Abstract
As part of the evaluation of porcine cells, tissues, and organs intended for transplantation into humans, we investigated the conditions required to induce expression and release of porcine endogenous retrovirus (PoEV) from primary cells. Pigs contain endogenous retroviral sequences encoding infectious retrovirus, yet little is known about the conditions required to activate the expression and release of PoEV from primary cells. We show here that mitogenic activation of peripheral blood mononuclear cells (PBMC) isolated from the National Institutes of Health (NIH) miniature pig and the Yucatan pig resulted in the activation and release of an infectious type C retrovirus. Coculture of activated porcine PBMC with pig or human cell lines resulted in the transfer and expression of PoEV-specific sequences and the establishment of a productive infection. Sequence comparison of portions of the PoEV pol gene expressed in pig cell lines productively infected with virus derived from NIH miniature pig and Yucatan pig PBMC revealed marked similarity, suggesting that one or a few loci may be capable of being activated to yield an infectious virus. These findings demonstrate that the presence of endogenous viruses in source animals needs to be carefully considered when the infectious disease potential of xenotransplantation is being assessed.
Recent progress in the field of immunosuppression and the shortage of human organs have led to renewed interest in the xenotransplantation of cells, tissues, and organs into humans for the purpose of providing permanent therapeutic benefit or as a bridging strategy until an appropriate human donor can be found. However, the source animals for xenotransplants are likely to carry infectious agents which may not be readily apparent or known to be pathogenic in the host species but which may be capable of infecting and progressing to disease in the recipient species. Of particular concern are endogenous infectious agents which may be latent and therefore not easily detected in the source animal. Extensive immunosuppression of the host, required to prevent xenograft rejection, may facilitate the spread of these agents, which would be eliminated in an immunocompetent individual.
Porcine xenografts are under evaluation for a variety of life-threatening and chronic diseases. Pig genomes contain endogenous retroviral sequences encoding infectious type C retroviral particles (2, 11, 15). Established porcine cell lines spontaneously release type C retroviral particles (1, 22, 26), although viral particles have not been observed in primary cultures of porcine cells (1). Initial reports suggested that the host range of porcine endogenous retrovirus was restricted to cells of porcine origin (11, 24), but recently, a type C retrovirus associated with the porcine kidney cell line PK-15, termed PERV-PK, was reported to infect human, mink, and porcine cell lines (15). Southern blot analysis suggested there may be up to 50 copies of this sequence in the pig genome (15). It is not known whether all of these loci encode infectious proviral sequences, and the conditions required for the activation and expression of infectious retrovirus from pig cells have not been determined. We therefore examined the conditions required for induction of infectious endogenous retrovirus from primary porcine cells. Our experiments were based on the observation that endogenous murine retroviruses can be activated during immune activation by a graft-versus-host reaction in vivo or by mixed lymphocyte reaction in vitro (5, 6, 9, 21). We find that mitogenic activation of porcine peripheral blood mononuclear cells (PBMC) results in the activation and release of an infectious retrovirus capable of infecting human cells.
MATERIALS AND METHODS
Isolation and activation of PBMC.
Acid-citrate dextrose-treated blood (450 ml) was obtained from National Institutes of Health (NIH) miniature pigs or Yucatan pigs maintained by the NIH animal facility at Poolesville, Md. PBMC were isolated by centrifugation over lymphocyte separation medium (Organon Teknika, Durham, N.C.). After three washes in phosphate-buffered saline (PBS), the PBMC were counted and cryopreserved in 90% fetal bovine serum and 10% dimethyl sulfoxide at 107 cells/ml. Thawed PBMC were mitogenically stimulated in the following medium: Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 μg of phytohemagglutinin (PHA) per ml, 10 ng of phorbol 12-myristate-13-acetate per ml, nonessential amino acids, 5 mM β-mercaptoethanol, 10 mM HEPES, 2 mM glutamine, 1 mM sodium pyruvate, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Where indicated, 0.2 μM A23187 (a calcium ionophore) was substituted for PHA in the medium.
Cell lines.
