Abstract
To better understand mechanisms of persistent rotavirus infections of cultured cells, we established independent, persistently infected cultures of MA104 cells, using rotavirus strain SA11. The cultures were either passaged when the cells reached confluence or supplemented with fresh medium every 7 days. Viral titers in culture lysates varied from 104 to 107 PFU per ml during 350 days of culture maintenance. Trypan blue staining indicated that 72 to 100% of cells in the cultures were viable, and immunocytochemical staining using a monoclonal antibody directed against viral protein VP6 demonstrated that 38 to 63% of the cells contained rotavirus antigen. We tested the capacity of rotaviruses isolated from the persistently infected cultures (PI viruses) to infect cells cured of persistent infection. Although wild-type (wt) and PI viruses produced equivalent yields in parental MA104 cells, PI viruses produced greater yields than wt virus in cured cells, which indicates that viruses and cells coevolve during persistent rotavirus infections of MA104 cells. To determine whether mutations in viruses and cells selected during these persistent infections affect viral entry, we tested the effect of trypsin treatment of the viral inoculum on growth of wt and PI viruses. Trypsin pretreatment is required for postattachment penetration of rotavirus virions into cells. In contrast to the case with wt virus, PI viruses produced equivalent yields with and without trypsin pretreatment in parental MA104 cells. However, PI viruses required trypsin pretreatment for efficient growth in cured cells. These results indicate that mutant viruses and cells are selected during maintenance of persistent rotavirus infections of MA104 cells and suggest that mutations in each affect trypsin-dependent steps in rotavirus entry.
Many cytolytic animal viruses, including rotavirus (13, 20), are capable of establishing persistent infections of cultured cells. In most cases, the mechanisms by which these persistent infections are maintained involve selection of mutant cells that do not efficiently support viral replication, selection of mutant viruses that are less cytopathic, or coevolution of mutant cells and viruses such that cellular resistance to viral replication is balanced by an enhanced capacity of the virus to infect the resistant cells (reviewed in reference 1). Viral entry has been shown to be a critical point of virus-cell interaction for the selection of mutations during persistent infections caused by several viruses, including coronavirus (11, 24), poliovirus (7, 31), and reovirus (17, 49). It is not known whether mutant viruses and cells are also selected during persistent rotavirus infections or whether such mutations affect rotavirus entry.
Rotaviruses are nonenveloped, icosahedral viruses that contain a genome consisting of 11 segments of double-stranded RNA (reviewed in reference 19). Rotavirus replication is initiated by attachment of the virus to cell surface receptors, which have not been identified with certainty. The attachment step is mediated by outer capsid protein VP4 (5, 16, 29, 32, 35, 38, 43), a spike protein that extends approximately 120 Å from the virion surface (41, 45, 52). The mechanism by which rotavirus enters cells is not precisely understood; however, current data suggest that rotavirus entry occurs by direct penetration of the virus into the cytosol. Rotavirus virions increase membrane permeability as measured by the release of radioactive chromium from cells (28) and the release of fluorescent dyes from liposomes (39) and isolated membrane vesicles (44). In addition, rotavirus virions induce fusion from without in cultured cells (22). Treatment of rotavirus with the intestinal protease trypsin significantly enhances viral infectivity (2, 3, 14, 18, 21), most likely by increasing the efficiency of membrane penetration (22, 28, 39, 44). Trypsin cleaves VP4 into two fragments, VP5* and VP8* (15, 18, 21, 34). VP5* is postulated to contain a fusion domain that facilitates rotavirus penetration into cells (36); VP8* contains sequences that mediate hemagglutination (23, 38) and likely participate in viral attachment to nucleated cells.
To better understand mechanisms of persistent rotavirus infections of cultured cells and to determine whether virus-cell coevolution involving viral entry occurs during persistent rotavirus infection, we characterized viruses and cells from MA104 cultures persistently infected with simian rotavirus strain SA11. These cultures produced high titers of infectious virus for prolonged periods and were cured by passage in medium containing anti-rotavirus antibodies. Viruses isolated from the persistently infected cultures and cells cured of persistent infection were tested to determine whether mutations in viruses and cells were selected during propagation of these persistent infections. Our findings suggest that persistent rotavirus infections are carrier cultures maintained by horizontal transmission of virus between cells and that the requirement for VP4 cleavage is a target for selection of mutant viruses and cells during maintenance of persistent rotavirus infection.
MATERIALS AND METHODS
Cells and viruses.
MA104 cells were obtained from Biowhittaker (Walkersville, Md.) and grown in a 5% CO2 atmosphere at 37°C in Eagle medium (Sigma Chemical Co., St. Louis, Mo.) that was supplemented to contain 10% fetal bovine serum (FBS) (Intergen, Purchase, N.Y.), 2 mM l-glutamine, 500 U of penicillin per ml, and 500 μg of streptomycin per ml (Irvine Scientific, Santa Ana, Calif.). Cells were passaged when confluent by using 1% trypsin with 5 mM EDTA (Irvine Scientific). Rotavirus strain SA11 was obtained from the American Type Culture Collection. Viruses isolated from persistently infected cultures of MA104 cells were designated PI viruses. Both wild-type (wt) and PI viruses were plaque purified twice in MA104 cells prior to preparation of lysate stocks (42). Virus stocks were grown in MA104 cells, which were maintained in Eagle medium without FBS and supplemented to contain 2 μg of porcine trypsin (Intergen) per ml. Purified virion preparations were made by infecting MA104 cells with trypsin-treated (20 μg of trypsin per ml, incubated at 37°C for 30 min) third-passage lysate stocks as previously described (10), with the exception that freon-extracted virus from the cell pellet and virus from the supernatant were collected by centrifugation through a 40% sucrose solution. To obtain purified rotavirus virions containing 35S-labeled proteins, MA104 cells were maintained in complete Eagle medium without FBS for 2 h after adsorption with trypsin-treated virus. The medium was removed, and methionine-free Eagle medium (Sigma) without FBS was added. Following incubation for 1 h, Easytag [35S]methionine (DuPont NEN, Boston, Mass.) (0.1 mCi per ml) was added to the medium and cells were incubated for an additional 1 h. After that time, the medium was removed and cells were maintained in complete Eagle medium without FBS and without trypsin for 20 h. The concentration of viral particles in preparations of purified virions was determined by using the conversion 1 optical density unit at A260 equals 2.1 × 1012 viral particles (46).
Establishment of persistently infected MA104 cell cultures.
Persistent rotavirus infections were established by infecting confluent MA104 cell monolayers (2 × 106 cells), at the 60th passage level, with independent third-passage lysate stocks of SA11 at a multiplicity of infection (MOI) of 0.1 PFU per cell (Table 1). The viral inocula were treated with 20 μg of porcine trypsin per ml at 37°C for 30 min prior to adsorption. The cultures were maintained in complete Eagle medium without FBS for the first 2 to 3 days, after which time the medium was supplemented to contain 10% FBS. Cells were either passaged when confluent or supplemented with fresh medium every seventh day if the cell density was not sufficient to permit passage. Trypsin was used to passage the cultures, but additional trypsin was not added to the medium. Cell culture lysates (5 ml per flask) were collected at each passage by two cycles of freezing and thawing (−70°C and 37°C).
TABLE 1.
