Abstract

Platinum-based chemotherapeutic agents are widely employed in cancer treatment because of their effectiveness in targeting DNA. However, this indiscriminate action often affects both cancerous and normal cells, leading to severe side effects and highlighting the need for innovative approaches in achieving precise drug delivery. Nanotechnology presents a promising avenue for addressing these challenges. Protein-based nanocarriers exhibit promising capabilities in the realm of cancer drug delivery with silk sericin nanoparticles standing out as a leading contender. This investigation focuses on creating a sericin-based nanocarrier (SNC) featuring surface charge reversal designed to effectively transport cisplatin (Cispt-SNC) into MCF-7 breast cancer cells. Utilizing AutoDock4.2, our molecular docking analyses identified key amino acids and revealed distinctive conformational clusters, providing insights into the drug–protein interaction landscape and highlighting the potential of sericin as a carrier for controlled drug release. The careful optimization and fabrication of sericin as the carrier material were achieved through flash nanoprecipitation, a straightforward and reproducible method that is devoid of intricate equipment. The physicochemical properties of SNCs and Cispt-SNCs, particularly concerning size, surface charge, and morphology, were evaluated using dynamic light scattering (DLS) and scanning electron microscopy (SEM). Chemical and conformational analyses of the nanocarriers were conducted using Fourier-transform infrared spectroscopy (FTIR) and circular dichroism (CD), and elemental composition analysis was performed through energy-dispersive X-ray spectroscopy (EDX). This approach aimed to achieve the smallest nanoparticle size for Cispt-SNCs (180 nm) and high drug encapsulation efficiency (84%) at an optimal sericin concentration of 0.1% (w/v), maintaining a negative net charge at a physiological pH (7.4). Cellular uptake and cytotoxicity were investigated in MCF-7 breast cancer cells. SNCs demonstrated stability and exhibited a pH-dependent drug release behavior, aligning with the mildly acidic tumor microenvironment (pH 6.0–7.0). Efficient cellular uptake of Cispt-SNC, along with DNA fragmentation and chromatin condensation, was found at pH 6, leading to cell apoptosis. These results collectively indicate the potential of SNCs for achieving controlled drug release in a tumor-specific context. Our in vitro studies reveal the cytotoxicity of both cisplatin and Cispt-SNCs on MCF-7 cells. Cisplatin significantly reduced cell viability at 10 μM concentration (IC50), and the unique combination of sericin and cisplatin showcased enhanced cell viability compared to cisplatin alone, suggesting that controlled drug release is indicated by a gradient decrease in cell viability and highlighting SNCs as promising carriers. The study underscores the promise of protein-based nanocarriers in advancing targeted drug delivery for cancer therapy.
Introduction
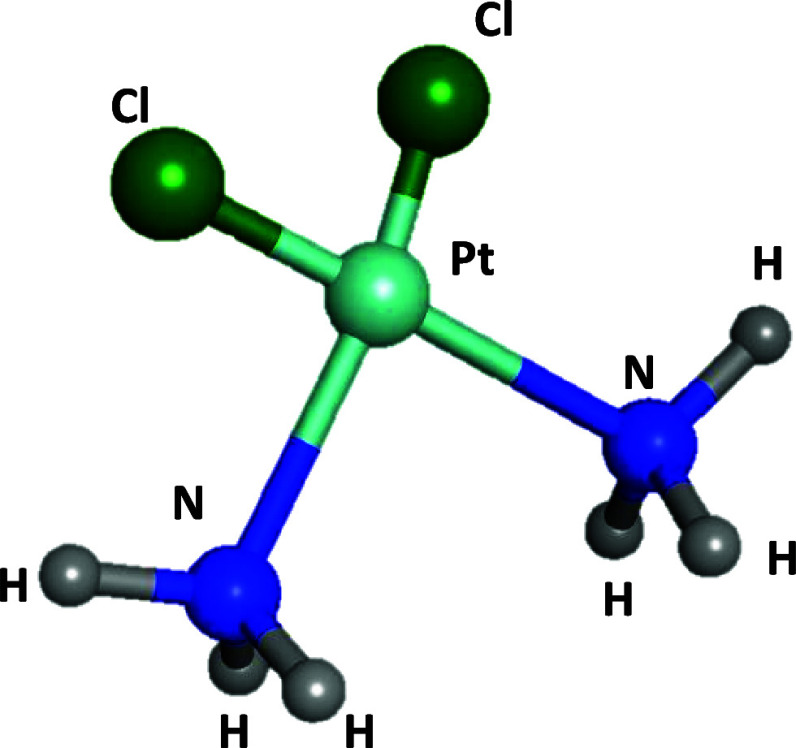
Breast cancer, a prevalent form of cancer, holds the distinguished position of being the most frequently diagnosed malignancy globally, accounting for the highest number of diagnosed cases (2.3 million, 24.5% of all cancers) and cancer-related deaths (684,996, 15.5% of all cancer deaths) in 2020.1 As a result, the estimated number of newly diagnosed invasive cancer cases among women in the United States in 2023 reveals that breast cancer alone represents 31% of the female cancer occurrences.2 Despite significant progress in comprehending and treating breast cancer in the past decade, the occurrence and fatality rate associated with it persistently escalate at a concerning pace. Current breast cancer treatment modalities, such as surgery, radiation, immunotherapy, gene therapy, photodynamic treatment, and chemotherapy, are effective in reducing the impact of breast cancer.3 However, their primary mode of action involves nonselective cell destruction, impacting both cancerous and noncancerous cells and leading to significant adverse effects. Common issues with conventional chemotherapy include the limited aqueous solubility, lack of selectivity toward cancer cells, and multidrug resistance, attributed to factors such as increased efflux pumps like P-glycoprotein.4 Multidrug resistance poses a major challenge, with some cancers exhibiting primary resistance, whereas others develop acquired resistance over time.4−6 Additionally, systemic chemotherapy faces limitations such as rapid clearance, low drug concentration at the tumor site, and considerable adverse effects beyond the tumor.6 Platinum-based drugs have become integral to cancer therapy, with approximately half of chemotherapy patients receiving them,7 The use of platinum agents in cancer treatment originated from the discovery of cisplatin’s antineoplastic properties by Rosenberg et al. in the 1960s.8 Cisplatin (Figure 1), also known as cis-diamminedichloroplatinum(II), is a metallic coordination compound with a square planar geometry and, along with oxaliplatin and carboplatin, constitutes a group of platinum-based anticancer agents sharing this geometric configuration. Cisplatin is highly effective against a range of cancers, including lung, ovarian, and testicular cancers. Carboplatin, less toxic but requiring higher doses, is used for ovarian, testicular, and small cell lung cancers. Oxaliplatin shows efficacy similar to cisplatin, particularly in esophageal and gastric cancers, and is used against colorectal and various other tumors, including those resistant to cisplatin and carboplatin.9 Cisplatin, as a cytotoxic drug, targets and eliminates cancer cells by causing DNA damage, inhibiting DNA synthesis and cell division, and inducing apoptosis.10 The mechanism of action of cisplatin and classical platinum agents involves four primary stages: (1) cellular uptake, (2) aquation/activation, and (3) DNA platination and cellular processing of Pt-DNA lesions, resulting in (4) either cell survival or apoptosis.11 The unaltered cisplatin can ingress tumor cells, predominantly through membrane diffusion,12 although recent research proposes active transportation facilitated by Cu-transporting proteins.13 Intracellularly, the relatively diminished chloride concentration (approximately 4–20 mM) instigates the substitution of one chloro ligand with water, generating a reactive, positively charged species that persists within the cell. In vitro investigations establish that this monoaquated platinum species is accountable for more than 98% of platinum binding to DNA in the cellular nucleus.14 It engages with a DNA base, commonly guanine, forming a monofunctional DNA adduct. Nanotechnology offer a promising solution to targeted drug delivery, reducing side effects of anticancer drugs, and controlled release, solving the mentioned critical issues of conventional cancer treatment methods.15 Recent advancements have introduced stimuli-responsive nanocarriers that effectively target tumor habitats, utilizing passive and active targeting approaches to overcome biological barriers. The unique physiological characteristics of the tumor microenvironment, such as pH, hypoxia, temperature, and enzyme upregulation, can trigger nanoparticle activation. This activation leads to accelerated drug release, enhanced cellular uptake, effective impact charge reversal action of nanocarriers, and improved drug distribution within the tumor volume.16 Protein-based charge reversal nanocarriers are a promising strategy that respond to changes in pH, particularly in the acidic microenvironment of tumors.17 They undergo a negative-to-positive charge transition, facilitating cellular uptake and interaction with negatively charged cell membranes.18 These nanocarriers possess desirable properties for breast cancer treatment. The investigation of protein nanocarriers as drug delivery vehicles that are generally recognized as safe (GRAS) is being conducted owing to their exceptional attributes.19 These attributes encompass biodegradability (when decomposed inside the body, they do not cause toxicity), nonantigenicity, the abundance of renewable sources, and the remarkable binding capacity toward diverse drugs. They also have the potential to minimize opsonization through the reticuloendothelial system (RES) with a steric barrier, lack immunostimulatory activity,20 and possess excellent functional properties like emulsification,21 gelation, foaming, and water binding.22 As a result, proteins are highly promising for developing safe drug delivery devices.23 Silk sericin (SS) is a pH-responsive protein that is ideal for creating charge-reversal nanocarriers. SS is a natural protein derived from the silk cocoon of the silkworm Bombyx mori.24 It can be obtained as industrial waste from the textile and silk industry, making it a sustainable and cost-effective source of protein.25 Silk sericin is a globular protein with molecular weight ranges from 10 to over 400 kDa,26 is synthesized in the silk glands of silkworms, and constitutes about 25% of the cocoon weight.27 It contains 18 amino acids, including both positively and negatively charged aromatic, polar, and nonpolar amino acids,28 with serine (40.51% of the polar amino acid side chains) being the most abundant polar amino acid. Sericin functions as an intrinsic natural biopolymer distinguished by the inclusion of acidic and basic amino acids, rendering it responsive to pH variations. This inherent quality positions sericin as a promising candidate for the design of reversible nanoparticles.29 Notably, following serine, sericin prominently features aspartic acid and glycine as the subsequent most abundant amino acids.25 The pivotal role played by these amino acids in influencing the charge reversal properties of sericin is of significant importance. Sericin is soluble and permeable to water due to its high content of hydrophilic amino acids (around 70%).28 It possesses various beneficial properties such as antioxidant,30 antibacterial, antityrosinase, anti-inflammatory, antitumor,31 and anti-GST activity. Additionally, sericin has the ability to form stable nanoparticles that can encapsulate different compounds. These nanoparticles provide prolonged circulation of the encapsulated compounds in the bloodstream, protect them from degradation, and allow targeted release at specific sites.32 The nanoprecipitation method offers a straightforward, gentle, and energy-efficient approach for manufacturing nanoparticles while also serving as an effective method for encapsulating both hydrophobic and hydrophilic substances.33 In antisolvent precipitation (ASP), a bottom-up technique, two fully miscible liquid solvents are utilized, with the protein solute being soluble in the first solvent but not in the second (antisolvent), enabling successful nanoencapsulation with control of its physicochemical characteristics.34,35 Hence, this study aimed to create a new protein-based nanocarrier using silk sericin through the nanoprecipitation method. This nanocarrier was utilized to transport cisplatin, as a platinum-based chemotherapy agent, to MCF-7 breast cancer cells. The nanoparticles were thoroughly examined for their physical and chemical properties, and the release of Cispt-SNC was evaluated at pH 7.4. Additionally, the impact of Cispt-SNC on the viability of MCF-7 cells was investigated at pH 7.4. The study also explored the uptake of Cispt-SNC within MCF-7 cells and assessed DNA fragmentation.
