Abstract
Introduction:
This study aims to explore machine learning (ML) methods for early prediction of Alzheimer’s disease (AD) and related dementias (ADRD) using the real-world electronic health records (EHRs).
Methods:
A total of 23,835 ADRD and 1,038,643 control patients were identified from the OneFlorida+ Research Consortium. Two ML methods were used to develop the prediction models. Both knowledge-driven and data-driven approaches were explored. Four computable phenotyping algorithms were tested.
Results:
The gradient boosting tree (GBT) models trained with the data-driven approach achieved the best area under the curve (AUC) scores of 0.939, 0.906, 0.884, and 0.854 for early prediction of ADRD 0, 1, 3, or 5 years before diagnosis, respectively. A number of important clinical and sociodemographic factors were identified.
Discussion:
We tested various settings and showed the predictive ability of using ML approaches for early prediction of ADRD with EHRs. The models can help identify high-risk individuals for early informed preventive or prognostic clinical decisions.
Keywords: Alzheimer’s disease (AD), Alzheimer’s disease and related dementias (ADRD), data-driven approach, machine learning, real-world data, risk prediction
1 |. INTRODUCTION
Alzheimer’s disease (AD) and related dementias (ADRD) is a class of complicated neurodegenerative disorders with symptoms ranging from short-term memory lapses to loss of bodily function until death.1 ADRD gradually diminishes the quality of life of the affected older population. An estimated 6.5 million Americans 65 years of age or older are living with AD—the sixth leading cause of death in 2022; and by 2050, 12.7 million people age 65 and older are projected to have AD.2 There is still no effective treatment for AD to date despite decades of investment.3–5 More than 99% of AD/ADRD-related clinical trials have failed to develop effective treatments and there have been only six U.S. Food and Drug Administration (FDA)–approved drugs (i.e., rivastigmine, galantamine, donepezil, memantine, manufactured combination of memantine and donepezil, and recently approved aducanumab) for alleviating the symptoms of ADRD since 1998.6,7 One potential reason for the high failure rate is that it may be too late to give the treatments to patients when the dementia is already symptomatic.3 Furthermore, underdiagnosis of ADRD is significant, especially in the primary care setting.2 Among older adults with probable dementia, 58.7% were either not diagnosed (39.5%) by a physician or unaware (19.2%) that they had the disease.8 Increasing evidence suggests that early recognition of ADRD is critical, not only to give patients with ADRD patients more time to prepare for the future, for example, referral to specialty care and to initiate timely preventive actions (e.g., diet and lifestyle changes) or early treatments of symptoms (e.g., diet and lifestyle changes), but also to help researchers better identify and characterize clinical trial participants.9 The neurodegenerative processes of ADRD start years before the onset of clinical symptoms and prior to diagnoses, leaving a wide time window of opportunities for early prediction and identification of risk factors.10
Many data sources have been explored for the prediction of ADRD,11,12 and various predictive modeling techniques including both statistical and machine learning (ML) models have been employed.13 For example, previous studies have investigated neuroimaging data such as those from the Alzheimer’s Disease Neuroimaging Initiative (ADNI),14,15 which demonstrated good performance,16 with many of these studies focused on predicting the conversion from mild cognitive impairment (MCI), already a clinical stage, to dementia due to ADRD.17–19 However, because they are expensive, neuroimaging procedures are normally ordered only for patients who either already have clinical symptoms or are at high-risk, making neuroimaging-based prediction methods impractical in real-world settings for early prediction.20,21
With the rapid adoption and continuous improvement of electronic health record (EHR) systems in the United States, large collections of longitudinal EHRs are becoming available for clinical research.22,23 Many risk factors of ADRD identified by previous studies (e.g., obesity, hypertension, high cholesterol) are routinely captured in patients’ EHRs.11 Recently, several studies have explored EHR data for ADRD risk prediction. Patient information such as diagnoses, medications, laboratory tests, and other medical procedures were investigated as predictors. For example, Nori et al. used de-identified administrative claims and EHR data from the OptumLabs and developed a predictive model using gradient boosting tree (GBT).24 Park et al. explored three ML algorithms including logistic regression (LR), support vector machines (SVMs), and random forests (RFs) using an administrative healthcare data set (e.g., claims and health check-ups) from South Korea.25 Both Nori et al. and Park et al. followed a data-driven strategy where all discrete diagnoses codes, medications, and procedures were used in building the predictive models. However, they did not consider existing knowledge of ADRD risk factors from domain experts or existing literature. Furthermore, Park et al. tested only two operational definitions of AD, that is, definite AD via diagnosed codes combined with dementia medication, and probable AD via only diagnosis codes;25 and the sample sizes were also small (i.e., n = 614 for definitive AD, and n = 2026 for probable AD). However, use of diagnostic codes alone is not accurate for identifying research-quality cohorts using EHRs,26–28 including for MCI29 and AD.30,31 This is why computable phenotypes (CPs, i.e., “clinical conditions, characteristics, or sets of clinical features that can be determined solely from EHRs and ancillary data sources.”) are needed.32–34 The National Institute on Aging (NIA)’s AD+ADRD Research Implementation Milestones also calls for “better electronic phenotyping of AD.”35 Nori et al. considered a broader definition of all AD-related dementias, including MCI, and used more complicated case definition rules (e.g., “a brain scan followed by a confirming diagnosis”),24 which may lead to a higher precision but limited sensitivity.
In contrast to previous studies, we comprehensively explored different prediction models and settings using large collections of real-world EHR data from the OneFlorida+ Clinical Research Consortium—a clinical research network that is part of the National Patient-Centered Clinical Research Network (PCORnet)36 funded by the Patient-Centered Outcomes Research Institute (PCORI), to create prediction models that can identify patients at risk of developing ADRD, including AD, vascular dementia (VaD), Lewy-body dementia (LBD), frontotem-poral dementia (FTD), or mixed dementia (i.e., patients with multiple ADRD subtypes). We tested four CP algorithms with different performances (in terms of specificity and sensitivity) to define the case group (i.e., patients with ADRD). Furthermore, we experimented with two different approaches to build the prediction models: (1) a knowledge-driven approach, where predictors were identified by domain experts based on existing literature, and (2) a data-driven approach, where all variables from the EHRs were used as predictors. In addition, we systematically explored different prediction windows: 0-year (i.e., all data before diagnosis), 1-year, 3-year, and 5-year using two widely used ML algorithms (i.e., LR and GBT). Our goal is to develop a robust prediction model that can identify patients at high risk for ADRD years before their initial diagnosis using routinely collected EHRs.
2 |. METHODS
2.1 |. Data source and study population
This study used data from the OneFlorida+ Clinical Research Consortium, a clinical research network contributing to the national PCORnet effort, with longitudinal EHRs linked with various other data sources (e.g., Medicaid and Medicare claims, vital statistics, and selected tumor registries, among others) for ≈16.8 million Floridians since 2012, and covering a wide range of patient characteristics including demographics, diagnoses, medications, procedures, vital signs, and lab tests, among others. This study was approved by the University of Florida Institutional Review Board under IRB201900182.
Figure 1(A) shows an overview of the study cohort construction process. Because ADRD is an age-associated disease, we are interested in predicting ADRD diagnosis in adults older than 50 years of age. As we needed to create a matched control cohort, we started with an initial cohort of 3,645,934 patients that (1) were 40 years of age and older as of January 1, 2012 (i.e., the starting date of the records in OneFlorida+); and (2) had at least 1 year of medical history in the database (i.e., at least two health care visits, where the last encounter of the patient observed in OneFlorida+ was at least 1 year from the first encounter).