Establishment of MA104 cell cultures persistently infected with rotavirus
| Third-passage stocka | Persistently infected cultureb | Cured cell culturec | PI virus straind |
|---|---|---|---|
| SA11-P3A | MA104/SA11-A | MX-S | |
| SA11-P3B | MA104/SA11-B | PI-SA11-B/135 | |
| SA11-P3C | MA104/SA11-C | ||
| SA11-P3D | MA104/SA11-D | PI-SA11-D/128 |
Independent twice-plaque purified stocks of wt rotavirus strain SA11 were passaged three times in MA104 cells to prepare stocks to establish independent, persistently infected cultures.
MA104-cell cultures persistently infected with rotavirus were established using trypsin-treated inocula of third-passage stocks of rotavirus strain SA11. Cells were maintained in medium without FBS or trypsin for the first 2 to 3 days, and then the medium was supplemented to contain 10% FBS but no trypsin. The cultures were either passaged with trypsin when cells reached confluence or supplemented with fresh medium every 7 days.
MA104 cells were cured of persistent infection by passage in medium containing a heat-inactivated, antirotavirus antiserum for a 35-day period. The resulting cured culture, termed MX-S, had nondetectable levels of infectious rotavirus in cell culture lysates, did not give rise to infectious centers, did not contain immunocytochemically detectable rotavirus antigen, and had nondetectable levels of rotavirus-specific RNA by RT-PCR.
PI virus strains were plaque purified twice from a lysate stock of a persistently infected cell culture.
Determination of viral titer in persistently infected cell culture lysates.
Viral titer in cell culture lysates was determined by plaque assay as previously described (42). Samples were treated with 10 μg of trypsin per ml, diluted serially 10-fold, and adsorbed to MA104-cell monolayers in duplicate in 6-well plates (Costar, Cambridge, Mass.). The MA104 cells were then overlaid with Eagle medium (Sigma) supplemented to contain 2 mM l-glutamine, 500 U of penicillin per ml, 500 μg of streptomycin per ml, 2 μg of trypsin per ml, and 1% agar (Difco, Detroit, Mich.) and incubated at 37°C. Plaques were counted on day 4 after staining with 1% neutral red (Fisher Scientific, Pittsburgh, Pa.).
Preparation of polyclonal antirotavirus antiserum.
Polyclonal rabbit antirotavirus antiserum was obtained by inoculating a New Zealand White rabbit with 100 μg of purified SA11 virions in complete Freund’s adjuvant, followed by booster doses of 50 μg of purified virions in incomplete Freund’s adjuvant at 2, 3, and 7 weeks after the initial inoculation (Cocalico, Reamstown, Pa.). Antiserum was heat-inactivated by incubation at 56°C for 30 min prior to use. A 1:5,120 dilution of antiserum was sufficient to achieve a 90% reduction in infectivity of SA11 as determined by plaque-reduction neutralization assay (51).
Infectious center assay.
Cells were washed three times in phosphate-buffered saline and enumerated. Cells were diluted serially 10-fold and deposited onto MA104-cell monolayers in duplicate in 6-well plates. After adsorption at room temperature for 2 h, cells were overlaid with complete Eagle medium containing 1% agar and processed according to the plaque assay technique (42).
Immunocytochemical staining with antirotavirus antibody.
Cells were grown in chambered glass slides (Lab Tek, Nunc, Naperville, Ill.) and fixed with 1:1 (vol/vol) acetone-methanol at 4°C for 5 min. Viral antigen was detected by using the VectaStain Elite ABC kit (Vector Laboratories, Burlingame, Calif.) according to the manufacturer’s instructions. The primary antibody was a 1:100 dilution of a rotavirus group A-specific monoclonal antibody (Serotec, Ebetsu, Japan) directed against VP6 (48). The chromogen was 3,3′-diaminobenzidine tetrahydrochloride (0.04%) in 0.05 M Tris-HCl (pH 7.4) and 0.025% H2O2.
Isolation of cells cured of persistent rotavirus infection.
MA104 cells were cured of persistent rotavirus infection by passage for 35 days in medium supplemented to contain 1% rabbit antirotavirus antiserum. Antibody-treated cells (designated MX) were tested for viral infection by plaque assay of cell culture lysates (42), infectious center assay, immunocytochemistry, reverse transcription-PCR amplification (RT-PCR) of rotavirus RNA, and electron microscopy.
Detection of rotavirus RNA by RT-PCR.
Parental MA104 cells, MA104 cells persistently infected with rotavirus, or MX cells cured of persistent infection (5 × 106 cells) were centrifuged to form a pellet, resuspended in 3.0 ml of lysis buffer (1% sodium dodecyl sulfate [SDS], 1 mM EDTA, and 50 mM Tris [pH 8.0]), and incubated at 37°C for 15 min. Lysates were extracted three times with 1:1 (vol/vol) phenol-chloroform after the addition of 60 μl 5 M NaCl. Nucleic acid was precipitated in 3 volumes of cold 100% ethanol after the addition of 1/10th volume of 3 M ammonium acetate (pH 5.2). Total cellular nucleic acid (2 μg) was used as template for RT-PCR (35 cycles of cDNA amplification). Nucleic acid was incubated in 10 μl 40% dimethyl sulfoxide at 95°C for 2 min and then added to ice-cold oligonucleotide primers and incubated on ice for 5 min prior to RT-PCR. Cycling parameters were set such that melting of cDNA template strands occurred at 95°C for 1 min, annealing of primers and cDNA template occurred at 45°C for 2 min, and synthesis of cDNA occurred at 72°C for 3 min. Oligodeoxynucleotide primers 5′-TAACTATTGTGCTCATAGAG and 5′-ATGTTCAAGATGGAGTCTAC corresponding to nucleotide sequences of rotavirus SA11 gene segment 7 (8) were used in these reactions. This primer pair is expected to amplify a 999-base-pair cDNA. Rotavirus genomic double-stranded RNA purified from SA11 virions and cellular nucleic acid purified from parental MA104 cells served as controls. RT-PCR products were resolved in a 1.0% agarose-Tris-borate-EDTA gel and visualized by ethidium bromide staining.
Electron microscopy.
Cells were centrifuged to form a pellet (1,000 × g for 3 min) and suspended in phosphate-buffered 2% glutaraldehyde. After primary fixation, cells were again centrifuged (1,000 × g for 3 min), resuspended in 1% osmium tetroxide, dehydrated in propylene oxide and increasing percentages of ethanol from 50% to 100%, and then embedded in an epoxy resin. Ultrathin sections were prepared with an LKB Ultratome III ultramicrotome and stained with lead citrate and uranyl acetate. Sections were examined in a Philips 300 electron microscope.
Growth of wt and PI rotaviruses in parental MA104 cells and cured MX cells.
Monolayers of parental MA104 cells or cured MX cells (105 cells) were infected with rotavirus strains at an MOI of 2 PFU per cell in 24-well plates (Costar). Prior to adsorption, the viral inoculum was either untreated or treated with 2.5 to 20 μg of trypsin per ml at 37°C for 30 min. Virus was adsorbed to cells at 4°C for 1 h, the inoculum was removed by washing twice with phosphate-buffered saline, and 1.0 ml of trypsin-free, fresh medium was added. After incubation at 37°C for 24 h, cells were frozen and thawed twice and virus in cell lysates was titrated on MA104 cell monolayers by plaque assay (42).
SDS-PAGE of rotavirus structural proteins.
