Abstract
Mason-Pfizer monkey virus (M-PMV), the prototype type D retrovirus, differs from most other retroviruses by assembling its Gag polyproteins into procapsids in the cytoplasm of infected cells. Once assembled, the procapsids migrate to the plasma membrane, where they acquire their envelope during budding. Because the processes of M-PMV protein transport, procapsid assembly, and budding are temporally and spatially unlinked, we have been able to determine whether cellular proteins play an active role during the different stages of procapsid morphogenesis. We report here that at least two stages of morphogenesis require ATP. Both procapsid assembly and procapsid transport to the plasma membrane were reversibly blocked by treating infected cells with sodium azide and 2-deoxy-d-glucose, which we show rapidly and reversibly depletes cellular ATP pools. Assembly of procapsids in vitro in a cell-free translation/assembly system was inhibited by the addition of nonhydrolyzable ATP analogs, suggesting that ATP hydrolysis and not just ATP binding is required. Since retrovirus Gag polyproteins do not bind or hydrolyze ATP, these results demonstrate that cellular components must play an active role during retrovirus morphogenesis.
Assembly and release of nascent retrovirus particles requires that the viral precursor polyproteins and genomic RNAs, and certain host cell tRNAs, migrate to the plasma membrane, where budding occurs. Two discrete intracellular transport pathways are utilized during the assembly of the infectious virion. The viral glycoproteins are synthesized on membrane-bound polysomes and are transported through the secretory pathway of the cell to the plasma membrane, where they colocalize with the immature capsid during the budding process (20). The major structural proteins of the viral capsid and the enzymatic proteins are synthesized in the cytoplasm on free polysomes and are transported to the underside of the plasma membrane (13, 36). While many of the details of the secretory pathway have been established, the mechanisms for intracytoplasmic protein transport are poorly understood.
The major structural polyprotein (Gag) of a nascent retrovirus capsid is encoded by the gag gene. Unlike most enveloped RNA viruses in which the viral glycoproteins mediate assembly by stabilizing the interactions between the capsid proteins and the viral membrane, retroviral Gag proteins can drive capsid assembly and budding in the absence of all the other viral gene products (19, 55, 58). As such, they contain all cis-acting information necessary for intracytoplasmic transport, capsid assembly, membrane binding, envelopment, and release from the cell surface. Assembly of the immature retrovirus capsid begins shortly after the Gag polyproteins are synthesized and modified by myristylation (15, 17, 40, 47–49). The Gag proteins of most retroviruses (the type C avian and mammalian viruses, lentiviruses, and human T-cell leukemia virus/bovine leukemia virus-related viruses) migrate directly to the plasma membrane, where they coalesce into spherical, immature capsids and simultaneously bud through the lipid bilayer, thereby acquiring their envelope. During or shortly after release, the Gag protein is cleaved by the viral protease into the internal structural (NH2-MA [matrix], CA [capsid], and NC [nucleocapsid]) proteins of the mature, infectious virion (22). In contrast, the Gag proteins of the mammalian and type B and D viruses (mouse mammary tumor virus [MMTV] and Mason-Pfizer monkey virus [M-PMV], respectively) accumulate in the cytoplasm, where they assemble into spherical structures in the absence of membranes. These nascent particles have been referred to as intracytoplasmic type A particles, but by analogy to other viruses and bacteriophages, we have redefined them as procapsids (55). Once assembled, procapsids are transported to the plasma membrane, from which they bud. Despite the different assembly strategies, the processes whereby Gag proteins assemble into procapsids are probably similar since a single amino acid change near the amino terminus of the Gag protein from M-PMV has been shown to convert it to the type C morphogenic pathway (41).
Genetic analyses of the gag genes from different retroviruses have shown that Gag proteins contain specific domains which are required for capsid formation. A membrane binding (M) domain has been located at the amino-terminal end of Gag of several retroviruses (31, 43, 60, 61). A late (L) domain functions during the budding and release. In Rous sarcoma virus (RSV) and M-PMV, the L domain is located between the MA and CA domains (57, 59). An equivalent domain in the lentiviruses has been found near the carboxy terminus of the Gag precursor (34). A third domain (I), located near the CA-NC junction, appears to be a region of interaction between Gag proteins (3, 56). Despite the lack of any extensive sequence similarities between different Gag proteins, there is functional conservation between assembly domains. Chimeric Gag proteins containing the M, L, and I domains from different retroviruses can assemble into capsid-like structures and mediate budding at the plasma membrane (3, 9, 10, 34).
The M-PMV Gag protein contains additional assembly elements which influence procapsid assembly, stability, and transport. This virus contains a region within Gag (known as p12) that is not found in either the type C viruses or lentiviruses. It has been suggested from biochemical data derived from studies with p12 deletion mutants that this domain assists in assembly by stabilizing intermolecular Gag associations (50). Protein stability and protein/procapsid transport depend on sequences in the MA domain which appear to be distinct from the M domain. As mentioned above, a single point mutation in MA at residue 55 results in a Gag protein that no longer assembles in the cytoplasm but rather assembles at the plasma membrane. This mutation lies within an 18-amino-acid region of the MA domain that has sequence similarity only to the type B retroviruses (41). The nuclear magnetic resonance-derived solution structure of a nonmyristylated M-PMV MA protein indicates that this region folds into a structured turn which is solvent accessible in the monomer and trimer models (8). Moreover, this structural feature is absent in human immunodeficiency virus (HIV), simian immunodeficiency virus, human T-cell leukemia virus, and bovine leukemia virus MA proteins (7, 18, 27–30, 37). It is reasonable, therefore, to suspect that this region contains a cytoplasmic protein transport signal which must interact with a cellular factor. In contrast, other mutations in either the myristic acid addition signal or at a variety of positions elsewhere in the MA coding region result in Gag proteins that fail to be released as virus-like particles despite assembling into procapsids in the cytoplasm (40, 43). Thus, the M-PMV Gag protein appears to contain a second cytoplasmic transport signal which normally directs assembled procapsids and not unassembled Gag proteins to the plasma membrane. It is implied in this model that the M-PMV Gag protein must utilize multiple cellular components during the different stages of assembly and release.
The type D retroviruses provide a useful system for studying morphogenic events since procapsid assembly, protein transport, and budding are temporally and spatially unlinked. We report here that in infected cells and an in vitro translation/assembly system, procapsid assembly and transport to the plasma membrane require ATP. Thus, cellular proteins do play an active role during at least two stages of M-PMV morphogenesis.
MATERIALS AND METHODS
Cells, viruses, and DNAs.