FIGURE 1.
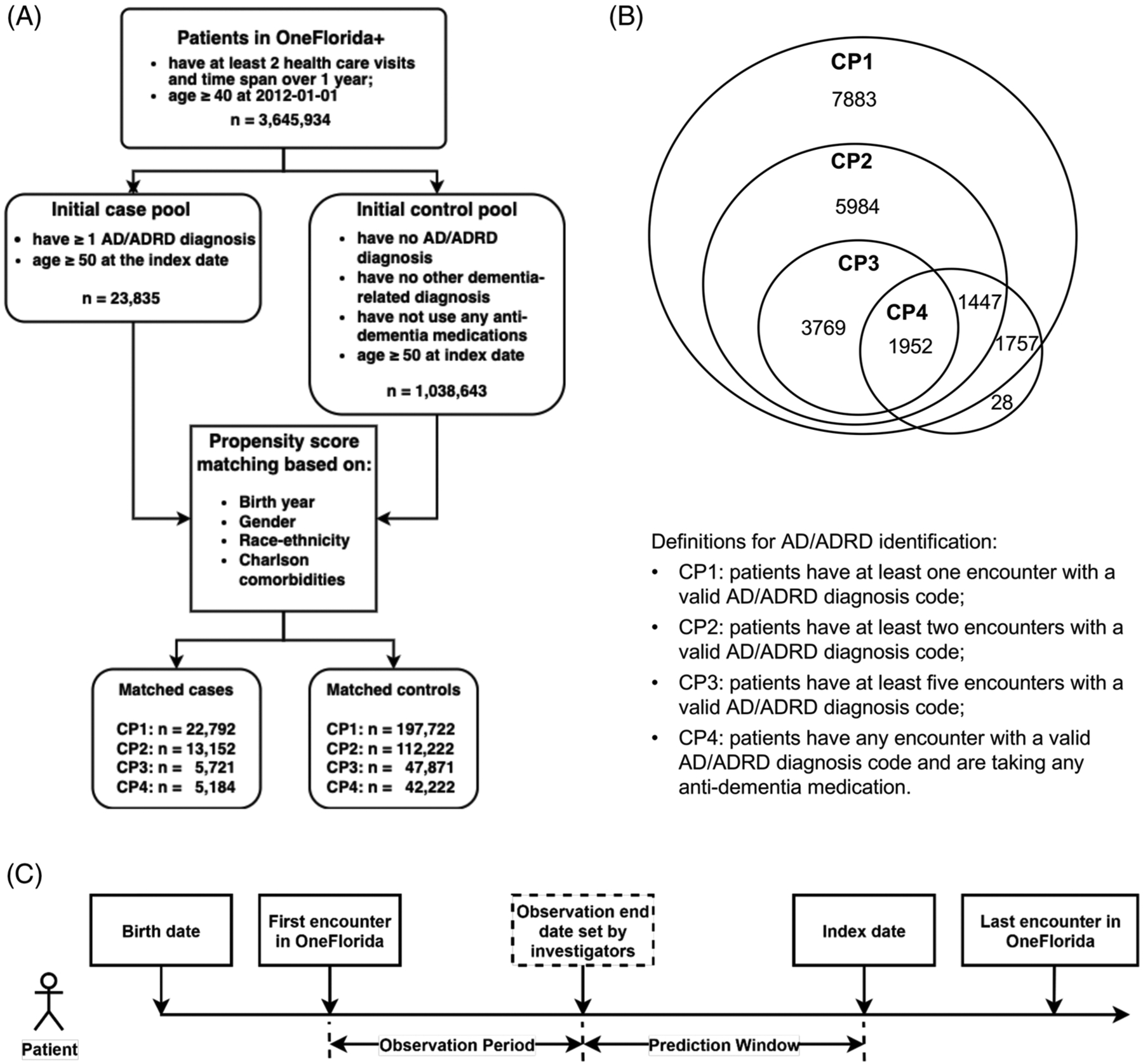
Overview of the study cohort extracted from the OneFlorida+ network. (A) Flowchart of patient selection. (B) Venn diagram of four computable phenotypes (CPs). (C) Patient timeline for the prediction task
2.2 |. Definition of cases and controls
Most previous studies on predicting ADRD-defined cases using International Classification of Diseases, Ninth/Tenth Revision (ICD-9/ICD-10) diagnosis codes including MCI and/or various subtypes of ADRD and prescribing of anti-dementia medications.24,25,37–40 In this study, we defined ADRD cases (i.e., AD, VaD, LBD, FTD, and mixed type) using a combination of diagnostic codes and anti-dementia medications (i.e., donepezil, galantamine, memantine, and rivastigmine), based on existing relevant computable phenotypes in the literature.41 There are currently six FDA-approved medications for AD treatment, that is, (rivastigmine, galantamine, donepezil, memantine, manufactured combination of memantine and donepezil, and aducanumab).7 We did not include aducanumab, as it was only recently approved in 2021, and the study data only covered patients in OneFlorida+ up to 2021. All the diagnostic and medication codes we used are included in the supplementary materials. Table 1 shows the four, rule-based CP algorithms we identified from the literature. Note that our focus is on leading causes of ADRD (AD, VaD, LBD, FTD, and mixed type), whereas some of the original CPs included AD only, or additional causes of dementias (see Table 1).
TABLE 1.
Performances of ADRD-related computable phenotypes
| Computable phenotype (CP) rules | Disease | Sensitivity | Specificity | PPV | NPV | F1 | Citation |
|---|---|---|---|---|---|---|---|
| CP1: patients have at least one encounter with a relevant ADRD diagnosis code | ADRD, other dementiasa | – | – | 0.825 | – | – | Wilkinson et al. (2019)39 |
| ADRD, other dementiasb | – | – | 0.73 | – | – | Fujiyoshi et al. (2017)40 | |
| AD | 0.7 | – | 0.77 | – | 0.73 | Tjandra et al. (2020)41 | |
| CP2: patients have at least two encounters with relevant ADRD diagnosis codes | ADRD, other dementiasa | – | – | 0.885 | – | – | Wilkinson et al. (2019)39 |
| AD | 0.24 | – | 0.74 | – | – | Wei et al. (2016)42 | |
| CP3: patients have at least five encounters with relevant AD/ADRD diagnosis codes | ADRD, other dementiasa | – | – | 0.907 | – | – | Wilkinson et al. (2019)39 |
| CP4: patients have at least one encounter with a relevant ADRD diagnosis code and at least one anti-dementia medication | AD | 0.04 | – | 0.8 | – | – | Wei et al. (2016)42 |
Includes Alzheimer’s disease (AD), vascular dementia, Lewy body dementia, frontotemporal dementia, senile dementia, Parkinson’s disease dementia, Huntington’s dementia, HIV-related dementia, alcohol-related dementia, and unspecified dementia.
Includes AD, vascular dementia, frontotemporal dementia, alcohol-related dementia, memory loss, and unspecified dementia;—indicates that the corresponding metric was not reported in the original study.