Rotavirus virions containing [35S]-labeled proteins were purified from infected MA104 cells (10), either untreated or treated with 1.25 to 10 μg of trypsin per ml at 37°C for 30 min, and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (33). In preparation for electrophoresis, virion samples were mixed 1:1 with 2× sample buffer (250 mM Tris [pH 6.8], 4% 2-mercaptoethanol, 2% SDS, 20% sucrose, and 0.02% bromophenol blue) and incubated at 65°C for 5 min. After incubation, samples were loaded into wells of an 8% polyacrylamide gel and electrophoresed at 200 V constant voltage for 45 min. Gels were fixed, dried onto filter paper under vacuum, and exposed to Kodak BiomaxMR film (Eastman Kodak Co., Rochester, N.Y.).
RESULTS
Establishment of persistently infected MA104 cell cultures using rotavirus strain SA11.
To determine whether rotavirus can establish persistent infections of MA104 cells, four independent cultures of these cells were infected with trypsin-treated inocula of independent, plaque-purified, third-passage lysate stocks of rotavirus strain SA11 (Table 1). The cultures were either passaged when cells reached confluence or supplemented with fresh medium every 7 days. Trypsin was used to passage the cultures, but additional trypsin was not added to the medium. Establishment of the persistently infected cultures was associated with intense cell crises in which only a few colonies of cells in the cultures survived. Recovery until first passage was prolonged, varying from 20 to 48 days, after which time no significant crises were observed, including times when the cultures were treated with trypsin to facilitate passage. Viral titers in lysates collected during passage of the four independent, persistently infected cultures varied from 104 to 107 PFU per ml for 350 days (data not shown). To assess cell viability, an aliquot of each culture was stained with trypan blue at several passages (Table 2 and data not shown). After the initial period of crisis, 72 to 100% of cells were viable as assessed by trypan blue exclusion. Therefore, MA104 cell cultures persistently infected with rotavirus can be established, and these cultures produce infectious virus for at least 350 days of culture maintenance.
TABLE 2.
Viability and antigen staining of cells in the persistently infected MA104/SA11-B culture
| Day of culture maintenance | Passage no. | Trypan blue exclusiona (%) | Immunocytochemical stainingb (%) |
|---|---|---|---|
| 90 | 6 | 72 | 63 |
| 144 | 10 | 100 | — |
| 153 | 11 | — | 52 |
| 163 | 12 | 95 | — |
| 173 | 13 | 85 | — |
| 194 | 15 | 86 | 42 |
| 210 | 16 | 87 | — |
| 248 | 20 | 100 | — |
| 330 | 28 | 96 | 38 |
Cells were mixed 1:4 (vol/vol) with 0.1% trypan blue, incubated at room temperature for 5 to 10 min, and enumerated using a hemacytometer. The percentage of cells not stained with trypan blue was determined by counting 100 cells. —, not done.
Viral antigen was detected with an immunoperoxidase assay using a monoclonal antibody directed against VP6. The percentage of cells containing rotavirus antigen was determined by counting approximately 1,000 cells.
To determine the percentage of cells in the persistently infected cultures containing rotavirus antigen, we used an immunoperoxidase assay to detect the rotavirus VP6 protein. The results show that 38 to 63% of cells contained VP6 antigen (Table 2 and data not shown), which indicates that a substantial percentage of cells in the cultures were infected with rotavirus. Viral antigen was not detected in uninfected MA104 cells or in persistently infected cells when the anti-VP6 monoclonal antibody was omitted from the immunocytochemistry procedure (data not shown). Infectious center assays performed in parallel demonstrated that >50% of cells contained infectious virus (data not shown).
In anticipation of studies with viruses obtained from persistent rotavirus infections of MA104 cells, PI viruses were isolated by plaque purification from the persistently infected MA104 cell cultures (Table 1). The particle-to-PFU ratio was determined for two of the PI virus isolates and was found to be 165/1 and 400/1 for PI-SA11-B/135 and PI-SA11-D/128, respectively, in comparison to 60/1 for wt SA11.
Cure of persistent rotavirus infection of MA104 cells.
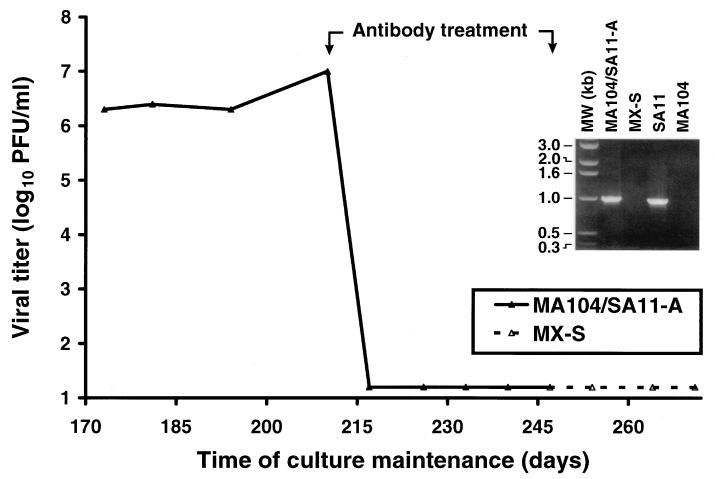
To assess whether persistent rotavirus infection of MA104 cells can be cured by passage in medium containing antirotavirus antibodies, the persistently infected MA104/SA11-A culture (subsequent to the 11th passage) was maintained in medium containing antirotavirus antiserum. Antibody treatment was continued for 35 days (4 passages), after which time the resulting culture, termed MX-S, was passaged in medium without antiserum. During the period of antibody treatment, viral titer decreased to less than 10 PFU per ml of culture lysate (the lower limit of detection) and remained undetectable for greater than 30 days after completion of antibody treatment (Fig. 1 and data not shown). We also tested whether the antibody-treated cells could give rise to infectious centers. In these experiments, no infectious centers were produced by the MX-S culture per 5 × 105 cells 24 days after completion of antibody treatment. In addition, antibody-treated cells did not contain immunocytochemically detectable rotavirus antigen 30 days after completion of antibody treatment (data not shown). Thus, antirotavirus antibody treatment of a persistently infected MA104 cell culture eradicates infectious virus and viral antigen from the culture.
FIG. 1.
Viral titer in cell culture lysates obtained from persistently infected MA104/SA11-A and cured MX-S cells. On the indicated day of culture maintenance, the titer of infectious virus in cell culture lysates was determined by plaque assay using MA104 cells. The period of antibody treatment (day 210 to day 245) is indicated. (Inset) RT-PCR to detect rotavirus-specific RNA in persistently infected and cured MA104 cell cultures. Total cellular nucleic acid was purified from each culture, and approximately 2 μg was used as template for RT-PCR (35 cycles of cDNA amplification). Oligodeoxynucleotide primers corresponding to the SA11 gene segment 7 sequence (8) were used in these reactions; genomic double-stranded RNA purified from SA11 virions and cellular nucleic acid purified from parental MA104 cells served as controls. RT-PCR products were resolved in a 1.0% agarose-Tris-borate-EDTA gel and visualized by ethidium bromide staining. Positions of DNA size markers are shown on the left in kilobase pairs. The primers used in these reactions are expected to amplify a 999-base-pair cDNA.