The following cell lines used in these studies were obtained from the American Type Culture Collection (ATCC) or as indicated: fetal swine testis ST-IOWA (passage 85 cells kindly provided by Richard Fister, Tufts University, or passage 117 cells from ATCC CRL-1746), 293 human embryonic kidney (ATCC CRL-1573), HeLa (ATCC CRL-2), mink lung fibroblast (ATCC CCL-64), bat lung fibroblast (ATCC CCL-88), rat2 fibroblast (ATCC CRL-1746), Mus dunni tail fibroblast (obtained from Olivier Danos), E36 Chinese hamster lung (obtained from Christine Kozak), and African green monkey kidney COS-7 (ATCC CRL-1650) cells. All cell lines were maintained in standard medium, i.e., Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum–2 mM glutamine–1 mM sodium pyruvate–100 U of penicillin per ml–100 μg of streptomycin per ml.
Infectivity assays.
Coculture assay for viral infectivity was performed as follows: 106 live or lethally irradiated (2,000 rads from a 137Cs source, Nordion GammaCell 100) virus producer cells were mixed with 5 × 105 target cells and plated in standard medium in 25-cm2 flasks. The following day, the cells were washed twice with PBS. Cultures were maintained by subpassaging 1 to 2 times/week as needed. As cultures reached confluence, culture medium was removed for reverse transcriptase assay, and in some cases, ≥106 cells were removed for RNA isolation and reverse transcription (RT)-PCR.
Reverse transcriptase assays.
Medium from cultured cells was precleared by centrifugation at 10,000 × g for 5 min. The supernatant was then assessed for reverse transcriptase activity as described previously (27), with modifications based on the method of Phan-Thanh et al. (16). The solubilized samples were incubated for 3 h at 37°C with 2× substrate buffer containing 50 mM Tris (pH 7.5), 10 mM dithiothreitol, 0.6 mM MnCl2, 10 μg of poly(rA) · poly(dT)12–18 (Pharmacia, Piscataway, N.J.) per ml, and 10 μCi of 3H[TTP] per 25 μl (22 Ci/mmol). All samples were assayed in triplicate.
Electron microscopy.
Cells were prepared for electron microscopy as follows. Cells were removed from flasks with trypsin-EDTA, washed in PBS, and fixed in 2% paraformaldehyde-2% glutaraldehyde in 0.10 M cacodylate buffer (pH 7.2 to 7.4) for 2 to 3 h at room temperature. Fixed cells were stored in PBS at 4°C until further processing. Samples were subsequently treated for 1 h with 2% osmium tetroxide, dehydrated with graded alcohols, and embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate and examined with a Zeiss EM 912 electron microscope.
Isolation and sequencing of PoEV cDNA.
A portion of the porcine endogenous retrovirus (PoEV) reverse transcriptase coding region was isolated from cDNA made from infected cells with degenerate oligonucleotide primers (Table 1), based on highly conserved regions among reverse transcriptase genes.
TABLE 1.
Primers used to isolate novel retroviral sequences
| Primer pool | Orientation | Sequence (5′ to 3′)a | Degeneracyb | Length (nt) |
|---|---|---|---|---|
| PQGWA | Sense | TACCAGTGGAACGTTCTGCCNCARGGNTGG | 32 | 30 |
| PQGMA | Sense | TACCAGTGGAACGTTCTGCCNCARGGNATG | 32 | 30 |
| PQGFA | Sense | TACCAGTGGAACGTTCTGCCNCARGGNTT | 32 | 29 |
| YMDDB | Antisense | GTAGTCTGATCCTACCAACADRTCRTCCATRTA | 24 | 33 |
N = A, T, C, G; R = A, G; D = A, G, T. Residues marked in bold and underlined represent the unique residues which code for either W, M, or F. These residues define the specificity of the oligonucleotide pool.
The number of different primers in the pool representing all codon possibilities at the degenerate positions indicated by N, R, or D.
Cellular RNA was isolated following guanidinium thiocyanate lysis and CsCl centrifugation. cDNA was prepared with avian myeloblastosis virus reverse transcriptase (10 U; Promega Corp.) in 1× avian myeloblastosis virus buffer containing 1 mM deoxynucleoside triphosphate, 30 U of RNasin, and 10 pmol of degenerate oligonucleotide YMDDB in a total volume of 30 μl. The samples were incubated for 1 h at 42°C under mineral oil. The cDNA templates (3 μl) were amplified with one of the degenerate oligonucleotide primer pools (50 pmol), PQGWA, PQGMA, or PQGFA, with the degenerate primer pool YMDDB (50 pmol). Hot-start amplifications were performed in 50 μl of 0.067 M Tris buffer (pH 8.8) containing 4 mM MgCl2, 16 mM (NH4)2SO4, 10 mM 2-mercaptoethanol, 0.1 mg of bovine serum albumin per ml (12), 100 μM each deoxynucleoside triphosphate, and Taq polymerase (1 U; Gibco/BRL) as follows: 35 cycles at 95°C for 1 min, at 55°C for 1 min, and at 72°C for 1 min. Following electrophoresis on a 2.5% agarose gel, PCR products were visualized by UV irradiation in the presence of ethidium bromide. The PCR products were cloned into pT7Blue (Novagen, Madison, Wis.) and sequenced with chain terminators (20).
