Abstract
The paramyxovirus genome, a nonsegmented, negative-polarity, single-stranded RNA of ∼15 kb, contains six transcription units flanked at the 3′ and 5′ ends by a short (∼ 50- to 60-nucleotide) extracistronic sequence, dubbed the positive and negative leader regions. These leader template regions, present at the 3′ end of the genome and the antigenome, have been shown to contain essential signals governing RNA replication activity. Whether they are sufficient to promote replication is still open to question. By using a series of Sendai virus defective interfering RNAs carrying a nested set of deletions in the promoter regions, it is shown here that for both the genomic and antigenomic promoters, a 3′-end RNA sequence of 96 nucleotides is required to allow replication. Sequence comparison of active and inactive promoters led to the identification of a set of three nucleotide hexamers (nucleotides 79 to 84, 85 to 90, and 91 to 96) containing a repeated motif RXXYXX [shown as 5′-3′ positive-strand]. Sequential mutation of each hexamer into its complementary sequence confirmed their essential role. The three hexamers are required, and their relative positioning is important, since displacing them by 6 nucleotides destroyed promoter function. RNAs carrying degenerate nucleotides in the three hexamers were used as replication templates. They led to the selection of actively replicating RNA species exclusively carrying the basic motif (GNNNNN)3 from nucleotides 79 to 96. These results clearly show that, apart from the region from nucleotides 1 to 31, previously identified as governing Sendai virus replication activity, a second element, spanning at the most nucleotides 79 to 96, appears essential. Thus, the paramyxovirus replication promoters are not confined to the leader template regions, as seems to be the case for the rhabdoviruses.
The genome of viruses belonging to the Paramyxovirinae subfamily is a nonsegmented single-stranded RNA (∼15 kb) of negative polarity, tightly assembled with more than 2,600 subunits of the nucleocapsid protein in a linear structure of helicoidal symmetry. The nucleocapsid is the biologically active viral genome. In general, it contains six transcription units, flanked at the 3′ and 5′ ends by two short extracistronic sequences, dubbed the positive and negative leader regions (the latter is also called the trailer region). During replication, the viral RNA is copied into its full complement, the antigenome, which is also in the form of a nucleocapsid. The leader template regions present at the 3′ end of the genome and the antigenome RNAs are known to contain essential signals governing genome replication and so represent genome and antigenome promoters for RNA replication. It is noteworthy that the promoter in the present context refers not only to the 3′-end nucleotides of the template RNA but also to the 5′-end nucleotides of the nascent RNA. This because the encapsidation efficiency of this nascent RNA is also a factor which governs replication (for a general review on paramyxovirus replication, see reference 30). The detailed participation of either RNA sequence still needs to be determined precisely, beyond the general assessment that the template 3′ end must be involved in binding the polymerase while the nascent RNA 5′ end could control replication elongation through encapsidation efficiency.
Sendai virus (SeV), a member of the Paramyxovirus genus in the Paramyxovirinae subfamily, matches these general features. Its genome is 15,384 nucleotides (nt) long with the positive and negative leaders containing 55 and 57 nt, respectively (see Fig. 1A). The six transcription units encode the six major structural proteins, from 3′ to 5′, the nucleocapsid protein (N), the phosphoprotein (P), the matrix protein (M), the envelope proteins (F and HN), and the large protein (L), and these genes represent the minimal essential genes of this virus subfamily. In many paramyxoviruses, the P gene codes for additional proteins from extra transcription units or from alternative expression mechanisms (30). The SeV P gene encodes as many as eight different polypeptides via overlapping open reading frames, mRNA editing, leaky ribosomal scanning, and a ribosomal shunt (11, 12, 24, 29). Among these extra proteins, the C proteins are a nested set of four polypeptides (called C′, C, Y1, and Y2), initiated at different translation start sites (in the 1+ frame relative to P) but having a common C-terminal end. These accessory C proteins have recently been shown to act on the replicating complex by increasing its selectivity for signals governing its function (43).
FIG. 1.
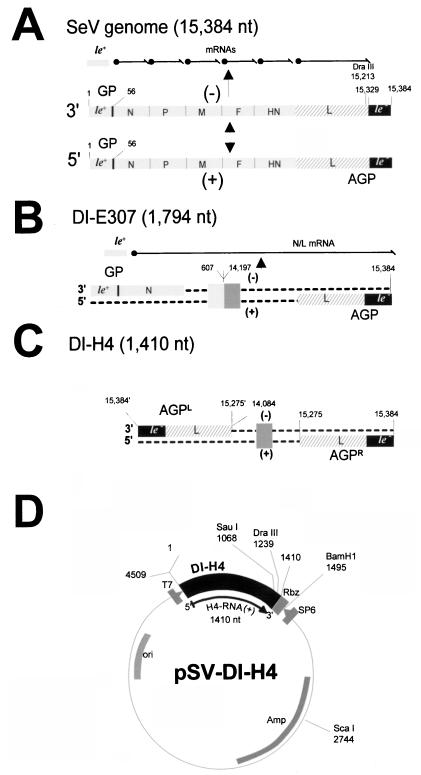
SeV RNAs and template plasmid structure. (A) The SeV genome (negative-strand RNA of 15,384 nt) is presented 3′ to 5′ as a grey rod, with the main transcription units indicated (not to scale). At each end, le+ and le− refer to the extracistronic regions, with the promoter for antigenome synthesis indicated (GP). The DraIII site, central to this study, is shown. Above, the transcription products include the le+ RNA and the six mRNAs, whose synthesis sequentially proceeds from the same GP region. Below, the antigenome is schematized exactly complementary to the full-length genome, with the AGP at its 3′ end. (B) The structure of the internal deletion DI RNA E307 is shown as negative and positive strands to emphasize the remnant GP and AGP promoter sequences, as in the SeV genome. A deletion from nt 607 to 14197 results in the fusion of the N and L genes, so that transcription produces on top of the le+ RNA a fused N/L mRNA. The E307 sequence is used in this study only as the donor of GP sequences. (C) The copy-back DI-H4 RNA structure emphasizes the presence, at the negative- strand 3′ end, of the same promoter sequence region (110 nt, nt 15275 to 15384) as that of the positive-strand 3′ end. Both strands contain in this way the same promoter, named AGPL and AGPR, respectively. No mRNA is transcribed from DI RNA. The box in the middle of the sequence scheme is there to specify the backbone sequence of the viral RNA, i.e., L sequence only for H4, N and L sequences for E307 (see panel B). Numbering for panels B and C corresponds to that of the full-length genome in panel A. (D) The H4 sequence is shown in the pSP65 plasmid, where it has been cloned along with the T7 promoter sequence. T7 transcription, which starts at the exact viral RNA 5′ end, produces an exact positive-strand DI RNA through endolytic cleavage at the 3′ end by the hepatitis delta virus ribozyme. Relevant restriction sites and positions are indicated. The numbering has been adjusted here to the H4 sequence inserted in plasmid pSP65.
