due to the small amount of amine on the surface used to immobilize the antibody, at the saturation point, there were insufficient antigens remaining for immobilization, and with increased concentration of antibody, the graph became stable. At the concentrations of 50 and 70 mol % APTES, the O.D. continued to increase with higher amounts of antibody due to the large number of amines that can enhance the immobilization efficiency of a higher amount antigen. Therefore, the one-step synthesis technique can be used to design and quantify amines on the surface of particles.
Scheme 2. Immobilization of Biomolecules on ASFNP Surface by Using Itaconic Acid Cross-Linker.
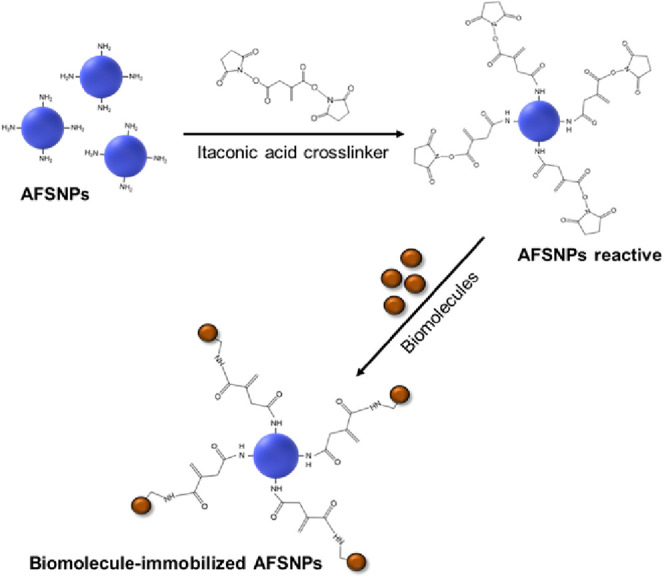
Figure 3.
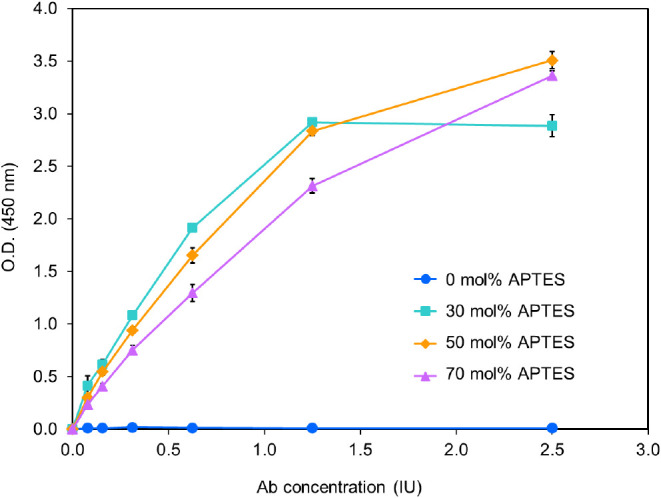
Efficiency determination of amine concentration on the surface of nanoparticles by ELISA technique.
Comparison of Synthesis Methods of AFSNPs to ELISA Signal
Furthermore, we determined and compared the biomolecular binding capacity of AFSNPs by conventional and one-step synthesis, as shown in Figure 4. It was found that the one-step synthesis method of AFSNPs resulted in higher ELISA signals for antirabies virus detection compared to the conventional method. Additionally, this method offers the advantage of using less APTES in the synthesis process. Although similar size nanoparticles were used in this study, the ELISA signal from the one-step synthesized nanoparticles was still strong due to the arrangement of the amine (−NH2) structure in the particles, making it feasible to immobilize between the nanoparticles and the antigens.35
Figure 4.
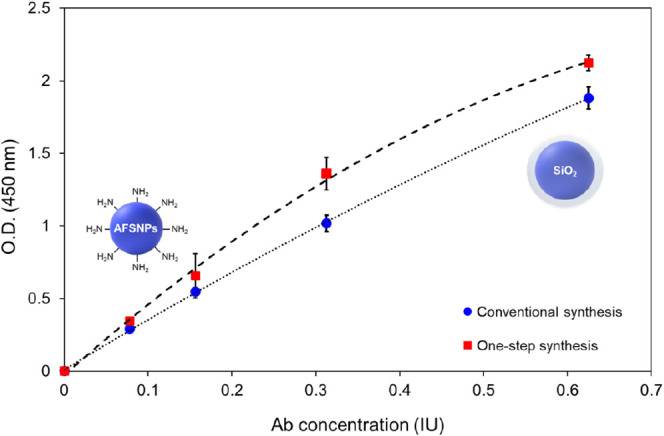
Comparison of biomolecules immobilization stability between conventional and one-step synthesis of AFSNPs.