CMMT cells, which produce infectious M-PMV, were initially established by cocultivating rhesus mammary tumor cells with rhesus monkey embryo cells (1, 6). Monolayers of CMMT cells were cultured in RPMI 1640 supplemented with 5% tryptose phosphate broth and 10% fetal bovine serum (regular growth medium). For ATP depletion studies, cells were treated with 10 mM sodium azide and 6 mM 2-deoxy-d-glucose in glucose-free RPMI 1640. Plasmid pTFCG (44) was used to program M-PMV Gag polyprotein translations in vitro.
Radiolabeling.
Confluent monolayers of CMMT cells in 100-mm-diameter dishes were incubated for 20 min in methionine- and cysteine-free RPMI 1640. Cells were then pulse-labeled for 30 min at 37°C with [35S]methionine-cysteine protein labeling mix (0.2 mCi/ml, 1,175 Ci/mmol; NEN). In pulse-chase experiments, pulse-labeled cells were chased for various times in either regular growth medium or glucose-free RPMI 1640 containing the metabolic inhibitors described above.
Cell lysis, immunoprecipitation, and gel electrophoresis.
The proportion of M-PMV Gag precursors that assembled into procapsids was monitored at various times after labeling with [35S]methionine-cysteine as described previously (40, 50). Briefly, radiolabeled CMMT cells were lysed on ice in TX-100 lysis buffer (0.25 M sucrose, 1.0 mM EDTA, 10 mM Tris-HCl [pH 7.5], 0.14 M NaCl, 0.5% Triton X-100, 0.25% deoxycholate [DOC]). Assembled M-PMV procapsids were separated from the bulk of the cellular proteins by centrifugation through a 20% sucrose cushion at 350,000 × g for 20 min at 4°C. The pellet was lysed in 1 ml of 1× lysis buffer B (0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% DOC, 0.15 M NaCl, 0.05 M Tris-HCl [pH 7.5]). The supernatants containing unassembled Gag precursor polyproteins were adjusted to 1× lysis buffer B by the addition of SDS and DOC. Gag proteins present in the supernatant and pelleted fractions were collected by immunoprecipitation using a rabbit anti-Pr78gag antiserum (44). Immunoprecipitates were dissolved in sample buffer (10% glycerol, 2.3% SDS, 63 mM Tris-HCl [pH 6.8], 5% β-mercaptoethanol, 0.01% bromophenol blue), boiled for 4 min, separated on an SDS–12% polyacrylamide gel (54), and visualized by fluorography. The band intensities on the resulting fluorograms were quantitated by using a high-performance digital imaging system and AlphaImager 2000 software (Alpha Innotech Corp., San Leandro, Calif.). The intensities of each of the Gag precursor proteins and their cleavage products (Pr95, p68, and p27 [CA]) were normalized to Pr78gag equivalents from their relative methionine content.
Measurement of ATP in cells.
The ATP levels in cells were measured as described by Parker et al. (35). Briefly, 3 × 106 CMMT cells were collected by centrifugation and incubated on ice for 10 min with 0.1 ml of ice-cold 0.5 M perchloric acid. The acid-insoluble material was removed by centrifugation at 12,000 × g for 2 min. The supernatant was removed and neutralized with 12.5 μl of 4 M KOH and 7.5 μl of 1 M potassium phosphate (pH 7.5). Insoluble material was removed by centrifugation at 12,000 × g for 20 min, and the supernatant was analyzed by high-pressure liquid chromatography (HPLC) using a Partisil-10 SAX anion-exchange column. The nucleotides were eluted with a 50-min linear gradient from 5 mM NH4H2PO4 (pH 2.8) to 750 mM NH4H2PO4 (pH 3.7) with a flow rate of 2 ml/min. The nucleotides were detected by their absorbance at 254 nm. The concentrations of ATP in the samples were calculated by comparison to known ATP standards.
Electron microscopy.
For analysis of procapsid assembly, CMMT cells were fixed for 1 h in 1% glutaraldehyde in phosphate-buffered saline (pH 7.0) at room temperature. The cells were removed from the plates, washed several times in phosphate-buffered saline, and then postfixed in 1% buffered osmium tetroxide for 1 h. The cells were rinsed again and then dehydrated with increasing concentrations of ethanol beginning with 50% and ending with 100%. The cells were rinsed three times with propylene oxide and then embedded in Polybed. Ultrathin sections were made by using a Rechert-Jung Ultra Cut E ultramicrotome. Sections were stained with uranyl acetate and lead acetate, examined, and photographed in a Hitachi-7000 transmission electron microscope.
Transcription, translation, and procapsid assembly in vitro.
Simultaneous transcription and translation reactions were performed from plasmid pTFCG in the TnT coupled reticulocyte lysate system (Promega). For analysis of completed in vitro assembly reactions by sucrose gradient analysis, lysates were diluted to a total volume of 100 μl with 30% (wt/wt) sucrose in 20 mM Tris (pH 8.0)–100 mM NaCl–100 μM dithiothreitol–0.1% Triton X-100 (gradient buffer) and loaded onto 2.2 ml continuous 30 to 55% (wt/wt) sucrose gradients in gradient buffer. Gradients were centrifuged in a TLS-55 rotor (Beckman Instruments) for 2 h at 55,000 rpm. Approximately 200-μl fractions were taken by hand from the top of the gradient. The pellet was suspended in 200 μl of 55% (wt/wt) sucrose in gradient buffer. Aliquots (10 μl) of each fraction were dissolved in SDS sample buffer and then loaded onto an SDS–12.5% polyacrylamide gel. After polyacrylamide gel electrophoresis (PAGE), radioactive bands were visualized by fluorography.
RESULTS
To determine whether M-PMV assembly, transport, and release are active events that utilize cellular components and not merely passive events such as diffusion and self-assembly, we asked whether viral morphogenesis would still occur under conditions where cell-specific processes are inhibited. Since Gag proteins are not known to bind or hydrolyze ATP, we specifically asked whether M-PMV procapsids could assemble and bud from infected cells in the absence of ATP.
Sodium azide and 2-deoxy-d-glucose reversibly deplete ATP levels in M-PMV-infected cells without affecting cell viability.