To determine whether rotavirus dsRNA was eliminated by antibody treatment of the persistently infected culture, we used an RT-PCR technique to detect rotavirus-specific nucleotide sequences. Cell lysates were prepared from the persistently infected and cured cultures, and oligodeoxynucleotide primers specific for rotavirus gene segment 7, which encodes nonstructural protein NSP3 (19), were used to prime cDNA synthesis (Fig. 1, inset). A band corresponding to the expected size for gene segment 7 was detected in the lysate prepared from the MA104/SA11-A culture but not in that from the MX-S culture (tested 24 passages after completion of antibody treatment). This technique can detect as few as 11 copies of gene 7 RNA per cell (data not shown). Therefore, the results suggest that rotavirus gene 7 sequences do not persist in MA104 cells after passage in medium containing antirotavirus antibodies.
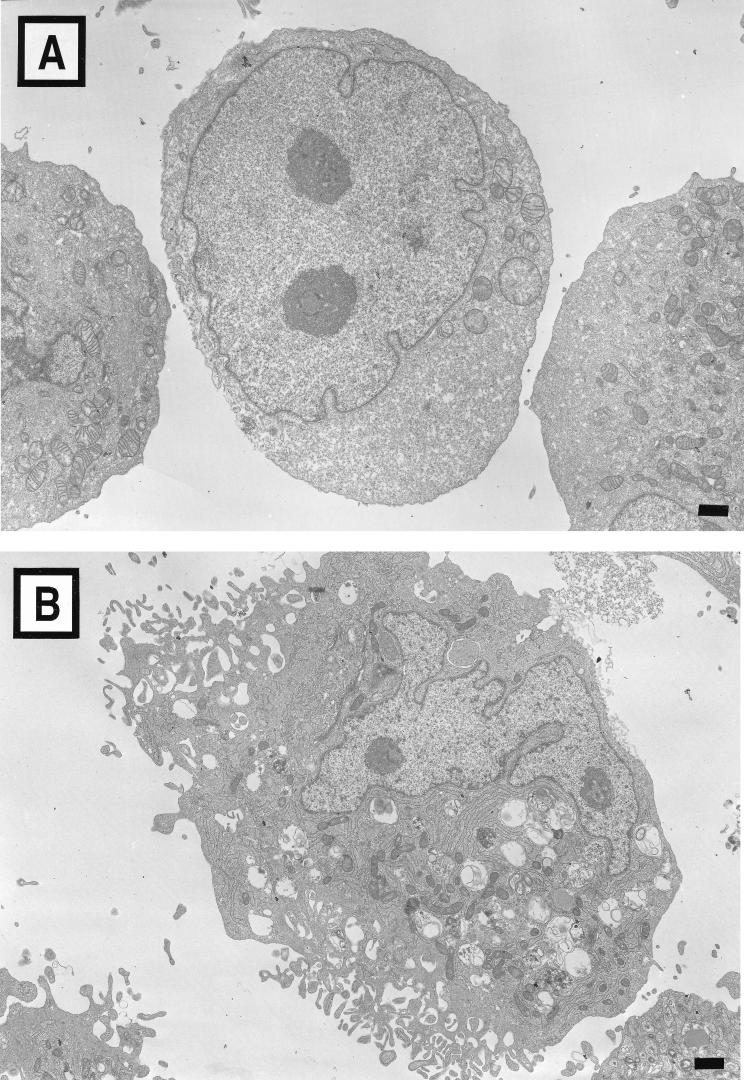
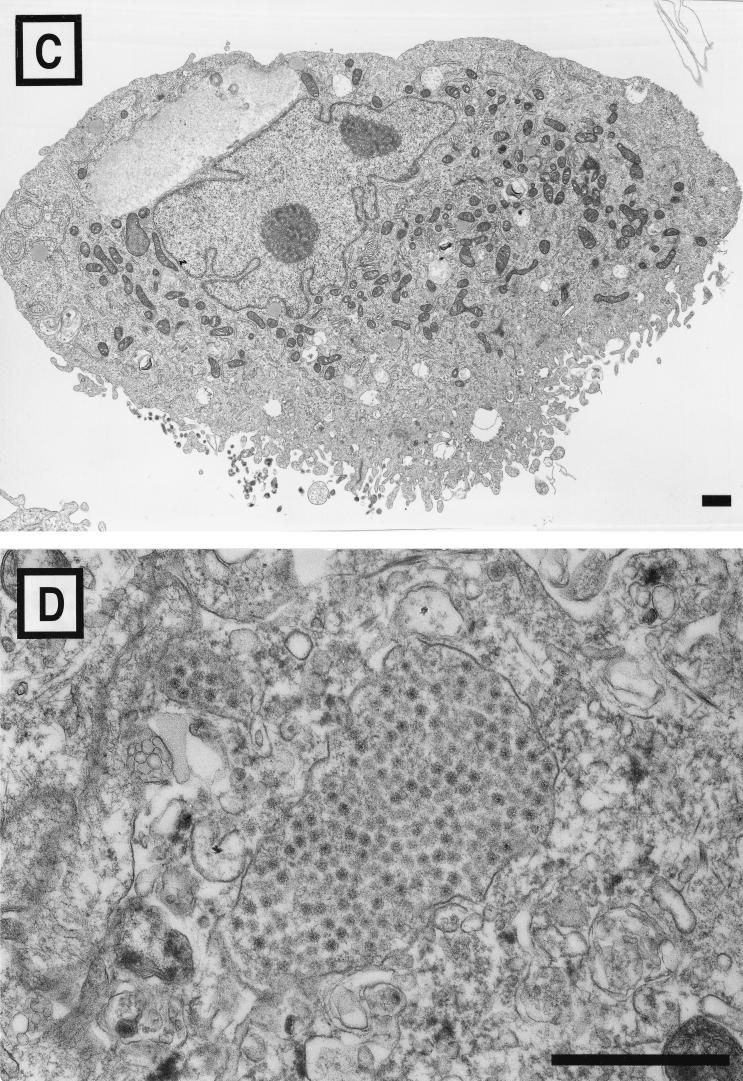
Ultrastructure of MA104 cells persistently infected with rotavirus and cured of persistent infection.
To define the location of virus in the persistently infected cells and to ascertain whether changes in cellular ultrastructure are associated with persistent rotavirus infection, we used electron microscopy to examine the morphology of uninfected MA104 cells (Fig. 2A), persistently infected MA104/SA11-A cells (Fig. 2B), and cured MX-S cells (Fig. 2C). In comparison to uninfected cells, both persistently infected and cured cells exhibited a polarized appearance: villouslike projections were observed on one surface, while the other surface was smooth. In addition, both types of cells contained increased numbers of mitochondria and vacuoles. These morphological features were observed in all thin sections examined. These features were not observed in uninfected MA104 cells passaged in medium containing antirotavirus antibodies for a 35-day period as a control (data not shown). Particles consistent with rotavirus virions were visualized in <5% of cells from the persistently infected cultures and were contained in membrane-bound structures (Fig. 2D). Rotavirus virions were not observed in cured MX-S cells.
FIG. 2.
Morphology of uninfected, persistently infected, and cured MA104 cells. (A) Uninfected MA104 cells. (B) Persistently infected MA104/SA11-A cells. (C) Cured MX-S cells. (D) Particles consistent with rotavirus virions enclosed in a membrane-bound structure in a persistently infected MA104/SA11-A cell. Bars, 1 μm.
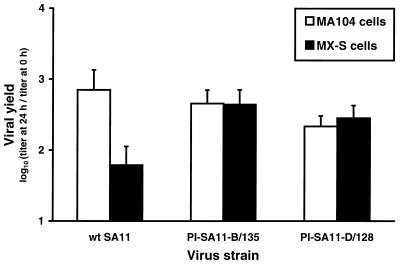
Growth of wt and PI rotaviruses in parental MA104 cells and cured MX cells.