Highly Enhanced ELISA Sensitivity and Specificity Using AFSNPs
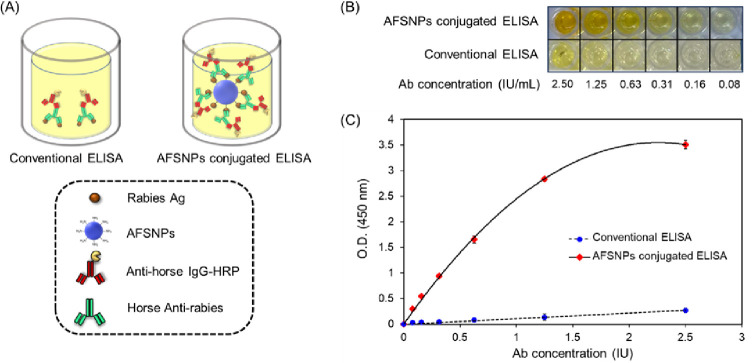
The highly sensitive ELISA based on the ASFNPs as nanocarriers of antirabies antigen by using an itaconic acid cross-linker in an indirect ELISA is shown in Figure 5. The indirect ELISA technique employs a two-step process for detection:36 first, a horse antirabies virus (primary antibody) specific to the rabies antigen binds to the target, and second, an antihorse IgG-HRP (labeled secondary antibody) binds to the primary antibody for detection, as shown in Figure 5A. The antirabies virus detection performance between conventional and AFSNPs-based ELISAs was also compared. It is expected that when AFSNPs are applied to ELISA, the amount of antigen immobilization and the ELISA signal can be increased. This is due to the assumption of increasing the surface area and functionality of nanoparticles. In Figure 5B, the developed reaction product color of the applied AFSNPs to ELISA showed significantly deeper yellow than the conventional ELISA. The absorbance was measured at 450 nm, and it was seen that the absorbance of AFSNP-conjugated ELISA showed a much higher signal than the conventional ELISA method as shown in Figure 5C. The limit of detection (LOD) of the coupled AFSNPs is about 0.05 IU, which is ten times more sensitive than the LOD of conventional ELISA (0.5 IU). The evidence showed their superiority in terms of detection sensitivity due to the efficient functionalization of ELISA with bioreceptors. AFSNPs can offer increased surface areas, which leads to greater capacity for biomolecule binding.37 Therefore, the AFSNPs can enhance the amplifying signal and the detection of the ELISA technique, which is useful in the early diagnosis of infection with very low concentrations of biomarkers.38
Figure 5.
(A) Schematic illustration of the roles of AFSNPs in improving the performance of conventional ELISA, (B) ELISA test, and (C) sensing performed in the presence and absence of AFSNPs.
Figure 6 shows the specificity of the proposed ELISA for antirabies virus detection. AFSNPs–rabies Ag was used to identify another sample, bovine serum albumin (BSA). The microplate was initially coated with the AFSNPs–rabies Ag. BSA did not result in a color change by demonstrating the same amount of the O.D. value as the negative control. Additionally, a negative control could confirm unimmobilized rabies Ag if the itaconic acid cross-linker was not used in the system. However, the O.D. value of antirabies was increased by 60-fold when compared to BSA, indicating that the AFSNPs-based enhanced ELISA has high specificity for antirabies virus detection.
Figure 6.
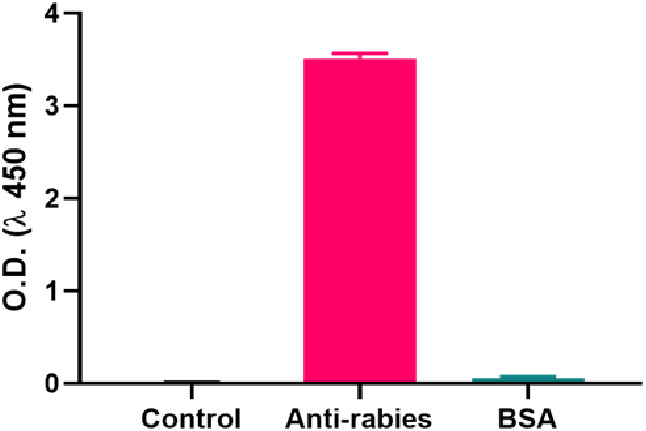
Specificity of the proposed ELISA for antirabies virus detection. The concentration of antirabies and BSA was 2.5 IU/mL and 1 mg/mL, respectively.