It has been shown that short-term exposure of certain cells to a combination of sodium azide and 2-deoxy-d-glucose can rapidly and reversibly deplete ATP pools without adversely affecting cell viability (25, 53). Given that the half times of procapsid assembly and release are ∼45 min and 3.5 h, respectively (42, 43), we initially determined whether these metabolic inhibitors were lethal to M-PMV-infected (CMMT) cells within this time frame. CMMT cells were incubated in either (i) normal growth medium, (ii) glucose-free, serum-free medium, or (iii) glucose-free, serum-free medium containing 10 mM sodium azide and 6 mM 2-deoxy-d-glucose. In each case, over 95% of the cells remained viable after 8 h when examined by a trypan blue exclusion assay. When the drugs were replaced after 4 h with normal growth medium and the cells were incubated for an additional 24 h, more than 95% of the cells were viable (data not shown).
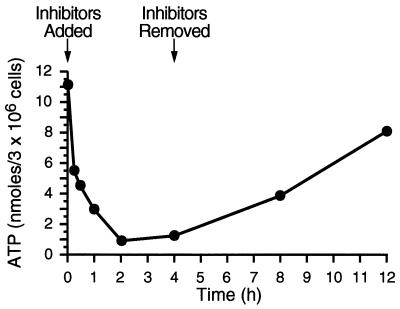
To determine whether ATP could be depleted prior to the onset of procapsid assembly and release, cells were treated with sodium azide and 2-deoxy-d-glucose for various times. Equal numbers of cells were harvested, washed, and lysed according to established protocols (35). The amount of ATP present in lysates was determined as described in Materials and Methods. As shown in Fig. 1, ATP levels dropped 4-fold within 1 h and more than 10-fold by 2 h. ATP remained at barely detectable levels until the metabolic inhibitors were removed at 4 h. Approximately 2 h after removal of the drugs, ATP levels began to rise, and by 12 h (8 h after inhibitors were removed), ATP levels had risen to within 70% of the mock-treated control sample levels.
FIG. 1.
Effects of metabolic inhibitors on the ATP levels in M-PMV-infected cells. CMMT cells were treated with 10 mM sodium azide and 6 mM 2-deoxy-d-glucose for increasing amounts of time. At various times, 3 × 106 cells were lysed and the amounts of ATP were measured by HPLC anion-exchange chromatography. The concentration of ATP in each sample was calculated based on the concentrations of known ATP standards.
ATP depletion inhibits procapsid assembly and blocks virus release.
Having shown that the cellular ATP pools were rapidly and reversibly depleted in the presence of the drugs, we asked whether assembly, transport, or release could occur in the absence of ATP. CMMT cells were pulse-labeled without drugs for 20 min. The rates at which these labeled Gag proteins were chased into procapsids and virus in either the absence or presence of the metabolic inhibitors were examined. CMMT cells produce three Gag-related polyproteins of 78, 95, and 180 kDa. The most abundant of these is the 78-kDa Gag polyprotein (Pr78gag). The 95- and 180-kDa proteins are the Gag-Pro (Pr95gag-pro) and Gag-Pro-Pol (Pr180gag-pro-pol) proteins which are synthesized via ribosomal frameshifting events. All three precursors are incorporated into procapsids and virions, but Pr180gag-pro-pol is made in very low amounts and is often difficult to detect by radiolabeling (see below). A smaller Gag-related protein of 68 kDa (p68) is also detected in M-PMV-infected cells. This protein originates from translational initiation from the second methionine codon in gag which is located at the end of the MA coding region (39).
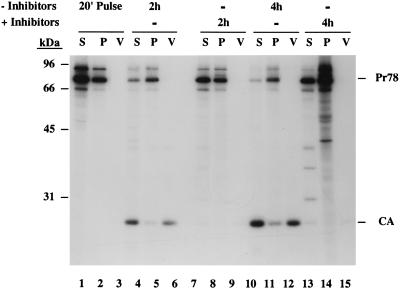
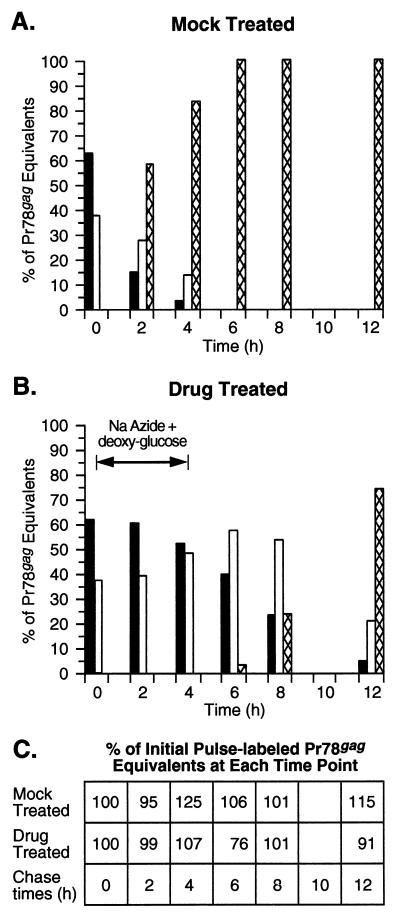
As shown in Fig. 2 (lanes 1 to 3), the Gag precursor polyproteins Pr78gag, Pr95gag-pro, and p68 were detected in both the soluble and pelleted fractions of pulse-labeled cells. These two fractions have been shown previously to contain unassembled Gag proteins and assembled procapsids, respectively (42). Of the total amount of these Gag precursors in pulse-labeled cells, 38% had already assembled into procapsids (Fig. 3A). No proteins were found in the virus pellet fraction at the end of the pulse-label. Following a 2-h chase in the absence of drugs, the total amount of cell-associated Gag precursors (both unassembled and assembled) had dropped to approximately 40% of that detected in the pulse (Fig. 3A). Of this amount, 65% was found in the procapsid-associated fraction. The decrease in cell-associated Gag proteins was primarily due to virus release rather than degradation since the total amount of Gag precursors and processed CA equivalents (normalized to Pr78gag equivalents based on methionine content) present in the cells and in virus is 95% of that in the pulse (Fig. 3C).
FIG. 2.
Kinetics of M-PMV procapsid assembly and virus release in the presence of the metabolic inhibitors. CMMT cells were pulse-labeled with [35S]methionine and cysteine for 20 min (lanes 1 to 3) and then chased for 2 h (lanes 4 to 9) and 4 h (lanes 10 to 15) in the absence (lanes 4 to 6 and 10 to 12) or presence of 10 mM sodium azide and 6 mM 2-deoxy-d-glucose (lanes 7 to 9 and 13 to 15). Cells were fractionated in TX-100 lysis buffer by centrifugation as described in Materials and Methods. Gag proteins present in the soluble (S [unassembled molecules]; lanes 1, 4, 7, 10, and 13) and pelleted fractions (P [assembled procapsids]; lanes 2, 5, 8, 11, and 14) and in virus (V; lanes 3, 6, 9, 12, and 15) were immunoprecipitated with anti-Pr78gag polyclonal serum. The positions of the molecular mass protein markers are indicated, as are the positions of Pr78gag and CA.