Figure 1.
3D structure of cisplatin.
Materials and Methods
Materials
Sericin Bombyx mori (silkworm) powder (S5201, reagent grade, ≥99%) and nonsolvent acetone (ACS reagent, ≥99.5%) were acquired from Sigma-Aldrich (St. Louis, USA). Cisplatin (CAS no. 15663-27-1, 99%, HPLC, MF-Cl2H6N2Pt; Figure 1) was obtained from Mylan (France). Ultrapure water (UP water) was produced using a Milli-Q (Millipore, Bedford, MA, USA) system.
Equipment Model
The following were used: Zetasizer instrument (Nano-ZS, Malvern Instruments Ltd., Worcestershire, UK), scanning electron microscope (SEM) (EM3200, KYKY Technology Co., China), FTIR spectrophotometer (IR Prestige-21, Shimadzu Co., Japan), circular dichroism spectrophotometer (J-1500, Jasco Co., Japan), spectrophotometric plate reader (ELx808, Lonza BioTek Co., Switzerland), and cytation cell imaging reader (Cytation 5, Biotek Co., USA)
Sequence Retrieval
The sequence of the sericin (P07856) protein was retrieved from the UniProt web server (https://www.uniprot.org/) in September 2023.36 The selection of this protein sequence was based on its close similarity in the number of amino acids to the target protein used in our research, facilitating meaningful comparative analyses.
Structure Prediction by I-TASSER
The structure prediction was performed using I-TASSER (Iterative Threading Assembly Refinement).37 The protein sequence was input into the I-TASSER,38 an online hierarchical approach utilized for predicting the structure and function of the protein.39 The tool is accessible at https://seq2fun.dcmb.med.umich.edu//I-TASSER/. The structure of sericin was predicted by employing the online I-TASSER tool.
Molecular Docking Analyses with AutoDock4.2
We conducted molecular docking analyses to elucidate the interactions between sericin and cisplatin, aiming to identify the critical amino acids involved in these bindings. The molecular docking analysis involved the selected sericin protein and the cisplatin molecule and was conducted using AutoDock4.2. The macromolecule preparation commenced with preprocessing the RBD protein through AutoDockTools1.5.7.40 This process encompassed water removal, the addition of Kollman charges, and polar hydrogens, culminating in the creation of a PDBQT format file. For ligand preparation, the 2D structures of the drug were initially sketched using ChemBioDrawUltra14.0 and subsequently converted into a 3D structure via Chembio3D Ultra14.0 followed by energy minimization and saving in PDB format. Given the substantial size of the structure, we partitioned the docking into three phases to enhance the computational efficiency. The protein was compartmentalized into three grid boxes. The first grid box had center coordinates at x = 71.963, y = 90.989, and z = 102.494; dimensions of 80 × 76 × 62 Å; and a spacing of 0.869 Å (Figure 2a). The second grid box centered at x = 114.997, y = 109.749, and z = 116.12 and featured dimensions of 60 × 52 × 92 Å with a 1 Å spacing (Figure 2b). The third grid box was positioned at x = 153.255, y = 129.716, and z = 134.526 nm, boasting dimensions of 125 × 82 × 52 Å and a 0.861 Å spacing (Figure 2c). This strategic division optimized the computational processing of the intricate structure. These grid boxes were strategically designed with a slight overlap to ensure comprehensive coverage and precision in the molecular docking analysis. The docking simulation was performed with the AutoDock Lamarckian Genetic Algorithm (LGA, runs 250) and 25 million energy evaluations per run to find the proper combination models. The selection of the most stable clusters was determined by considering the most stable or lowest bonding energy, and further confirmation was derived from the histogram and repetition of amino acids.
Figure 2.

(A) The first grid box with center coordinates at x = 71.963, y = 90.989, and z = 102.494; dimensions of 80 × 76 × 62 Å; and a spacing of 0.869 Å. (B) The second grid box with a center at x = 114.997, y = 109.749, and z = 116.12, featuring dimensions of 60 × 52 × 92 Å with a 1 Å spacing. (C) The third grid box with center coordinates at x = 153.255, y = 129.716, and z = 134.526 nm; dimensions of 125 × 82 × 52 Å; and a 0.861 Å spacing.
Preparation of Protein-Based Nanocapsules
To determine the optimal protein concentration, a previous study investigated different concentrations of sericin protein using the anti-solvent precipitation method.41 In our study, we built upon a prior investigation that explored various concentrations of sericin protein (0.1, 0.2, 0.3, 0.5, and 1% (w/v)) to determine the optimal formulation.41 Our work aimed to contribute by selecting the 0.1% concentration, which was identified as the most suitable for preparing protein-based nanocapsules in terms of achieving small, spherical, and uniform particles with high cisplatin encapsulation. The use of flash nanoprecipitation, as adapted from Nelemans et al., serves as a robust and reproducible method that ensures the accessibility and practicality of nanocarrier fabrication.42 Consequently, sericin was directly dispersed in ultrapure water at the concentration of 0.1% (w/v) with gentle stirring at room temperature. Subsequently, a protein-based nanocapsule was synthesized utilizing the nanoprecipitation or anti-solvent precipitation (ASP) method with minor adjustments.43 Briefly, 20 μL of silk sericin solution (0.1% (w/v)) was gradually added dropwise (10 μL/5 s) to an organic phase consisting of a nonsolvent (acetone) under vigorous stirring (1500 rpm) at room temperature. During this process, water and acetone were evaporated completely, resulting in the formation of sericin-based nanocapsules.
Drug Loading to Sericin-Based Nanocapsules
In this study, drug loading to sericin-based nanocapsules was achieved through the direct system of dissolution into the aqueous phase with 1 mg of cisplatin that was solved in 1 mL of ultrapure water. The next step involved adding the 200 μL (containing 0.2 mg of cisplatin) of dissolved Cisplatin and sericin aqueous phase mixture into the acetone organic phase under forceful stirring (1500 rpm) and pH 7.4. This process was done without any tools or devices, only with an insulin Hamilton syringe for greater accuracy and control, one drop per 5 s, under intense stirring. The SNCs containing the drug (Cispt-SNC) were recovered by completely evaporating water and acetone. Formerly, intracellular delivery of cisplatin utilizing pH-responsive SNCs for anticancer therapeutics was achieved. In addition to assessing the physical appearance and storage stability, the Cispt-SNC was tested for variations in quality features when added to cell culture media.
Basic Physicochemical Properties of Cispt-SNC
The dynamic light scattering technique using the Zetasizer instrument (Nano-ZS, Malvern Instruments Ltd., Worcestershire, UK) at 25 °C was employed to determine the polydispersity index (PDI), particle surface charge (zeta potential), and particle size of SNC and Cispt-SNC. Additionally, other essential features of SNC and Cispt-SNC such as aggregation, morphology, size, and shape were evaluated using the scanning electron microscope (SEM) (EM3200, KYKY Technology Co., China) at an operating voltage of 25 kV.
Fourier Transform Infrared Spectroscopy (FTIR)
FTIR spectroscopy was utilized to analyze the chemical composition, molecular properties, and surface adsorption of nanoparticles’ functional groups. To do so, 1 to 2 mg aliquots of pure cisplatin, lyophilized blank SNC (B-SNC), and Cispt-SNC were mixed and crushed together with 100 mg of spectroscopy-grade potassium bromide (Sigma-Aldrich). The mixture was then placed in the FTIR sample holder and pressed followed by recording the spectra over a scanning range of 400–4000 cm–1 with a spectral resolution of 4 cm–1 using an FTIR spectrophotometer (IR Prestige-21, Shimadzu Co., Japan).
Conformational Analysis by Circular Dichroism (CD)
Circular dichroism spectrophotometry was used to assess the secondary structures of sericin B-SNC for their dispersion properties. A quartz cell with a path length of 1 mm was used, and the examination was carried out using a Jasco Co. circular dichroism spectrophotometer (J-1500). The examination involved scanning the samples three times using ultraviolet light between 180 and 250 nm, with a scan rate of 100 nm/min.
Energy-Dispersive X-ray Spectroscopy (EDX)
Energy-dispersive X-ray spectroscopy (EDX) was performed to investigate the elemental composition of cisplatin in SNCs for potential cancer treatment. The analysis aimed to determine the composition of the Cispt-SNCs and the extent of cisplatin encapsulation within the nanoparticles. Samples were prepared and analyzed using an instrument equipped with an energy-dispersive X-ray detector, allowing for elemental mapping and point analysis on selected regions of interest to determine the presence and relative abundance of cisplatin.
Determination of Entrapment Efficiency (EE%) and Drug Loading (DL%)
The ultracentrifugation method was used to evaluate the entrainment efficiency and drug loading.44 The Cispt-SNC solution was ultracentrifuged at 18,200 rpm for 30 min using Optima L-90K equipment from Beckman Coulter Co, USA, to separate the unbound drug from the Cispt-SNCs. UV–vis spectrophotometry was used to evaluate the unentrapped cisplatin by assessing 1 mL of the supernatant at 210 nm. A standard calibration curve was created using various concentrations of cisplatin ranging between 0.1 and 100 μg/mL, which was a straight line with an r2 of 0.98. The free drug amounts were determined using this curve.45 Finally, the entrapment efficiency (EE) (eq 1) and drug loading (DL) (eq 2) of cisplatin on Cispt-SNC were calculated using the following formula:
| 1 |
| 2 |
In pursuit of optimizing drug loading and encapsulation efficiency, researchers utilize protein-based carriers to achieve several key objectives. Protein nanocarriers enhance drug stability and solubility while enabling controlled drug release, thereby improving efficacy and reducing administration frequency.46 Moreover, these carriers contribute to enhanced safety by minimizing the toxic effects and immunogenicity. Furthermore, the engineered nature of protein nanocarriers enables targeted delivery to specific tissues or cells, reducing off-target effects and enhancing drug accumulation at tumor sites.47 Additionally, the encapsulation of drugs in proteins proves beneficial for enhancing oral bioavailability with each protein exhibiting a tendency to encapsulate either hydrophobic or hydrophilic molecules. Notably, silk proteins exhibit higher encapsulation efficiency for hydrophilic substances.48
In Vitro Drug Release
In vitro release is increasingly garnering attention as a reliable surrogate test for assessing product performance.49 The study analyzed the release of cisplatin from Cispt-SNC at pH 7.4 through in vitro testing using a dialysis bag with a 20 kDa molecular weight cutoff from Sigma-Aldrich.50 Initially, 5 mL of Cispt-SNC was placed into the dialysis bag and immersed in a fixed volume of release medium consisting of 80 mL of PBS with a pH of 7.4. The bottles were then subjected to incubation for 55 h at 37 ± 0.5 °C at 300 rpm in an orbital mixer from Benchmark Scientific. Samples were taken at set intervals of time (0.5, 1, 2, 4, 24, 30, 48, and 65 h), where 1 mL of PBS dialysate was extracted and replaced with a fresh medium of the same volume. The samples were analyzed using UV–vis spectrophotometry (Mini 1240, Shimadzu Co., Japan) at a λmax corresponding to 210 nm. In essence, the design is easy to perform, simple, and available in all pharmaceutical laboratories under the same setup.51
In Vitro Biological Evaluation of Cispt-SNC
Cell Culture Model
The study utilized a human breast adenocarcinoma cell line known as MCF-7 (NCBI C135 Pasteur Institute of Iran, Karaj, Iran) as an in vitro model. MCF-7 cells possess several features of differentiated mammary epithelium such as functional estrogen receptor expression,52 responsiveness to estradiol, and the ability to invade and metastasize, making it an ideal model for the study.53 Additionally, MCF-7 is a luminal A subtype (HER2-negative and progesterone receptor-positive) cell line.54 The cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic–antimycotic solution (100 μg/mL streptomycin, 100 U/mL penicillin, and 0.25 μg amphotericin B) and placed in an atmosphere of 5% CO2, 37 °C, and humidified air. The culture medium was refreshed on a daily basis.