SeV, in common with many viruses (for a review, see reference 22), very readily produces deleted versions of the genome during rapid multiplication. These can be amplified by the replication complex expressed by the nondeleted (ND) genome, and since they replicate better than the ND genome, they are referred to as defective interfering (DI) RNAs. These molecules have been valuable in the development of reverse genetics of the negative-stranded RNA viruses. Due to their small size, they were the first viral RNA templates to be successfully replicated when they were transcribed from DNA plasmids in a system in which the replicating complex (the L, P, and N proteins) was itself expressed from plasmids (for a review, see reference 9). Two DI RNAs have been instrumental in the study of SeV. The first is an internal deletion DI RNA (see Fig. 1B) (DI-E307), which retained the ND RNA ends (i.e., the genomic [GP] and antigenomic [AGP] promoters). E307, as expected from its primary structure, was shown to have retained its transcription potential (18). The second is a copy-back DI RNA (DI-H4), which lacks transcription potential through the loss of the genomic promoter. This is replaced by 110 nt identical to those present at the plus-strand 3′ end, so that DI-H4 contains antigenomic promoters on both the minus (AGPL) and the plus (AGPR) strands (see Fig. 1C) (4). When the two RNAs were compared, DI-H4 was found to replicate 20-fold better than E307 (6). Since the two RNAs have different promoters at the minus-strand 3′ end, this difference in replication was postulated to be due to the possession of the stronger AGP promoter on both the positive and negative-strands of DI-H4. Experiments involving reciprocal sequence exchanges between the two promoters supported this conclusion, since there was a direct correlation between the amount of either promoter present and the replication ability of the chimeric RNAs: the larger the AGP sequence, the greater the replication and vice versa (6). In a further study, it was shown that the first 31 nt of AGP are responsible for the high replication property (10- to 20-fold) (44). Other features of the promoters were unravelled by using these RNA derivatives, including the absence of an interference of transcription on the replication ability (interference that could have explained the lower replication ability of a transcribing DI RNA [6]) and the lack of influence of the extent of RNA terminal complementarity (44), which had been postulated to positively influence replication (47).
The reciprocal sequence exchanges made to create these RNA derivatives resulted in the juxtaposition of sequence from each promoter, making it difficult to dissect the relative contributions of the components of the “hybrid” promoters to their activity. In fact, the preparation of derivatives with exact reciprocal exchanges required this complementation between promoter components. Therefore, this approach did not provide firm information about the minimal sequence length required for an active replication promoter. This minimal length was originally predicted to be that of the leader template regions, since these do not participate in the coding capacity. This prediction was recently verified for vesicular stomatitis virus (VSV), a virus of the Rhabdoviridae family having many features in common with the Paramyxoviridae (32). In the case of the paramyxoviruses, this question is still open, and there are some indications that the paramyxoviruses may differ in that respect from the rhabdoviruses. First, the primary structure of the naturally obtained defective RNAs shows that the shortest end sequence remnant of the nondefective genome is 94 nt (i.e., extending past the leader regions), with a range between 94 and 168 nt (4, 17, 31, 37, 41). This contrasts with VSV, for which at least one copy-back DI RNA has been shown to contain only the negative leader template region (45 nucleotides [38]), with a range varying between 45 and 150 nt (22). Second, a sequence homology between the first 3′-end nucleotides of SeV and a stretch of 15 to 20 nt further inward than the leader template region (nt 75 through 90) led to the speculation that these nucleotides could be part of the promoter (BB3 box [1, 10]). Third, in addition to the demonstration that AGP nt 1 to 31 confer high replication efficiency in SeV, a second region, broadly defined as extending from nt 48 to 98 (i.e., extending past the leader template region) was found to participate in the modulation of replication efficiency (44). Fourth, the inhibition of replication resulting from an 18- or 12-nt insertion between nt 47 and 67 of GP of an E307 derivative was interpreted as the forbidden displacement of a putative promoter element located downstream of nt 67 (36). These results were supported by a recent study, in which a 6-nt insertion at this position was found to obliterate replication under defined conditions (43). Finally, it may be significant that all the artificial paramyxovirus minigenomes that were created and reported to successfully replicate when expressed from DNA plasmids contained promoters of more than 100 nucleotides (7, 8, 15, 42, 48).
The present study shows that the minimal length of the genomic and antigenomic promoters of SeV extends past the leader template regions to nt 91, at the least. A motif, consisting of 5′-G79(N)5-G85(N)5-G91(N)5-3′ (shown as plus-strand DNA) is identified, and its position cannot be displaced by 6 nt to either side. This identification was made by site-directed mutagenesis and by sequence analysis of RNAs selected by efficient replication from a pool of template RNAs containing degenerate nucleotides at these critical positions.
MATERIALS AND METHODS
Virus and cells.
Cells (A549 and HeLa) were grown under a 5% CO2 atmosphere in regular minimal essential medium (MEM) supplemented with 5% fetal calf serum. Recombinant vaccinia virus expressing the T7 RNA polymerase, vTF7-3, a gift from Bernard Moss (National Institutes of Health, Bethesda, Md.) was described by Fuerst et al. (20) and used as specified. vTF7-3 stocks were grown in HeLa cells with titers ranging from 5 × 108 to 1 × 109 PFU/ml.
Sequence and plasmids.
The complete SeV RNA primary sequence (15,384 nt) is taken from the work of Shioda et al. (39, 40) and Neubert et al. (35). The sequence is written as DNA according to standard convention, 5′ to 3′ from left to right. The plasmids expressing SeV N (pGem-N), P plus C (pGem-P/C, here referred to as PCwt), and L (pGem-L) under the control of the T7 RNA polymerase promoter have been described previously (11, 14, 23, 24).
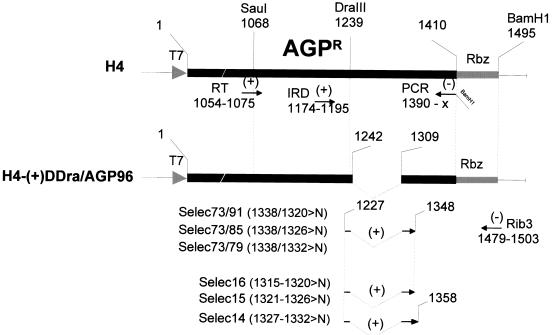
The structures of the SeV DI-H4 and E307 RNAs are presented in Fig. 1B and C, in relation to the nondefective RNA (Fig. 1A). The cloning of these DI RNAs in pSP65 (respectively pSV-DI-H4 and pSV-E307) under the control of the T7 RNA polymerase promoter has been described previously (4, 6, 18), and a schematic representation of pSV-DI-H4 is shown in Fig. 1D. As before (6, 44), the 3′ end of the viral genome is called the genomic promoter (GP) and the 3′ end of the antigenome is called the antigenomic promoter (AGP). In the case of H4 RNA, which contains the AGP on both the minus- and plus-strand RNA, the distinction between the two promoters is made by using AGPL (left) or AGPR (right). In all the derivatives used in this study, only the 3′ end of the H4 positive- strand RNA (AGPR) has been modified. H4-AGP(+)57 was obtained by replacing the DraIII-BamHI fragment pSV-DI-H4 (see Fig. 2) with that of H4-AGP(−/+)57 (44). The DI-H4s of the ΔDra/GP series (see Fig. 3 and 4) were constructed by replacement of the same H4 DraIII-BamHI fragment with DraIII-BamHI fragments obtained by PCR amplification from either H4-AGP(−)120/(+)98 (44) or H4-AGP(−/+)57 with DraIII carrying oligonucleotides adjusted to match the deletions and Rib3 as amplimers (see Fig. 2). The DI-H4s of the ΔDra/AGP series carrying deletions (see Fig. 3 and 6), site-directed mutations (see Fig. 5), or degenerate positions (see Fig. 7 and 8) were constructed by replacement of the H4 DraIII-BamHI fragment with a PCR product obtained with primer Rib3 and the corresponding primer spanning the DraIII site carrying the appropriate sequence, with pSV-E307 as the template (see Fig. 2, lower part, where only the primers for the degenerate positions are shown). For H4-AGP(+)57 (see Fig. 3), in which the first 57 nt of the H4 plus-strand 3′ end (AGPR) was replaced by 57 nt from E307 GP, the previously used nomenclature applies (detailed in reference 44). For all the other H4 derivatives, a description of their structure is shown in the figures, along with their names. It is noteworthy that all the derivatives used in this study contain a total number of nucleotides that is a multiple of 6 to comply with the rule of six (5).