FIG. 3.
Graphic representation of the effects of metabolic inhibitors on M-PMV morphogenesis. The fluorograms shown in Fig. 2 and 4 were quantitated as described in Materials and Methods, and the band intensities of the Gag precursor polyproteins Pr95gag-pro, Pr78gag, and p68 as well as the processed p27 (CA) were normalized to Pr78gag equivalents with respect to their methionine content. The percentages of the total Pr78gag equivalents present in the soluble (solid bars) and pelleted fractions (open bars) and in virus pellets (cross-hatched bars) at each time point during the mock treatment (A) and drug treatment (B) are shown. (C) The amounts of Pr78gag equivalents in the soluble and pelleted lysate fractions and in the viral pellets at each time point were combined and are reported as the percentage of the initial pulse-labeled Gag proteins.
At the end of the 2-h chase, significant amounts of the 27-kDa mature CA were found in both cell lysates and virus. The CA protein is derived from Gag following the action of the viral protease. This processing of Gag takes place during or shortly after budding at the plasma membrane. Mutations that block transport to or budding from the plasma membrane prevent Gag processing (40, 43). The presence of CA in the cell-associated fraction is therefore a measure of virus budding and probably represents virus particles that have budded from the plasma membrane but remain cell associated during the fractionation protocol. Finding the majority of the cell-associated CA in the soluble fraction is common since cleaved Gag proteins in mature virions are easily solubilized with detergent (51). After the 4-h chase, only 17% of the pulse-labeled Gag precursors remained cell associated; of this amount, the majority (80%) fractionated with procapsids (Fig. 2, lanes 10 to 12; Fig. 3A). The reduction in total cell-associated Gag precursors and the increase in assembled procapsids were accompanied by increasing amounts of CA associated with cells and virus pellets. The kinetics of M-PMV procapsid assembly and release in CMMT cells (half-lives of 1 h and 3 to 4 h, respectively) are consistent with that previously observed in COS-1 and HeLa cells (43).
In contrast to normal virus morphogenesis, procapsid assembly and virus release were dramatically inhibited in cells treated with sodium azide and 2-deoxy-d-glucose. After 2 h of drug treatment, 99% of the Gag precursors present in the pulse-labeled cells could still be precipitated from the cell lysates (combined soluble and pelleted fractions) with the polyclonal antiserum (Fig. 3C). As was seen in the pulse-labeled cell extracts, approximately 40% of the radiolabeled Gag proteins had assembled into procapsids (Fig. 2, lanes 7 and 8; Fig. 3B). In contrast to the results observed after the 2-h chase in the absence of drugs, no Gag cleavage products were detected in either the cell lysates or virus pellets (Fig. 2, lanes 7 to 9). The absence of CA in these fractions suggests that those procapsids that had assembled during the pulse and the early time points of drug treatment did not reach the stage of budding from the plasma membrane. After 4 h of drug treatment, virtually 100% of the pulse-labeled Gag precursors could still be detected in the cell lysates (Fig. 3C). After subtracting the nonspecific and abnormally high background from the pelleted fraction (Fig. 2, lane 14), we found that the amount of cell-associated Gag which had assembled into procapsids had risen slightly to 48% (Fig. 3B). These procapsids, however, were not released from the cell as virus particles (Fig. 2, lane 15). Since Gag proteins are not known to bind or hydrolyze ATP, these results suggest that procapsid assembly and either protein transport to the plasma membrane or budding/release rely on cellular proteins that utilize ATP.
Restoration of assembly and release by removing the metabolic inhibitors.
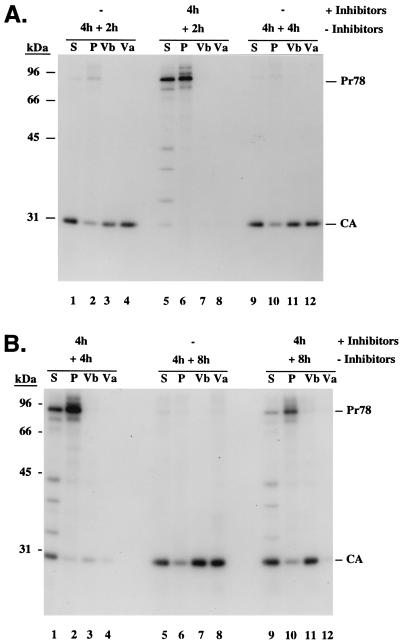
If the hypothesis that procapsid assembly, transport, and/or membrane associations is ATP dependent is correct, it would be reasonable to expect that these processes would resume if the cells were allowed to regenerate ATP. To examine this, parallel monolayers of pulse-labeled cells were chased in the absence or presence of drugs as described above except that after 4 h, the metabolic block was removed by replacing the drug-containing medium with normal growth medium. Cells were fractionated, and virus particles that had accumulated during the initial 4-h chase (Fig. 4, lanes Va) and during the following chase (Vb) were analyzed. After a 6-h chase in the absence of the drugs, more than 95% of the pulse-labeled Gag proteins had been processed (Fig. 3A), and approximately 50% of these were released as free virus particles (Fig. 4A, lanes 1 to 4). Mature virions remained associated with cells even after a 12-h chase, when 32% of the CA protein was found in the cell lysates (Fig. 4B, lanes 5 to 8).
FIG. 4.
Restoration of M-PMV morphogenesis by removing the metabolic block. Plates of CMMT cells identical to those used for Fig. 2 were pulse-labeled for 20 min and then chased in the absence (A, lanes 1 to 4 and 9 to 12; B, lanes 5 to 8) or presence of 10 mM sodium azide and 6 mM 2-deoxy-d-glucose (A, lanes 5 to 8; B, lanes 1 to 4 and 9 to 12) for 4 h. The culture media were replaced with normal growth medium without drugs, and the cells were further incubated for either 2 h (A, lanes 1 to 8), 4 h (A, lanes 9 to 12; B, lanes 1 to 4), or 8 h (B, lanes 5 to 12). Gag-specific proteins present in virus that had accumulated in the culture media during the initial 4-h chase (lanes Va) and the subsequent chase (Vb) and in the soluble (S) and pelletable (P [assembled procapsids]) fractions of cell lysates were immunoprecipitated with anti-Pr78gag polyclonal serum.