We defined the controls as patients who (1) had no ADRD-related diagnoses (i.e., AD, VaD, LBD, and FTD); (2) had no other conditions associated with or causally related to dementia (e.g., MCI, Parkinson’s disease, Huntington’s disease, HIV, alcoholism, etc.); and (3) had no exposure to dementia-related medications. Each case was matched with up to 10 patients from the control group without replacement based on propensity scores, controlling for patients’ birth years, gender, race-ethnicity, and Charlson comorbidity index (dementia-related conditions were excluded). Note that we tested four different CPs; thus each case cohort defined by each CP has a corresponding matched control cohort, as shown in Figure 1 (A, B).
2.3 |. Observation period and prediction window
Figure 1(C) shows a patient timeline for the prediction task using the OneFlorida+ database. We defined the index date for the case as the first encounter with an ADRD diagnosis or a dementia-related medication prescription, whichever came first. For the controls, we identified a similar “index date” according to their corresponding cases. We required that the control must have an age within 1 year of the case and must have encounters within 6 months of the index date of its corresponding case. We then selected the encounter date of the control closest to the index date of its corresponding case as reference “index date” for the control.
We split the patient timeline before the index date into two segments: (1) the prediction window: we defined various prediction windows, including 0 years, 1 year, 3 years, and 5 years before the index date (i.e., recorded ADRD diagnosis or treatment), and (2) the observation period: defined as from the first encounter recorded in the OneFlorida+ database to the beginning of the defined prediction window. Only data from the observation period were used for prediction and we used different prediction windows to assess how far in advance we could predict ADRD. We included only patients who had at least a 1-year observation period in the predictive models.
2.4 |. Machine learning algorithms and features
We first tested various ML algorithms (e.g., random forest, support vector machine, etc.) on a smaller subset of the entire population and chose two algorithms for the rest of the experiments: logistic regression as the baseline, and GBTs,42 which had the best performance in our small-scale experiments.
We adopted two feature engineering strategies, including a knowledge-driven approach and a data-driven approach. For the knowledge-driven approach, we used risk factors identified from the existing literature43,44 and by domain experts on the study team including: (1) medical diagnoses of obesity, diabetes, hyperlipidemia, hypertension, heart disease, stroke, depression, anxiety, concussion, sleep disorders, periodontitis, smoking, and alcohol use; (2) medication exposures, including nonsteroidal anti-inflammatory drugs (NSAIDs), statins, anticholinergics, hormone replacement therapies, antihypertensives, benzodiazepines, and proton pump inhibitors; and (3) the most recent vital signs and lab test results, including body mass index (BMI), systolic/diastolic blood pressure, total cholesterol, high-density lipoprotein, glucose, and hemoglobin A1C (HbA1c) in the observation period. Diagnoses and medication histories were coded as binary variables in the models. BMIs, blood pressures, and lab result values were categorized based on the reference normal range (e.g., abnormally low, normal, or abnormally high).
For the data-driven approach, we used all variables captured by the EHRs (including and beyond those listed for the knowledge-driven approach) with minimal pre-processing in the prediction models. More specifically, we categorized the demographic and behavioral variables, such as age, gender, race, ethnicity, marital status, and smoking status. We also included all discrete diagnosis codes, all medication RxNorm codes (and National Drug Codes [NDCs] are mapped to RxNorm), and all procedure codes recorded in patients’ EHRs as categorical features. To reduce the sparsity of the features, we adopted a code grouping/merging strategy similar to that described in our previous study.45 For diagnoses, we mapped a total of 30,064 unique ICD-9/ICD-10 codes to a number of 1912 PheWas (Phenome-wide association studies) groups.46 For medication, we mapped all the RxNorm codes for clinical drugs and branded drugs (22,492 unique codes) to ingredient-level codes (3159 unique codes). We aggregated all 11,880 unique procedure codes (including Current Procedural Terminology [CPT] and ICD-9/ICD-10 procedure codes) into 228 unique CCS (Clinical Classification Software) groups. We encoded all categorical features using the one-hot encoding scheme. Same as the knowledge-driven approach, we treated the real-valued variables (i.e., BMI, systolic and diastolic blood pressures, and lab test results) as categorical variables in the model. Because a patient can have multiple findings and/or lab test measurements from different encounters, we used the values from the most recent encounter before the end of the observation period for categorization. For lab test results, we used the abnormal indicator available in the PCORnet common data model (CDM) and then lab results were converted into normal, abnormal high, critical high, abnormal low, and critical low levels.
2.5 |. Experiments and evaluation
We split data into a training set (80%) and a testing set (20%) using stratified sampling. We optimized ML models using the training set via 5-fold cross validation and optimized hyperparameters using randomized search. We then applied the best model on the test set for evaluation. For the evaluation metric, we used the area under the receiver-operating characteristic (ROC) curve (AUC). We also calculated the sensitivity, specificity, positive-predicted value (PPV), and negative-predicted value (NPV) determined by optimizing the maximum value of Youden’s index. We conducted bootstrapping with 100 iterations to obtain point estimate and 95% confidence intervals (Cis) for each evaluation metric.
2.6 |. Identification of important risk factors
We adopted the SHAP (SHapley Additive exPlanations) method47 to identify the key risk factors contributed to the prediction and estimated the associations between the risk factors and the outcomes. We created SHAP bar plots and summary plots for the top 20 risk factors for the GBT models (as they are the best performing models) for both knowledge-driven and data-driven approaches. Positive SHAP values indicated features that can increase the probability of developing ADRD, whereas negative SHAP values indicated features that can decrease the risk. The absolute SHAP value of each feature indicates its importance.
3 |. RESULTS
Because our CP1 (i.e., have at least one encounter with a valid ADRD diagnosis code) has the most relaxed rules, Table 2 shows the descriptive statistics of the case and control groups identified using CP1. Descriptive statistics of the other three CPs are detailed in the Tables S1–S3 (descriptive statistics in case and control groups for CP2, CP3, and CP4). Table 3 shows the prediction performance measure by AUC for knowledge-driven and data-driven approaches.
TABLE 2.
Descriptive statistics in case and control groups for CP1
| Case N = 22,792 | Control N = 197,722 | |||||
|---|---|---|---|---|---|---|
| Variable | Sub-categories | N or mean | % or SD | N or mean | % or SD | p-value |
| Demographics | ||||||
| Age | at index date | 77.9 | 10.5 | 64.6 | 10.0 | N/A |
| Gender | Female | 13,844 | 60.7% | 117,055 | 59.2% | N/A |
| Male | 8,948 | 39.3% | 80,667 | 40.8% | N/A | |
| Race and ethnicity | Hispanic | 4,461 | 19.6% | 26,212 | 13.3% | N/A |
| Non-Hispanic White | 11,841 | 52.0% | 119,052 | 60.2% | N/A | |
| Non-Hispanic Black | 4,413 | 19.4% | 33,806 | 17.1% | N/A | |
| Non-Hispanic Other | 609 | 2.7% | 6,095 | 3.1% | N/A | |
| Unknown | 1,468 | 6.4% | 12,557 | 6.4% | N/A | |
| Length of medical history in OneFlorida+ | ||||||
| Number of years, range | [1, 2) | 5,139 | 22.5% | 50,205 | 25.4% | <0.001a |
| [2, 4) | 7,461 | 32.7% | 64,663 | 32.7% | ||
| [4, 6) | 5,839 | 25.6% | 48,074 | 24.3% | ||
| [6, +) | 4,353 | 19.1% | 34,780 | 17.6% | ||
| Number of encounters | All types of encounters | 270.3 | 508.7 | 100.5 | 228.4 | <0.001c |
| Ambulatory-encounter | 65.7 | 100.9 | 49.7 | 83.2 | <0.001c | |
Pearson chi-square test.