FIG. 2.
Primers. The primers (RT and PCR), used for the RT-PCR amplification of a fragment covering the AGPR region of DI RNA derivatives in verification of the selection experiment results (see Fig. 7 and 8), are shown below the schematized H4 sequence, along with the IRD primer used for the sequencing. Below the H4-(+)ΔDra/AGP96 schematized sequence, the series of oligonucleotides used to construct the plasmids containing the degenerate nucleotides are outlined, with Rib3 commonly used. Numbering refers to positions in the pSV-DI-H4 plasmid (Fig. 1D).
FIG. 3.
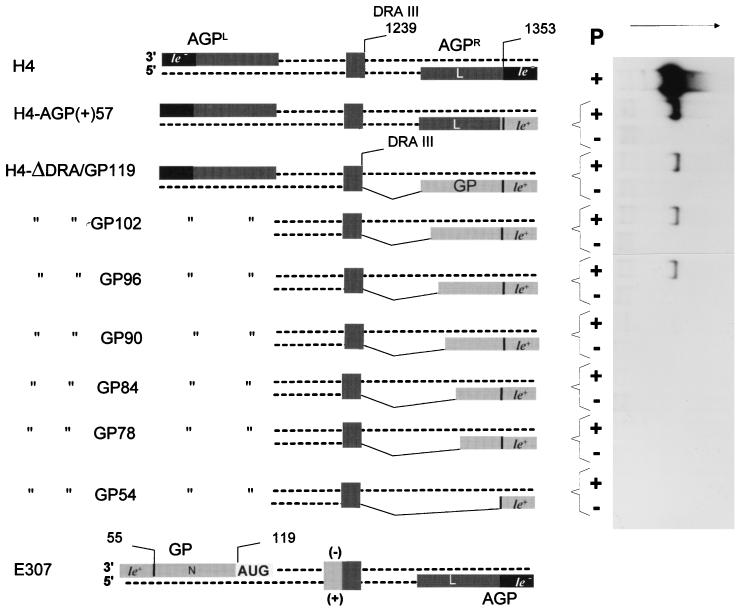
Minimal genomic promoter length. The H4-ΔDra/GP series (obtained as described in Materials and Methods) in which various GP lengths have been fused to the DraIII site, replacing the original AGPR sequence, are shown with (from top to bottom) decreasing lengths of the GP sequence. The promoter on the 3′ end of the H4 negative strand (AGPL) is the same for all the derivatives. H4-AGP(+)57 is particular in that the GP 57 nt exactly replaces the AGPR 57 nt, introducing an le+ template region in place of le− one. E307 RNA is shown for general DI RNA structure information and because it is the original source of the GP sequences. To the right-hand side, a Northern blot analysis of the encapsidated viral RNAs recovered from in vivo replication assays (see Materials and Methods) primed with the RNAs described to the left is shown. The P + or − lanes refer, respectively, to fully competent replicating assays and to assays done in the absence of the P protein as negative controls. The RNA probe (5′ ex) (44) is of positive polarity to score the RNA species complementary to the T7 RNA transcript.
FIG. 4.
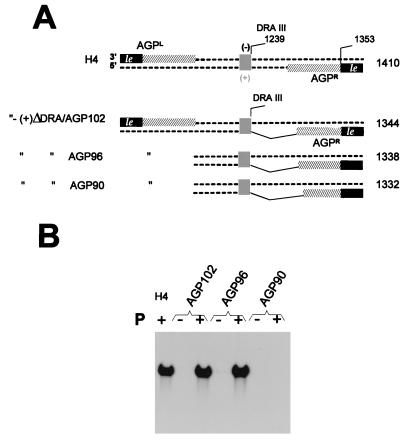
Minimal AGP length. (A) As in Fig. 3, except that the proper AGPR region of DI-H4 RNA has been increasingly truncated from the DraIII site position toward the plus-strand 3′ end, leaving the number of nucleotides indicated in the denomination of the derivatives (102, 96, and 90 nt). The numbers to the right refer to the total number of nucleotides in the RNA. (B) Northern blot as in Fig. 3, with the RNA derivatives with AGP truncations shown in panel A.
FIG. 6.
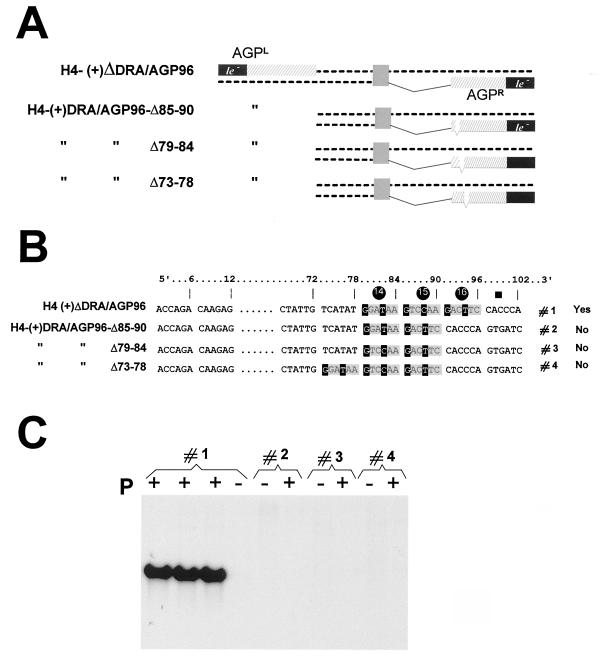
The three hexamers are required for competent replication. (A) H4(+)ΔDra/AGP96 derivatives carrying, in AGPR, deletions of one hexamer as indicated. (B) Outline of the relevant sequence in the derivatives schematically presented in panel A. Note that in derivatives 2 and 3, the sequence in hexamer 14 is different. In derivative 4, the three hexamers are present but displaced toward the end (left) by 6 nt. (C) Replication ability of the RNAs presented in panels A and B measured by Northern blot analysis, as in Fig. 3C. For derivative 1, the result is shown in triplicate.
FIG. 5.
Importance of the postulated motif in hexamers 14, 15, and 16. (A) SeV AGP and GP are aligned (as positive-strand DNA from 5′ to 3′) in groups of 6 nt (hexamers), to emphasise a sequence similarity repeated in the three hexamers 14, 15, and 16. The four sequences presented below the AGP and GP sequences originate from modified versions of GP, which led to successful (competent Yes) or unsuccessful (competent No) replication (reference 33 and unpublished data). The postulated motif is the nucleotide sequence predicted to be required for competent replication in the three hexamers. (B) H4(+)ΔDra/AGP96 derivatives carrying, in AGPR, substitutions introduced at positions marked with an asterisk. Every mutation consists of the replacement of a nucleotide(s) by its (their) complement(s). Yes and No refer to the ability of these RNA derivatives to be replicated, as shown in panel C. (C) Northern blot analysis of the encapsidated RNAs described in panel B recovered from replication assays as described in the legend to Fig. 3.
FIG. 7.