RT-PCR assay for detection of PoEV.
Primers based on the sequence described by Tristem et al. (25) were used to obtain additional sequences of the reverse transcriptase coding region. RNA from ≥106 cells (isolated with RNA stat-60; Tel-test “B,” Inc., Friendswood, Tex.) was converted to cDNA with 2.5 μM random hexamers and Superscript II (Life Technologies, Gaithersburg, Md.). cDNA templates were amplified with the PoEV-derived primers PB905 (sense) (5′ CCGCAGGGATGGGTTTGGCAAAGCA 3′) and PB906 (antisense) (5′ ACGTACTGGAGGAGGGTCACCTGA 3′) for 30 cycles at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 1 min. To obtain sequence information, the PCR products were cloned into the TAII vector (Invitrogen Corporation, San Diego, Calif.). Multiple clones were sequenced to verify nucleotide differences. To detect expressed PoEV sequences in cells, the amplification products were separated on a 1% agarose gel, transferred to Nytran (Schleicher & Schuell, Keene, N.H.), and hybridized with a 32P-labelled probe (nick translated). The PoEV-specific probe was an amplified product obtained with PB903, 5′ GACGGGTAACCCACTCGTTTCTGGT 3′, and PB906 with the amplification conditions described above. The membrane was hybridized at 42°C in hybrisol I (Oncor, Gaithersburg, Md.) and stringently washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 65°C. To assess cDNA synthesis, all cDNA preparations were examined for their ability to amplify G3PDH (GenBank accession no. M32599).
Nucleotide sequence accession numbers.
The GenBank accession number for the ST-NIH sequence is AF033259, and that for sequence ST-Yucatan is AF033260.
RESULTS
Mitogenic stimulation of porcine PBMC releases infectious type C retrovirus.
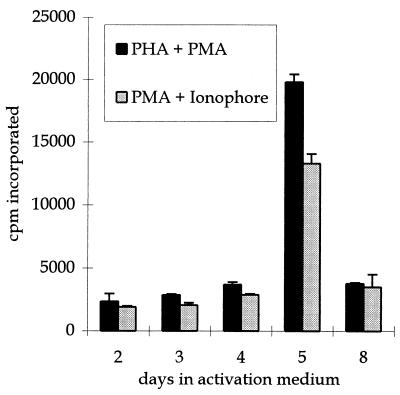
To investigate the conditions which would activate PoEV(s), primary PBMC isolated from the blood of an NIH miniature pig were exposed to two different combinations of mitogens. Activity of retroviral reverse transcriptase was measured in the medium of stimulated cells. Five days after stimulation, the level of reverse transcriptase activity showed a sharp increase in activity, which returned to baseline levels by 8 days (Fig. 1). Treatment of PBMC with PHA and phorbol myristate acetate (PMA) or with PMA and the calcium ionophore A23187 resulted in comparable increases in reverse transcriptase activity with similar kinetics (Fig. 1). PHA and PMA treatment of PBMC isolated from blood of a different strain of pig, the Yucatan, also showed a similar increase in reverse transcriptase activity (data not shown).
FIG. 1.
Porcine PBMC cultured with two combinations of mitogens release reverse transcriptase activity. Cell culture medium was removed at each of the days indicated after porcine PBMC were exposed to either PHA and PMA (black bars) or PMA and calcium ionophore A23187 (grey bars), as described in Materials and Methods. The culture medium was precleared of cell and cellular debris and then assayed for reverse transcriptase activity. The mean values ± standard deviations of the mean for counts per minute (cpm) of [3H]TTP incorporated are shown on the y axis.