Two-sample t-test.
Wilcoxon rank–sum test.
TABLE 3.
Area under the curve (AUC) comparison of knowledge-driven and data-driven feature selection using GBT and LR for 0, 1, 3, and 5 years
| Feature strategy | CP | Model | 0-year AUC (95% CI) |
1-year AUC (95% CI) |
3-year AUC (95% CI) |
5-year AUC (95% CI) |
|---|---|---|---|---|---|---|
| Knowledge-driven | CP1 | LR | 0.892 (0.892, 0.893) | 0.870 (0.87, 0.87) | 0.855 (0.854, 0.856) | 0.841 (0.84, 0.842) |
| GBT | 0.894 (0.894, 0.895) | 0.871 (0.871, 0.872) | 0.856 (0.855, 0.857) | 0.839 (0.838, 0.84) | ||
| CP2 | LR | 0.898 (0.898, 0.899) | 0.878 (0.877, 0.878) | 0.868 (0.867, 0.869) | 0.849 (0.847, 0.851) | |
| GBT | 0.899 (0.899, 0.9) | 0.878 (0.877, 0.879) | 0.868 (0.867, 0.869) | 0.845 (0.843, 0.847) | ||
| CP3 | LR | 0.900 (0.899, 0.901) | 0.879 (0.878, 0.88) | 0.872 (0.87, 0.873) | 0.851 (0.847, 0.854) | |
| GBT | 0.900 (0.899, 0.901) | 0.879 (0.878, 0.88) | 0.867 (0.866, 0.869) | 0.849 (0.845, 0.852) | ||
| CP4 | LR | 0.877 (0.876, 0.878) | 0.856 (0.855, 0.858) | 0.846 (0.844, 0.848) | 0.830 (0.826, 0.834) | |
| GBT | 0.877 (0.876, 0.878) | 0.854 (0.853, 0.855) | 0.841 (0.839, 0.843) | 0.830 (0.826, 0.835) | ||
| Data-driven | CP1 | LR | 0.917 (0.918, 0.917) | 0.884 (0.885, 0.884) | 0.856 (0.857, 0.855) | 0.827 (0.828, 0.826) |
| GBT | 0.931 (0.932, 0.931) | 0.9 (0.9, 0.899) | 0.876 (0.877, 0.876) | 0.854 (0.855, 0.853) | ||
| CP2 | LR | 0.922 (0.922, 0.921) | 0.887 (0.888, 0.886) | 0.863 (0.864, 0.862) | 0.826 (0.828, 0.824) | |
| GBT | 0.936 (0.936, 0.935) | 0.904 (0.905, 0.903) | 0.884 (0.885, 0.883) | 0.858 (0.86, 0.856) | ||
| CP3 | LR | 0.919 (0.92, 0.918) | 0.882 (0.883, 0.881) | 0.856 (0.858, 0.855) | 0.817 (0.82, 0.813) | |
| GBT | 0.939 (0.94, 0.938) | 0.906 (0.908, 0.906) | 0.884 (0.885, 0.882) | 0.853 (0.856, 0.85) | ||
| CP4 | LR | 0.891 (0.892, 0.89) | 0.851 (0.853, 0.85) | 0.82 (0.822, 0.818) | 0.785 (0.79, 0.78) | |
| GBT | 0.911 (0.912, 0.91) | 0.876 (0.877, 0.875) | 0.854 (0.856, 0.852) | 0.831 (0.835, 0.828) |
As shown in Table 3, the GBT model consistently outperformed the LR model in terms of AUC in all four cohorts using either the knowledge-driven or the data-driven feature selection approach. The GBT model with data-driven feature selection achieved the best AUC scores of 0.939, 0.906, 0.884, and 0.854 for predicting ADRD at 0, 1, 3, and 5 years before initial diagnosis, respectively, where the best performing 0-, 1-, and 3-year models were from the CP3 cohort and the best performing 5-year model was from the CP2 cohort. Consistently, the AUC decreased as the prediction window became larger; the CIs grew larger as the prediction window increased from 0 years to 5 years; and the data-driven approach using all EHR data elements significantly outperformed the knowledge-driven approach with features derived from domain knowledge. Similar results are obtained for other performance metrics including specificity, sensitivity, PPV, and NPV (see Tables S4–S7).
Figure 2 shows the SHAP summary plots cross-comparing 1-year versus 5-year and CP1 versus CP3 models under the knowledge-driven approach. We only show CP1 and CP3 in the main text because, as case definition algorithms, (1) CP1 captures the most patients (see Figure 1(B)) but with high false positives, and (2) CP3 has the highest PPV (i.e., the lowest misclassification errors, although with significantly higher false negatives). The SHAP summary plots for all other models are included in the Supplementary Material (see Figures S1–S4). As shown in Figure 2, age, history of stroke, history of depression, history of diabetes, history of diabetes, and history of heart disease are the most important features to predict the risk of ADRD. The risk increases with age, stroke, and depression, whereas BMI in range (18.5, 23] reduced the risk of ADRD. BMI gradient boosting tree 30 is also negatively associated with the risk of ADRD.
FIGURE 2.