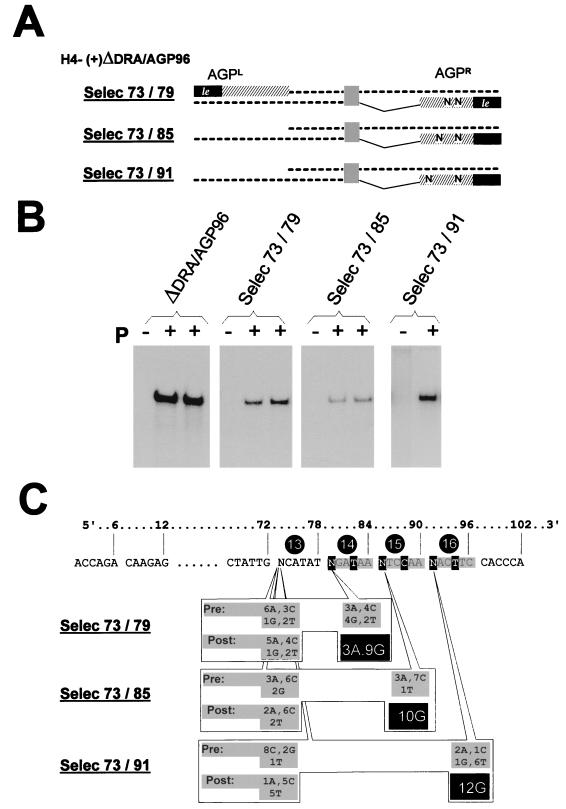
Selection applied to the first nucleotide of each of the three hexamers. Three oligonucleotides were created to contain two degenerate positions; one common to all three at position 73, and the other at position 79, 85, or 91, respectively, corresponding to the first position of each of hexamers 14, 15, and 16 (see the outlined sequence in panel B). PCR products, obtained with each of these three degenerate oligonucleotides with, as the other amplimer, the Rib3 primer (Fig. 2), were cloned into the DraIII and BamHI sites of H4(+)ΔDra/AGP96 to prepare the Selec 73/79, Selec 73/85, and Selec 73/91 plasmids with modified AGPR sequences. Pooled plasmid preparations were obtained from bacterial colonies scraped from a large LA petri dish (diameter, 13 cm) plated with 9/10 of a transformation reaction mixture; the remaining 1/10 was plated to pick individual colonies, allowing the sequencing of individual species (10 to 15 clones analyzed) to characterize the starting pool of plasmids (Pre distribution in panel C). After replication, using as the template plasmid each of the three pools, an RT-PCR fragment covering the AGPR region was obtained and cloned (see Materials and Methods) and 10 to 15 individual clones were sequenced to characterize the nucleotide (Post) present at the original degenerate position. (A) Schematized representation of the RNAs, with the degenerate positions indicated by N. (B) Northern blot analysis of the replicated encapsidated RNAs (as in Fig. 3), shown in duplicate for H4(+)ΔDra/AGP96, Selec 73/79, and Selec 73/85. (C) Nucleotide distribution at the positions of interest. The number accompanying the nucleotide designation refers to the number of clones containing this base at the indicated position. Pre, nucleotide distribution in the original plasmid preparation; Post, nucleotide distribution in the RT-PCR products after replication.
FIG. 8.
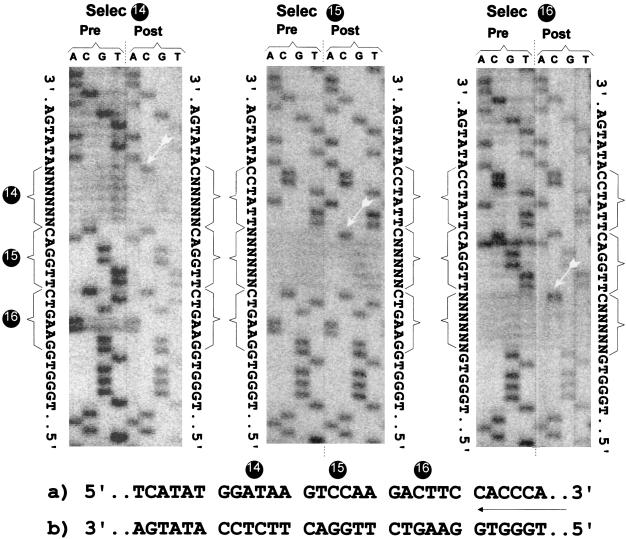
Selection applied to the 6 nt of each of the three hexamers. Three oligonucleotides were designed to contain six degenerate positions corresponding to hexamers 14, 15, and 16. Pooled plasmid preparations were obtained, as in Fig. 7. This time, Pre sequence characterization was done by sequencing the pooled plasmid preparation. After replication, RT-PCR products were obtained as in Fig. 7, and Post sequence characterization was done by directly sequencing the RT-PCR products of the replicated RNA. At the sides of each sequence panel, the Pre and Post sequences are outlined. At the bottom is shown the AGPR sequence from 5′ to 3′ (a) and the sequence in the sequencing gel (complement of that in a) (b).
Replication system and recovery of the encapsidated RNA.
The replication system used has been described previously (6). It has been used here with changes in the procedure of replicated RNA recovery. In brief, A549 cells (about 107) were infected with vTF7-3 (multiplicity of infection, 3). At 1 h later, the cells were transfected with 5 μg of pGem-N, 5 μg of pGem-PCwt, 1.5 μg of pGem-L, or 5 μg of the plasmid carrying the DI-RNA sequence, by incubation of the cells (3 h at 33°C) with 3 ml of MEM containing the plasmid mixture and 15 μl of Transfectace (Gibco BRL). Then 7 ml of MEM–5% fetal calf serum was added. At 40 h later, the cells were disrupted in 500 μl of lysis buffer (0.6% Nonidet P-40, 20 mM Tris-HCl [pH 8.0], 10 mM NaCl) and the clarified cytoplasmic extracts (3,000 × g for 15 min at 4°C) supplemented with 300 μl of TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing 3 mM CaCl2 were incubated with 2.5 μg of micrococcal nuclease (Biofinex) for 15 min at 37°C. After nuclease inactivation by addition of 20 mM EDTA and 4 mM EGTA (final concentrations), encapsidated RNA, resistant to nuclease digestion, was treated with proteinase K (1 μg/μl for 10 min at 55°C; Boehringer Mannheim) in the presence of 1% Sarkosyl. After chloroform-phenol extraction, the RNA was separated from residual DNA by pelleting it through a 5.7 M CsCl cushion (12 h at 32,000 rpm in a Beckman SW55 at 12°C) and resuspended in TE buffer.
Analysis of the encapsidated RNA.
One-fifth of the replicated RNA was analyzed by Northern blotting with an RNA probe of the same polarity as the T7 DI RNA transcript (plus strand) to detect the minus-strand RNA resulting from replication (6, 43, 44). When required, the remaining four-fifths of the RNA was used to reverse transcribe a 5′-end portion of the minus-strand RNA (extremity complementary to the end carrying the AGPR sequence) with Moloney murine leukemia virus reverse transcriptase (Promega) as specified by the manufacturer, with 100 U of enzyme in a 50-μl reaction mixture (60 min at 42°C with primer RT [Fig. 2]). One-tenth of this reaction mixture was then PCR amplified with Taq DNA polymerase (Gibco BRL) as specified by the manufacturer (100-μl reaction mixture subjected to 25 cycles with RT and PCR primers [Fig. 2]). After purification on columns (QIAquick PCR purification kit [Qiagen]), two-thirds of the PCR product was sequenced with a LiCor automated sequencing device (MWG-Biotech AG) as specified by the manufacturer (with the IRD primer [Fig. 2]). When not directly sequenced, the RT-PCR product was subcloned into the DraIII and BamHI sites of pSV-DI-H4 plasmid to be sequenced as individual clones.