Conclusions
This study proposes an amine-modified surface on SNPs via a one-step synthesis, a novel immobilization of biomolecules on nanoparticle surfaces, and its application to biomolecular detection. The one-step synthesis was developed to overcome the complex and time-consuming steps of the conventional synthesis approach. In addition, a newly developed cross-linker between the N-terminal and N-terminal was used to bind the proteins onto the AFSNPs. The findings demonstrated that the ELISA exhibited a substantially higher sensitivity for detecting antirabies virus as compared to the conventional method. It is worth noting that this improved signal was achieved despite using only one-fifth of the quantity of APTES, which is a critical component in the synthesis process. Furthermore, we conducted a comparison between conventional ELISA and AFSNPs-based ELISA for the detection of antirabies virus. Our findings revealed that the AFSNPs-based ELISA exhibited ten times higher sensitivity than the conventional ELISA. Thus, the presence of AFSNPs has played a crucial role in enhancing signal amplification and improving the detection effectiveness of the ELISA technique. This notable improvement holds considerable value, particularly in the early diagnosis of infections with remarkably low concentrations of biomarkers.
Experimental Section
Chemicals and Materials
Tetraethyl orthosilicate (TEOS) (98.0%), (3-aminopropyl)triethoxysilane (APTES) (99%), ethyl alcohol (ACS reagent, ≥ 99.5%), ammonium hydroxide solution (30%), ethylcarbodiimide hydrochloride (EDC) (98%), N-hydroxysuccinimide (NHS) (98%), and antihorse IgG (whole molecule)-peroxidase antibody produced in rabbit were purchased from Sigma-Aldrich. Itaconic acid (99%) was purchased from Thermo Scientific. Rabies virus antigen and TRCS antirabies serum were purchased from Biovalys Co., Ltd. BlockPRO1 min protein-free blocking buffer was purchased from Energenesis Biomedical Co., Ltd. Washing buffer 20×, 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate, and stop solution were purchased from Peprotech. Deionized water (18 MΩ) was used throughout the experiment.
Synthesis of AFSNPs
The uniform-sized AFSNPs were prepared by a one-step synthesis of modified Stöber as described in a previous study.10 Briefly, TEOS and APTES were added to a solvent mixture of ethanol and distilled water in the presence of ammonium hydroxide as catalysts for hydrolysis and condensation. The mixture was stirred for 6 h at room temperature. The washing process with centrifugation and redispersion was repeated three times. The final products were collected and lyophilized for the next use.
Immobilization of Biomolecules on AFSNPs
Cross-linking reactions were carried out for 30 min at room temperature in a 1.5 mL Eppendorf tube with a mixture of itaconic acid, distilled water, EDC, and NHS. Then, AFSNPs and rabies virus antigen were added to the mixture at the same time. AFSNPs were incubated with rabies virus antigen (concentration of 0.4, 0.2, 0.1, 0.05, 0.025, and 0.0125 μg, respectively) for 1 h at 25 °C. The functionalized NPs were then centrifuged at 4000 rpm for 10 min, and the precipitated NPs were resuspended in wash buffer three times to remove the excess antigen. The supernatant was collected for antigen quantification.
Characterizations
The size and surface charge of the AFSNPs in ethanol were determined by using a laser particle analyzer system (NANO ZS, Malvern instruments Ltd., UK). A He–Ne laser was used as the light source. The incident wavelength was 633 nm, and the measurement angle was 173°. ζ-potential measurements were performed at 25 °C using highly diluted colloidal dispersions. The morphologies of the AFSNPs were investigated by using scanning electron microscopy (SEM, JSM-IT800). To determine the elemental composition of the particles studied during the electron microscopy measurements, an energy-dispersive X-ray spectroscopy (EDS) detector was used together with the SEM. For the measurement in combination with SEM, diluted colloidal dispersions were dropped onto a copper tape on an aluminum substrate (cleaned with ethanol) to produce well-separated particles on the surface. The chemical bond structure of the AFSNPs was confirmed by Fourier transform infrared spectroscopy (FTIR). The spectra from 500 to 4000 cm–1 were recorded by an FTIR spectrometer (Thermo Scientific, USA). The samples were scanned in the attenuated total reflection (ATR) mode.