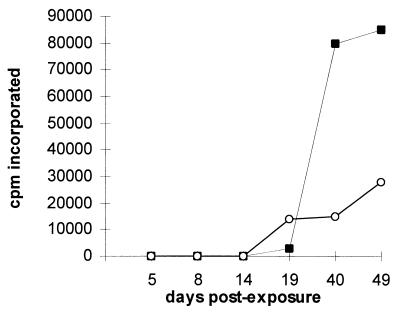
To determine whether the increase in reverse transcriptase activity following mitogenic stimulation correlated with the presence of infectious retrovirus, stimulated PBMC, which were isolated either from an NIH miniature pig or a Yucatan pig, were cocultured with ST-IOWA, a porcine cell line previously shown to be susceptible to infection by PoEV (11). As shown in Fig. 2, reverse transcriptase activity began to increase in ST cells at 14 days postexposure to PBMC from both strains of pigs and continued to increase throughout the duration of the experiment. The increase in reverse transcriptase activity observed over time indicates that mitogenic activation of porcine PBMC released an infectious retrovirus capable of spreading through the ST-IOWA cell culture.
FIG. 2.
Infectious virus is isolated after coculture of activated PBMC from the NIH miniature pig or the Yucatan with ST-IOWA cells. PBMC isolated from the NIH miniature pig or Yucatan pig were cultured with PHA and PMA for 5 days prior to coculture with ST-IOWA cells. Cell culture medium was removed at each of the days indicated and assayed for reverse transcriptase activity. The values for counts per minute of [3H]TTP incorporated for cells exposed to NIH miniature pig PBMC are shown by solid black squares, and those for cells exposed to Yucatan pig PBMC are shown by open circles. Background values for uninfected ST-IOWA cells have been subtracted from the values shown. cpm, counts per minute.
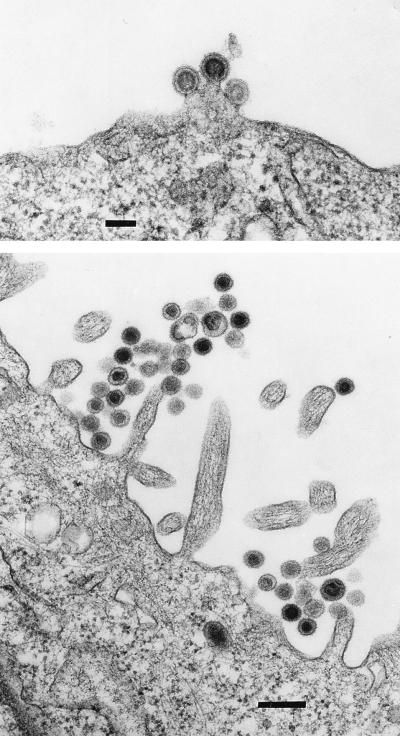
Retroviruses released by porcine cells were previously shown to have a type C morphology (1, 13). We examined the ST-IOWA cells productively infected with virus from the NIH miniature pig or the Yucatan pig (ST-NIH or ST-Y, respectively) by electron microscopy. As shown in Fig. 3, the morphology of the virus associated with both cell lines is typical of a type C retrovirus (23). No such particles were observed in control uninfected ST-IOWA cells. The PoEV obtained from these productively infected ST cell lines is referred to as PoEV-NIH or PoEV-Y.
FIG. 3.
ST-IOWA cells productively infected with PoEV release type C particles. ST-IOWA cells productively infected with PoEV from the Yucatan pig (top; bar = 100 nm) and the NIH miniature pig (bottom; bar = 0.25 μm).
PBMC-derived PoEVs are related to but distinct from pig kidney cell line-derived PERV-PK.