RESULTS
The SeV promoters extend past the leader template regions.
The copy-back DI-H4 RNA (Fig. 1C) was used as the backbone for all the RNA template derivatives tested for replication in this study. The replication system was identical to that used previously (5, 6, 43, 44). Cells in culture were transfected with a series of four plasmids carrying genes encoding, for three of them, the functions required for encapsidation of the RNA template (protein N) and for replication (proteins P and L). The pGem-P plasmid (PCwt) also expresses the four accessory C proteins (13). The fourth plasmid expresses the template RNA (as plus-strand RNA) from the exact 5′ to the exact 3′ viral nucleotides (Fig. 1D). The four plasmids were transcribed by T7 RNA polymerase expressed from the recombinant vaccinia virus, vTF7-3, infecting the transfected cells. The replicated RNAs were purified as a nuclease-resistant fraction, and the extent of replication was monitored by Northern blotting with an RNA probe of positive polarity to score the RNA complementary to the template RNA produced by the T7 polymerase. Dependence on the SeV RNA polymerase was controlled by omitting the P protein (or P and L).
In a previous study, a reciprocal exchange between E307 and H4 RNAs had produced an H4 RNA derivative in which AGPR had been replaced with the first 57 GP nucleotides [H4-AGP(+)57 (Fig. 3; the figure legend gives the nomenclature of the RNA derivatives)]. Out of its natural context, the 57 GP nucleotides allowed H4-AGP(+)57 to replicate (44). Such a construct, however, could not be used to assess the minimal length of GP, since it is difficult to rule out the participation of the adjacent AGP sequence (from nucleotide 58 inward) in creating the active promoter. To avoid a possible complementation with AGP sequences, GP sequences had to be fused to the H4 plus strand sufficiently far from the 3′ end. A DraIII site, located 171 nt from the AGPR extremity (in the 3′-end open reading frame [ORF] of the L gene, ∼30 nt before the stop codon), was then chosen to generate H4 derivatives with various GP sequence lengths incorporated at AGPR (Fig. 3, H4-ΔDRA/GP series). This location was considered sufficiently remote from the end because numerous natural paramyxovirus copy-back DI RNAs replicate efficiently with the minus-strand promoter (inverted repeat, AGPL [Fig. 1C]) close to 100 nt long (e.g., 110 nt in SeV DI-H4 [see also Introduction]), indicating that the promoter sequence is unlikely to extend past this limit. In the H4-ΔDRA/GP series, the GP 119 nt (H4-ΔDRA/GP119) correspond to the sequence at the genome 3′ end extending to the start codon of the first transcription unit, the N gene. H4-ΔDRA/GP54, on the other hand, contains just the leader positive template region. The derivatives in between contain increasing amount of sequence (78 to 102 nt) all ending in the 5′ noncoding region of the N gene. Figure 3 shows the replication ability of these RNAs. This has to be compared with the P (−) assay, which represents the negative control. Replication of derivatives H4-ΔDRA/GP119 to H4-ΔDRA/GP96 was clearly positive (P+ lanes), while for H4-ΔDRA/GP90 to 54, no signal over the background was detected. Positive replication reached a much lower level than that of H4 RNA, consistent with the replacement of the strong AGP promoter with sequence derived from the weak GP promoter (6). A DraIII-ScaI fragment (containing the GP sequence, the ribozyme, and part of the vector plasmid [Fig. 1D]) was excised from the H4-ΔDRA/GP90 and H4-ΔDRA/GP54 plasmids (expressing nonreplicating RNAs) and replaced with the same fragment derived from the original pSV-DI-H4 to re-create an integer H4-RNA. This was found to replicate at the H4 level (data not shown), excluding the possibility that an unexpected modification had occurred in the region that had not been deliberately modified (and therefore not sequenced) during construction of the derivative. All the RNA transcripts of the H4-ΔDRA/GP series contained the same 5′ end, making it unlikely that lower encapsidation ability of some of the T7 RNA transcripts could account for the differences in replication observed. A Northern blot analysis of the encapsidated T7 RNA transcripts produced in a P and L (−) assay, using a minus-strand RNA probe, confirmed the equivalence of encapsidation of all the derivative RNAs (data not shown). When the replication of the functional H4-ΔDRA/GP RNAs (RNAs 119, 102, and 96) is compared to that of H4-AGP(+)57, it is noteworthy that the former RNAs replicated on average 10-fold less efficiently. If all four constructs share the first 57 GP nucleotides, the H4-AGP(+)57 sequence then diverges into the AGP sequence. The implication of these facts will be addressed in Discussion.
Similar derivatives were constructed with AGP sequences of various lengths fused to the DraIII site (Fig. 4A, H4-ΔDRA/AGP series). Replication was observed with 102 and 96 AGP nt but not with 90 nt (Fig. 4B). It is noteworthy that positive replication reached the level of H4, confirming that none of the sequence deleted between nt 97 and the DraIII site (nt 170) is required. In conclusion, for both GP and AGP, the replication promoter is shown to extend past the leader template regions down to nt 96 at the most.
Three hexamers of importance for effective replication.
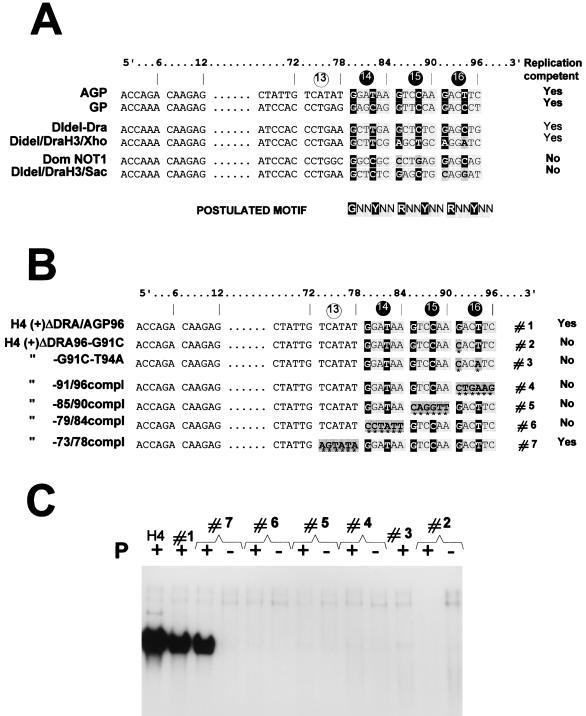
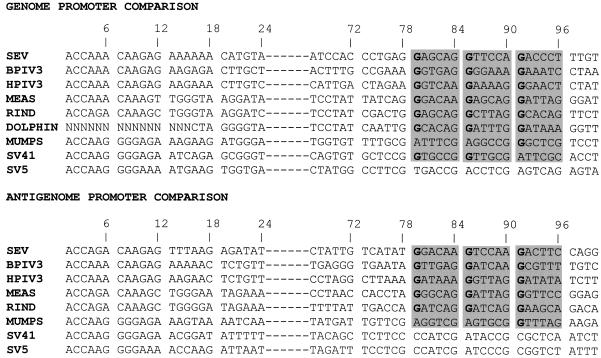
Examination of the sequences beyond the leader template regions highlighted a similarity in the region spanning nt 75 to 90 (1, 10). An alignment of the AGP and GP sequences in that region revealed a possible G1NNYNN6 motif repeated three times between nt 79 and 96 (36). The alignment was done by comparing blocks of 6 nt (hexamers), based on the rationale that the template RNA is likely to be seen by the RNA polymerase in this context. This conclusion was drawn from data suggesting (i) that each N protein subunit covers exactly 6 nt (5, 16) and (ii) that the six contact points of the N subunit with the 6 nt are not equivalent (36). The phase context of each nucleotide was then postulated to be an important feature of the signals recognized by the replicating (transcribing?) complex (28, 36). A limited comparison with other GP promoters (generated for other purposes, some of which are reported by Mottet et al. [33]), competent or not competent for replication, appeared to support the presence of a purine in position 1 of each hexamer, so that the postulated conserved motif can be better written R1NNYNN6, with a preference for G in position 1 (Fig. 5A). The presence of this motif would argue for the importance of the three hexamers 14 (nt 79 to 84), 15 (nt 85 to 90), and 16 (nt 91 to 96) in defining the active promoter.