Cell Viability and Cytotoxicity
To evaluate cell viability/cytotoxicity for MCF-7 cells,55 the growth-inhibiting impact of cisplatin and Cispt-SNCs (0.4, 0.6, 1, 2, 5, 10, 15, 25, and 50 μM) at pH 7.4 was measured. This was done using a colorimetric MTT assay kit following a previously described protocol.56 The cells were initially seeded in 96-well plates at a density of 3 × 103 cells/well and were then treated with cisplatin and Cispt-SNCs at the aforementioned concentrations using the media at pH 7.4 for 24 and 48 h. After this treatment period, 10 μL of the MTT dye (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide, 5 mg/mL in phosphate-buffered saline) was added to each well,57 and the plates were incubated at 37 °C for 4 h. Next, the media were aspirated, and 100 μL of a solubilization solution (10% SDS in 0.01 M HCl) was added to each well to dissolve the formant crystals. The absorbance of the plates was measured at a wavelength of 570 nm by using a spectrophotometric plate reader (ELx808, Lonza BioTek Co., Switzerland). Subsequently, the percentage of viable cells was calculated using the below formula (eq 3), enabling the determination of the IC50:
| 3 |
Intracellular Uptake and DAPI Staining
To examine how the fabricated nanoparticle is taken up by cells, intracellular uptake and DAPI staining were performed.54,56 Initially, a 1 mg/mL suspension of Cispt-SNCs was mixed with an equal volume of FITC (5 μg/mL) and left to stir for 10 h in the dark at room temperature. The resulting suspension was then centrifuged at 13,000 rpm for 10 min and washed three times with PBS. Following this, MCF-7 cells were seeded in six-well culture plates at a density of 2 × 105 cells per well. After the medium was removed, the cells were washed twice with PBS and then treated with 1000 μg/mL of stained Cispt-SNC at pH 6 for 8 h. The cells were then washed three times with PBS and fixed using 4% paraformaldehyde (100 μL/well) for 15 min, and the nucleus was stained using DAPI (4′,6-diamidino-2-phenylindole; 40 μM) staining. After 24 h of incubation, the cells were washed with PBS, fixed with 4% (v/v) paraformaldehyde for 30 min, and rinsed twice with PBS. DAPI was then added for 20 min in the dark to stain the nucleus. Finally, the cells were washed with PBS three times, and the well (n = 3) was analyzed using a Cytation cell imaging reader (Cytation 5, Biotek Co., USA) to evaluate the results of the study.
Statistical Analysis
The data collected underwent statistical analysis using SPSS version 23.0 for Windows and GraphPad Prism 8 software. To identify any significant differences between the groups, one-way analysis of variance (ANOVA) was utilized. The experiments were conducted three times, and the mean values with their corresponding standard deviations (SDs) were presented. A p value below 0.05 was deemed statistically significant.
Results
Molecular Docking Analyses
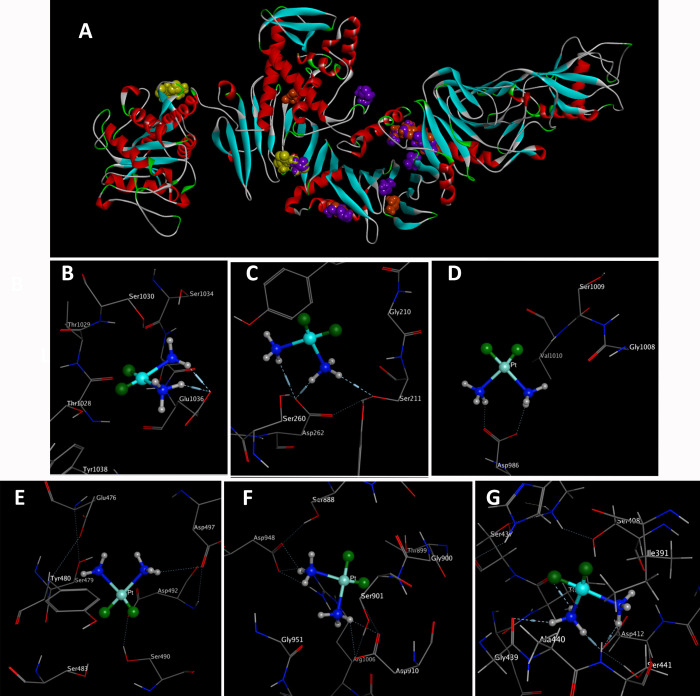
The analysis uncovered distinctive conformational clusters across 250 runs, employing an RMSD tolerance of 0.5 Å for all three grid boxes. The self-docking calculations exhibited outstanding performance across all cases, with mean RMSD values consistently ≤1.0. Particularly noteworthy were the lower RMSD values associated with poses of the highest affinity.58 According to the results (Figure 3a), key amino acids pivotal in the drug–protein interaction encompassed Asp 412, 910, 497, 262, 209, 371, 492, 986, 890, 931, 364, 1007, 948, and 287; Glu 1036, 491, and 476; Ser 103, 211, 490, 483, 1009, 822, 493, 471, 474, 917, and 901; Gly 501, 1008, 824, and 439; Leu 500 and 835; Tyr 362 and 480; Thr 1028, 899, 867, and 436; and Ile 493 and 498. Consequently, blind docking revealed the identification of active regions and amino acids within sericin that can interact with cisplatin. This refers to the central regions of the sericin structure visible in certain sections of grid box 2 and those overlapping with the western part of grid box C, as illustrated in (Figure 3a). The clustering histogram provided valuable insights into the distribution of binding energies within the identified clusters. Specifically, within grid box A, cluster 1 (Figure 3b) demonstrated the lowest binding energy of −5.22 kcal/mol, involving Glu 1036, whereas cluster 2 exhibited the lowest binding energy of −4.58 kcal/mol, involving Asp 262 and Serine 211 (Figure 3c). In grid box B, the most stable clusters displayed the lowest binding energies, with one involving Asp 986 at −5.72 kcal/mol (Figure 3d) and another involving Asp 497 at −5.88 kcal/mol (Figure 3e). Within grid box C, the most stable cluster exhibited the lowest binding energy of −6.03 kcal/mol, involving Asp 910 and 948 (Figure 3f). The molecular docking results highlighted the predominant and most stable clusters formed with negatively charged amino acids, particularly aspartic acid and glutamate. This interaction has implications for influencing the surface charge of our nanocarrier. The I-TASSER analysis identified significant ligand binding sites, namely, Ser 406, 407, and 408; Ala 411; Asp 412; Lys 413; Asp 414; Thr 422; and Ser 375. Consequently, we introduced a novel grid box positioned at x = 124.34, y = 110.184, and z = 96.655 with dimensions of 60 × 60 × 60 employing an RMSD tolerance of 0.5. This investigation unveiled a stable cluster characterized by the lowest binding energy of −5.51 kcal/mol, which included Asp 412 as identified by the I-TASSER analysis. Additionally, Gly 439 and Thr 436 (Figure 3g) highlighted their robust stability and prevalence in the binding interaction. The distribution pattern across the histogram emphasized the diversity of energetically favorable conformations, highlighting the significance of these findings in comprehending the molecular landscape of the investigated system. The initial docking interaction between sericin and cisplatin revealed notable hydrogen bond formation with the glutamate (Glu) residue at position 1036 in the receptor (Figure 3b). Specifically, the ligand atom N1 (donor atom) established a hydrogen bond with O– (acceptor atom) of Glu 1036 at a distance of 2.77 Å, displaying a strong interaction with an energy of −7.4 kcal/mol. Additionally, ligand atom N2 (donor atom) formed another hydrogen bond with the same O– (acceptor atom) of Glu 1036, albeit at a slightly longer distance of 3.17 Å, with an interaction energy of −3.7 kcal/mol. In the docking interaction (Figure 3c), the ligand atom N1 engaged in a hydrogen bond with the hydroxyl group of serine 211 in the receptor, manifesting at a distance of 3.03 Å and yielding an associated binding energy of −2.2 kcal/mol. Notably, ligand atom N1 established an additional hydrogen bond with the oxygen atom O– of Asp 262, which is characterized by a distance of 2.78 Å and a substantial binding energy of −6.2 kcal/mol. Simultaneously, ligand atom N2 participated in a distinct hydrogen bond with the same Asp 262 residue at the position, showcasing a distance of 3.40 Å and a binding energy of −2.3 kcal/mol. In the docking interactions depicted in Figure 3d–g, various hydrogen bonding interactions between sericin and cisplatin were observed. In Figure 3e, ligand atom N1 formed a hydrogen bond with the oxygen atom (O) of aspartic acid Asp 497 at a distance of 3.55 Å, resulting in a binding energy of −1.7 kcal/mol. Meanwhile, in Figure 3d, ligand atoms N1 and N2 established hydrogen bonds with the same Asp residue at position 986, with each interaction occurring at a distance of 2.50 Å and exhibiting a significant binding energy of −5.7 kcal/mol. Turning to Figure 3f, cisplatin atoms N1 and N2 engaged in hydrogen bonding as hydrogen donors with Asp residues at positions 910 and 948 in the receptor. Specifically, ligand atoms N1 and N2 formed hydrogen bonds with the oxygen atoms of Asp 910 and 948 at distances of 2.63 and 3.21 Å, respectively, accompanied by binding energies of −7.5 and −3.2 kcal/mol. In Figure 3g, N1 forms a hydrogen bond with the oxygen atom of Asp 412 at a distance of 2.58 Å, resulting in a substantial binding energy of −13.4 kcal/mol. Additionally, ligand atom N1 engages in hydrogen bonding interactions with the oxygen atoms (O) of Thr 436 and Gly 439 residues, observed at distances of 2.66 and 3.30 Å, respectively. The significance of these hydrogen bonds is underscored by their specific roles in fostering stability within the sericin–cisplatin complex with a particular emphasis on the involvement of Asp 910 and 412.
Figure 3.
Panel A illustrates the docking of sericin with cisplatin, revealing possible binding sites of cisplatin shown in boxes 1, 2, and 3, highlighted in yellow, purple, and orange, respectively. Notably, box 2 exhibits a higher likelihood of cisplatin binding compared to the other boxes. The most stable clusters in terms of energy and reproducibility, along with the most important amino acids involved in interactions, are shown in panels B, C, D, E, F, and G.