To test whether mutant viruses and cells are selected during persistent rotavirus infections of MA104 cells, independent PI viruses were tested for growth in parental MA104 cells and cured MX-S cells. Monolayers of parental and cured cells at approximately the same passage number were infected with trypsin-treated inocula of wt SA11, PI-SA11-B/135, and PI-SA11-D/128 at an MOI of 2 PFU per cell (Fig. 3). After 24 h of viral growth in cured cells, the PI viruses produced 10- to 25-fold greater yields than that of wt SA11, which indicates that mutant viruses capable of enhanced growth were selected during propagation of the persistently infected cultures. In addition, wt SA11 produced 10-fold greater yields in parental cells than in cured cells, which demonstrates that mutant cells manifesting a block to rotavirus replication were also selected during these persistent infections. Thus, these results indicate that mutations arise in both viruses and cells during maintenance of MA104 cell cultures persistently infected with rotavirus.
FIG. 3.
Growth of wt and PI viruses in parental MA104 cells and cured MX cells. Monolayer cultures of MA104 cells and MX-S cells (5 × 105 cells) were infected with wt SA11, PI-SA11-B/135, and PI-SA11-D/128 at an MOI of 2 PFU per cell. Prior to adsorption, each viral inoculum was treated with 10 μg of trypsin per ml at 37°C for 30 min. After 1 h of adsorption, the inoculum was removed, 1.0 ml of fresh trypsinfree medium was added, and cells were incubated at 37°C for 24 h. Virus in cell lysates was titrated by plaque assay using MA104 cells. The results are presented as the mean viral yields (titer at 24 h divided by titer at 0 h) for four independent experiments. Error bars indicate standard deviations of the means.
Growth of PI rotaviruses with and without trypsin pretreatment.
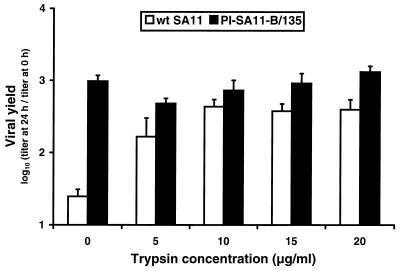
To determine whether mutations in viruses and cells selected during persistent rotavirus infections of MA104 cells affect viral entry, we tested the capacity of wt and PI SA11 viruses to infect MA104 cells in the absence of trypsin pretreatment. For these experiments, virus stocks were prepared without trypsin supplementation of the medium. Monolayers of parental MA104 cells were infected with wt SA11 and PI-SA11-B/135 at an MOI of 2 PFU per cell (Fig. 4). Prior to adsorption, the viral inoculum was either untreated or treated with trypsin at concentrations of from 5 to 20 μg per ml. In the absence of trypsin pretreatment, PI-SA11-B/135 produced approximately 50-fold greater yields than wt SA11 after 24 h of viral growth. As the concentration of trypsin used to treat the viral inoculum was increased, yields of wt SA11 approached those of PI-SA11-B/135 in the absence of trypsin pretreatment. However, increasing concentrations of trypsin did not increase yields of PI-SA11-B/135. Therefore, wt and PI rotaviruses differ substantially in the requirement for trypsin pretreatment to efficiently infect MA104 cells, suggesting that PI viruses have acquired mutations that alter steps in viral entry.
FIG. 4.
Growth of wt and PI viruses with and without trypsin pretreatment. Monolayer cultures of MA104 cells (105 cells) were infected with wt SA11 and PI-SA11-B/135 at an MOI of 2 PFU per cell. Prior to adsorption, each viral inoculum was treated with trypsin at the concentrations shown at 37°C for 30 min. After 1 h of adsorption, the inoculum was removed, 1.0 ml of fresh trypsinfree medium was added, and cells were incubated at 37°C for 24 h. Virus in cell lysates was titrated by plaque assay using MA104 cells. The results are presented as the mean viral yields (titer at 24 h divided by titer at 0 h) for four independent experiments. Error bars indicate standard deviations of the means.
Evolution of the capacity of PI rotaviruses to infect MA104 cells without trypsin pretreatment.
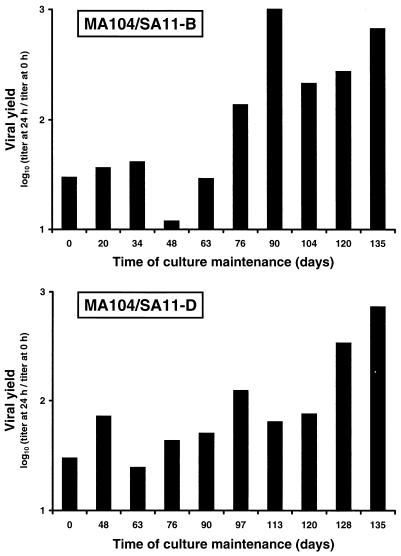
To determine when in the course of persistent infection the capacity for trypsin-independent viral growth evolved, we measured viral yields after infections were initiated with untreated cell culture lysate stocks collected over time from the persistently infected MA104/SA11-B and MA104/SA11-D cultures (Fig. 5). The capacity of viruses in the persistently infected cultures to grow in MA104 cells without trypsin pretreatment was not a property of the virus stocks used to establish persistent infection but instead was gradually selected in these cultures by passage day 100. Thus, the property of trypsin-independent viral growth evolves during maintenance of persistent rotavirus infection of MA104 cells.
FIG. 5.
Effect of day of cell culture maintenance on the capacity of viruses from the persistently infected MA104 cell cultures to grow without trypsin pretreatment. Monolayers of MA104 cells (105 cells) were infected with cell culture lysates (0.2 ml of lysate stock) obtained from the persistently infected MA104/SA11-B and MA104/SA11-D cultures. Prior to adsorption, each viral inoculum was not treated with trypsin. After 1 h of adsorption, the inoculum was removed, fresh trypsinfree medium was added, and cells were incubated at 37°C for 24 h. Virus in cell lysates was titrated by plaque assay using MA104 cells. The results are presented as the mean viral yields (titer at 24 h divided by titer at 0 h) for two independent experiments.
SDS-PAGE analysis of structural proteins of wt and PI rotaviruses.
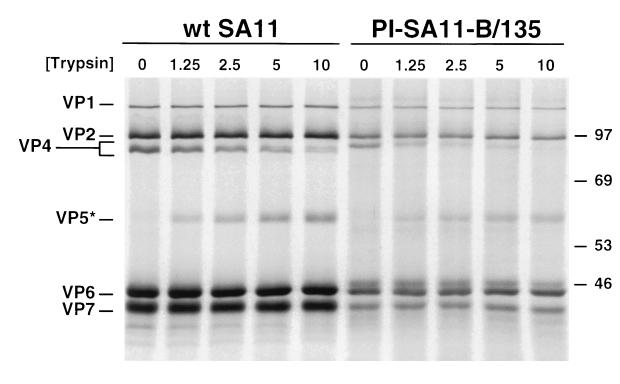
Our finding that PI rotaviruses are capable of trypsin-independent growth prompted us to test whether these viruses contain cleaved VP4 proteins. Radiolabeled virions of wt SA11 and PI-SA11-B/135 were purified from lysate stocks prepared without trypsin and subjected to SDS-PAGE and autoradiography (Fig. 6). Prior to electrophoresis, virions were either untreated or treated with trypsin at concentrations of from 1.25 to 10 μg per ml. In the absence of trypsin treatment, bands corresponding to uncleaved VP4 were observed for both wt and PI viruses, indicating that wt and PI viruses prepared without trypsin contain intact VP4 proteins. Using these electrophoresis conditions, the VP4 protein of PI-SA11-B/135 migrated slightly slower than that of wt SA11. After trypsin treatment of purified virions of wt and PI viruses, bands of equivalent electrophoretic mobility corresponding to trypsin-cleavage fragment VP5* were observed. In these experiments, there was no discernible difference between wt and PI viruses in the relative decrease in VP4 band intensity with increasing concentrations of trypsin. Thus, PI virus VP4 proteins are uncleaved in stocks prepared without trypsin but remain susceptible to trypsin-mediated proteolysis.