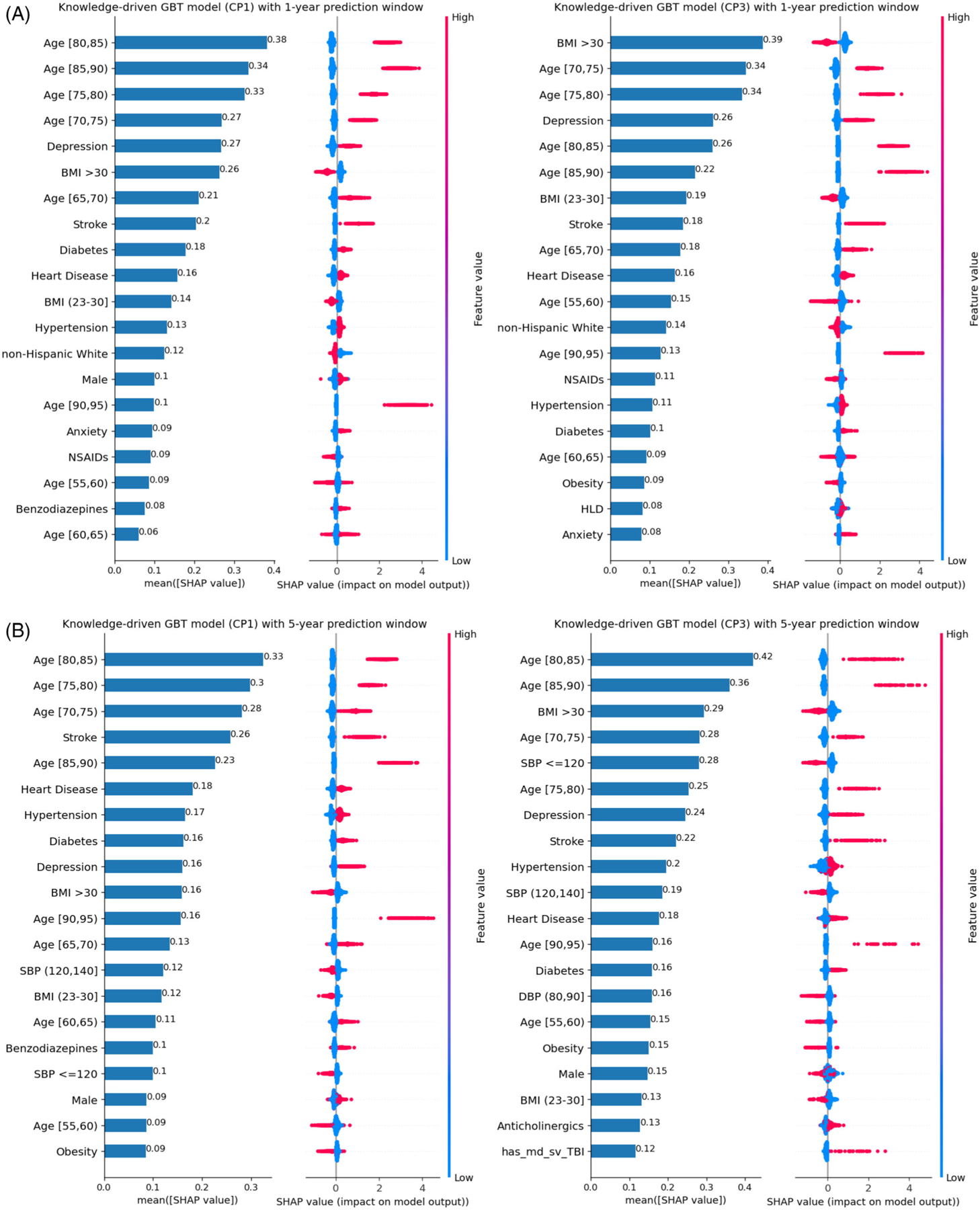
Under the knowledge-drive approach, SHAP plot of the top-20 features for GBT models based on CP1 and CP3 algorithms: (A) 1-year prediction window and (B) 5-year prediction windows
Similarly, Figure 3 examined the top-20 important data-driven features identified using SHAP values for 1-year and 5-year and CP1 versus CP3 models. The SHAP plots for all other data-driven models are included in the supplementary material (see Figures S5–S8). As shown in Figure 3, age, cerebrovascular disease, mood disorders, diabetes mellitus, malaise, and fatigue are the most important features for predicting the risk of ADRD. Age ≥70, history of cerebral ischemia, and history of cerebrovascular disease increase the risk of AD/ADRD. Age < 60 decreases the diagnosis risk for all models. BMI > 30 is also negative associated with the risk of ADRD. Black and African Americans, debility, contusion, and epilepsy are positively associated with the risk of ADRD.
FIGURE 3.
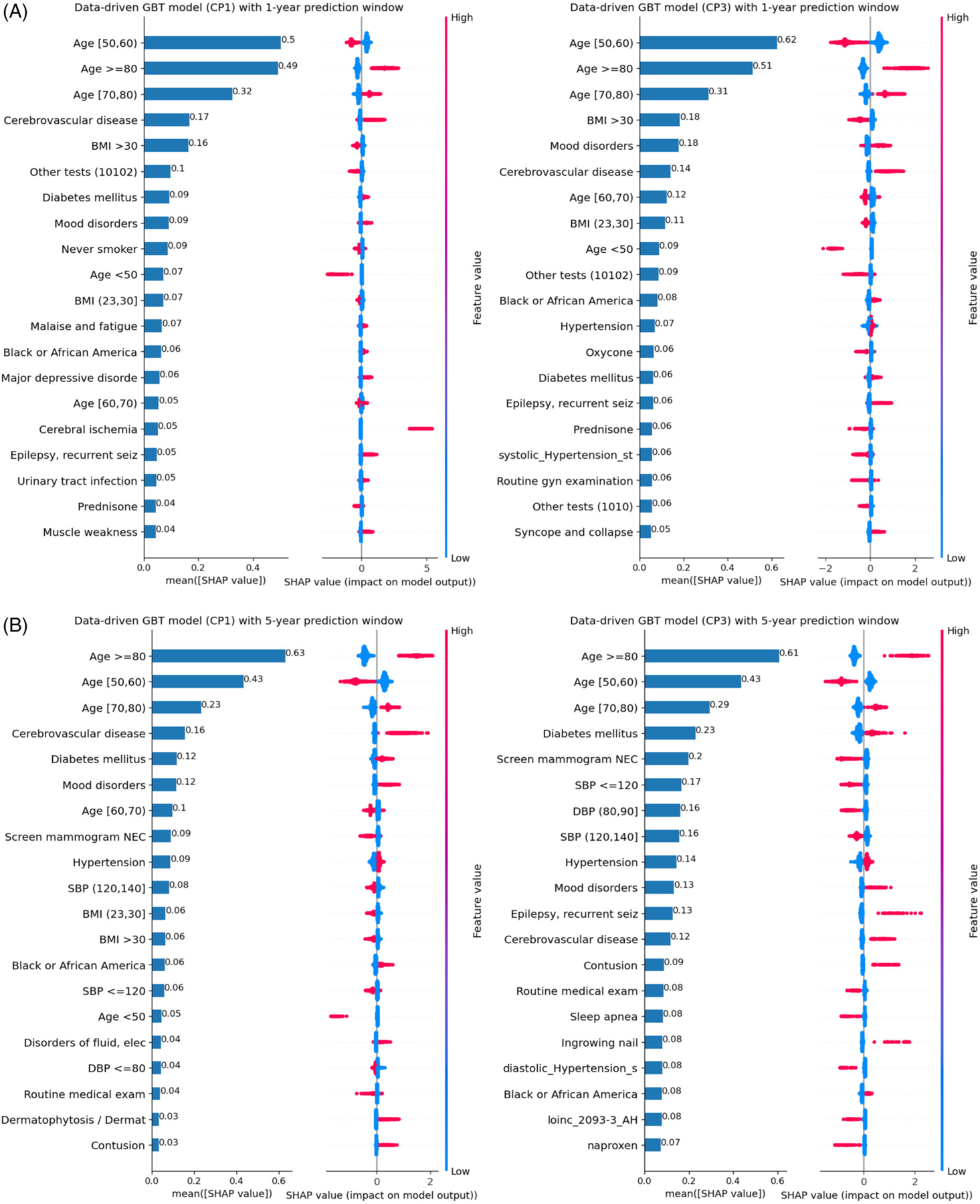
Under the data-driven approach, SHAP plot of the top-20 features for GBT models based on CP1 and CP3 algorithms: (A) 1-year prediction window and (B) 5-year prediction window. *SBP: systolic blood pressure
Results of all the models and settings that we have tested are included in the supplementary materials.