Development and Characterization of Protein-Based Nanocapsules
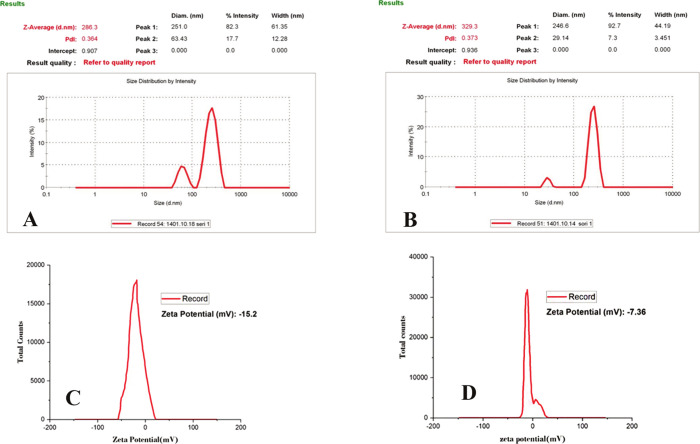
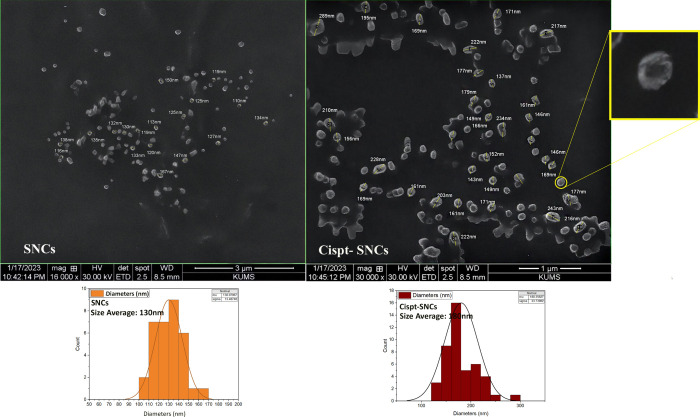
The nanoprecipitation method was used to synthesize these nanocarriers.34,42 Based on these studies, the 0.1% (w/v) sericin formulation was used throughout the experiments. DLS measurements of SNCs were relatively homogeneous with moderate size variability (286.3 nm, PDI = 0.364; Figure 4a). Loading cisplatin into SNCs resulted in a slight increase in size distribution, with a z-average size of 329.3 nm and size distribution of PDI = 0.373 (Figure 4b), indicating that the encapsulation process led to a slightly larger particle size compared to the SNC formulation. The zeta potential analysis indicated −15.2 mV for SNCs at pH 7.4 (Figure 4c). Subsequently, upon loading with cisplatin, the zeta potential of Cispt-SNCs dropped to −7.36 mV (Figure 4d). The scanning electron microscopy (SEM) analysis revealed a uniform distribution of Cispt-SNCs without aggregation, showcasing a spherical shape with an average diameter of approximately 180 nm. Additionally, it is noteworthy that the average diameter of SNCs determined by SEM was found to be 130 nm (Figure 5). The amount of EE (%) in single Cispt-SNCs was 84%. The cisplatin was loaded in the SNC, with a DL (%) of approximately 24%. The data presented in this study demonstrate the successful fabrication of self-assembled SNCs with a spherical morphology and a narrow size distribution.
Figure 4.
Size distribution analysis of (A) SNCs and (B) Cispt-SNCs. Zeta potential analysis of (C) SNCs and (D) Cispt-SNCs
Figure 5.
SEM analysis of SNCs and Cispt-SNCs with the average size of 130 and 180 nm, respectively.
FTIR
FTIR analysis (Figure 6) of cisplatin showed prominent peaks at 3460 and 3417 cm–1, indicating N–H stretching vibrations and the presence of NH3 ligands. Aquation processes replace chloride ligands with aqua ligands, confirmed by the peak at 3417 cm–1 corresponding to the O–H stretching vibrations. Minor peaks at 474 and 615 cm–1 suggest Pt–Cl stretching vibrations and the inclusion of chloride ligands in cisplatin’s coordination sphere before the aquation process. Spectra from sericin powder and Blank-SNC displayed similar peaks; therefore, only one spectrum was shown in the FTIR diagram for clarity. The stretching vibrations of N–H bonds at 3388 cm–1, corresponding to amine groups in sericin, were detected, along with the stretching vibrations of O–H bonds, indicating the presence of hydroxyl groups in the protein structure. The presence of proteins is indicated by the detection of an individual amide absorption band at 1654 cm–1 corresponding to the amide I band. This peak represents the stretching vibrations of C=O bonds. Two peaks observed at 1539 and 1525 cm–1 correspond to the amide II band, which arises from N–H bending vibrations. Additionally, a range of peaks between 1259 and 1384 cm–1, known as the amide III region (C–N stretching and N- H bending), indicate the presence of a random coil structure in sericin. Another peak observed at 1384 and 1460 cm–1 can be attributed to C–H and O–H bending vibrations as well as C–OH stretching vibrations of hydroxyl amino acid side chains, such as serine. This peak further confirms the presence of sericin residues within the sericin structure. The findings of FTIR analysis of CisPt-SNCs revealed that the incorporation of cisplatin into the sericin structure results in shifting peaks, new peaks, and altered intensities. In particular, the shifting of prominent peaks in the spectrum, such as those related to NH stretching and CH stretching, suggests changes in the vibrational modes due to the presence of cisplatin. In this spectrum, the shifting of prominent peaks from 3388 to 3402 cm–1 (NH stretching), 2924 to 2927 cm–1 (CH stretching), 1654 to 1656 cm–1 (C=O stretching), 1539 to 1541 cm–1 (N–H bending), 1384 to 1386 cm–1 (C–N stretching), and 615 to 621 cm–1 (Pt- ligand bending) suggests changes in the vibrational modes due to the presence of cisplatin.These shifts can be attributed to the formation of intermolecular hydrogen bonds between the NH and OH groups of sericin with cisplatin. The appearance of new peaks at 2742, 2677, 2627, and 2495 cm–1 further indicates the incorporation of cisplatin into the sericin structure. The increased intensity of peaks related to amide linkages suggests the formation of new amide linkages in Cispt SNCs, reflecting the interaction between sericin and cisplatin. Comparing the loaded sample to SNCs, notable changes are observed with some peaks removed while a new peak was observed. Peaks at 1109 and 1076 cm–1 present in the blank spectrum are removed, whereas a shoulder peak is observed at 1041 cm–1, indicating the presence of new vibrations in the Cispt-SNC spectrum. The findings suggest that the incorporation of cisplatin into sericin can enhance molecular interactions and the potential for controlled release and delivery of cisplatin using sericin-based carriers.
Figure 6.
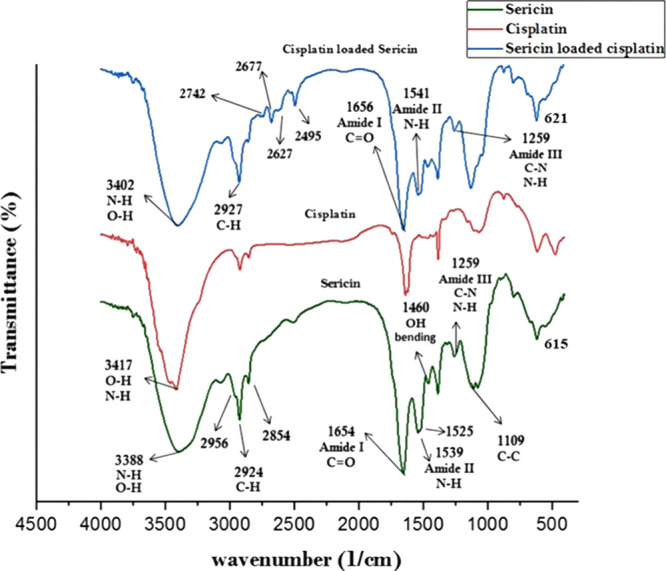
FTIR spectra of SNC, cisplatin, and Cispt-SNC.
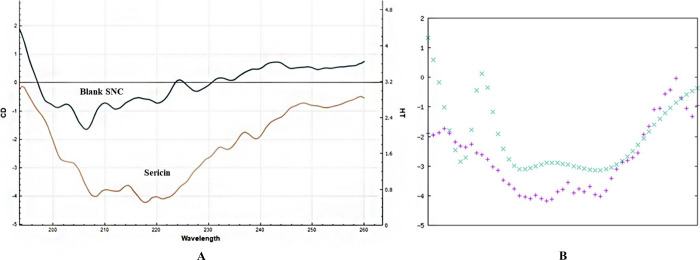
Circular Dichroism (CD)
The utilization of circular dichroism (CD) as a sensitive biophysical technique allowed for the detailed examination of protein conformational changes in both this current research and our previous study.41 In this study, CD was used to investigate the secondary structure of sericin, following its formation of nanocapsules through the nanoprecipitation method. By measuring the difference in absorption of right and left circularly polarized light, CD was able to determine the secondary structure of the protein (Figure 7a), which has a molecular weight of 198.6 kDa and 1758 amino acid residues. The CD spectra showed a weak negative band at 218 nm, indicative of the β-sheet structure in sericin. Additionally, the CD spectra of the sericin nanocapsules showed a good match to the native sericin spectrum with a weak negative band at 208 nm. Using the K2D3 software (Figure 7b), it was estimated that intact sericin had 10.27% β-sheet and 4.6% α-helix in its secondary structure. Overall, the results of this study suggest that the nanoprecipitation method successfully preserved both the primary and the secondary structures of the protein.
Figure 7.
(A) CD spectra of sericin and B-SNC. Sericin displayed a weak negative band at 218 nm, indicating a β-sheet structure. (B) The secondary structure was assessed using the K2D3 software.
Energy-Dispersive X-ray Analysis (EDX)
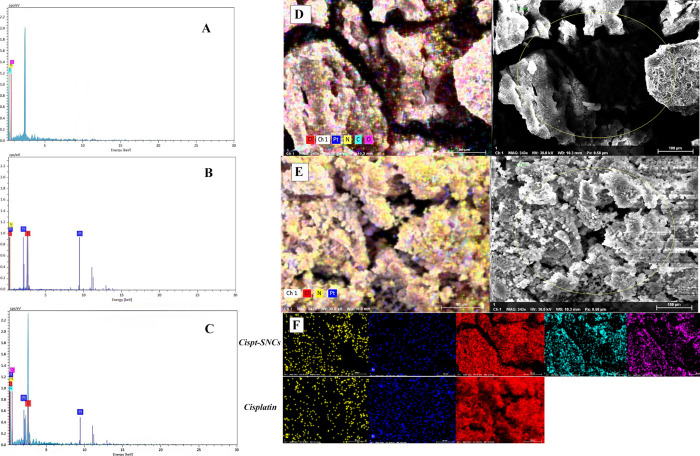
The element composition of SNC, Cispt-SNCs, and cisplatin was further analyzed through EDX. The EDX spectra captured during SEM showed the presence of Cl and Pt peaks for Cispt-SNCs (Figure 8a), which correlates with the FTIR spectra of the Cispt-SNCs that indicated the presence of cisplatin. Furthermore, when comparing EDX spectra from regions (Figure 8b,c) cisplatin with the same weight percentage for both Cispt-SNC and cisplatin samples, the peaks corresponding to Pt and Cl in the Cispt-SNCs are greatly reduced, and there was a 40% reduction in the wt % values calculated for these elements. To further examine the element composition and distribution of cisplatin in the SNCs, SEM and EDX mapping analysis from the surface was carried out (Figure 8d,e). The data shown illustrate that the content of Pt atom and the atomic ratio of platinum to nitrogen (Pt/N) of cisplatin are higher than those of the Cispt-SNCs. In the EDX mapping photos of Cispt-SNCs, platinum is navy blue, nitrogen is yellow, chlorine is red, carbon is pale blue, and oxygen is purple (Figure 8f). In this comparison, the amount of Pt on the surface of the synthesized sample of Cispt-SNCs is decreased significantly compared with the amount of Pt in the sample of cisplatin. Based on the EDX mapping photos. These results indicate that cisplatin was successfully incorporated within the SNCs, and notably, it was well distributed throughout the particle structure.
Figure 8.
EDX spectra of (A) SNCs (B) cisplatin (C) and Cispt-SNCs. EDX mapping images of the (D) element distribution of Cispt-SNCs, (E) cisplatin SEM-EDX, and (F) colorized elemental distribution (platinum is navy blue, nitrogen is yellow, chlorine is red, carbon is pale blue, and oxygen is purple).