Enzyme-Linked Immunosorbent Assays (ELISA)
The 96-well ELISA plates (Immune; Nunc) were coated with antigen-immobilized AFSNPs (0.01 mg mL–1) at 4 °C for overnight. Then, they were washed with washing buffer three times and were blocked with blocking buffer for 6 min. After the blocking buffer was removed, the diluted solution of antibody (100 μL) with desired concentrations (2.50, 1.25, 0.63, 0.31, 0.16, and 0.08 IU/mL) was then added to each well and incubated for 1 h at room temperature. Next, the well plate was washed with washing buffer for seven times, followed by the addition of rabbit antihorse IgG (whole molecule) – HRP 100 μL/well at optimized dilution of 1:3000 and incubated for 1 h at room temperature. The well plate was washed again with washing buffer for seven times. Later, the TMB substrate was added to each well and incubated for 15 min at room temperature. Finally, the reaction was stopped by adding stop solution and the absorbance of the product was measured at wavelengths of 450 and 570 nm using a microplate reader (Infinite M Plex, Tecan Trading AG, Switzerland).
Acknowledgments
This research has received funding support from the NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation (B05F640218), National Higher Education Science Research and Innovation Policy Council, Thailand Science Research and Innovation (TSRI) (71698), and was also supported by the National Research Council of Thailand (NRCT) and Chiang Mai University (N42A650185).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07548.
EDS mapping images, EDS spectrum, and FTIR of SNPs and AFSNPs (PDF)
Author Contributions
# S.U. and K.K. equally contributed to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- LaFleur L.; Yager P.. Chapter II.5.13 - Medical Biosensors. In Biomaterials Science, 3rd ed.; Ratner B. D.; Hoffman A. S.; Schoen F. J.; Lemons J. E.; Academic Press, 2013; pp 996–1006.. [Google Scholar]
- Farka Z.; Juřík T.; Kováǎ D.; Trnková L.; Skládal P. Nanoparticle-Based Immunochemical Biosensors and Assays: Recent Advances and Challenges. Chem. Rev. 2017, 117 (15), 9973–10042. 10.1021/acs.chemrev.7b00037. [DOI] [PubMed] [Google Scholar]
- Martín-Gracia B.; Martín-Barreiro A.; Cuestas-Ayllón C.; Grazú V.; Line A.; Llorente A.; de la Fuente J. M.; Moros M. Nanoparticle-based biosensors for detection of extracellular vesicles in liquid biopsies. J. Mater. Chem. B 2020, 8 (31), 6710–6738. 10.1039/D0TB00861C. [DOI] [PubMed] [Google Scholar]
- Špringer T.; Ermini M. L.; Špačková B.; Jabloňků J.; Homola J. Enhancing Sensitivity of Surface Plasmon Resonance Biosensors by Functionalized Gold Nanoparticles: Size Matters. Anal. Chem. 2014, 86 (20), 10350–10356. 10.1021/ac502637u. [DOI] [PubMed] [Google Scholar]
- Altug H.; Oh S.-H.; Maier S. A.; Homola J. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 2022, 17 (1), 5–16. 10.1038/s41565-021-01045-5. [DOI] [PubMed] [Google Scholar]
- Gergeroglu H.; Yildirim S.; Ebeoglugil M. F. Nano-carbons in biosensor applications: An overview of carbon nanotubes (CNTs) and fullerenes (C60). SN Appl. Sci. 2020, 2 (4), 603. 10.1007/s42452-020-2404-1. [DOI] [Google Scholar]
- Giner-Casares J. J.; Henriksen-Lacey M.; Coronado-Puchau M.; Liz-Marzán L. M. Inorganic nanoparticles for biomedicine: Where materials scientists meet medical research. Mater. Today 2016, 19 (1), 19–28. 10.1016/j.mattod.2015.07.004. [DOI] [Google Scholar]
- Park Y.; Jeong S.; Kim S. Medically translatable quantum dots for biosensing and imaging. J. Photochem. Photobiol., C 2017, 30, 51–70. 10.1016/j.jphotochemrev.2017.01.002. [DOI] [Google Scholar]