A series of derivatives of H4-ΔDRA/AGP96, the one carrying the shortest AGP active promoter, was then produced in an attempt to verify the requirement for these hexamers and eventually the importance of the motif they contain. First, each hexamer was serially converted into its complement sequence (Fig. 5B, constructs 4, 5, and 6), with hexamer 13, not predicted to contain a particularly required sequence, taken as a control (construct 7). The absence of replication of the first three, in contrast to the quasinormal replication ability of the latter (Fig. 5C), pointed to the importance of the wild-type sequence in each of the three hexamers. It also indicated that three hexamers are required together since two hexamers remained in each of the nonreplicating constructs. The requirement for the three hexamers was further confirmed by the absence of replication of derivatives in which hexamer 15 or 14 had been deleted, leaving two adjacent hexamers with wild-type sequence (Fig. 6B and C, constructs 2 and 3). Note, in Fig. 6, that the deletion of hexamer 13 (construct 4), shown not to be important in itself, abolished replication. Interestingly, this shows that it is not enough for the three hexamers to be present and integer; they also cannot be displaced by 6 nt toward the template 3′ end (see Discussion).
The motif is rather simple.
In a preliminary attempt to test the validity of the motif, the G1 or the G1 and T4 of hexamer 16 were changed into their complement nucleotides (Fig. 5B and C, derivatives 2 and 3). The abolition of replication by the single transversion in derivative 2 strongly reinforced the possibility that the postulated motif could be important. A more systematic verification of the importance of the G1 in each hexamer was then undertaken as follows. Starting with the plasmid carrying H4-(+)ΔDRA/AGP96, three new constructs were prepared with degenerate nucleotides (A, C, G, and T) replacing G1 of hexamers 14, 15, and 16 (nt 79, 85, and 91, respectively). In each new construct, another degenerate site was created at position 1 of hexamer 13 (nt 73), a position predicted not to be critical (Fig. 5, construct 7). Each plasmid preparation (described as Selec 73/79, Selec 73/85, and Selec 73/91 [Fig. 7A]) contained a mixture of 16 different sequences. After transfection, the encapsidated RNA which had efficiently replicated was purified and the region spanning the AGPR was amplified by RT-PCR, cloned, and sequenced to determine the nucleotide present at the original degenerate positions. Figure 7B shows that each plasmid preparation led to a significant replication above the background. Figure 7C summarizes the sequencing results. At position 73, the distribution of the nucleotides in the RT-PCR products appeared to be random (with the limitation attached to the small number of clones sequenced), suggesting that replication can take place with any nucleotide at this position. In contrast, an almost absolute bias for G was observed in position 1 of hexamers 14, 15, and 16 in the RT-PCR products, a bias that the distribution in the original clones cannot account for. Note that this bias for G79, G85, or G91 was observed concomitant with the absence of preference for any nucleotide at position 73, which served as the internal control in each case. There appears, then, to be a strong selection for the RNAs carrying a G at position 1 of each hexamer.
The analysis of the sequence requirement was extended in a similar experiment in which all the six nucleotides in each hexamer were degenerate, creating the plasmid preparations Selec 14, Selec 15 and Selec 16. The transfections were thus made with potentially a mixture of 4,096 different sequences (46). The sequence analysis of either the initial plasmid preparations and of the RT-PCR products amplified from the replicated RNAs (data not shown) was done this time without cloning, for obvious reasons. Figure 8 shows the printout image of the sequences obtained from the automatic sequencer. The results are identical for the three hexamers. Sequencing of the transfected plasmid pools showed a degenerate sequence of six nucleotides at the expected positions. After replication, the degenerate positions were still observed but were truncated by one nucleotide where, in all three cases, a C was now visible; these C’s correspond to the G’s of the sequenced plus-strand templates. The conclusions are twofold. First, this second selection experiment confirmed the bias for the presence of G at position 1 of each of the three hexamers. Second, they ruled out the strict requirement of any particular nucleotide in the five other positions, under the limitation of the selective pressure applied here, i.e., productive replication only.
DISCUSSION
The first 31 nt of the SeV replication promoters have been previously shown to represent an essential element, since they adjust the efficiency of replication of the DI RNAs in the same replication assay as that used here (6, 44). These promoters now extend past the leader template regions, since, for genome as well as for antigenome, they contain 96 nt at the most. A second element has been identified in this extension as confined in the three hexamers 14, 15, and 16 and as consisting of AGP in the basic motif 5′-G79NNNNN-G85NNNNN-G91NNNNN-3′. In fact, the experiments demonstrated the requirement for three G’s interspaced with 5 nt, so that the motif could end at nt 91 with G91, the most remote feature, which is absolutely required. In retrospect, the H4-ΔDRA/GP and H4-ΔDRA/AGP RNAs containing 90 nt of the promoter linked to the DraIII site could have been functional if the next nucleotide (nt 91) had been a G. In both cases, however, the fusion with the DRA site inserted a C at this position, confirming the detrimental effect of C91 as in H4-ΔDRA/AGP-G91C (construct 2, Fig. 5B). The absence of the minimal motif in the other derivatives of the same series that failed to replicate was also confirmed in retrospect (data not shown). The requirement for the presence of three interspaced G’s appears absolute, since any combination of only two remaining is nonfunctional. Moreover, the position of the motif appears rigid, since displacement by 6 nt toward the end is not permitted. It is the distance to the end that is measured here.
Because of the identical requirement for a minimal length and the presence of the same basic motif, it is likely that these features are also those of GP. A previous study of GP examined the consequence of insertions of one, two, or three hexamers between nt 47 and 67. Under conditions which corresponded to those of the experiments presented here, these insertions, including that of a single hexamer, were not permitted (43). In view of the present results, it is likely that the insertions eliminated replication by causing inward displacement of the motif. Together, these results argue for a requirement for the exact positioning of the motif, which cannot be displaced by one hexamer to either side.
By image reconstruction of negatively stained electron micrographs, the SeV nucleocapsid has been described as a left-handed helix counting 13 N subunits per turn, with each subunit predicted to contact 6 nt (16), a prediction which was later corroborated by discovery of the rule of six (5). It may be significant that in this structure, the three hexamers, 14, 15, and 16, are positioned on the same face of the helix, exactly stacked above hexamers 1, 2, and 3, a stacking that cannot be skewed in either direction by addition or deletion of one hexamer. In this way, the first three hexamers are in proximity to the second element, although they are separated by 79 nt. This spatial arrangement suggests that the two elements are seen by the same surface of the replicating complex, although in the absence of any structural information, such considerations should remain speculative.