In Vitro Drug Release
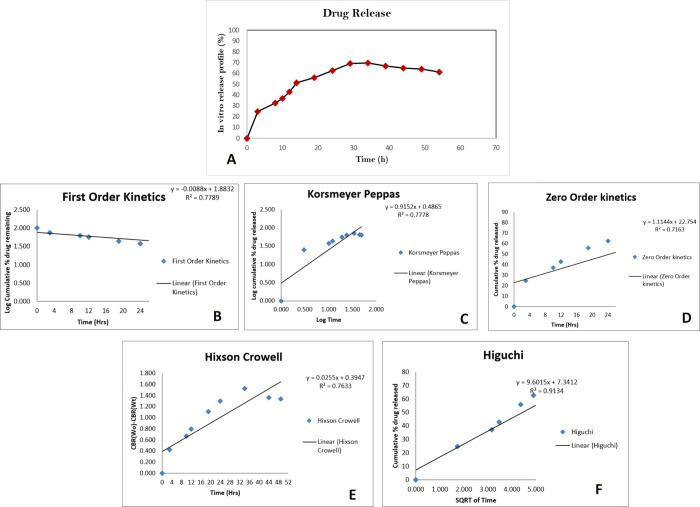
The in vitro release of cisplatin from Cispt-SNC was examined by the dialysis bag method under a pH value of 7.4 for up to 55 h (Figure 9a). No significant drug release was observed at the beginning, indicating the stability of the Cispt-SNCs. However, an initial burst release of 24.89% was detected after 3 h. The cumulative drug release gradually increased over time, with sustained drug release observed at 8 h (32.89%), 10 h (37.2%), and 12 h (43.05%). The release rate continued to rise, reaching 51.66% at 14 h and 55.97% at 19 h. At 24 h, the cumulative release of cisplatin reached 62.74%, and the release percentage gradually increased to 69.2% at 29 h and 69.82% at 34 h. From 39 to 54 h, the release percentage remained stable, ranging from 61.51 to 66.74%. The sustained release behavior indicates the ability of SNCs to maintain drug concentration within a therapeutic range over an extended period. The release kinetics of cisplatin was analyzed using various mathematical models, including first-order kinetics (R2=0.77, Figure 9b), Korsmeyer–Peppas (R2=0.77, Figure 9c), zero-order kinetics (R2=0.71, Figure 9d), Hixson–Crowell (R2=0.76, Figure 9e), and Higuchi (R2=0.91, Figure 9f). The cumulative percentage of drug release was monitored over time, and the data were fitted to these kinetic models. Among the tested models, Higuchi exhibited the highest coefficient of determination (R2 = 0.91), indicating a strong correlation between the observed and predicted values (Figure 9f). Therefore, the release of cisplatin from the formulation was found to follow Higuchi kinetics, suggesting a diffusion-controlled release mechanism. The Higuchi model, describing the release of drugs from an insoluble matrix as the square root of a time-dependent process based on the Fickian diffusion equation (Q = KHt1/2), aligns with the observed data. This model, derived by Professor Takeru Higuchi, provides valuable insights into drug release mechanisms and facilitates device optimization. The direct proportionality between the cumulative amount of drug released and the square root of time, as demonstrated by the Higuchi equation, holds significance in understanding and optimizing drug delivery systems. This analysis contributes to a comprehensive understanding of drug release behavior and aids in formulating effective controlled-release strategies.59
Figure 9.
(A) The in vitro release of cisplatin from SNCs at a pH value of 7.4. The release kinetics of cisplatin was analyzed using various mathematical models including first-order kinetics (B), Korsmeyer–Peppas (C), zero-order kinetics (D), Higuchi (F), and Hixson–Crowell (E).
In Vitro Cytotoxicity
To assess the cytotoxic effects in cell cultures, SNCs were tested against MCF-7 cells. The test results indicated that SNCs exhibited minimal cytotoxicity toward MCF-7 cells even when the concentration was raised to 1000 μg/mL. This finding highlights the great potential of SNCs in acting as an effective drug carrier for chemotherapy treatment.
Cell Viability
To evaluate the potential of cisplatin and CisPt-SNCs (at concentrations of 0.4, 0.6, 1, 2, 5, 10, 15, 25, and 50 μM) on the viability of MCF-7 cells at pH 7.4, the MTT assay was performed (Figure 10). After 24 h, a significant decrease in cell viability was observed between the control group and those treated with cisplatin (IC50 = 10 μM) and Cispt-SNCs (IC50 = 25 μM). Cells treated with Cispt-SNCs displayed a dose-dependent inhibition. At a concentration of 0.4 μM, there was an 80% reduction in cell viability in both cisplatin and CisPt-SNCs groups compared to the control group (p value of 0.0001%). However, no significant difference was observed in each concentration separately. Compared with free cisplatin, Cispt-SNCs had similar cytotoxicity at low cisplatin concentrations up to 5 μM. Notably, Cispt-SNCs acted with a gentle gradient decreasing cell viability, which represents the controlled release of cisplatin as confirmed in the release test. At 10 μM concentration, cisplatin achieved its IC50 and showed significant superiority (p value of 0.0001%) over both control and cisplatin loaded in SNCs, whereas the viability of cells treated with Cispt-SNCs was 67%. At a concentration of 25 μM, CisPt-SNCs reached their IC50, showcasing a significant difference (p value of 0.0001%) in comparison to cisplatin at the same concentration. As the concentration increased from 10 μM onward, significant differences were observed, particularly at concentrations of 15 and 50 μM, where significant variations were found between cisplatin and CisPt-SNCs with p values of 0.001 and 0.01%. By loading cisplatin into SNCs, the cell viability increased to 30% from 20% observed when treated with cisplatin alone at a final concentration of 50 μM. Collectively, the data showcases that at pH 7.4, Cispt-SNCs efficiently enable controlled release, successfully delivering drug doses to cancer cells, demonstrating enhanced cell viability compared to cisplatin alone, and indicating promising potential as carriers due to the gradient decrease in cell viability, signifying controlled drug release.
Figure 10.
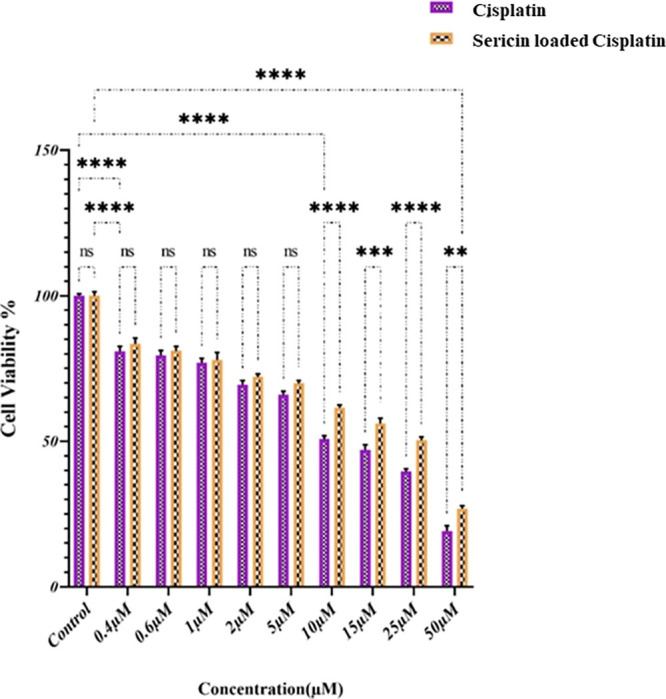
MCF-7 cell viability after treatment with various concentrations of Cispt-SNCs and cisplatin. Results are mean ± SD, *p ≤ 0.05 compared to respective controls. Results are reported in five replicates.
Intracellular Uptake and DAPI Staining
A photograph taken using fluorescence microscopy shows the morphology of MCF-7 cells treated with FITC-labeled Cispt-SNCs at the IC50 concentration (25 μM) for 4 h exposure at pH 6 (Figure 11). In our prior research, we conducted tests on sericin at varying pH levels to evaluate drug release and cell viability. The findings revealed that the highest drug release from this nanocarrier transpires at pH 6, albeit significantly compromising cell viability.41 Hence, we selected pH 6 for cellular uptake studies. The FITC-labeled Cispt-SNCs were taken up by the cells, potentially through endocytosis, and were found to be distributed in the cellular cytoplasm by interacting with the cytoskeleton fibers. The presence of cells stained by DAPI was also observed, which bind to DNA A-T rich regions and show nuclear condensation and DNA fragmentation. The captured images showed that the Cispt-SNC-treated cells underwent significant DNA fragmentation, chromatin condensation, and apoptosis.60 The merged fluorescent DAPI and FITC images also confirmed this result. Cancer cells display negatively charged surfaces directly linked to the secretion of lactic acid, an exclusive metabolic feature stemming from their high glycolysis rate.61 Additionally, these cells possess highly negative surface charges attributed to the accumulation of glycoproteins rich in sialic acid,62 allowing only positively charged nanoparticles to attach to them.63 By altering the nanocarrier surface charge from negative to positive through a charge reversal process, the FITC-labeled Cispt-SNCs were able to attach to the MCF-7 membrane easily and were picked up by these cells via transcytosis. The DAPI results revealed chromatin condensation and DNA fragmentation in MCF-7 cells, indicating DNA damage that leads to apoptosis. Therefore, the encapsulation of cisplatin in SNCs promotes MCF-7 cell apoptosis.60
Figure 11.
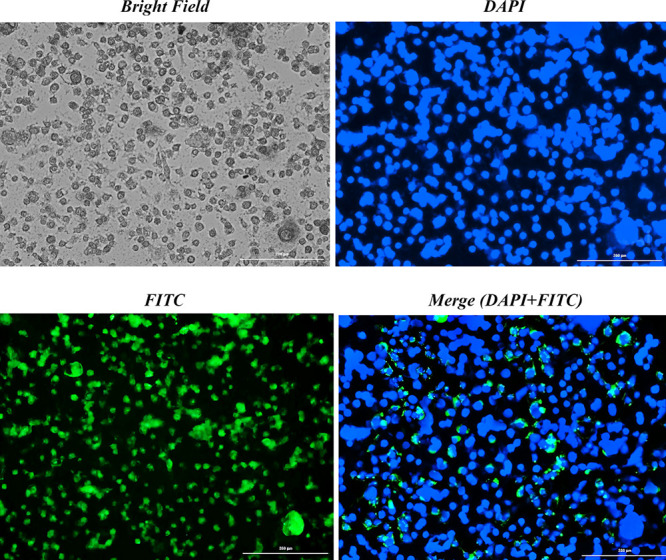
Intracellular uptake of FITC-labeled Cispt-SNCs at pH 6 alongside DAPI staining in MCF-7 cells, displayed with a bar scale of 200 nm.