In order to identify and characterize the PoEV(s) induced by mitogenic activation of porcine PBMC, we used a PCR assay developed to identify unknown retroviruses (see Materials and Methods). This method uses pools of degenerate oligonucleotide primers containing the sequence possibilities for two regions of highly conserved amino acid sequences within the reverse transcriptase genes of different retroviruses. A similar assay was used to identify unknown herpesviruses by targeting the DNA polymerase gene (18). cDNA synthesis and PCRs were performed on RNA isolated from ST-NIH cells with a single downstream degenerate primer pool, YMDDB, and one of three different upstream primer pools, each of which targets different retroviral reverse transcriptase sequences (Table 1). A 150-bp product was observed from the porcine sample when the upstream primer PQGFA (based on sequence found in human T-cell leukemia virus type 1) was used but not when the PQGMA or PQGWA upstream primer (based on sequence identified in mouse mammary tumor virus or lentiviruses, respectively) was used (Table 1). The 150-bp PCR product was cloned and sequenced. Computer-based sequence comparisons with BLAST demonstrated that this sequence was highly related to the reverse transcriptase genes of other retroviruses and showed a high degree of sequence conservation to that previously reported for an endogenous retrovirus present in the pig genome (25). Oligonucleotide primers based on this published sequence were used to amplify a larger, 483-bp portion of the reverse transcriptase coding region from cDNA made from RNA of ST-NIH and ST-Y cells. Only one nucleotide difference was detected between the 483-bp sequences from PoEV-NIH and PoEV-Y (99.8% nucleotide identity; Table 2). These sequences varied by either one or two nucleotides, respectively, from the published PERV-MP sequence, obtained from the virus produced in the MPK miniature-pig kidney cell line (15). Neither nucleotide change altered the encoded amino acid sequence. Comparison of PoEV-NIH and PoEV-Y nucleotide sequences with the sequence of the virus identified in the PK(15) pig kidney cell line, PERV-PK, showed greater nucleotide differences, with only 95% nucleotide identity with the PERV-PK sequence from this region (15). Ninety-four percent nucleotide identity was found in a comparison of this region of PoEV-NIH and PoEV-Y with the same sequence characterized in the pig genome (25).
TABLE 2.
Two nucleotide positions distinguish PoEV from the NIH miniature pig and the Yucatan pig sequences from PERV-MP in the reverse transcriptase coding region of expressed sequences
PoEV-NIH and PoEV-Y display tropism for human cells.
To explore the species tropism of PoEV, cell lines representative of different species were exposed to PoEV-NIH or PoEV-Y and assessed for evidence of infection by reverse transcriptase assay (Table 3). Reverse transcriptase activity was detectable in porcine, mink, and human cell lines after exposure to PoEV-Y, whereas porcine and human cell lines, but not mink or any of the other cell lines examined, expressed reverse transcriptase activity after exposure to PoEV-NIH.
TABLE 3.
Reverse transcriptase activity in cells exposed to PoEV isolated from the NIH miniature pig or the Yucatan pig
| Target cell origin | Cpm 3H[TTP] incorporateda
|
|
|---|---|---|
| ST - NIHb | ST - Yc | |
| Mus dunni tail fibroblast | Bkgd | NDe |
| Rat fibroblast (Rat2) | Bkgd | ND |
| Mink lung (CCL-65) | Bkgd | 4,026 |
| Bovine (CCL-22, MDBK) | ND | Bkgdd |
| Hamster lung (E36) | Bkgd | Bkgd |
| Swine testis (ST-IOWA) | 11,515 | 10,919 |
| Human cervical cancer (HeLa) | 5,382 | ND |
| Human embryonic kidney (293) | 18,432 | 27,622 |
Samples of cell culture supernatant were removed 22 days after initiation of coculture and assayed for reverse transcriptase activity, as described in Materials and Methods. Values represent the counts per minute of 3H[TTP] incorporated in coculture sample, with subtraction of background values from matched cultures not exposed to ST-NIH miniature-pig cells.
ST-NIH miniature-pig cells were lethally irradiated and cocultured with target cells as described in Materials and Methods.
Target cells were exposed to culture supernatant from productively infected ST-Y cells.
Bkgd, value measured in coculture sample was less than twofold greater than background value.
ND, not determined.