FIG. 6.
SDS-PAGE of structural proteins of wt and PI viruses. Purified [35S]methionine-labeled virions of wt SA11 and PI-SA11-B/135 at a concentration of 2 × 1011 particles per ml were treated with trypsin at the concentrations shown (in micrograms per milliliter) at 37°C for 30 min. Equal volumes of samples (2 × 109 particles) were loaded into wells of an 8% SDS-polyacrylamide gel. After electrophoresis, gels were prepared for fluorography and exposed to film. Viral proteins are labeled on the left, and positions of molecular size markers are shown on the right (in kilodaltons).
Effect of trypsin pretreatment on growth of PI rotaviruses in cured MX cells.
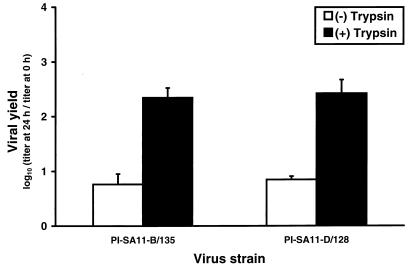
Since PI viruses produced equivalent yields in parental MA104 cells with and without trypsin treatment of the viral inoculum, we tested the effect of trypsin pretreatment on growth of PI viruses in cured MX cells. Monolayers of MX-S cells were infected with PI-SA11-B/135 and PI-SA11-D/128 at an MOI of 2 PFU per cell with and without trypsin pretreatment (Fig. 7). After 24 h of viral growth, trypsin-treated PI viruses produced approximately 40-fold greater yields than untreated viruses. Therefore, PI viruses require trypsin pretreatment to efficiently infect cured MX cells but not parental MA104 cells.
FIG. 7.
Effect of trypsin pretreatment on growth of PI viruses in cured MX cells. Monolayer cultures of MX-S cells (105 cells) were infected with PI-SA11-B/135 and PI-SA11-D/128 at an MOI of 2 PFU per cell. Prior to adsorption, each viral inoculum was either untreated or treated with 10 μg of trypsin per ml at 37°C for 30 min. After 1 h of adsorption, the inoculum was removed, 1.0 ml of fresh trypsinfree medium was added, and cells were incubated at 37°C for 24 h. Virus in cell lysates was titrated by plaque assay using MA104 cells. The results are presented as the mean viral yields (titer at 24 h divided by titer at 0 h) for four independent experiments. Error bars indicate standard deviations of the means.
DISCUSSION
MA104 cells can be persistently infected with rotavirus.
In this report, we show that MA104 cell cultures infected with rotavirus strain SA11 can support persistent infection for prolonged periods. After an initial period of crisis in which only a few cells survived, the cultures stabilized and produced substantial titers of infectious virus for 350 days of continuous passage. Treatment of persistent rotavirus infection of MA104 cells with antirotavirus antibodies resulted in cure of viral infection, which suggests that horizontal transmission of virus between cells is required for propagation of persistent rotavirus infection. During maintenance of the persistently infected cultures, mutant cells were selected that do not support efficient growth of wt virus and mutant viruses were selected that can infect the resistant cells. Furthermore, mutant viruses selected during these persistent infections do not require trypsin pretreatment to productively infect parental MA104 cells but do so to infect MA104 cells cured of persistent infection. These observations suggest that mutations in viruses and cells affecting a trypsin-sensitive step in rotavirus entry coevolve during maintenance of persistent infection of MA104 cells.
In comparison to persistent rotavirus infections established previously using AU-BEK bovine fetal kidney cells (13) and RK13 rabbit kidney cells (20), persistently infected MA104 cell cultures produce higher titers of infectious virus and a greater percentage of cells contain viral antigen. However, the vast majority of MA104 cells in the persistently infected cultures were viable after an initial period of crisis. By electron microscopy, cells in the persistently infected MA104 cell cultures exhibited significant morphological changes in comparison to parental cells; however, few cells in the cultures contained detectable viral particles. These observations suggest that viruses in the persistently infected cultures can exit the cell without producing lysis. This model would explain the occurrence of high viral titers in culture lysates and a large percentage of antigen-positive cells in the absence of large numbers of detectable viral particles by electron microscopy and significant alterations in cell viability. In support of this hypothesis, a nonlytic mechanism of viral egress has been shown for rotavirus infection of cultured Caco-2 intestinal epithelial cells (27).
Persistent rotavirus infections of MA104 cells are carrier cultures in which viruses and cells coevolve.
The finding that MA104 cell cultures persistently infected with rotavirus can be cured by treatment with neutralizing antirotavirus antibodies suggests that these cultures are carrier cultures maintained by horizontal transmission of virus between cells (17, 37). Neutralizing antibodies are believed to block early steps in viral replication, such as attachment, penetration, or disassembly (reviewed in reference 50). Rotavirus infectivity is neutralized by antibodies directed against outer capsid proteins VP4 and VP7 (6, 25, 26, 30, 47), and monoclonal antibodies directed against VP4 block viral attachment (43) and penetration (28). These steps would be required to maintain horizontal transmission of virus in a persistently infected culture. Therefore, the capacity of neutralizing antibodies to eradicate persistent infection suggests that lateral viral spread is required to maintain viral infection in these cultures.
During maintenance of persistent rotavirus infections of MA104 cells, mutations are selected in both viruses and cells: PI viruses grow better than wt virus in cured MX cells, and cured MX cells are less permissive for growth of wt virus than parental MA104 cells. These observations support a model of persistent infection which holds that during the initial rounds of viral replication in a persistently infected culture, cells manifesting resistance to viral replication are selected for their capacity to survive increasing viral titers (i.e., those cells surviving crisis). As the persistent infection is maintained, mutant viruses are selected that exhibit an augmented capacity to infect the resistant cells by bypassing the cellular block to viral replication. For these persistent infections to survive, an equilibrium must be reached between viral cytopathicity and cellular resistance in which ongoing viral replication is not sufficient to completely lyse the culture. Thus, these persistent infections also can be described as chronic infections in which lysis is restricted to a subset of cells.
Our results indicate that mutant MA104 cells and PI rotaviruses coevolve mutations that affect the requirement for trypsin treatment of the viral inoculum prior to initiation of infection. Treatment of rotavirus virions with trypsin increases the efficiency of viral penetration into the cell (22, 28, 39, 44). Therefore, it is likely that mutations in viruses and cells selected during persistent rotavirus infections of MA104 cells affect viral entry. Furthermore, selection of mutant viruses and cells altered in viral entry also supports a model of horizontal viral transmission for propagation of these persistent infections since entry steps would be required if viruses were transmitted horizontally in the cultures. Virus-cell coevolution affecting viral entry has been documented previously in studies of persistent coronavirus (11) and reovirus (17, 49) infections; however, a novel mechanism involving proteolysis of the viral attachment protein appears to be required for the maintenance of persistent rotavirus infection.