4 |. DISCUSSION
In this study, we systematically examined the prediction of ADRD using ML models and four different CP algorithms to identify ADRD patients from the OneFlorida+ real-world data (RWD). For each ML algorithm, we compared knowledge-driven and data-driven approaches for feature selection and tested the performance using different prediction window sizes including 0, 1, 3, and 5 years before ADRD diagnosis. The GBT model with data-driven features selection significantly outperformed the LR models and/or knowledge-driven approaches and achieved the best AUC scores of 0.939, 0.906, 0.884, and 0.854 for the prediction of 0, 1, 3, and 5 years before ADRD diagnosis, respectively, indicating that ML models can predict ADRD years before diagnosis using real-world clinical data. Early prediction of ADRD in real-world clinical settings can help providers, patients, and their caregivers plan ahead of time (e.g., lifestyle changes such as increase muscle strength, as muscle weakness is a risk factor revealed by our study associated with an increase future ADRD risk) and hopefully soon, to apply potential effective pharmacological disease-modifying treatments. All the models had high NPVs (≈0.9), making them effective in pinpointing people who will not develop ADRD, but relatively low PPVs (≈0.4–0.5), indicating that the models will identify a high proportion of false positives (see Table S4). Nevertheless, the ADRD prediction models using EHR can target higher risk patients for confirmatory biomarker studies.
The performance of the models varied across different prediction windows and cohorts identified using different CP rules. It is understandable that the model performance dropped as the size of the prediction window increased from 0 years to 5 years; and that the models were more confident in predicting ADRD onset as more data points were available closer to the initial diagnoses. For example, the CIs of these models became smaller when the prediction window reduced to 0 years. Note that 0-year prediction (i.e., predicting ADRD right before the onset dates) can be significantly biased and have low utility as the models may well be predicting clinicians’ diagnostic processes rather than the disease development itself. Nevertheless, the 0-year models provide us the upper bounds of the models’ predictive ability, where the best AUC score for the 0-year model is 0.939, 10.9% higher than the 5-year model (i.e., an AUC score of 0.856). An interesting finding is that the performance varied among the different models built with the four different CPs (i.e., used to define ADRD cases as the outcomes). The cohort identified by CP3 (i.e., patients have at least five unique encounters with ADRD codes) had the best prediction performance followed by cohorts identified by CP2, CP1, and CP4, in general with minor variations. Most previous studies predicting ADRD often identified cases using only diagnosis codes without considering other information such as medications to develop more complex CP algorithms to reduce the misclassification errors when identifying cases.
We compared knowledge-driven and data-driven feature selection in ML models and the experimental results showed that the data-driven feature selection outperformed knowledge-driven across all models, demonstrating that ML methods are capable of exploring a large feature space and identifying features that human experts might have missed. For example, preventive care such as routine medical exam, mammogram screening, and routine gynecology examination are negatively associated with the risk of ADRD under the data-driven approach. In particular, mammogram screening is one of the top-20, important features in several settings. Preventive care may be a surrogate marker for social determinants of health. Women who receive preventive health care are more likely to be highly educated, wealthier, and have health insurance. They are more likely to adopt other healthy lifestyle habits (not measured by EHR, such as physical exercise, healthy eating, lifelong learning, taking vitamins, and so on) that are associated with reduced risk of ADRD. However, we want to emphasize that associations are not causations, and predictive models are not causal models. The underlying causal mechanisms about how mammogram screening and routine gynecology examination reduce the risk of ADRD is still not clear. Nevertheless, our prediction models pinpoint the factors and directions of interest that warrant further investigations (e.g., patients who frequently use preventive services are generally healthier than those who do not).
Furthermore, the advantage of data-driven approach over knowledge-driven approach diminished as the size of the prediction window increases (e.g., an AUC of 0.9 vs an AUC of 0.871% to 5.7% improvement for the 1-year CP1 models, whereas 0.856 vs 0.841% to 1.8% improvement for the 5-year CP1 models, comparing data-driven vs knowledge-driven approaches, respectively). This is potentially because the knowledge-driven features are derived from established scientific literature (that summarized randomized control trials and large cohort studies among others) that capture the underlying causal mechanisms of the disease etiology and development process, whereas the data-driven approach may capture more spurious correlations between some of the additional trivial features and disease outcome, which can still improve the predictive ability of the corresponding models.
We identified important features of each model by calculating their SHAP values (see Figures S1–S8). We observed that different ML models often utilized different combinations of features, possibly due to the variations in model assumptions of the classification algorithms. For example, the analysis of top-10 features across the four different cohorts using data-driven and knowledge-driven approaches for feature selection showed that the NL models tend to use different combinations of features for different prediction windows, indicating that the feature importance changed relative to the progress of disease (i.e., some factors are important for long-time prediction such as 5-year model and others are important for short-term prediction such as 1-year model). This is understandable, as prior evidence suggests that different factors may influence the development of ADRD at different stages of the patient’s life course,48 and there exists heterogeneity in ADRD progression pathways (e.g., faster progression or with the different clinical syndrome).49,50 In addition, there are differences between the data-driven and knowledge-driven approaches. The top-10 features of the four different CP rules of the knowledge-driven approach are very similar for the same prediction windows. Age, BMI range (18.5, 23], history of stroke, and history of depression were the most important features under the knowledge-driven approaches. However, for the data-driven approaches, the top-10, important features are slightly different across different CP rules, even for the same prediction window. Although some of the features identified by the data-driven approach, such as mood disorders, malaise, and fatigue, could be part of the preclinical ADRD, they may be extremely useful for identifying asymptomatic individuals at high risk of ADRD and for allowing the search for new drugs or other methods such as vaccines or genetic manipulations that might slow or prevent the progress of ADRD (e.g., identify participants at high ADRD risk with preclinical risk factors into these trials).51 Nevertheless, age ≥70, and cerebrovascular disease, mood disorders, and diabetes mellitus are positively associated with the risk of ADRD across all cohorts. BMI >30 is negatively associated with the risk of ADRD in both data-driven and knowledge-driven approaches.
ADRD is a significant cause of morbidity and mortality in the elderly. Long-term cognitive decline due to ADRD can affect the diagnosis and treatment of comorbidities and generate complex management.52 Although AD is the most common diagnosis, other ADRDs often share biologically and clinically similar features with AD, making them difficult to distinguish from. There exists a substantial level of underdiagnosis of AD, especially in the primary care settings. Early recognition of potential ADRD as a group through automated prediction models is critical in clinical settings, so that the patients with high risks can be referred to specialized clinics and guide subsequent care of these patients. Furthermore, to build clinical decision support based on these prediction models, it is not that a single prediction model will rule it all but likely that a combination of tailored prediction models will have to be used. For example, along a patient’s life course, the first step may require models to identify ADRD risk years in advance (e.g., our 5-year model); and as the disease progresses, a short-term model (e.g., the 1-year model) can be used, followed by the need to identify the risk of subtypes. Moreover, ADRD prediction models cannot only give ADRD patients more time to prepare for the future, but also be used in other settings, such as to help researchers better identify and characterize clinical trial participants.9
Our study does have several limitations. First, our results were not validated with an external EHR data set. Although OneFlorida+ is a large, real-world clinical database, the study population may not be representative of the general ADRD population. Second, our study was a case-control study that considered a cohort of patients ≥ 50 years of age (i.e., a cohort of patients at risk for ADRD). Compared to cohort studies, case-control studies are relatively simple to implement, but they are not as good at showing causality and are more prone to bias.53 Third, this study is a secondary analysis of observational EHR data, which comes with its own pitfalls,54 including data quality issues55 such as complex missingness situation and documentation errors (that can leave to a number of analytical challenges, e.g., misclassification errors), selection bias of the population captured by EHRs, and confounding and informed presence bias56 (e.g., information captured in EHR is not random, but tied to patients’ encounters with the health system when they are ill), among many other issues. For example, our study did not consider clinical texts in EHRs that may contain additional information about patients’ status. Nevertheless, EHR-based study is a critical new tool to generate real-world evidence as promoted by the FDA.57,58 Our attempts to use CP phenotypes is one way of dealing with these limitations to minimize misclassification errors.