The first promoter element (nt 1 to 31) was functionally defined as containing, by itself, a signal regulating the efficiency of replication (6, 44). Whether the different replication efficiency reflects a different binding affinity for the RNA polymerase is not known. A difference in elongation efficiency following the entry of an identical RNA polymerase has to be envisaged as well. Since elongation is a step proposed to depend on the rate of nascent chain encapsidation (25, 26, 46), encapsidation initiation can also control replication efficiency. This complex and still unresolved mechanism of control of replication leaves ample space for a role for the second promoter element identified in the present study. The higher replication efficiency of H4-AGP(+)57 RNA than of H4-ΔDRA/GP RNA (Fig. 3), two RNAs which differ in the second element, suggests that the one on AGP can be functionally different from the one on GP. This second element could then constitute the feature previously identified in AGP as exerting a positive effect on replication, a minor effect compared to that of the nt 1 to 30 element and apparent only in the absence of this latter [as in H4-AGP(+)57 RNA (44)]. This interpretation is not immediately consistent with the definition of the nt 79 to 91 element as being (GNNNNN)3, a basic motif identical in GP and AGP. It is noteworthy, however, that the experiments performed to define this motif selected for the ability to replicate but not for the ability to differentially replicate.
The motif (GNNNNN)3 in a nucleocapsid structure where each N subunit is predicted to contact 6 nt is reminiscent of the motif (GNN)6 found in tobacco mosaic virus, a rodlike virus where each coat protein subunit contacts three nucleotides. This trinucleotide motif of tobacco mosaic virus constitutes the site of the initiation of virus assembly (45), and the definition of the structure of TMV at atomic resolution (34) has provided insight into the mechanism of this assembly. When a G is present in position 1 of the trinucleotide, it can form two additional hydrogen bonds to the coat protein, a feature that makes the presence of G at this position particularly significant for the nucleation site of assembly, where six GNN motifs are found. By analogy, it is tempting to speculate that the three hexamers 14, 15, and 16 could represent the nucleation site for SeV nucleocapsid assembly (a site postulated to be within the first 12 to 18 nt, based on assembly experiments of the VSV nucleocapsid [2, 3] before it was known that the two viruses differ in the extent of promoter sequence). If this were grounded, the initiation site for SeV encapsidation would be located past the initiation site for the first transcription unit (nt 56). This could argue against the transcription-to-replication-switch model which postulates that the nascent RNA encapsidation is effective before the polymerase reaches the site where the switch must occur (25, 26). If the encapsidation site were past the transcription initiation site, it would be difficult to explain how a polymerase that cannot replicate due to too low an N protein content could be switched into transcription. It is noteworthy in this respect that for respiratory syncytial virus, an increased N expression was shown not to alter the balance between transcription and replication, although replication was stimulated (19), indicating that in this particular example the two processes were not linked.
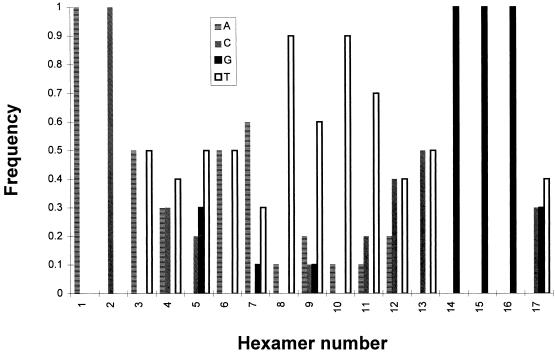
The (GNNNNN)3 motif is located such that, for three times in a row, the G is positioned as the first of the six nucleotides contacting the N protein (hexamer phase context 1). This may constitute another example of the importance of the hexamer phase in defining the signals addressed to the RNA polymerase. An alignment analysis of the motif present at the mRNA start site of nine paramyxoviruses (for which the total number of nucleotides is a multiple of 6) showed a remarkable degree of conservation of the phase context with a significant preference for starting at hexamer positions 1 and 2 (28). The conservation of the phase was also noticed for the motif involved in the P gene mRNA editing, a AnGn purine run which directs a number of G insertions in the P mRNA provoking a frameshift to access an internal ORF (V ORF [reviewed in reference 27]). Since the number of G insertions varies among different virus groups to mirror their requirements to switch between the in-frame and out-of-frame ORFs, it is particularly significant that the conservation is the strongest in each virus group (28). When the GP and AGP of the nine paramyxoviruses are aligned, a strong conservation of the (GNNNNN)3 in hexamers 14, 15, and 16 is again apparent. Of 17 promoters, 11 contain the motif at the right position, a number that increases to 14 of 17 if A, the other purine, is tolerated in place of G in one or two hexamers (Fig. 9). Computation of the frequency with which a particular nucleotide occupies the first position in the first 17 hexamers of the promoters for which the viral RNA is a multiple of 6 nt highlights the significant higher representation of G for hexamers 14, 15, and 16 (Fig. 10). It is noteworthy at this point that an insertion of 18 nt in the middle of the GP motif (at nt 80), unexpectedly re-creating a (GNNNNN)3 motif but shifted by 1 nt toward the end, so that the G’s are now in hexamer position 6 (G6 NNNNNG6 NNNNNG6 N), failed to yield a viable recombinant SeV (21), in contrast to a 18-nt insertion at nt 119. This could suggest that a change of one position in the phase context is not permitted, although in changing the phase, the distance to the end (to the first element) is similarly affected, so that the two parameters are concomitantly affected.
FIG. 9.
Prevalence of (GNNNNN)3 among paramyxoviruses. The GP and AGP sequences (as positive-strand DNA from 5′ to 3′) of nine paramyxoviruses are aligned to emphasize the sequence similarity in the three hexamers under focus here. SeV, BPIV3 (bovine parainfluenza type 3), and HPIV3 (human parainfluenza type 3) are closely related in the SeV subgroup. MEAS (measles virus), RIND (rinderpest virus), and DOLPHIN belong to the subgroup of the morbilliviruses. MUMPS, SV41, and SV5 (simian virus 41 and 5) belong to the subgroup of the rubulaviruses. The AGP sequence of the dolphin morbillivirus is not available.
FIG. 10.
Frequency of the presence of each nucleotide in the first hexamer position. The frequency with which a particular nucleotide is found in the first hexamer position is plotted as a function of the hexamer number in the GP and AGP of the SeV and morbillivirus subgroups (Fig. 9), which are known to contain a viral RNA with the total number of nucleotides being a 6n + 0 number.
In conclusion, a clear demonstration of the minimal promoter length is provided with the confirmation that both GP and AGP extend past the leader template regions. A basic motif of (GNNNNN)3 with a necessary positioning in hexamers 14, 15, and 16 with respect to the end and with, most probably, a necessary hexamer phase context of 1 has been defined. This constitutes the second element of SeV promoters. Whether another sequence between these two elements (in nt 31 to 78) participates in the replication promoter definition is open to question. An H4-(+)ΔDra/AGP96 derivative with all the nucleotides between nt 32 and 78 turned into their complements failed to replicate (preliminary data [not shown]). Whether this failure argues for the presence of still another element in between the two described or results in the insertion of a poisoned sequence is under investigation through selection experiments of the kind presented here.
ACKNOWLEDGMENTS
We are indebted to Stephane Hausmann and Dominique Garcin for discussions and technical advice (to C.T.) and to Nathalie Fouillot-Coriou for general supervision of D.M. L.R. thanks Dave Rowlands for editing of the manuscript and for his efforts to put it in Shakespearean language.