Mutations in PI viruses confer trypsin-independent growth.
Since trypsin-mediated proteolysis of VP4 is required for efficient entry of rotavirus into cells (22, 28, 39, 44), we reasoned that viruses having altered requirements for trypsin pretreatment might be selected during maintenance of the cultures. We found that PI viruses produce substantially greater yields than wt virus in parental MA104 cells in the absence of trypsin pretreatment. The trypsin-independent phenotype was selected in both persistently infected cultures tested and required approximately 100 days of culture maintenance to become fully manifest. These findings suggest that a protease requirement for efficient viral growth is a point of balance for viral growth and cellular survival during maintenance of persistent infection.
Several types of mutations might confer the trypsin-independent phenotype selected during persistent rotavirus infection of MA104 cells. Trypsin cleaves outer capsid protein VP4, resulting in the generation of particle-associated cleavage fragments VP5* and VP8* (15, 18, 21, 34). It is possible that mutations selected during persistent infection alter the VP4 cleavage site, rendering the protein susceptible to cleavage by other host proteases, such as those present on the cell surface (reviewed in reference 12). Alternatively, mutations selected during persistent infection might alter the fusion domain of VP5* to allow it to mediate membrane penetration in the absence of trypsin-mediated VP4 cleavage. It seems unlikely that progeny virions exiting infected cells contain cleaved VP4 proteins, as experiments using SDS-PAGE to assess viral structural proteins indicate that progeny virions contain intact VP4. However, the electrophoretic mobility of PI virus VP4 protein is altered in comparison to that of wt virus, which suggests that mutations were selected in PI virus VP4 during persistant infection. It is conceivable that mutations in other rotavirus proteins that interact with VP4, such as outer capsid protein VP7 or inner capsid protein VP6 (45, 52), might confer trypsin-independent viral growth. In support of this idea, heterologous VP4-VP7 pairings in rotavirus reassortants can affect the stability of rotavirus virions (10), alter antibody-binding domains of VP4 (9), and influence VP4-mediated viral attachment to cells (35).
The observation that PI viruses do not require trypsin pretreatment to produce high titers of viral progeny in parental MA104 cells, but do so to produce high titers in cured MX cells, establishes a strong link between trypsin-independent viral growth and maintenance of persistent infection. These findings suggest that a cellular mutation selected during persistent infection alters the requirement for VP4 proteolysis. Our results are consistent with a model in which establishment of persistent rotavirus infection of MA104 cells is associated with selection of viruses capable of utilizing a protease expressed at the cell surface in lieu of trypsin to cleave VP4. During maintenance of persistent infection, this protease might be down-regulated by MA104 cells to allow cell survival in a carrier culture. Thus, PI viruses would be predicted to grow well without trypsin pretreatment in parental cells in which the protease is expressed, but not in mutant cells in which the protease is down-regulated, consistent with the growth of PI viruses in this study. In support of this model, decreased expression of cell surface molecules required for viral entry has been demonstrated for other types of persistent infections. Mutant cells selected during persistent coronavirus (11), encephalomyocarditis virus (40), and poliovirus (7, 31) infections express decreased numbers of viral receptors, which results in attenuation of viral cytopathicity in cultures persistently infected with these viruses. Alternatively, it is possible that mutations selected in PI rotaviruses facilitate viral uptake into cells by receptor-mediated endocytosis, which is normally a nonproductive pathway for rotavirus entry (4). In this scenario, viral mutations affecting the requirement for VP4 proteolysis would lead to a dependence on proteases contained in the endocytic compartment and these proteases, rather than those expressed at the cell surface, would be the targets for mutations leading to cellular resistance to viral replication. In either case, our findings suggest that mutations in both viruses and cells selected during persistent rotavirus infections of MA104 cells lead to altered requirements for proteolysis to facilitate early steps in rotavirus replication. Ongoing studies of mutant viruses and cells selected during these persistent infections should increase our understanding of how rotavirus enters cells.
ACKNOWLEDGMENTS
We thank Mary Estes and Frank Ramig for essential discussions and Geoff Baer, Jim Chappell, and Frank Ramig for reviews of the manuscript. We also thank Sue Crawford and Sharon Tollefson for expert technical advice.
This research was supported by a postdoctoral fellowship from the International Pediatric Research Foundation (J.Z.M.), by Public Health Service awards AI05050 (P.F.W.) and AI32539 (T.S.D.) from the National Institute of Allergy and Infectious Diseases, and by the Elizabeth B. Lamb Center for Pediatric Research.
REFERENCES
- 1.Ahmed R, Morrison L A, Knipe D M. Persistence of viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 219–249. [Google Scholar]
- 2.Almeida J D, Hall T, Banatvala J E, Totterdell B M, Chrystie I L. The effect of trypsin on the growth of rotavirus. J Gen Virol. 1978;40:213–218. doi: 10.1099/0022-1317-40-1-213. [DOI] [PubMed] [Google Scholar]
- 3.Babiuk L A, Mohammed K, Spence L, Fauvel M, Petro R. Rotavirus isolation and cultivation in the presence of trypsin. J Clin Microbiol. 1977;6:610–617. doi: 10.1128/jcm.6.6.610-617.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass D M, Baylor M, Chen C, Upadhyayula U. Dansylcadaverine and cytochalasin D enhance rotavirus infection of murine L cells. Virology. 1995;212:429–437. doi: 10.1006/viro.1995.1500. [DOI] [PubMed] [Google Scholar]
- 5.Bass D M, Mackow E R, Greenberg H B. Identification and partial characterization of a rhesus rotavirus binding glycoprotein on murine enterocytes. Virology. 1991;183:602–610. doi: 10.1016/0042-6822(91)90989-o. [DOI] [PubMed] [Google Scholar]
- 6.Birch C J, Heath R L, Gust I D. Use of serotype-specific monoclonal antibodies to study the epidemiology of rotavirus infection. J Med Virol. 1988;24:45–53. doi: 10.1002/jmv.1890240107. [DOI] [PubMed] [Google Scholar]
- 7.Borzakian S, Couderc T, Barbier Y, Attal G, Pelletier I, Colbere-Garapin F. Persistent poliovirus infection: establishment and maintenance involve distinct mechanisms. Virology. 1992;186:398–408. doi: 10.1016/0042-6822(92)90005-a. [DOI] [PubMed] [Google Scholar]
- 8.Both G W, Bellamy A R, Siegman L J. Nucleotide sequence of the dsRNA genomic segment 7 of simian 11 rotavirus. Nucleic Acids Res. 1984;12:1621–1626. doi: 10.1093/nar/12.3.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D, Burns J W, Estes M K, Ramig R F. Phenotypes of rotavirus reassortants depend upon the recipient genetic background. Proc Natl Acad Sci USA. 1989;86:3743–3747. doi: 10.1073/pnas.86.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Ramig R F. Determinants of rotavirus stability and density during CsCl purification. Virology. 1992;186:228–237. doi: 10.1016/0042-6822(92)90077-3. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Baric R S. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol. 1996;70:3947–3960. doi: 10.1128/jvi.70.6.3947-3960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W T. Membrane proteases: roles in tissue remodeling and tumour invasion. Curr Opin Cell Biol. 1992;4:802–809. doi: 10.1016/0955-0674(92)90103-j. [DOI] [PubMed] [Google Scholar]