- Yang Y.; Zhang M.; Song H.; Yu C. Silica-Based Nanoparticles for Biomedical Applications: From Nanocarriers to Biomodulators. Acc. Chem. Res. 2020, 53 (8), 1545–1556. 10.1021/acs.accounts.0c00280. [DOI] [PubMed] [Google Scholar]
- Baipaywad P.; Kim Y.; Wi J.-S.; Paik T.; Park H. Size-controlled synthesis, characterization, and cytotoxicity study of monodisperse poly(dimethylsiloxane) nanoparticles. J. Ind. Eng. Chem. 2017, 53, 177–182. 10.1016/j.jiec.2017.04.023. [DOI] [Google Scholar]
- Wagner J.; Gößl D.; Ustyanovska N.; Xiong M.; Hauser D.; Zhuzhgova O.; Hočevar S.; Taskoparan B.; Poller L.; Datz S.; Engelke H.; Daali Y.; Bein T.; Bourquin C. Mesoporous Silica Nanoparticles as pH-Responsive Carrier for the Immune-Activating Drug Resiquimod Enhance the Local Immune Response in Mice. ACS Nano 2021, 15 (3), 4450–4466. 10.1021/acsnano.0c08384. [DOI] [PubMed] [Google Scholar]
- Baipaywad P.; Wi J.-S.; Park H.; Paik T. Tailoring cubic and dodecagonal quasicrystalline mesophases of mesoporous organosilica nanoparticles and core/shell structure. Mater. Sci. Eng., C 2019, 98, 666–674. 10.1016/j.msec.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Lee J. Y.; Kim M. K.; Nguyen T. L.; Kim J. Hollow Mesoporous Silica Nanoparticles with Extra-Large Mesopores for Enhanced Cancer Vaccine. ACS Appl. Mater. Interfaces 2020, 12 (31), 34658–34666. 10.1021/acsami.0c09484. [DOI] [PubMed] [Google Scholar]
- Selvarajan V.; Obuobi S.; Ee P. L. R. Silica Nanoparticles—A Versatile Tool for the Treatment of Bacterial Infections. Front. Chem. 2020, 8, 602. 10.3389/fchem.2020.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjua T. I.; Cao Y.; Yu C.; Popat A. Clinical translation of silica nanoparticles. Nat. Rev. Mater. 2021, 6 (12), 1072–1074. 10.1038/s41578-021-00385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayl A. A.; Abd-Elhamid A. I.; Aly A. A.; Bräse S. Recent progress in the applications of silica-based nanoparticles. RSC Adv. 2022, 12 (22), 13706–13726. 10.1039/D2RA01587K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira L. F.; Bouchmella K.; de Almeida Gonçalves K.; Bettini J.; Kobarg J.; Cardoso M. B. Functionalized Silica Nanoparticles as an Alternative Platform for Targeted Drug-Delivery of Water Insoluble Drugs. Langmuir 2016, 32 (13), 3217–3225. 10.1021/acs.langmuir.6b00214. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Sun Y.; Wang J.; Yang Y.; Li Y.; Yuan Y.; Liu C. Charge-Reversal APTES-Modified Mesoporous Silica Nanoparticles with High Drug Loading and Release Controllability. ACS Appl. Mater. Interfaces 2016, 8 (27), 17166–17175. 10.1021/acsami.6b05370. [DOI] [PubMed] [Google Scholar]
- Miller P. J.; Shantz D. F. Covalently functionalized uniform amino-silica nanoparticles. Synthesis and validation of amine group accessibility and stability. Nanoscale Adv. 2020, 2 (2), 860–868. 10.1039/C9NA00772E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widyasari D. A.; Kristiani A.; Randy A.; Manurung R. V.; Dewi R. T.; Andreani A. S.; Yuliarto B.; Jenie S. N. A. Optimized antibody immobilization on natural silica-based nanostructures for the selective detection of E. coli. RSC Adv. 2022, 12 (33), 21582–21590. 10.1039/D2RA03143D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talreja K.; Chauhan I.; Ghosh A.; Majumdar A.; Butola B. S. Functionalization of silica particles to tune the impact resistance of shear thickening fluid treated aramid fabrics. RSC Adv. 2017, 7 (78), 49787–49794. 10.1039/C7RA09834K. [DOI] [Google Scholar]
- Samanta D.; Sarkar A. Immobilization of bio-macromolecules on self-assembled monolayers: Methods and sensor applications. Chem. Soc. Rev. 2011, 40 (5), 2567–2592. 10.1039/c0cs00056f. [DOI] [PubMed] [Google Scholar]