Discussion
Platinum-based drugs, owing to their soft nucleophilic character, have a high affinity for binding peptides and proteins that contain sulfur residues of cysteine or methionine. Notably, these drugs tend to preferentially bind with glutathione, an antioxidant peptide. Despite this, their predominant targets are the nucleus and DNA.64 Although platinum-based compounds serve as effective chemotherapeutic agents, their impact on both cancerous and normal cells results in significant side effects. Unfortunately, the drugs also concentrate in other fast-growing tissues, causing unwanted toxicity. There are over 40 specific side effects associated with platinum-based drugs, which can be broadly grouped into seven categories, including nephrotoxicity, ototoxicity, neurotoxicity, cardiotoxicity, hematological toxicity, hepatotoxicity, and gastrointestinal toxicity, making them challenging to use.65 The application of nanotechnology stands as a promising solution toward alleviating the aforementioned challenges and increasing the effectiveness of cancer therapy through targeted drug delivery mechanisms to malignant cells.66,67 Of recent progress, nanomedicine denotes an ever-emerging domain of cancer diagnosis and treatment. Furthermore, proteins, characterized by their repeatable amino acid sequences, feature multiple functional groups, rendering them facile in their interaction with other biopolymers to produce nanocarriers, thereby improving the solubility, biocompatibility, nontoxicity, nonimmunogenicity, and bioavailability of bioactive agents.68 Protein nanocarriers have enormous potential in the field of oncology and can be an effective avenue for delivering bioactive substances. Encapsulating small-sized drugs in nanostructures can improve their pharmacokinetics and mitigate potential side effects.69 Specifically, we sought to explore the use of a sericin nanocarrier for delivering cisplatin in the form of Cispt-SNC and examine its potential to synergistically combat breast cancer in the MCF-7 cell line. In our molecular docking analyses, we aimed to unravel the intricate interactions between sericin and cisplatin with a focus on identifying the pivotal amino acids governing these bindings. Our research focused on elucidating the specific amino acids and active domains of sericin engaged in interactions with cisplatin, particularly emphasizing the influence on negatively charged amino acids, including aspartic and glutamic acid. These interactions are pivotal in modulating the zeta potential of the nanocarrier. Notably, amino acids Asp 412, Asp 910, Asp 497, and Glu1036 emerged as key contributors, underscoring their significant roles in orchestrating these interactions through the formation of hydrogen bonds. To determine the optimal protein concentration for preparing protein-based nanocapsules, five different sericin concentrations were tested (0.1, 0.2, 0.3, 0.5, and 1% w/v) through investigational experimentation. It is worth noting that the concentration of protein in these nanocarriers can influence their size,70 with higher protein concentrations leading to a wider range of nanoparticle sizes.71 Based on our findings, a 0.1% sericin concentration was selected for subsequent experiments. Our finding is in line with a previous study where the lowest concentration of sericin (0.1%)41 showed the appropriate nanoparticle size for SNCs (130 nm) and Cispt-SNCs (180 nm) with spherical morphology and the highest release efficiency of 69.82% at 34 h. Sericin, particularly at a low concentration of 0.1%, demonstrates excellent solubility,72 making it a highly promising candidate for antisolvent precipitation in nanoparticle production. This characteristic aligns with the properties of highly water-soluble proteins that have garnered significant interest for their application in antisolvent precipitation-based methods.43 In antisolvent precipitation, a soluble protein in a solvent is introduced to a nonsolvent (antisolvent), resulting in the precipitation of nanoparticles.73 Notably, sericin exhibits solubility in one phase (the solvent) while being insoluble in the other (the nonsolvent), showcasing its suitability for the antisolvent precipitation technique. It is generally accepted that small particles (<500 nm) can avoid the reticuloendothelial system (RES),74 resulting in a longer circulation time. Therefore, for proper cellular uptake and escape from RES, the size of the nanocarrier must be in this range. The zeta potential analysis of SNCs revealed a value of −15.2 mV at pH 7.4, indicating the robust stability of the nanocarriers and minimizing the risk of undesirable clearance by the reticuloendothelial system (RES).75 Also, nanoparticles that are negatively charged76 evoke a lower immune response compared to neutral or positively charged counterparts.77 The zeta potential of Cispt-SNCs decreased to −7.36 mV upon cisplatin loading. Cisplatin, being considered neutral, undergoes a ligand exchange in aqueous physiological environments, replacing the negatively charged chloro ligands with neutral aqua ligands. Thus, in physiological environments, cisplatin can acquire a positive charge,11,78 which can contribute to the alteration of the zeta potential of the Cispt-SNCs. These findings align with the outcomes derived from molecular docking, indicating that cisplatin’s interaction with negatively charged amino acids, such as aspartic acid and glutamic acid, resulted in the formation of the most stable clusters (Asp 412, 910, and 497 and Glu1036). Zeta potential alterations are influenced by the occurrence of individual functional groups contingent upon the ionic characteristics of the polyelectrolyte. Specifically, negatively charged groups such as carboxylic groups diminish the zeta potential, whereas positively charged groups like amino groups result in an elevation of the zeta potential value.79 This correlation suggests a potential link between these interactions and the observed decrease in the negative zeta potential of the nanocarrier (from −15.2 to −7.36) upon cisplatin encapsulation. The apparent affinity of cisplatin for negatively charged amino acids potentially contributes to the alteration of the overall charge of the nanocarrier. Notably, nanoparticles with a surface potential between −10 and +10 mV80 are less prone to phagocytosis and nonspecific interactions.81 SEM analysis depicted the uniform distribution of Cispt-SNCs without aggregations, presenting a spherical shape with an average diameter of around 180 nm. SEM, providing dry particle size, yielded an average diameter smaller than the Zetasizer, which measures in solutions and accounts for particle hydration-induced size increase.82 The SNCs, as analyzed through CD analysis, displayed a secondary structure comprising 10.27% β-sheet and 4.6% α-helix. β-Sheets,41 because of their greater flexibility and accessible binding sites, offer advantages for protein encapsulation compared to α-helix.83 The EDX mapping photos, through elemental analysis, confirmed the effective incorporation of cisplatin within the SNCs. This was evidenced by the presence of Pt and Cl elements evenly dispersed throughout the entire particle structure. Nanoparticle uptake by cells involves endocytosis, comprising phagocytosis and pinocytosis. Phagocytosis occurs in macrophages and polymorphonuclear neutrophils for large particles, whereas pinocytosis, involving mechanisms like macropinocytosis and clathrin-mediated endocytosis, caveolae-mediated endocytosis, and clathrin- and caveolae independent endocytosis, occurs in all cell types for smaller particles.84 The process includes binding to the cell membrane and internalization. Physicochemical characteristics, especially surface charge, influence binding, with positively charged nanoparticles favorably interacting with the negatively charged cell membrane.85 The tumor microenvironment plays a crucial role in determining the effectiveness of nanocarriers as drug delivery systems. Solid tumors typically exhibit pH values ranging from 6.0 to 7.0 due to carbonic anhydrase activity and lactic acid production resulting from anaerobic glycolysis. This acidic environment can trigger the protonation of functional groups on nanocarriers, causing a shift in their surface charge from negative/neutral to positive, which may lead to the collapse of the carrier and release of its drug cargo.86 Silk sericin, containing a high proportion of serine and aspartic acid, has the potential to self-assemble. The carboxyl groups on aspartic acid give sericin a net negative charge at neutral pH. Under the mild acidic conditions found in the cancer microenvironment, ionizable groups in sericin may undergo protonation, causing a charge reversal. This reversal not only disrupts electrostatic interactions between sericin’s amino acid side chains and facilitates the intracellular release of cisplatin (a drug that remains stable at low pH values) but also promotes the favorable interaction of positively charged SNCs with the negatively charged cell membrane.87 This stability in an acidic environment can potentially aid in the diffusion of SNC.88 This incorporation enhanced the cellular uptake by our nanocarrier. Our previous research that demonstrated that drug release from the SNCs was pH-dependent also showed high cell toxicity and drug release at pH 6.41 Furthermore, DAPI results in MCF-7 cells demonstrated chromatin condensation and DNA fragmentation, indicative of DNA damage and subsequent apoptosis. Hence, the encapsulation of cisplatin in SNCs actively promotes apoptosis in MCF-7 cells. At the 24 h mark, cisplatin significantly reduced MCF-7 cell viability, particularly at a concentration of 10 Μm. Cispt-SNC also displayed a decrease in cell viability mainly at concentrations of 25 and 50 μM. On the other hand, sericin exhibited remarkable antioxidant activity by preventing lipid peroxidation and oxidative stress.89 At lower doses, sericin demonstrated antioxidant and protective effects on both normal and tumor cells. However, at higher doses, its toxicity toward cancer cells becomes evident, which is consistent with previous observations.67 Additionally, the cytotoxic effects of Cispt-SNC were observed at concentrations of 25 and 50 μM on the MCF-7 cell viability, in line with the charge-reversal property of Cispt-SNC at low pH levels.
As a suggestion for future research, it is recommended that detailed molecular investigations be conducted into the intracellular mechanisms of CisPt-SNCs. Additionally, considering a consistent computational study could inspire potential mutational investigations on the carrier, with the aim of enhancing the efficiency of drug encapsulation and loading. These suggestions aim to contribute to the continuous improvement of protein-based nanocarrier studies.
Conclusions
This study explores the potential of SNCs for controlled cisplatin delivery in solid breast cancer therapy. We developed a pH-sensitive nanocarrier by using a straightforward and reproducible nanoprecipitation method. Cisplatin was encapsulated in this nanocarrier, exhibiting a charge-reversal property that switches its surface charge from negative to positive in mildly acidic environments. Optimizing the sericin concentration at 0.1% produced SNCs with small sizes and high drug encapsulation, crucial for efficient cellular uptake. Our analysis of SNCs and Cispt-SNCs confirmed their suitable size, surface charge, and morphology, supporting their potential for effective drug delivery. Chemical and conformational analyses via FTIR and EDX further validated the successful cisplatin encapsulation in sericin nanocarriers. The substantial drug release from SNCs over 54 h highlights their effectiveness in controlled drug delivery. Controlled drug release is indicated by a gradient decrease in cell viability, highlighting SNCs as promising carriers. In vitro experiments reveal the cytotoxic effects of cisplatin and Cispt-SNCs on MCF-7 cells, resulting in decreased cell viability at their respective IC50 concentrations of 10 and 25 μM. Interaction between Cispt-SNCs and MCF-7 cells induces apoptosis, emphasizing the system’s potential. This study underscores the promise of protein-based nanocarriers in refining cancer therapy precision, providing impetus for further exploration in oncology. The unique sericin–cisplatin combination represents a significant advancement in targeted drug delivery, highlighting natural protein based-nanotechnology’s role in cancer treatment.
Acknowledgments
This study was supported financially by Kermanshah University of Medical Sciences.
The authors declare no competing financial interest.