In the drug-treated plates, the block to procapsid assembly and release was relieved by removing the metabolic inhibitors. The amount of the labeled Gag precursors that remained soluble (40%) 2 h after the drugs were removed was less than that seen (52%) after 4 h of continuous drug treatment. This decrease in soluble Gag was accompanied by an increase in procapsid assembly but not virus budding (Fig. 4A, lanes 5 to 8; Fig. 3B). Four hours after the drugs were removed, the amount of unassembled Gag (23%) had decreased and there was a significant increase in the cell-associated and virus-associated CA protein (Fig. 4B, lanes 1 to 4). These changes are consistent with the rise in ATP levels after the drugs were removed (Fig. 1). By the point at which ATP levels had risen to within 70% of that of mock-treated cells (8 h after removal of the drugs), only 26% of pulse-labeled Gag precursors were found to be cell associated. The remainder of the Gag proteins had been processed into CA; of these, >70% were found in the virus pellets (Fig. 4B, lanes 9 to 12; Fig. 3B). The relative difference in the amount of Gag proteins present in the various fractions was not due to protein degradation but rather was due to renewed procapsid assembly and virion release since the total amount of labeled Pr78gag equivalents that was immunoprecipitated with the polyclonal antisera varied by <25% of that detected after the initial pulse-labeling (Fig. 3C).
In vitro assembly of M-PMV procapsids is dependent on ATP.
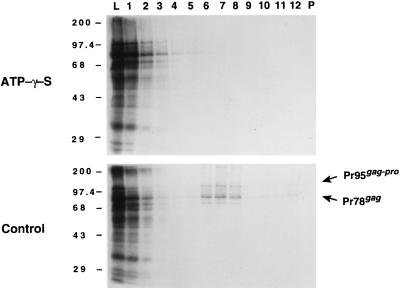
To further confirm our observation that M-PMV procapsid assembly requires ATP, we assayed whether Pr78gag could assemble into procapsids in vitro in the presence of a nonhydrolyzable ATP analog, ATPγS. We have shown previously that Pr78gag and Pr95gag-pro proteins can assemble in vitro into procapsid-like structures which are indistinguishable in density and appearance from authentic procapsids produced in mammalian cells (44). Furthermore, since we have shown that anti-Gag monoclonal antibodies can inhibit assembly, this system is suitable for testing potential inhibitors of this process. Coupled transcription and translation of Pr78gag and Pr95gag-pro was initiated in reticulocyte lysates and allowed to proceed for 30 min at 30°C to allow enough time for sufficient protein synthesis. At that time, cycloheximide and either ATPγS or water were added to the reaction mixture. The lysate was then incubated for an additional 90 min, after which the reactions were analyzed for procapsid assembly by gradient analysis and SDS-PAGE (Fig. 5). The absence of Pr78gag and Pr95gag-pro in the middle of the density gradient shows that the addition of ATPγS completely inhibited procapsid assembly (Fig. 5, top panel). In contrast, ∼50% of the Pr78gag and Pr95gag-pro precursors had assembled into procapsids that sedimented to a density of ∼1.2 g/ml in control samples (Fig. 5, bottom panel). Identical results were obtained with the similar nonhydrolyzable ATP analog AMP-PNP (data not shown). These data demonstrate that ATP hydrolysis and not just ATP binding is required for procapsid assembly.
FIG. 5.
Sucrose gradient analysis of M-PMV gag and gag-pro programmed in vitro translations. Equal volumes of gradient fractions were separated by SDS-PAGE (10% polyacrylamide gel) and visualized by fluorography. Lane numbers represent the gradient fractions beginning from the top (lane 1). Lane L contained an equivalent aliquot of the translation reaction mixture which was loaded onto each gradient; lane P contained the material which had pelleted in the tube. The electrophoretic positions of the molecular mass protein markers are indicated in kilodaltons on the left, and the positions of Pr78gag and Pr95gag-pro are shown on the right. The translation reactions which were treated with ATPγS (top panel) or H2O (control; bottom panel) are indicated.
Analysis of procapsid transport and membrane associations by electron microscopy.
While the in vitro data demonstrated that procapsid assembly requires ATP, the data from the pulse-chase experiments in the presence of the metabolic inhibitors indicated that a postassembly step was also dependent upon ATP. However, it was not possible to distinguish whether (i) transport of assembled procapsids to the plasma membrane, (ii) stable associations between the procapsids and the membrane, or (iii) budding was the ATP-dependent step. To discriminate between these possibilities, mock-treated and drug-treated cells were directly examined by electron microscopy. If procapsid transport was inhibited in the presence of the drugs, then one would expect to find a majority of the capsids in the cytoplasm and not associated with membranes. If either membrane association or budding was inhibited and procapsid transport was not, the majority of the procapsids would be expected to localize near the plasma membrane.