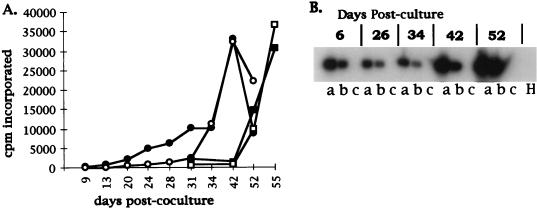
Although ST-IOWA cells do not produce retroviral particles, as shown by electron microscopy analysis and infectivity assays, they do express PoEV-specific sequences as detected by RT-PCR (data not shown). Thus, the observation that virus produced in ST-NIH and ST-Y cells infects human cell lines could be explained by the presence of a virus generated either by recombination of PoEV-NIH or PoEV-Y viral sequences with endogenous elements present in ST or by pseudotyping of PoEV-NIH or PoEV-Y viral proteins with envelope proteins expressed in ST cells. Therefore, to determine whether the virus released from activated porcine PBMC could directly infect human cells, we directly cocultured NIH miniature pig PBMC, either live or lethally irradiated 6 days after stimulation with PHA and PMA, with either ST-IOWA, human embryonic kidney 293, or African green monkey COS-7 cells. For each cell line, a control culture not exposed to porcine PBMC was also examined. Infection was monitored by reverse transcriptase assay and, where indicated, by PoEV-specific RT-PCR assay (described in Materials and Methods). Reverse transcriptase activity released from ST-IOWA cells exposed to either live or lethally irradiated PBMC was initially detectable approximately 2 weeks after coculture (Fig. 4A). ST-IOWA cells could not be monitored by RT-PCR assay because of expression of endogenous sequences present in control cultures (data not shown). Reverse transcriptase activity was detected in 293 cells at day 31 and increased by day 55 to the levels observed in the PBMC-ST-IOWA cell coculture (Fig. 4A). RT-PCR assay of 293 cells exposed to either live (lane a) or irradiated (lane b) porcine PBMC demonstrated that expression of PoEV-specific sequences could be observed as early as 6 days (Fig. 4B). No evidence of infection was observed in any of the cocultures with COS-7 cells with either reverse transcriptase or RT-PCR over the course of the experiment (55 days; data not shown). The results for 293 cells clearly demonstrate that PoEV can be directly transferred to human cells from porcine PBMC. Further, the increasing levels of reverse transcriptase activity observed in the 293 cell cultures represent a spreading and productive infection by PoEV in human cells.
FIG. 4.
Infectious virus isolated by coculture of activated PBMC from NIH miniature pig with ST-IOWA or human 293 cells. PBMC isolated from the NIH miniature pig were activated with PHA and PMA. Six days after activation, the cells were cocultured with ST-IOWA cells or human 293 cells. (A) Reverse transcriptase activity in cell culture medium removed at each of the days indicated. The mean values for counts per minute of [3H]TTP incorporated are shown on the y axis. The values for ST-IOWA cells cocultured with live PBMC are shown by closed circles, and those for ST-IOWA cells cocultured with lethally irradiated PBMC are shown by open circles. The results for the coculture of 293 cells with live PBMC are shown by closed squares, and those for 293 cells cocultured with lethally irradiated PBMC are shown by open squares. cpm, counts per minute. (B) Assay by RT-PCR followed by Southern blot analysis (described in Materials and Methods) of RNA obtained from 293 cells at the days indicated after coculture (numbers across the top) with either live PBMC (a), lethally irradiated PBMC (b), or control 293 (c) cultures. H, water only in lieu of cDNA template, a negative control for the PCR.
DISCUSSION
We have demonstrated that mitogenic activation of primary PBMC from two strains of pig, the NIH miniature pig and the Yucatan pig, results in the release of type C retrovirus that infects human as well as porcine cells. Limited host range analysis suggested that these viruses may be distinct, as the Yucatan-derived virus infected mink cells but the NIH miniature pig-derived virus did not. Although there is a previous report of a pig kidney cell line, PK(15), which spontaneously expresses type C particles with a human host range (15), this is the first demonstration that an endogenous retrovirus that is directly infectious to human cells can be mobilized from primary porcine cells.
The pig genome has been estimated to contain approximately 50 loci of endogenous retroviral sequences, based on hybridization with a probe derived from the pol region (15). Endogenous retroviral sequences are typically defective, representing partial, deleted, or otherwise disabled genomes incapable of encoding infectious retrovirus. Whether the loci which do encode infectious virus are common to all strains of pigs or unique to the strains tested is not known. The origins of the NIH miniature pig and the Yucatan pig are obscure. The Yucatan pig is derived from a herd which roamed wild in the Yucatan peninsula of Mexico (14). The NIH miniature pig is derived from crosses which included both feral and domesticated breeds found within the United States and elsewhere (4, 19). Sequence comparison of the reverse transcriptase coding regions revealed a high degree of nucleotide identity (482 of 483 nucleotides) of these viruses to each other as well as to PERV-MP, a porcine retrovirus which was reported to infect porcine cells, but not human cells, while some sequence divergence was observed when this region of PoEV-NIH and PoEV-Y was compared to the PERV-PK virus, which infects human cell lines (15). The divergence from PERV-PK may reflect the presence of more than one locus in the pig genome which encodes a virus capable of infecting human cells or the genetic heterogeneity of pig retroviral loci in different breeds. Sequence comparison of other regions of the retroviral genome such as the long terminal repeat or envelope may help to elucidate the relationship among these viruses. Indeed, it was recently reported that at least two classes of envelope are expressed in cells infected with PK(15)-derived virus (8).