5 |. CONCLUSION
Early prediction of ADRD is challenging as multiple and complex mechanisms are involved in pathogenesis. This study systematically explored four different cohorts of patients with ADRD identified using four CP rules from relaxed to strict and examined feature selection using knowledge-driven and data-driven approaches. Our study identified the prediction variance among different cohorts and different prediction windows and further assessed the important features of different cohorts using SHAP values. The presented prediction models could help identify patients at a higher risk of ADRD ahead of onset, identify important factors for early prediction (and early prevention), help reveal research directions (i.e., why certain risk factors are significant), and facilitate recruitment of individuals at the early stage of probable ADRD before into clinical trials. Future work shall focus on (1) advanced analytical methods that can account for the data and analytical challenges with real-world EHR data, and (2) validating the prediction models built with OneFlorida+ data using external data sets (both EHR and non-EHR data sources) that cover different populations, given the known heterogeneity of ADRD.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic Review: We reviewed the literature using traditional methods and sources (e.g., Google Scholar, PubMed). Research suggests that early prediction of Alzheimer’s disease and related dementia (ADRD) in real-world clinical settings can help providers, patients, and their caregivers plan ahead of time (e.g., lifestyle changes) and it is hoped soon, to apply potential effective pharmacological disease-modifying treatments.
Interpretation: The study demonstrated the feasibility of using machine learning approaches for early prediction of ADRD diagnosis (as a proxy of ADRD onset).
Future Directions: Further work is necessary to identify the best strategy to implement ADRD prediction algorithms like the ones in this study into the clinical workflow. A promising direction would be a shared decision-making app that helps patient-physician dyads assess a patient’s personalized risk, understand the limitations of the risk prediction, and decide on the best course of action from evidence-based risk factor modifications.
ACKNOWLEDGMENTS
This study was partially supported by a Patient-Centered Outcomes Research Institute (PCORI®) Award (ME-2018C3-14754); grants from National Institute on Aging R56AG069880, R21AG068717, K01AG058781, and R01AG076234; a grant from the National Cancer Institute, R01CA246418 R01; a grant from Centers for Disease Control and Prevention (CDC) U18DP006512-01; a grant from the Ed and Ethel Moore Alzheimer’s Disease Research Program of the Florida Department of Health (FL DOH #9AZ14), and a Momentum Teaming grant from the University of Pittsburgh. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding institutions.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing interests.
Author disclosures are available in the supporting information.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.2022 Alzheimer’s disease facts and figures. Alzheimers Dement. 2022;18(4):700–789. doi: 10.1002/alz.12638 [DOI] [PubMed] [Google Scholar]
- 3.Mehta D, Jackson R, Paul G, Shi J, Sabbagh M. Why do trials for Alzheimer’s disease drugs keep failing? A discontinued drug perspective for 2010–2015. Expert Opin Investig Drugs. 2017;26(6):735–739. doi: 10.1080/13543784.2017.1323868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berk C, Paul G, Sabbagh M. Investigational drugs in Alzheimer’s disease: current progress. Expert Opin Investig Drugs. 2014;23(6):837–846. doi: 10.1517/13543784.2014.905542 [DOI] [PubMed] [Google Scholar]
- 5.Slomski A Another Amyloid-beta Blocker Fails to Halt Dementia. JAMA. 2019;321(24):2396–2396. doi: 10.1001/jama.2019.7821 [DOI] [PubMed] [Google Scholar]
- 6.Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther. 2014;6(4):37. doi: 10.1186/alzrt269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.How Is Alzheimer’s Disease Treated? National Institute on Aging. Published July 8, 2021. Accessed December 31, 2022. https://www.nia.nih.gov/health/how-alzheimers-disease-treated [Google Scholar]
- 8.Amjad H, Roth DL, Sheehan OC, Lyketsos CG, Wolff JL, Samus QM. Underdiagnosis of dementia: an observational study of patterns in diagnosis and awareness in US older adults. J Gen Intern Med. 2018;33(7):1131–1138. doi: 10.1007/s11606-018-4377-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mclinden K, Tools for earlier detection of cognitive impairment and dementia. National Institute on Aging. Accessed December 31, 2022. https://www.nia.nih.gov/research/blog/2020/12/tools-earlier-detection-cognitive-impairment-and-dementia [Google Scholar]
- 10.Crous-Bou M, Minguillón C, Gramunt N, Molinuevo JL. Alzheimer’s disease prevention: from risk factors to early intervention. Alzheimers Res Ther. 2017;9(1):71–71. doi: 10.1186/s13195-017-0297-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Simon G, Yu F. Advancing Alzheimer’s research: a review of big data promises. Int J Med Informatics. 2017;106:48–56. doi: 10.1016/j.ijmedinf.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang EYH, Harrison SL, Errington L, et al. Current Developments in Dementia Risk Prediction Modelling: an Updated Systematic Review. PLoS One. 2015;10(9):e0136181–e0136181. doi: 10.1371/journal.pone.0136181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R, Herskovits EH. Machine-learning techniques for building a diagnostic model for very mild dementia. Neuroimage. 2010;52(1):234–244. doi: 10.1016/j.neuroimage.2010.03.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s & Dementia. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR Jr, Bernstein MA, Fox NC, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): mRI methods. J Magn Reson Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini E, Ballerini L, Hernandez M, del CV, et al. Machine learning of neuroimaging for assisted diagnosis of cognitive impairment and dementia: a systematic review. Alzheimers Dement (Amst). 2018;10:519–535. doi: 10.1016/j.dadm.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends Neurosci. 2011;34(8):430–442. doi: 10.1016/j.tins.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hojjati SH, Ebrahimzadeh A, Khazaee A, Babajani-Feremi A. Predicting conversion from MCI to AD by integrating rs-fMRI and structural MRI. Comput Biol Med. 2018;102:30–39. doi: 10.1016/j.compbiomed.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 19.Moscoso A, Silva-Rodríguez J, Aldrey JM, et al. Prediction of Alzheimer’s disease dementia with MRI beyond the short-term: implications for the design of predictive models. Neuroimage Clin. 2019;23:101837–101837. doi: 10.1016/j.nicl.2019.101837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cedazo-Minguez A, Winblad B. Biomarkers for Alzheimer’s disease and other forms of dementia: clinical needs, limitations and future aspects. Exp Gerontol. 2010;45(1):5–14. doi: 10.1016/j.exger.2009.09.008 [DOI] [PubMed] [Google Scholar]
- 21.Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. Neuroimage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beam AL, Kohane IS. Big Data and Machine Learning in Health Care. JAMA. 2018;319(13):1317–1318. doi: 10.1001/jama.2017.18391 [DOI] [PubMed] [Google Scholar]