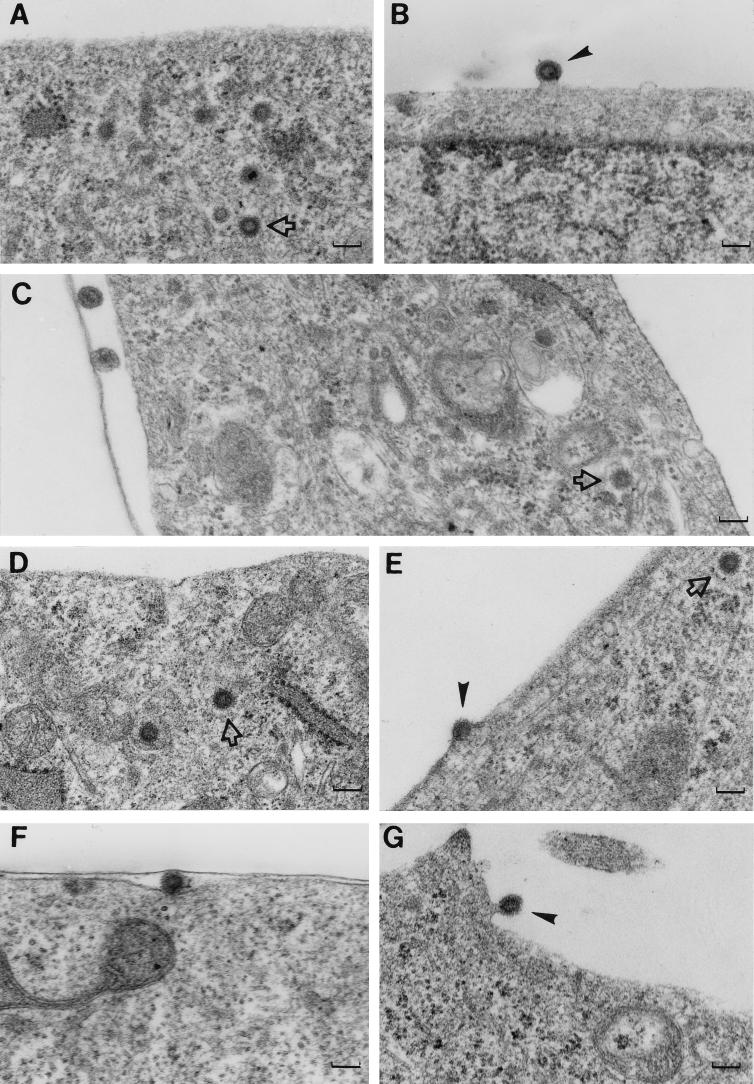
Numerous procapsids clustered in the cytoplasm and associated with the plasma membrane were detected in mock-treated cells (Fig. 6A and B). The cytoplasmic procapsids and budding particles clearly display the electron-dense doughnut shape characteristic of immature retrovirus cores. These structures were easily identified in essentially every cell examined. In contrast, detection of cytoplasmic procapsids and especially membrane-associated procapsids in drug-treated cells became progressively harder the longer the cells were treated. In cells that were treated for 1.5 h (Fig. 6C), several cytoplasmic procapsids were detected in approximately half of the cells (n ≈ 100 cells) that were examined. However, no membrane-associated forms were found. As shown, a few cell-free virus particles were found in these samples, but these displayed a morphology characteristic of mature particles (i.e., electron-dense cores). These particles were more than likely released from the cell prior to ATP depletion since the pulse-chase data demonstrated that particle release and Gag processing were inhibited in the presence of the inhibitors. After 3 h of drug treatment, ∼40% of the cells (n ≈ 200 cells) contained cytoplasmic procapsids. Out of all of these cells, only one membrane-associated procapsid was found (Fig. 6E). In contrast, multiple membrane-associated procapsids were found in cells that had been treated with drugs for 3 h and then incubated in their absence for 90 min (Fig. 6F and G). These data are consistent with the pulse-chase experiments (i.e., absence of cell-associated CA in the drug-treated cell lysates), and they indicate that transport of assembled procapsids to the plasma membrane is ATP dependent.
FIG. 6.
Electron micrographs of CMMT cells treated with the metabolic inhibitors. Thin sections of CMMT cells which had been mock treated or drug treated were examined by electron microscopy to determine at which stage processed assembly or release was arrested. (A and B) Sections of mock-treated cells; (C to G) sections of cells treated with drugs for 1.5 h (C), treated with drugs for 3 h (D and E), and treated with drugs for 3 h and then without drugs for 1.5 h (F and G). Arrow, cytoplasmic procapsids; arrowheads, budding virions. Magnification, ×51,000; bars, approximately 100 nm.
DISCUSSION
According to current models, release of a type D retrovirus particle requires at least six distinct posttranslational events: (i) transport of nascent Gag proteins from their sites of synthesis to the defined assembly domains in the cytoplasm, (ii) assembly of the procapsid, (iii) transport of the assembled procapsid through the cytoplasm to the plasma membrane, (iv) membrane binding, (v) capsid envelopment and budding, and (vi) membrane fusion to release the virus particle. Based on genetic evidence, some of these events are thought to be dependent on host cell factors. The ability of a single point mutation in the MA domain (R55W) to alter the site of assembly from the cytoplasm to the plasma membrane suggests that wild-type Gag proteins arrive at the cytoplasmic assembly sites by a specific transport mechanism (41). Certain point mutations elsewhere in the MA domain result in the accumulation of procapsids in the cytoplasm (40, 43). Transport of these mutant procapsids to the plasma membrane is apparently blocked. Taken together, the phenotypes of these mutants suggest that transport to and transport from the cytoplasmic assembly sites are both active processes driven by cellular components rather than a passive process such as diffusion. The data presented in this report not only support the concept of active transport through the cytoplasm but also provide evidence that cellular factors play an active role during procapsid assembly.
Pulse-chase experiments in conjunction with cell fractionation studies which separate unassembled (soluble) Gag proteins from assembled procapsids indicate that procapsid assembly can be inhibited with the metabolic inhibitors sodium azide and 2-deoxy-d-glucose. During the pulse-labeling, a significant proportion of the nascent Gag molecules had been incorporated into procapsids that could be pelleted by centrifugation. This was expected since previous work had shown that in HeLa cells and COS-1 cells which express the M-PMV Gag polyprotein, half of the newly synthesized Gag proteins are incorporated into procapsids within 45 min (42, 43). In the absence of drugs, there was a steady increase in procapsid-associated and virus-associated Gag during the chases. In contrast, the amount of pulse-labeled Gag that fractionated with assembled procapsids in the presence of drugs rose only slightly, and none was found associated with released virus. In fact, the block to assembly occurred within the amount of time it took for the drugs to deplete the ATP pools approximately fourfold. The ability to block assembly in vitro with nonhydrolyzable ATP analogs demonstrates that assembly itself requires ATP hydrolysis. Since these experiments focus on the assembly process itself, it is entirely possible that transport of newly synthesized Gag proteins to the assembly sites also requires ATP.
Because chaperonins facilitate protein folding in an ATP-dependent manner (16, 52), the requirement for ATP during M-PMV procapsid assembly raises the possibility that cytosolic chaperonins play an active role during procapsid morphogenesis. Such a role for chaperone-assisted capsid assembly has been demonstrated with hepatitis B virus (HBV). Lingappa et al. (24) found that HBV core proteins translated in vitro could assemble into structures which closely resembled authentic HBV cores. They also demonstrated, using sucrose gradients and coimmunoprecipitation experiments, that a significant portion of the HBV core proteins colocalized with CC60 (a TCP-1-related chaperone) in an ATP-dependent manner. TCP-1 is a subunit of a large hetero-oligomeric ring complex (CCT/TRiC) consisting of two rings of eight 55-kDa subunits (14, 16). Like other chaperones, TRiC mediates protein folding and requires ATP for the release of its substrate. Clearly the association of a substrate protein with TRiC would result in a complex that could be easily pelleted by centrifugation. In fact, HBV core proteins associated with CC60 were found at the bottom of sucrose gradients. Also, the HBV core protein could be chased out of the pellet (CC60 complex) and into fractions containing either HBV capsids or unassembled core proteins by manipulating the energy substrates (i.e., by addition of nucleoside triphosphates or apyrase) of the in vitro extracts. We have also found in vitro-translated M-PMV Gag proteins in the pellets of sucrose gradients (44). However, unlike the HBV core protein, the pelleted M-PMV protein appears to assemble into aberrant, dead-end structures that bear no resemblance to M-PMV capsids or TRiC ring-like structures. Since ATP is required to release substrate polypeptides from the TRiC complex, it would be expected that under limited ATP conditions, there would be an increase in the substrate-TRiC complex. To this end, addition of nonhydrolyzable ATP analogs to the in vitro translation resulted in an increase of material at the top of the gradient and not in the pellet. Thus, it is unlikely that Gag interacts with TCP-1. Other candidate cytosolic chaperones include the Hsp70/Hsp90 family. Determination of whether these cellular proteins assist in procapsid assembly awaits further experimentation.