This work was supported by a grant from the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Blumberg B M, Chan J, Udem S A. Function of paramyxovirus 3′ and 5′ end sequences in theory and practice. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 235–247. [Google Scholar]
- 2.Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg B M, Leppert M, Kolakofsky D. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell. 1981;23:837–845. doi: 10.1016/0092-8674(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 4.Calain P, Curran J, Kolakofsky D, Roux L. Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology. 1992;191:62–71. doi: 10.1016/0042-6822(92)90166-m. [DOI] [PubMed] [Google Scholar]
- 5.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calain P, Roux L. Functional characterisation of the genomic and antigenomic promoter of the Sendai virus. Virology. 1995;211:163–173. doi: 10.1006/viro.1995.1464. [DOI] [PubMed] [Google Scholar]
- 7.Collins P L, Mink M A, Hill III M G, Camargo E, Grosfeld H, Stec D S. Rescue of a 7502-nucleotide (49.3% of full-length) synthetic analog of respiratory syncytial virus genomic RNA. Virology. 1993;195:252–256. doi: 10.1006/viro.1993.1368. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conzelmann K-K. Genetic manipulation of non-segmented negative-strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 10.Crowley J C, Dowling P C, Menonna J, Sliverman J I, Schuback D, Cook S D, Blumberg B M. Sequence variability and function of measles virus 3′ and 5′ ends and intercistronic regions. Virology. 1988;164:498–506. doi: 10.1016/0042-6822(88)90564-8. [DOI] [PubMed] [Google Scholar]
- 11.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curran J, Kolakofsky D. Scanning independant ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J. 1989;8:521–526. doi: 10.1002/j.1460-2075.1989.tb03406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran J, Kolakofsky D. Sendai virus P gene produces multiple proteins from overlapping open reading frames. Enzyme. 1990;44:244–249. doi: 10.1159/000468762. [DOI] [PubMed] [Google Scholar]
- 14.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 15.Dimock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelman E H, Wu S-S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enami M, Kohama T, Sugiura A. A measles virus subgenomic RNA: structure and generation mechanism. Virology. 1989;171:427–433. doi: 10.1016/0042-6822(89)90611-9. [DOI] [PubMed] [Google Scholar]
- 18.Engelhorn M, Stricker R, Roux L. Molecular cloning and characterization of a Sendai virus internal deletion defective RNA. J Gen Virol. 1993;74:137–141. doi: 10.1099/0022-1317-74-1-137. [DOI] [PubMed] [Google Scholar]
- 19.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasan M K, Kato A, Shioda T, Sakai Y, Yu D, Nagai Y. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J Gen Virol. 1997;78:2813–2820. doi: 10.1099/0022-1317-78-11-2813. [DOI] [PubMed] [Google Scholar]
- 22.Holland J J. Defective interfering rhabdoviruses. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Publishing Corp.; 1987. pp. 297–360. [Google Scholar]
- 23.Kast W M, Roux L, Curran J, Blom H J J, Voordouw A C, Meloen R H, Kolakofsky D, Melief C J M. Protection against lethal Sendai virus infection by in vivo priming of virus-specific cytotoxic T lymphocytes with a free synthetic peptide. Proc Natl Acad Sci USA. 1991;88:2283–2287. doi: 10.1073/pnas.88.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene J D, Chien I M, Lazzarini R A. Vesicular stomatitis defective interfering particle containing a mutated, internal leader RNA gene. Proc Natl Acad Sci USA. 1981;78:2090–2094. doi: 10.1073/pnas.78.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kingsbury D W. The molecular biology of paramyxoviruses. Med Microbiol Immunol. 1974;160:73–83. doi: 10.1007/BF02121714. [DOI] [PubMed] [Google Scholar]
- 26.Kolakofsky D, Blumberg B M. A model for the control of non-segmented negative strand virus genome replication. In: Mahy B W J, Minson A C, Darby G K, editors. Virus persistence. Cambridge, United Kingdom: Cambridge University Press; 1982. pp. 203–213. [Google Scholar]
- 27.Kolakofsky D, Curran J, Pelet T, Jacques J P. Paramyxovirus P gene mRNA editing, chapter 5. In: Benne R, editor. RNA editing. Chichester, United Kingdom: Ellis Horwood; 1993. pp. 1–21. [Google Scholar]
- 28.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolakofsky D, Vidal S, Curran J. Paramyxovirus RNA synthesis and P gene expression. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 215–233. [Google Scholar]
- 30.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1204. [Google Scholar]
- 31.Leppert M, Kort L, Kolakofsky D. Further characterization of Sendai virus DI-RNAs: a model for their generation. Cell. 1977;12:539–552. doi: 10.1016/0092-8674(77)90130-1. [DOI] [PubMed] [Google Scholar]
- 32.Li T, Pattnaik A K. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: indentification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 33.Mottet G, Mühlemann A, Tapparel C, Hoffmann F, Roux L. A Sendai virus vector leading to the efficient expression of mutant M proteins interfering with virus particle budding. Virology. 1996;221:159–171. doi: 10.1006/viro.1996.0362. [DOI] [PubMed] [Google Scholar]
- 34.Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic-acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by X-ray fiber diffraction. J Mol Biol. 1989;208:307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 35.Neubert W J, Eckerskorn C, Homann H E. Sendai virus NP gene codes for a 524 amino acid NP protein. Virus Genes. 1991;5:25–32. doi: 10.1007/BF00571728. [DOI] [PubMed] [Google Scholar]
- 36.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 37.Re G G. Deletion mutants of paramyxoviruses. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 275–298. [Google Scholar]
- 38.Schubert M, Keen J D, Lazzarini R A. A specific internal RNA polymerase recognition site of VSV RNA is involved in the generation of DI particles. Cell. 1979;18:749–757. doi: 10.1016/0092-8674(79)90128-4. [DOI] [PubMed] [Google Scholar]
- 39.Shioda T, Hidaka Y, Kanda T, Shibuta H, Nomoto A, Iwasaki K. Sequence of 3′687 nucleotides from the 3′ end of Sendai virus genome RNA and the predicted amino acid sequences of viral NP, P and C proteins. Nucleic Acids Res. 1983;11:7317–7330. doi: 10.1093/nar/11.21.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shioda T, Iwasaki K, Shibuta H. Determination of the complete nucleotide sequence of the Sendai virus genome RNA and the predicted amino acid sequences of the F, HN, and L proteins. Nucleic Acids Res. 1986;14:1545–1563. doi: 10.1093/nar/14.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidhu M S, Crowley J, Lowenthal A, Karcher D, Mennona J, Cook S, Udem S A, Dowling P. Defective measles virus in human subacute sclerosing panencephalitis brain. Virology. 1994;202:631–641. doi: 10.1006/viro.1994.1384. [DOI] [PubMed] [Google Scholar]
- 42.Sidhu M S, Kaelin K, Spielhofer P, Radecke F, Schneider H, Masurekar M, Dowling P C, Billeter M A, Udem S A. Rescue of a synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- 43.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 45.Turner D R, Joyce L E, Butler P J. The tobacco mosaic virus assembly origin RNA: functional characteristics defined by direct mutagenesis. J Mol Biol. 1988;203:531–547. doi: 10.1016/0022-2836(88)90190-8. [DOI] [PubMed] [Google Scholar]
- 46.Vidal S, Kolakofsky D. Modified model for the switch from Sendai virus transcription to replication. J Virol. 1989;63:1951–1958. doi: 10.1128/jvi.63.5.1951-1958.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Q, Hardy R, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]