References
- Becerra-Tomás N.; Balducci K.; Abar L.; Aune D.; Cariolou M.; Greenwood D. C.; Markozannes G.; Nanu N.; Vieira R.; Giovannucci E. L.; Gunter M. J.; Jackson A. A.; Kampman E.; Lund V.; Allen K.; Brockton N. T.; Croker H.; Katsikioti D.; McGinley-Gieser D.; Mitrou P.; Wiseman M.; Cross A. J.; Riboli E.; Clinton S. K.; McTiernan A.; Norat T.; Tsilidis K. K.; Chan D. S. M. Postdiagnosis dietary factors, supplement use and breast cancer prognosis: Global Cancer Update Programme (CUP Global) systematic literature review and meta-analysis. Int. J. Cancer 2023, 152 (4), 616–634. 10.1002/ijc.34321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R. L.; Miller K. D.; Wagle N. S.; Jemal A. Cancer statistics, 2023. Ca Cancer J. Clin 2023, 73 (1), 17–48. 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- Gunasekaran G.; Bekki Y.; Lourdusamy V.; Schwartz M. Surgical treatments of hepatobiliary cancers. Hepatology 2021, 73, 128–136. 10.1002/hep.31325. [DOI] [PubMed] [Google Scholar]
- Chidambaram M.; Manavalan R.; Kathiresan K. Nanotherapeutics to overcome conventional cancer chemotherapy limitations. Journal of pharmacy & pharmaceutical sciences 2011, 14 (1), 67–77. 10.18433/J30C7D. [DOI] [PubMed] [Google Scholar]
- Yao Y.; Zhou Y.; Liu L.; Xu Y.; Chen Q.; Wang Y.; Wu S.; Deng Y.; Zhang J.; Shao A. Nanoparticle-based drug delivery in cancer therapy and its role in overcoming drug resistance. Front. Mol. Biosci. 2020, 7, 193. 10.3389/fmolb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davodabadi F.; Sajjadi S. F.; Sarhadi M.; Mirghasemi S.; Nadali Hezaveh M.; Khosravi S.; Kamali Andani M.; Cordani M.; Basiri M.; Ghavami S. Cancer chemotherapy resistance: Mechanisms and recent breakthrough in targeted drug delivery. Eur. J. Pharmacol. 2023, 958, 176013 10.1016/j.ejphar.2023.176013. [DOI] [PubMed] [Google Scholar]
- Galanski M.; Jakupec M. A.; Keppler B. K. Update of the preclinical situation of anticancer platinum complexes: novel design strategies and innovative analytical approaches. Curr. Med. Chem. 2005, 12 (18), 2075–2094. 10.2174/0929867054637626. [DOI] [PubMed] [Google Scholar]
- Rosenberg B.; Vancamp L.; Trosko J. E.; Mansour V. H. Platinum compounds: a new class of potent antitumour agents. nature 1969, 222 (5191), 385–386. 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- Szefler B.; Czeleń P. Will the Interactions of Some Platinum (II)-Based Drugs with B-Vitamins Reduce Their Therapeutic Effect in Cancer Patients? Comparison of Chemotherapeutic Agents such as Cisplatin, Carboplatin and Oxaliplatin—A Review. International Journal of Molecular Sciences 2023, 24 (2), 1548. 10.3390/ijms24021548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari S.; Bernard Tchounwou P. Cisplatin in cancer therapy: molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovec T. Cisplatin and beyond: molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiology and oncology 2019, 53 (2), 148–158. 10.2478/raon-2019-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gately D.; Howell S. Cellular accumulation of the anticancer agent cisplatin: a review. British journal of cancer 1993, 67 (6), 1171–1176. 10.1038/bjc.1993.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S.; Lee J.; Thiele D. J.; Herskowitz I. Uptake of the anticancer drug cisplatin mediated by the copper transporter Ctr1 in yeast and mammals. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (22), 14298–14302. 10.1073/pnas.162491399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. S.; Berners-Price S. J.; Hambley T. W. Slowing of cisplatin aquation in the presence of DNA but not in the presence of phosphate: improved understanding of sequence selectivity and the roles of monoaquated and diaquated species in the binding of cisplatin to DNA. Inorganic chemistry 2000, 39 (25), 5603–5613. 10.1021/ic000847w. [DOI] [PubMed] [Google Scholar]
- Patra J. K.; Das G.; Fraceto L. F.; Campos E. V. R.; Rodriguez-Torres M. d. P.; Acosta-Torres L. S.; Diaz-Torres L. A.; Grillo R.; Swamy M. K.; Sharma S.; Habtemariam S.; Shin H. S. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018, 16 (1), 1–33. 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J.; Lane L. A.; Nie S. Stimuli-responsive nanoparticles for targeting the tumor microenvironment. J. Controlled Release 2015, 219, 205–214. 10.1016/j.jconrel.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.; Chen D.; Li L.; Sun K. Charge reversal nano-systems for tumor therapy. J. Nanobiotechnol. 2022, 20, 1–27. 10.1186/s12951-021-01221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D.; Xu Z.; Hu Z.; Hu B.; Yang M.; Zhu L. pH-triggered charge-reversal silk sericin-based nanoparticles for enhanced cellular uptake and doxorubicin delivery. ACS Sustainable Chem. Eng. 2017, 5 (2), 1638–1647. 10.1021/acssuschemeng.6b02392. [DOI] [Google Scholar]
- Kariduraganavar M. Y.; Heggannavar G. B.; Amado S.; Mitchell G. R.. Protein nanocarriers for targeted drug delivery for cancer therapy. In Nanocarriers for Drug Delivery; Elsevier, 2019; pp 173–204. [Google Scholar]
- Narwade M.; Gajbhiye V.; Gajbhiye K. R.. Protein nanocapsules as a smart drug delivery platform. In Stimuli-Responsive Nanocarriers; Elsevier, 2022; pp 393–412. [Google Scholar]
- Elzoghby A. O.; Abo El-Fotoh W. S.; Elgindy N. A. Casein-based formulations as promising controlled release drug delivery systems. J. Controlled Release 2011, 153 (3), 206–216. 10.1016/j.jconrel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Chen L.; Remondetto G. E.; Subirade M. Food protein-based materials as nutraceutical delivery systems. Trends in Food Science & Technology 2006, 17 (5), 272–283. 10.1016/j.tifs.2005.12.011. [DOI] [Google Scholar]
- Elzoghby A. O.; Samy W. M.; Elgindy N. A. Albumin-based nanoparticles as potential controlled release drug delivery systems. Journal of controlled release 2012, 157 (2), 168–182. 10.1016/j.jconrel.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Lamboni L.; Gauthier M.; Yang G.; Wang Q. Silk sericin: A versatile material for tissue engineering and drug delivery. Biotechnology advances 2015, 33 (8), 1855–1867. 10.1016/j.biotechadv.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Seo S.-J.; Das G.; Shin H.-S.; Patra J. K. Silk Sericin Protein Materials: Characteristics and Applications in Food-Sector Industries. International Journal of Molecular Sciences 2023, 24 (5), 4951. 10.3390/ijms24054951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S. C.; Dash B. C.; Dash R.; Kaplan D. L. Natural protective glue protein, sericin bioengineered by silkworms: Potential for biomedical and biotechnological applications. Prog. Polym. Sci. 2008, 33 (10), 998–1012. 10.1016/j.progpolymsci.2008.08.002. [DOI] [Google Scholar]
- Shitole M.; Dugam S.; Tade R.; Nangare S.. Pharmaceutical applications of silk sericin. In Annales pharmaceutiques francaises; Elsevier, 2020, Vol. 78, pp 469–486. [DOI] [PubMed] [Google Scholar]
- Wu J.-H.; Wang Z.; Xu S.-Y. Preparation and characterization of sericin powder extracted from silk industry wastewater. Food chemistry 2007, 103 (4), 1255–1262. 10.1016/j.foodchem.2006.10.042. [DOI] [Google Scholar]
- Vepari C.; Kaplan D. L. Silk as a Biomaterial. Prog. Polym. Sci. 2007, 32 (8–9), 991–1007. 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Q. Applications of natural silk protein sericin in biomaterials. Biotechnology advances 2002, 20 (2), 91–100. 10.1016/S0734-9750(02)00003-4. [DOI] [PubMed] [Google Scholar]
- Zhaorigetu S.; Yanaka N.; Sasaki M.; Watanabe H.; Kato N. Inhibitory effects of silk protein, sericin on UVB-induced acute damage and tumor promotion by reducing oxidative stress in the skin of hairless mouse. Journal of Photochemistry and Photobiology B: Biology 2003, 71 (1–3), 11–17. 10.1016/S1011-1344(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Takechi T.; Wada R.; Fukuda T.; Harada K.; Takamura H. Antioxidant activities of two sericin proteins extracted from cocoon of silkworm (Bombyx mori) measured by DPPH, chemiluminescence. ORAC and ESR methods. Biomedical Reports 2014, 2 (3), 364–369. 10.3892/br.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez Rivas C. J.; Tarhini M.; Badri W.; Miladi K.; Greige-Gerges H.; Nazari Q. A.; Galindo Rodríguez S. A.; Román R. Á.; Fessi H.; Elaissari A. Nanoprecipitation process: From encapsulation to drug delivery. Int. J. Pharm. 2017, 532 (1), 66–81. 10.1016/j.ijpharm.2017.08.064. [DOI] [PubMed] [Google Scholar]
- Reverchon E. Supercritical antisolvent precipitation of micro-and nano-particles. journal of supercritical fluids 1999, 15 (1), 1–21. 10.1016/S0896-8446(98)00129-6. [DOI] [Google Scholar]
- Ramos R.; Koh K.; Gabryelczyk B.; Chai L.; Kanagavel D.; Yan X.; Ganachaud F.; Miserez A.; Bernard J. Nanocapsules Produced by Nanoprecipitation of Designed Suckerin-Silk Fusion Proteins. ACS Macro Lett. 2021, 10 (5), 628–634. 10.1021/acsmacrolett.1c00171. [DOI] [PubMed] [Google Scholar]
- UniProt: a hub for protein information. Nucleic Acids Res. 2015, 43 (D1), D204–D212. 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A.; Kucukural A.; Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols 2010, 5 (4), 725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A.; Xu D.; Poisson J.; Zhang Y. A protocol for computer-based protein structure and function prediction. J. Visualized Exp. 2011, 57, e3259 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A.; Yang J.; Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic acids research 2012, 40 (W1), W471–W477. 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry 2009, 30 (16), 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghaz F.; Asadi Z.; Sajadimajd S.; Kashfi K.; Arkan E.; Rahimi Z. Codelivery of resveratrol melatonin utilizing pH responsive sericin based nanocarriers inhibits the proliferation of breast cancer cell line at the different pH. Sci. Rep. 2023, 13 (1), 11090. 10.1038/s41598-023-37668-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans L. C.; Buzgo M.; Simaite A.. Optimization of protein precipitation for high-loading drug delivery systems for immunotherapeutics. In Proceedings; MDPI, 2020, Vol. 78, p 29. [Google Scholar]
- Joye I. J.; McClements D. J. Production of nanoparticles by anti-solvent precipitation for use in food systems. Trends in Food Science & Technology 2013, 34 (2), 109–123. 10.1016/j.tifs.2013.10.002. [DOI] [Google Scholar]
- Kanoujia J.; Faizan M.; Parashar P.; Singh N.; Saraf S. A. Curcumin loaded sericin nanoparticles: Assessment for biomedical application. Food Hydrocolloids for Health 2021, 1, 100029 10.1016/j.fhfh.2021.100029. [DOI] [Google Scholar]
- Lv Y.; He H.; Qi J.; Lu Y.; Zhao W.; Dong X.; Wu W. Visual validation of the measurement of entrapment efficiency of drug nanocarriers. International journal of pharmaceutics 2018, 547 (1–2), 395–403. 10.1016/j.ijpharm.2018.06.025. [DOI] [PubMed] [Google Scholar]
- Wang B.; Xu X.; Li B.; Wei Z.; Lu S.; Li J.; Liu K.; Zhang H.; Wang F.; Yang Y. Protein-based nanocarriers for efficient Etoposide delivery and cancer therapy. Nano Res. 2023, 1–5. 10.1007/s12274-023-5841-5. [DOI] [Google Scholar]