- 13.Chiarini A, Arista S, Giammanco A, Sinatra A. Rotavirus persistence in cell cultures: selection of resistant cells in the presence of foetal calf serum. J Gen Virol. 1983;64:1101–1110. doi: 10.1099/0022-1317-64-5-1101. [DOI] [PubMed] [Google Scholar]
- 14.Clark S M, Barnett B B, Spendlove R S. Production of high-titer bovine rotavirus with trypsin. J Clin Microbiol. 1979;9:413–417. doi: 10.1128/jcm.9.3.413-417.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark S M, Roth J R, Clark M L, Barnett B B, Spendlove R S. Trypsin enhancement of rotavirus infectivity: mechanisms of enhancement. J Virol. 1981;39:816–822. doi: 10.1128/jvi.39.3.816-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y J, Estes M K. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J Virol. 1994;68:5942–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dermody T S, Nibert M L, Wetzel J D, Tong X, Fields B N. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J Virol. 1993;67:2055–2063. doi: 10.1128/jvi.67.4.2055-2063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espejo R T, López S, Arias C. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J Virol. 1981;37:156–160. doi: 10.1128/jvi.37.1.156-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1625–1655. [Google Scholar]
- 20.Estes M K, Graham D Y. Establishment of rotavirus persistent infection in cell culture. Arch Virol. 1980;65:187–192. doi: 10.1007/BF01317330. [DOI] [PubMed] [Google Scholar]
- 21.Estes M K, Graham D Y, Mason B B. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falconer M M, Gilbert J M, Roper A M, Greenberg H B, Gavora J S. Rotavirus-induced fusion from without in tissue culture cells. J Virol. 1995;69:5582–5591. doi: 10.1128/jvi.69.9.5582-5591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiore L, Greenberg H B, Mackow E R. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology. 1991;181:553–563. doi: 10.1016/0042-6822(91)90888-i. [DOI] [PubMed] [Google Scholar]
- 24.Gallagher T M, Escarmis C, Buchmeier M J. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg H B, Valdesuso J, Wyke K V, Midthun K, Walsh M, McAuliffe V, Wyatt R G, Kalica A R, Flores J, Hoshino Y. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J Virol. 1983;47:267–275. doi: 10.1128/jvi.47.2.267-275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshino Y, Sereno M M, Midthun K, Flores J, Kapikian A Z, Chanock R M. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci USA. 1985;82:8701–8704. doi: 10.1073/pnas.82.24.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jourdan N, Maurice M, DeLautier D, Quero A M, Servin A L, Trugnan G. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol. 1997;71:8268–8278. doi: 10.1128/jvi.71.11.8268-8278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaijot J-K T, Shaw R D, Rubin D H, Greenberg H B. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J Virol. 1988;62:1136–1144. doi: 10.1128/jvi.62.4.1136-1144.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalica A R, Flores J, Greenberg H B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983;125:194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 30.Kalica A R, Greenberg H B, Wyatt R G, Flores J, Sereno M M, Kapikian A Z, Chanock R M. Genes of human (strain Wa) and bovine (strain UK) rotaviruses that code for neutralization and subgroup antigens. Virology. 1981;112:385–390. doi: 10.1016/0042-6822(81)90285-3. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan G, Levy A, Racaniello V R. Isolation and characterization of HeLa cell lines blocked at different steps in the poliovirus life cycle. J Virol. 1989;63:43–51. doi: 10.1128/jvi.63.1.43-51.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitaoka S, Fukuhara N, Suzuki H, Sato T, Konno T, Ebina T, Ishida N. Characterization of monoclonal antibodies against human rotavirus hemagglutinin. J Med Virol. 1986;19:313–323. doi: 10.1002/jmv.1890190404. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.López S, Arias C F, Bell J R, Strauss J H, Espejo R T. Primary structure of the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. Virology. 1985;144:11–19. doi: 10.1016/0042-6822(85)90300-9. [DOI] [PubMed] [Google Scholar]
- 35.Ludert J E, Feng N, Yu J H, Broome R L, Hoshino Y, Greenberg H B. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J Virol. 1996;70:487–493. doi: 10.1128/jvi.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dant M N, Greenberg H B. Characterization of the rhesus rotavirus VP3 gene: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahy B W J. Strategies of viral persistence. Br Med Bull. 1985;41:50–55. doi: 10.1093/oxfordjournals.bmb.a072024. [DOI] [PubMed] [Google Scholar]
- 38.Méndez E, Arias C F, López S. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J Virol. 1993;67:5253–5259. doi: 10.1128/jvi.67.9.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandi P, Charpilienne A, Cohen J. Interaction of rotavirus particles with liposomes. J Virol. 1992;66:3363–3367. doi: 10.1128/jvi.66.6.3363-3367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardoe I U, Grewal K K, Baldeh M P, Hamid J, Burness A T. Persistent infection of K562 cells by encephalomyocarditis virus. J Virol. 1990;64:6040–6044. doi: 10.1128/jvi.64.12.6040-6044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad B V V, Burns J W, Marietta E, Estes M K, Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional electron microscopy. Nature. 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 42.Ramig R F. Isolation and genetic characterization of temperature-sensitive mutants of simian rotavirus SA11. Virology. 1982;120:93–105. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 43.Ruggeri F M, Greenberg H B. Antibodies to the trypsin cleavage peptide VP8* neutralize rotavirus by inhibiting binding of virions to target cells in culture. J Virol. 1991;65:2211–2219. doi: 10.1128/jvi.65.5.2211-2219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz M C, Alonso-Torre S R, Charpilienne A, Vasseur M, Michelangeli F, Cohen J, Alvarado F. Rotavirus interaction with isolated membrane vesicles. J Virol. 1994;68:4009–4016. doi: 10.1128/jvi.68.6.4009-4016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw A L, Rothnagel R, Chen D, Ramig R F, Chiu W, Prasad B V V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith R E, Zweerink H J, Joklik W K. Polypeptide components of virions, top components and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi K, Urasawa T, Morita Y, Greenberg H B, Urasawa S. Direct serotyping of human rotavirus in stools using serotype 1-, 2-, 3-, and 4-specific monoclonal antibodies to VP7. J Infect Dis. 1987;155:1159–1166. doi: 10.1093/infdis/155.6.1159. [DOI] [PubMed] [Google Scholar]
- 48.Urasawa S, Urasawa T, Taniguchi K, Morita Y, Sakurada N, Saeki Y, Morita O, Hasegawa S. Validity of an enzyme-linked immunosorbent assay with serotype-specific monoclonal antibodies for serotyping human rotavirus in stool specimens. Microbiol Immunol. 1988;32:699–708. doi: 10.1111/j.1348-0421.1988.tb01431.x. [DOI] [PubMed] [Google Scholar]
- 49.Wetzel J D, Chappell J D, Fogo A B, Dermody T S. Efficiency of viral entry determines the capacity of murine erythroleukemia cells to support persistent infections by mammalian reoviruses. J Virol. 1997;71:299–306. doi: 10.1128/jvi.71.1.299-306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitton J L, Oldstone M B A. Immune responses to viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 345–374. [Google Scholar]
- 51.Wyatt R G, Greenberg H B, James W D, Pittman A L, Kalica A R, Flores J, Chanock R M, Kapikian A Z. Definition of human rotavirus serotypes by plaque reduction assay. Infect Immun. 1982;37:110–115. doi: 10.1128/iai.37.1.110-115.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeager M, Berriman J A, Baker T S, Bellamy A R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]