Live porcine cells are used for therapeutic purposes in a growing number of clinical trials. Cases in which tissues or organs are minimally manipulated are likely to transfer leukocytes with the xenograft into a human recipient. Our findings suggest that subsequent mitogenic activation of the transferred porcine lymphocytes may release a retrovirus infectious for the recipient. Further analysis of the biological characteristics of this virus are needed to determine its tissue-specific infection properties. One strain of PoEV, PERV-PK, is reported to infect human cell lines of kidney, lung, muscle, and B- and T-cell origin (15), suggesting that PoEV(s) may infect a broad range of human tissues in vivo. Additional information is needed to determine whether other primary porcine tissues spontaneously release or can be induced to release infectious retrovirus.
Although it is difficult to predict the clinical outcome of human infection by PoEV, type C retroviruses closely related to PoEV have been associated with disease in nonhuman primates. Three of 10 immunosuppressed nonhuman primates developed lymphoma and died after exposure to a retroviral vector contaminated with high doses of replicating murine leukemia virus as well as defective endogenous murine retroviral genomes (3, 17). This experiment inadvertently models one aspect of xenotransplantation, that is, chronic exposure to a retrovirus in an immunosuppressed recipient. A second relevant example is the development of chronic myelogenous leukemia in normal juvenile gibbon apes after intraperitoneal inoculation with gibbon ape leukemia virus SEATO (7), which may have originated from an endogenous retrovirus of Asian wild mice (10). This represents another precedent for transmission and disease induction in a higher-order primate by a type C retrovirus endogenous to a nonprimate mammalian species. Preliminary infectivity studies of African green monkey (COS-7) and Rhesus kidney (FRhK) (15) cell lines suggest that PoEV may not infect monkey cells, and therefore nonhuman primates may not be suitable for assessing the safety risk associated with this virus. The absence of a nonhuman primate animal model and the finding that primary porcine lymphocytes release retrovirus infectious for human cells underscore the need for careful screening of porcine xenograft recipients for transfer of PoEV and potential development of disease. In this regard, the PoEV-specific RT-PCR assay which we have developed may be useful.
ACKNOWLEDGMENTS
We thank Robin Weiss, Jonathan Stoye, and Maribeth Eiden for helpful discussions, Eda Bloom and Dennis Klineman for critical evaluation of the manuscript, Marilyn Lundquist for technical assistance in the preparation of samples for electron microscopy, and Jackie Newell and David Sachs for providing background information on miniature pig breeds.
REFERENCES
- 1.Armstrong J A, Porterfield J S, Madrid A T D. C-type virus particles in pig kidney cell lines. J Gen Virol. 1971;10:195–198. doi: 10.1099/0022-1317-10-2-195. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste R E, Todaro G J. Evolution of type C viral genes: preservation of ancestral murine type C viral sequences in pig cellular DNA. Proc Natl Acad Sci USA. 1975;72:4090–4094. doi: 10.1073/pnas.72.10.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donahue R E, Kessler S W, Bodine D, McDonagh K, Dunbar C, Goodman S, Agricola B, Byrne E, Raffeld M, Moen R, Bacher J, Zsebo K M, Nienhuis A W. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176:1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.England D C, Panepinto L M. Conceptual and operational history of the development of miniature swine. In: Tumbleson M E, editor. Swine in biomedical research. 1st ed. Vol. 1. New York, N.Y: Plenum Press; 1985. pp. 17–22. [Google Scholar]
- 5.Hirsch M S, Black P H, Tracy G S, Leibowitz S, Schwartz R S. Leukemia virus activation in chronic allogeneic disease. Proc Natl Acad Sci USA. 1970;67:1914–1917. doi: 10.1073/pnas.67.4.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch M S, Phillips S M, Solnik C, Black P H, Schwartz R S, Carpenter C B. Activation of leukemia viruses by graft-versus-host and mixed lymphocyte reactions in vitro. Proc Natl Acad Sci USA. 1972;69:1069–1072. doi: 10.1073/pnas.69.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakami T G, Kollias G V, Jr, Holmberg C. Oncogenicity of gibbon type-C myelogenous leukemia virus. Int J Cancer. 1980;25:641–646. doi: 10.1002/ijc.2910250514. [DOI] [PubMed] [Google Scholar]