- Masud M. K.; Na J.; Younus M.; Hossain M. S. A.; Bando Y.; Shiddiky M. J. A.; Yamauchi Y. Superparamagnetic nanoarchitectures for disease-specific biomarker detection. Chem. Soc. Rev. 2019, 48 (24), 5717–5751. 10.1039/C9CS00174C. [DOI] [PubMed] [Google Scholar]
- Gloag L.; Mehdipour M.; Chen D.; Tilley R. D.; Gooding J. J. Advances in the Application of Magnetic Nanoparticles for Sensing. Adv. Mater. 2019, 31 (48), 1904385. 10.1002/adma.201904385. [DOI] [PubMed] [Google Scholar]
- Udomsom S.; Mankong U.; Paengnakorn P.; Theera-Umpon N. Novel Rapid Protein Coating Technique for Silicon Photonic Biosensor to Improve Surface Morphology and Increase Bioreceptor Density. Coatings 2021, 11 (5), 595. 10.3390/coatings11050595. [DOI] [Google Scholar]
- Bootz A.; Vogel V.; Schubert D.; Kreuter J. Comparison of scanning electron microscopy, dynamic light scattering and analytical ultracentrifugation for the sizing of poly(butyl cyanoacrylate) nanoparticles. Eur. J. Pharm. Biopharm. 2004, 57 (2), 369–375. 10.1016/S0939-6411(03)00193-0. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Sun Y.; Mao Y.; Chen K.; Cao Z.; Qi D. Controllable synthesis of raspberry-like PS–SiO2 nanocomposite particles via Pickering emulsion polymerization. RSC Adv. 2018, 8 (7), 3910–3918. 10.1039/C7RA13086D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto K.Infrared and Raman Spectra of Inorganic and Coordination Compounds, Part B: Applications in Coordination, Organometallic, and Bioinorganic Chemistry; Wiley, 2009. [Google Scholar]
- Suteewong T.; Sai H.; Bradbury M.; Estroff L. A.; Gruner S. M.; Wiesner U. Synthesis and Formation Mechanism of Aminated Mesoporous Silica Nanoparticles. Chem. Mater. 2012, 24 (20), 3895–3905. 10.1021/cm301857e. [DOI] [Google Scholar]
- Karnati S. R.; Oldham D.; Fini E. H.; Zhang L. Application of surface-modified silica nanoparticles with dual silane coupling agents in bitumen for performance enhancement. Constr. Build. Mater. 2020, 244, 118324. 10.1016/j.conbuildmat.2020.118324. [DOI] [Google Scholar]
- Lin-Vien D.; Colthup N. B.; Fateley W. G.; Grasselli J. G.. The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules; Elsevier Science, 1991. [Google Scholar]
- Koike N.; Chaikittisilp W.; Shimojima A.; Okubo T. Surfactant-free synthesis of hollow mesoporous organosilica nanoparticles with controllable particle sizes and diversified organic moieties. RSC Adv. 2016, 6 (93), 90435–90445. 10.1039/C6RA22926C. [DOI] [Google Scholar]
- Saptarshi S. R.; Duschl A.; Lopata A. L. Interaction of nanoparticles with proteins: Relation to bio-reactivity of the nanoparticle. J. Nanobiotechnol. 2013, 11 (1), 26. 10.1186/1477-3155-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeppel K. M.; Zheng X. S.; Cui X. T. Enhancing surface immobilization of bioactive molecules via a silica nanoparticle based coating. J. Mater. Chem. B 2018, 6 (19), 3058–3067. 10.1039/C8TB00408K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Zhou Y.; Chandrawati R. Metal and Metal Oxide Nanoparticles to Enhance the Performance of Enzyme-Linked Immunosorbent Assay (ELISA). ACS Appl. Nano Mater. 2020, 3 (1), 1–21. 10.1021/acsanm.9b02003. [DOI] [Google Scholar]
- Lin A. V.Indirect ELISA. In ELISA: Methods and Protocols, Hnasko R., Eds.; Springer New York: New York, NY, 2015; pp 51–59.. [Google Scholar]
- Xing Z. C.; Chang Y.; Kang I. K. Immobilization of biomolecules on the surface of inorganic nanoparticles for biomedical applications. Sci. Technol. Adv. Mater. 2010, 11 (1), 014101. 10.1088/1468-6996/11/1/014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L.; Wu W. B.; Yang X. X.; Huang C. Z. Gold nanoparticle-based enhanced ELISA for respiratory syncytial virus. New. J. Chem. 2014, 38 (7), 2935–2940. 10.1039/C4NJ00253A. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.