- Cheng Z.; Li Y.; Zhao D.; Zhao W.; Wu M.; Zhang W.; Cui Y.; Zhang P.; Zhang Z. Nanocarriers for intracellular co-delivery of proteins and small-molecule drugs for cancer therapy. Frontiers in Bioengineering and Biotechnology 2022, 10, 994655 10.3389/fbioe.2022.994655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarhini M.; Greige-Gerges H.; Elaissari A. Protein-based nanoparticles: From preparation to encapsulation of active molecules. International journal of pharmaceutics 2017, 522 (1–2), 172–197. 10.1016/j.ijpharm.2017.01.067. [DOI] [PubMed] [Google Scholar]
- D’Souza S. A Review ofIn VitroDrug Release Test Methods for Nano-Sized Dosage Forms. Advances in Pharmaceutics 2014, 2014, 1–12. 10.1155/2014/304757. [DOI] [Google Scholar]
- Motiei M.; Kashanian S. Novel amphiphilic chitosan nanocarriers for sustained oral delivery of hydrophobic drugs. European journal of pharmaceutical sciences 2017, 99, 285–291. 10.1016/j.ejps.2016.12.035. [DOI] [PubMed] [Google Scholar]
- Abdel-Mottaleb M. M.; Lamprecht A. Standardized in vitro drug release test for colloidal drug carriers using modified USP dissolution apparatus I. Drug development and industrial pharmacy 2011, 37 (2), 178–184. 10.3109/03639045.2010.502534. [DOI] [PubMed] [Google Scholar]
- Geyer F. C.; Rodrigues D. N.; Weigelt B.; Reis-Filho J. S. Molecular classification of estrogen receptor-positive/luminal breast cancers. Advances in Anatomic Pathology 2012, 19 (1), 39–53. 10.1097/PAP.0b013e31823fafa0. [DOI] [PubMed] [Google Scholar]
- Yu S.; Kim T.; Yoo K. H.; Kang K. The T47D cell line is an ideal experimental model to elucidate the progesterone-specific effects of a luminal A subtype of breast cancer. Biochem. Biophys. Res. Commun. 2017, 486 (3), 752–758. 10.1016/j.bbrc.2017.03.114. [DOI] [PubMed] [Google Scholar]
- Wright P. K.; Jones S. B.; Ardern N.; Ward R.; Clarke R. B.; Sotgia F.; Lisanti M. P.; Landberg G.; Lamb R. 17β-estradiol regulates giant vesicle formation via estrogen receptor-alpha in human breast cancer cells. Oncotarget 2014, 5 (10), 3055. 10.18632/oncotarget.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath N.; Vassell R.; Chattopadhyay M.; Kogan M.; Kashfi K. Nitro-aspirin inhibits MCF-7 breast cancer cell growth: effects on COX-2 expression and Wnt/β-catenin/TCF-4 signaling. Biochemical pharmacology 2009, 78 (10), 1298–1304. 10.1016/j.bcp.2009.06.104. [DOI] [PubMed] [Google Scholar]
- Ahmadian S.; Barar J.; Saei A. A.; Fakhree M. A. A.; Omidi Y. Cellular toxicity of nanogenomedicine in MCF-7 cell line: MTT assay. J. Visualized Exp. 2009, 26, e1191 10.3791/1191-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M.; Kodela R.; Nath N.; Dastagirzada Y. M.; Velázquez-Martínez C. A.; Boring D.; Kashfi K. Hydrogen sulfide-releasing NSAIDs inhibit the growth of human cancer cells: a general property and evidence of a tissue type-independent effect. Biochemical pharmacology 2012, 83 (6), 715–722. 10.1016/j.bcp.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Castro-Alvarez A.; Costa A. M.; Vilarrasa J. The performance of several docking programs at reproducing protein–macrolide-like crystal structures. Molecules 2017, 22 (1), 136. 10.3390/molecules22010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paarakh M. P.; Jose P. A.; Setty C. M.; Peterchristoper G. V. Release kinetics–concepts and applications. Int. J. Pharm. Res. Technol. 2019, 8 (1), 12–20. 10.31838/ijprt/08.01.02. [DOI] [Google Scholar]
- Badran A.; Tul-Wahab A.; Zafar H.; Mohammad N.; Imad R.; Ashfaq Khan M.; Baydoun E.; Choudhary M. I. Antipsychotics drug aripiprazole as a lead against breast cancer cell line (MCF-7) in vitro. PLoS One 2020, 15 (8), e0235676 10.1371/journal.pone.0235676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le W.; Chen B.; Cui Z.; Liu Z.; Shi D. Detection of cancer cells based on glycolytic-regulated surface electrical charges. Biophysics Reports 2019, 5, 10–18. 10.1007/s41048-018-0080-0. [DOI] [Google Scholar]
- Li Z.; Ruan J.; Zhuang X. Effective capture of circulating tumor cells from an S180-bearing mouse model using electrically charged magnetic nanoparticles. J. Nanobiotechnol. 2019, 17 (1), 1–9. 10.1186/s12951-019-0491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Le W.; Wang Y.; Li Z.; Wang D.; Lin L.; Cui S.; Hu J. J.; Hu Y.; Yang P.; Ewing R. C.; Shi D.; Cui Z. Targeting negative surface charges of cancer cells by multifunctional nanoprobes. Theranostics 2016, 6 (11), 1887. 10.7150/thno.16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone T. C.; Suntharalingam K.; Lippard S. J. The next generation of platinum drugs: targeted Pt (II) agents, nanoparticle delivery, and Pt (IV) prodrugs. Chem. Rev. 2016, 116 (5), 3436–3486. 10.1021/acs.chemrev.5b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oun R.; Moussa Y. E.; Wheate N. J. The side effects of platinum-based chemotherapy drugs: a review for chemists. Dalton transactions 2018, 47 (19), 6645–6653. 10.1039/C8DT00838H. [DOI] [PubMed] [Google Scholar]
- Farokhzad O. C.; Langer R. Impact of nanotechnology on drug delivery. ACS Nano 2009, 3 (1), 16–20. 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- Ammar H. O.; Shamma R. N.; Elbatanony R. S.; Khater B. Antioxidants in cancer therapy: recent trends in application of nanotechnology for enhanced delivery. Scientia Pharmaceutica 2020, 88 (1), 5. 10.3390/scipharm88010005. [DOI] [Google Scholar]
- Wen C.; Zhang J.; Zhang H.; Duan Y. New perspective on natural plant protein-based nanocarriers for bioactive ingredients delivery. Foods 2022, 11 (12), 1701. 10.3390/foods11121701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqosaibi A. I. Nanocarriers for anticancer drugs: Challenges and perspectives. Saudi J. Biol. Sci. 2022, 29 (6), 103298 10.1016/j.sjbs.2022.103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi Kajani A.; Haghjooy Javanmard S.; Asadnia M.; Razmjou A. Recent advances in nanomaterials development for nanomedicine and cancer. ACS Applied Bio Materials 2021, 4 (8), 5908–5925. 10.1021/acsabm.1c00591. [DOI] [PubMed] [Google Scholar]
- Sandra F.; Khaliq N. U.; Sunna A.; Care A. Developing protein-based nanoparticles as versatile delivery systems for cancer therapy and imaging. Nanomaterials 2019, 9 (9), 1329. 10.3390/nano9091329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao G.; Cai R.; Wang Y.; Liu L.; Zuo H.; Zhao P.; Umar A.; Mao C.; Xia Q.; He H. Bioinspired design of AgNPs embedded silk sericin-based sponges for efficiently combating bacteria and promoting wound healing. Materials & Design 2019, 180, 107940 10.1016/j.matdes.2019.107940. [DOI] [Google Scholar]
- Khan S. A.; Schneider M. Improvement of nanoprecipitation technique for preparation of gelatin nanoparticles and potential macromolecular drug loading. Macromol. Biosci. 2013, 13 (4), 455–463. 10.1002/mabi.201200382. [DOI] [PubMed] [Google Scholar]
- Kommareddy S.; Amiji M. Poly (ethylene glycol)–modified thiolated gelatin nanoparticles for glutathione-responsive intracellular DNA delivery. Nanomedicine: nanotechnology, biology and medicine 2007, 3 (1), 32–42. 10.1016/j.nano.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K.; Li Y.; Luo J.; Lee J. S.; Xiao W.; Gonik A. M.; Agarwal R. G.; Lam K. S. The effect of surface charge on in vivo biodistribution of PEG-oligocholic acid based micellar nanoparticles. Biomaterials 2011, 32 (13), 3435–3446. 10.1016/j.biomaterials.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Zhang R.; Xu Z. P. Nanoparticle-based nanomedicines to promote cancer immunotherapy: recent advances and future directions. Small 2019, 15 (32), 1900262 10.1002/smll.201900262. [DOI] [PubMed] [Google Scholar]
- Arvizo R. R.; Miranda O. R.; Moyano D. F.; Walden C. A.; Giri K.; Bhattacharya R.; Robertson J. D.; Rotello V. M.; Reid J. M.; Mukherjee P. Modulating pharmacokinetics, tumor uptake and biodistribution by engineered nanoparticles. PloS one 2011, 6 (9), e24374 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corinti D.; Coletti C.; Re N.; Piccirillo S.; Giampa M.; Crestoni M. E.; Fornarini S. Hydrolysis of cis-and transplatin: structure and reactivity of the aqua complexes in a solvent free environment. RSC Adv. 2017, 7 (26), 15877–15884. 10.1039/C7RA01182B. [DOI] [Google Scholar]
- Ostolska I.; Wiśniewska M. Application of the zeta potential measurements to explanation of colloidal Cr(2)O(3) stability mechanism in the presence of the ionic polyamino acids. Colloid Polym. Sci. 2014, 292 (10), 2453–2464. 10.1007/s00396-014-3276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernsting M. J.; Murakami M.; Roy A.; Li S.-D. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. Journal of controlled release 2013, 172 (3), 782–794. 10.1016/j.jconrel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.; Bhatia S. Nanoparticles types, classification, characterization, fabrication methods and drug delivery applications. Natural Polymer Drug Delivery Systems: Nanoparticles, Plants, and Algae 2016, 33–93. 10.1007/978-3-319-41129-3_2. [DOI] [Google Scholar]
- Liu M.; Zhang X.; Yang B.; Deng F.; Ji J.; Yang Y.; Huang Z.; Zhang X.; Wei Y. Luminescence tunable fluorescent organic nanoparticles from polyethyleneimine and maltose: facile preparation and bioimaging applications. RSC Adv. 2014, 4 (43), 22294–22298. 10.1039/c4ra03103b. [DOI] [Google Scholar]
- Kopeć K.; Pędziwiatr M.; Gront D.; Sztatelman O.; Sławski J.; Łazicka M.; Worch R.; Zawada K.; Makarova K.; Nyk M.; Grzyb J. Comparison of α-helix and β-sheet structure adaptation to a quantum dot geometry: Toward the identification of an optimal motif for a protein nanoparticle cover. ACS Omega 2019, 4 (8), 13086–13099. 10.1021/acsomega.9b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Byrne J. D.; Napier M. E.; DeSimone J. M. More effective nanomedicines through particle design. small 2011, 7 (14), 1919–1931. 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest V.; Pourchez J. Preferential binding of positive nanoparticles on cell membranes is due to electrostatic interactions: A too simplistic explanation that does not take into account the nanoparticle protein corona. Materials Science and Engineering: C 2017, 70, 889–896. 10.1016/j.msec.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Mo R.; Sun Q.; Xue J.; Li N.; Li W.; Zhang C.; Ping Q. Multistage pH-responsive liposomes for mitochondrial-targeted anticancer drug delivery. Advanced materials 2012, 24 (27), 3659–3665. 10.1002/adma.201201498. [DOI] [PubMed] [Google Scholar]
- Li Z.; Gao Y.; Li W.; Li Y.; Lv H.; Zhang D.; Peng J.; Cheng W.; Mei L.; Chen H. Charge-reversal nanomedicines as a smart bullet for deep tumor penetration. Smart Mater. Med. 2022, 3, 243–253. 10.1016/j.smaim.2022.01.008. [DOI] [Google Scholar]; Das G.; Shin H.-S.; Campos E. V. R.; Fraceto L. F.; del Pilar Rodriguez-Torres M.; Mariano K. C. F.; de Araujo D. R.; Fernández-Luqueño F.; Grillo R.; Patra J. K. Sericin based nanoformulations: a comprehensive review on molecular mechanisms of interaction with organisms to biological applications. J. Nanobiotechnol. 2021, 19, 1–22. 10.1186/s12951-021-00774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbownik A.; Szałek E.; Urjasz H.; Głęboka A.; Mierzwa E.; Grześkowiak E. The physical and chemical stability of cisplatin (Teva) in concentrate and diluted in sodium chloride 0.9%. Contemp. Oncol./Współczesna Onkol. 2012, 16 (5), 435–439. 10.5114/wo.2012.31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel G. A.; Álvarez-López C.. Extraction and antioxidant activity of sericin, a protein from silk. Brazilian Journal of Food Technology 2020, 23. [Google Scholar]