The requirement for ATP during M-PMV procapsid assembly is not inconsistent with previous results which demonstrated that other retroviral Gag proteins can be purified and assembled into capsid-like structures under appropriate conditions without an apparent need for ATP. Campbell and Vogt have shown that RSV and HIV Gag proteins can be overexpressed in and purified from bacteria in a soluble form (4, 5). By manipulating the pH and ionic strength and by including RNA in the assembly reaction, it was shown that these proteins could self-assemble into spherical capsid-like structures. Since the RSV and HIV Gag proteins remained soluble in bacteria rather than accumulating in inclusion bodies, it is entirely possible that these proteins had been properly folded into assembly-competent forms by bacterial chaperone proteins prior to their purification. Thus, the requirement for ATP had already been met. In contrast, overexpression of the M-PMV Gag protein in bacteria results in the accumulation of capsid-like structures in inclusion bodies (21). Purification of Gag from the insoluble inclusion bodies required denaturation using 8 M urea. However, such denatured Gag proteins could be renatured and assembled into capsid-like structures by slowly removing the urea under the correct pH and ionic strength. This slow, controlled renaturation of Gag could conceivably supersede the proposed requirement for ATP-dependent (chaperone-assisted) protein folding.
ATP-dependent assembly is not unique to the type D retroviruses. Lingappa et al. (23) demonstrated that HIV type 1 (HIV-1) Gag proteins translated in vitro can assemble into capsids in a wheat germ extract but only in the presence of ATP. They further showed that assembly is dependent on both a detergent-insoluble and a detergent-soluble component present in the extracts. However, since capsid assembly and budding occur concomitantly during lentivirus morphogenesis and probably during assembly in vitro, it is difficult to distinguish at which step these cellular factors are being utilized. In contrast, we have shown directly that ATP hydrolysis is a prerequisite for the assembly of precursor proteins into the procapsid shell. In addition, we present several lines of evidence that assembled M-PMV procapsids arrive at the plasma membrane by an active process rather than by passive diffusion. While a portion of the nascent Gag proteins had assembled into procapsids during the pulse-labeling, these proteins failed to be released as virus particles upon ATP depletion. Based on the absence of cell-associated CA during the drug treatment and on the absence of membrane-associated procapsids in the electron micrographs, the block to particle release appears to occur prior to membrane binding. To this end, the phenotype of the procapsids during the drug treatment resembles that of the transport-defective mutant, A18V (43). Cells expressing this mutant contain an array of procapsids in the cytoplasm but none in association with the plasma membrane. Furthermore, the absence of cell-associated CA in A18V-expressing cells or in cells expressing a variety of other mutants that are incapable of budding from the plasma membrane indicates that the viral protease becomes active after binding the plasma membrane, perhaps during either envelopment or release. Based on the recent nuclear magnetic resonance-derived solution structure of a nonmyristylated M-PMV MA protein, the arginine at residue 18 lies within an alpha helix which has been proposed to be located on the membrane-proximal surface of the MA monomer and trimer models (8). Provided that a similar structural motif exists in the context of the myristylated Gag polyprotein, this region would be solvent accessible and therefore available to associate with the cellular transport machinery. This phenotypic similarity between the transport-defective procapsids and what was observed with wild-type procapsids during ATP depletion provides strong evidence that transport to the plasma membrane is an active process requiring recognition of procapsid structural elements by cellular components.
It has been suggested that elements of the cytoskeleton may provide a motive force for delivering certain Gag molecules to the site of assembly or for budding. Actin has been found in purified HIV-1 and MMTV virions (2, 32), and cell fractionation studies of HIV-1 and murine leukemia virus-infected cells have shown that Gag proteins cosediment with detergent-insoluble material which contains cytoskeletal proteins and membrane components (11, 12, 26, 38). Furthermore, the efficiency of HIV-1, MMTV, and Moloney murine leukemia virus particle release can be diminished (but not completely blocked) pharmacologically by using cytochalasins, colchicine, and wortmannin (26, 33, 38, 45, 46). These data suggest that the cytoskeleton may play a role during capsid morphogenesis. However, a specific interaction between the cytoskeleton and Gag that is relevant to protein/procapsid transport or assembly has not yet been demonstrated. Based on the electron micrographs presented in this report as well as on numerous others of M-PMV (both published and unpublished), we have not observed procapsids in close association with either cytoskeletal elements or membranes other than the plasma membrane. Nonetheless, we are exploring whether microtubule or microfilament destabilization has any effect on the efficiency or the rate of M-PMV procapsid assembly and release.
ACKNOWLEDGMENTS
We are grateful to Eugene Arms and Dale Abrahamson at the UAB Comprehensive Cancer Center Electron Microscopy Core Facility for excellent assistance with electron microscopy and to Lucy M. Rose at the Southern Research Institute (Birmingham, Ala.) for assistance in ATP quantitations. We also thank Sally Weldon and John West for critical reading of the manuscript.
This work was supported by Public Health Service grants AI09054 to R.A.W., AI29157 to W.B.P., AI093001 to M.S., and CA-27834 to E.H.