- 8.Le Tissier P, Stoye J P, Takeuchi Y, Patience C, Weiss R A. Two sets of human-tropic pig retroviruses. Nature. 1997;389:681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- 9.Levy J A, Datta S K, Schwartz R S. Recovery of xenotropic virus but not ecotropic virus during graft-versus-host reaction in mice. Clin Immunol Immunopathol. 1977;7:262–268. doi: 10.1016/0090-1229(77)90053-8. [DOI] [PubMed] [Google Scholar]
- 10.Lieber M, Scherr C J, Todaro G J, Benveniste R E, Callahan R, Coon H G. Isolation from the Asian mouse Mus Caroli of an endogenous type C virus related to infectious primate type C viruses. Proc Natl Acad Sci USA. 1975;72:2315–2319. doi: 10.1073/pnas.72.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieber M M, Sherr C J, Benveniste R E, Todaro G J. Biologic and immunologic properties of porcine type C viruses. Virology. 1975;66:616–619. doi: 10.1016/0042-6822(75)90234-2. [DOI] [PubMed] [Google Scholar]
- 12.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 13.Moennig V, Frank H, Hunsmann G, Ohms P, Schwarz H, Schafer W. C-type particles produced by a permanent cell line from a leukemic pig. Virology. 1974;57:179–188. doi: 10.1016/0042-6822(74)90119-6. [DOI] [PubMed] [Google Scholar]
- 14.Panepinto L M, Kroc R L. History, genetic origins, and care of Yucatan miniature and micro pigs. Lab Anim. 1995;June:31–34. [Google Scholar]
- 15.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 16.Phan-Thanh L, Kaeffer B, Bottreau E. Porcine retrovirus: optimal conditions for its biochemical detection. Arch Virol. 1992;123:255–265. doi: 10.1007/BF01317262. [DOI] [PubMed] [Google Scholar]
- 17.Purcell D F J, Broscius C M, Vanin E F, Buckler C E, Nienhuis A W, Martin M A. An array of murine leukemia virus-related elements is transmitted and expressed in a primate recipient of retroviral gene transfer. J Virol. 1996;70:887–897. doi: 10.1128/jvi.70.2.887-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C-C, Bosch M L. Identification of two homologs of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachs D H, Leight G, Cone J, Schwarz S, Stuart L, Rosenberg S. Transplantation in miniature swine. I. Fixation of the major histocompatibility complex. Transplantation. 1976;22:559–567. doi: 10.1097/00007890-197612000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherr C J, Lieber M M, Todaro G J. Mixed splenocyte cultures and graft versus host reactions selectively induce an “S-tropic” murine type C virus. Cell. 1974;1:55–58. [Google Scholar]
- 22.Strandstrom H, Veijalainen P, Moennig V, Hunsmann G, Schwarz H, Schafer W. C-type particles produced by a permanent cell line from a leukemic pig. I. Origin and properties of the host cells and some evidence for the occurrence of type-C-like particles. Virology. 1974;57:175–178. doi: 10.1016/0042-6822(74)90118-4. [DOI] [PubMed] [Google Scholar]
- 23.Teich N. Taxonomy of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 25–308. [Google Scholar]
- 24.Todaro G J, Benveniste R E, Lieber M M, Sherr C J. Characterization of a type C virus released from the porcine cell line PK(15) Virology. 1974;58:65–74. doi: 10.1016/0042-6822(74)90141-x. [DOI] [PubMed] [Google Scholar]
- 25.Tristem M, Kabat P, Lieberman L, Linde S, Karpas A, Hill F. Characterization of a novel murine leukemia virus-related subgroup within mammals. J Virol. 1996;70:8241–8246. doi: 10.1128/jvi.70.11.8241-8246.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tumilowicz J J, Hung C-L, Kramarsky B. Concurrent replication of a papovavirus and a C-type virus in the CCL 33 porcine cell line. In Vitro. 1979;15:922–928. doi: 10.1007/BF02618050. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C A, Eiden M V, Marsh J W. Quantitative micro p30 and reverse transcriptase assays for Moloney murine leukemia virus. J Virol Methods. 1994;48:109–118. doi: 10.1016/0166-0934(94)90093-0. [DOI] [PubMed] [Google Scholar]