REFERENCES
- 1.Ahmed M, Mayyasi S A, Chopra H C, Zelljadt I, Jensen E M. Mason-Pfizer monkey virus isolated from spontaneous mammary carcinoma of a female monkey. I. Detection of virus antigens by immunodiffusion, immunofluorescent, and virus agglutination techniques. J Natl Cancer Inst. 1971;46:1325–1334. [PubMed] [Google Scholar]
- 2.Arthur L O, Bess J, Jr, Sowder II R C, Benveniste R E, Mann D L, Chermann J C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 3.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra H C, Mason M M. A new virus in a spontaneous mammary tumor of a rhesus monkey. Cancer Res. 1970;30:2081–2086. [PubMed] [Google Scholar]
- 7.Christensen A M, Massiah M A, Turner B G, Sundquist W I, Summers M F. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J Mol Biol. 1996;264:1117–1131. doi: 10.1006/jmbi.1996.0700. [DOI] [PubMed] [Google Scholar]
- 8.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deminie C A, Emerman M. Incorporation of human immunodeficiency virus type 1 Gag proteins into murine leukemia virus virions. J Virol. 1993;67:6499–6506. doi: 10.1128/jvi.67.11.6499-6506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deminie C A, Emerman M. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J Virol. 1994;68:4442–4449. doi: 10.1128/jvi.68.7.4442-4449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and retrovirus assembly. Virology. 1983;130:415–426. doi: 10.1016/0042-6822(83)90096-x. [DOI] [PubMed] [Google Scholar]
- 12.Edbauer C A, Naso R B. Cytoskeleton-associated Pr65gag and assembly of retrovirus temperature-sensitive mutants in chronically infected cells. Virology. 1984;134:389–397. doi: 10.1016/0042-6822(84)90306-4. [DOI] [PubMed] [Google Scholar]
- 13.Eisenman R N, Vogt V M, Diggelmann H. Synthesis of avian RNA tumor virus structural proteins. Cold Spring Harbor Symp Quant Biol. 1974;39:1067–1075. doi: 10.1101/sqb.1974.039.01.122. [DOI] [PubMed] [Google Scholar]
- 14.Frydman J, Nimmesgern E, Erdjument-Btomage H, Wall J S, Tempst P, Hartl F. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type I. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 17.Henderson L E, Krutzsch H C, Oroszlan S. Myristyl amino-terminal acylation of murine retrovirus proteins: an unusual post-translational protein modification. Proc Natl Acad Sci USA. 1983;80:339–443. doi: 10.1073/pnas.80.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 20.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–254. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 21.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leis J, Baltimore D, Bishop J M, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald H R, Kock C J. Energy metabolism and T-cell-mediated cytolysis. J Exp Med. 1977;146:698–709. doi: 10.1084/jem.146.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldarelli F, King N W, Jr, Yagi M J. Effects of cytoskeletal disrupting agents on mouse mammary tumor virus replication. Virus Res. 1987;7:281–295. doi: 10.1016/0168-1702(87)90043-8. [DOI] [PubMed] [Google Scholar]
- 27.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 28.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, Kingsman S, Kingsman A, Campbell I. Structural similarity between the p17 matrix protein of HIV-1 and interferon-gamma. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 29.Matthews S, Barlow P, Clark N, Kingsman S, Kingsman A, Campbell I. Refined solution structure of p17, the HIV matrix protein. Biochem Soc Trans. 1995;23:725–729. doi: 10.1042/bst0230725. [DOI] [PubMed] [Google Scholar]
- 30.Matthews S, Mikhailov M, Burny A, Roy P. The solution structure of the bovine leukemia virus matrix protein and similarity with lentiviral matrix proteins. EMBO J. 1996;15:3267–3274. [PMC free article] [PubMed] [Google Scholar]
- 31.Nelle T D, Wills J W. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panem S. Cell cycle-dependent inhibition of Kirsten murine sarcoma-leukemia virus release by cytochalasin B. Virology. 1977;76:146–151. doi: 10.1016/0042-6822(77)90291-4. [DOI] [PubMed] [Google Scholar]
- 34.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker W B, Shaddix S C, Chang C H, White E L, Rose L M, Brockman R W, Shortnacy A T, Montgomery J A, Secrist J, Bennett L., Jr Effects of 2-chloro-9-(2-deoxy-2-fluoro-beta-d-arabinofuranosyl)adenine on K562 cellular metabolism and the inhibition of human ribonucleotide reductase and DNA polymerases by its 5′-triphosphate. Cancer Res. 1991;51:2386–2394. [PubMed] [Google Scholar]
- 36.Purchio A F, Jovanovich S, Erikson R L. Sites of synthesis of viral proteins in avian sarcoma virus-infected chicken cells. J Virol. 1980;35:629–636. doi: 10.1128/jvi.35.3.629-636.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 38.Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 39.Rhee, S. S., and E. Hunter. Unpublished results.
- 40.Rhee S S, Hunter E. Myristylation is required for intracellular transport but not for assembly of D-type retrovirus capsids. J Virol. 1987;61:1045–1053. doi: 10.1128/jvi.61.4.1045-1053.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 42.Rhee S S, Hunter E. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J Virol. 1990;64:4383–4389. doi: 10.1128/jvi.64.9.4383-4389.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhee S S, Hunter E. Amino acid substitutions within the matrix protein of type D retroviruses affect assembly, transport, and membrane-association of a capsid. EMBO J. 1991;10:535–546. doi: 10.1002/j.1460-2075.1991.tb07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasaki H, Nakamura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satake M, Luftig R B. Microtubule-depolymerizing agents inhibit Moloney leukemia virus production. J Gen Virol. 1982;58:339–349. doi: 10.1099/0022-1317-58-2-339. [DOI] [PubMed] [Google Scholar]
- 47.Schultz A, Henderson L E, Oroszlan S. Fatty acylation of proteins. Annu Rev Cell Biol. 1988;4:611–647. doi: 10.1146/annurev.cb.04.110188.003143. [DOI] [PubMed] [Google Scholar]
- 48.Schultz A M, Rabin E H, Oroszlan S. Post-translational modification of Rauscher leukemia virus precursor polyproteins encoded by the gag gene. J Virol. 1979;30:255–266. doi: 10.1128/jvi.30.1.255-266.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz A M, Rein A. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J Virol. 1983;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sommerfelt M A, Rhee S S, Hunter E. Importance of p12 protein in Mason-Pfizer monkey virus assembly and infectivity. J Virol. 1992;66:7005–7011. doi: 10.1128/jvi.66.12.7005-7011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart L, Schatz G, Vogt V M. Properties of avian retroviral particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd M J, Viitanen P V, Lorimer G H. Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 53.Trinchieri G, De Marchi M. Antibody-dependent cell-mediated cytotoxicity in humans. II. Energy requirement. J Immunol. 1975;115:256–260. [PubMed] [Google Scholar]
- 54.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weldon R A, Jr, Hunter E. Molecular requirements for retrovirus assembly. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1996. pp. 381–410. [Google Scholar]
- 56.Weldon R A, Jr, Wills J W. Characterization of a small (25-kDa) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 59.Yasuda, J., and E. Hunter. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 60.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]