- 23.Goldstein BA, Navar AM, Pencina MJ, Ioannidis JPA. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 2016;24(1):198–208. doi: 10.1093/jamia/ocw042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nori VS, Hane CA, Crown WH, et al. Machine learning models to predict onset of dementia: a label learning approach. Alzheimers Dement (N Y). 2019;5:918–925. doi: 10.1016/j.trci.2019.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JH, Cho HE, Kim JH, et al. Machine learning prediction of incidence of Alzheimer’s disease using large-scale administrative health data. NPJ Digit Med. 2020;3(1):46. doi: 10.1038/s41746-020-0256-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shivade C, Raghavan P, Fosler-Lussier E, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH. Advances in Electronic Phenotyping: from Rule-Based Definitions to Machine Learning Models. Annu Rev Biomed Data Sci. 2018;1:53–68. doi: 10.1146/annurev-biodatasci-080917-013315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–54. doi: 10.1136/amiajnl-2012-000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edmonds EC, Delano-Wood L, Jak AJ, Galasko DR, Salmon DP, Bondi MW. “Missed” Mild Cognitive Impairment: high False-Negative Error Rate Based on Conventional Diagnostic Criteria. J Alzheimers Dis. 2016;52(2):685–691. doi: 10.3233/JAD-150986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponjoan A, Garre-Olmo J, Blanch J, et al. How well can electronic health records from primary care identify Alzheimer’s disease cases? Clin Epidemiol. 2019;11:509–518. doi: 10.2147/CLEP.S206770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin PJ, Kaufer DI, Maciejewski ML, Ganguly R, Paul JE, Biddle AK. An examination of Alzheimer’s disease case definitions using Medicare claims and survey data. Alzheimers Dement. 2010;6(4):334–341. doi: 10.1016/j.jalz.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 32.Tasker RC. Why Everyone Should Care About Computable Phenotypes. Pediatr Crit Care Med. 2017;18(5):489–490. doi: 10.1097/PCC.0000000000001115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. doi: 10.1186/s13073-015-0166-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015;22(6):1220–1230. doi: 10.1093/jamia/ocv112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.NIA. Milestone 1.H. Published 2018. Accessed March 29, 2021. http://www.nia.nih.gov/research/milestones/population-studies-precision-medicine-health-disparities/milestone-1-h
- 36.Forrest CB, McTigue KM, Hernandez AF, et al. PCORnet® 2020: current state, accomplishments, and future directions. J Clin Epidemiol. 2021;129:60–67. doi: 10.1016/j.jclinepi.2020.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Miled Z, Haas K, Black CM, et al. Predicting dementia with routine care EMR data. Artif Intell Med. 2020;102(101771):101771. doi: 10.1016/j.artmed.2019.101771 [DOI] [PubMed] [Google Scholar]
- 38.Venugopalan J, Tong L, Hassanzadeh HR, Wang MD. Multimodal deep learning models for early detection of Alzheimer’s disease stage. Sci Rep. 2021;11(1):3254. doi: 10.1038/s41598-020-74399-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo H, Lau KK, Wong GHY, et al. Predicting dementia diagnosis from cognitive footprints in electronic health records: a case-control study protocol. BMJ Open. 2020;10(11):e043487. doi: 10.1136/bmjopen-2020-043487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albrecht JS, Hanna M, Kim D, Perfetto EM. Predicting diagnosis of Alzheimer’s disease and related dementias using administrative claims. J Manag Care Spec Pharm. 2018;24(11):1138–1145. doi: 10.18553/jmcp.2018.24.11.1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson T, Schnier C, Bush K, et al. Identifying dementia outcomes in UK Biobank: a validation study of primary care, hospital admissions and mortality data. Eur J Epidemiol. 2019;34(6):557–565. doi: 10.1007/s10654-019-00499-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ke G, Meng Q, Finley T. LightGBM: a Highly Efficient Gradient Boosting Decision Tree. In: Guyon I, Luxburg UV, Bengio S, eds. Advances in Neural Information Processing Systems. Curran Associates, Inc.; 2017:3146–3154. http://papers.nips.cc/paper/6907-lightgbm-a-highly-efficient-gradient-boosting-decision-tree.pdf [Google Scholar]
- 43.Xu W, Tan L, Wang HF, et al. Meta-analysis of modifiable risk factors for Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2015;86(12):1299–1306. doi: 10.1136/jnnp-2015-310548 [DOI] [PubMed] [Google Scholar]
- 44.Baumgart M, Snyder HM, Carrillo MC, Fazio S, Kim H, Johns H. Summary of the evidence on modifiable risk factors for cognitive decline and dementia: a population-based perspective. Alzheimers Dement. 2015;11(6):718–726. doi: 10.1016/j.jalz.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Gong Y, Waheed N, et al. Identifying cancer patients at risk for heart failure using machine learning methods. AMIA Annu Symp Proc. 2019;2019:933–941. https://www.ncbi.nlm.nih.gov/pubmed/32308890 [PMC free article] [PubMed] [Google Scholar]
- 46.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26(9):1205–1210. doi: 10.1093/bioinformatics/btq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg SM, Lee SI, A Unified Approach to Interpreting Model Predictions. In: Proceedings of the 31st International Conference on Neural Information Processing Systems. NIPS’17. Curran Associates Inc.; 2017:4768–4777. [Google Scholar]
- 48.Pujades-Rodriguez M, Assi V, Gonzalez-Izquierdo A, et al. The diagnosis, burden and prognosis of dementia: a record-linkage cohort study in England. PLoS One. 2018;13(6):e0199026. doi: 10.1371/journal.pone.0199026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goyal D, Tjandra D, Migrino RQ, Giordani B, Syed Z, -Wiens J Characterizing heterogeneity in the progression of Alzheimer’s disease using longitudinal clinical and neuroimaging biomarkers. Alzheimers Dement (Amst). 2018;10(1):629–637. doi: 10.1016/j.dadm.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam B, Masellis M, Freedman M, Stuss DT, Black SE. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res Ther. 2013;5(1):1. doi: 10.1186/alzrt155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nitrini R Preclinical diagnosis of Alzheimer’s disease: prevention or prediction? Dement Neuropsychol. 2010;4(4):259–261. doi: 10.1590/S1980-57642010DN40400002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manemann SM, Chamberlain AM, Bielinski SJ, Jiang R, Weston SA, Roger VL. Predicting Alzheimer’s disease and related dementias in heart failure and atrial fibrillation. Am J Med. 2022. doi: 10.1016/j.amjmed.2022.11.010. Published online December 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song JW, Chung KCp. Observational studies: cohort and case-control studies. Plast /Reconstr Surg. 2010;126(6):2234–2242. doi: 10.1097/PRS.0b013e3181f44abc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Data M Secondary Analysis of Electronic Health Records. 1st ed. Springer International Publishing; 2016. doi: 10.1007/978-3-319-43742-2 [DOI] [Google Scholar]
- 55.Bian J, Lyu T, Loiacono A, et al. Assessing the practice of data quality evaluation in a national clinical data research network through a systematic scoping review in the era of real-world data. J Am Med Inform Assoc. 2020;27(12):1999–2010. doi: 10.1093/jamia/ocaa245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haneuse S, Daniels M. A general framework for considering selection bias in EHR-based studies: what data are observed and why? EGEMS (Wash, DC). 2016;4(1):1203. doi: 10.13063/2327-9214.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Concato J, Corrigan-Curay J. Real-world evidence - where are we now? N Engl J Med. 2022;386(18):1680–1682. doi: 10.1056/NEJMp2200089 [DOI] [PubMed] [Google Scholar]
- 58.Sherman RE, Anderson SA, Dal Pan GJ, et al. Real-world evidence - what is it and what can it tell us? N Engl J Med. 2016;375(23):2293–2297. doi: 10.1056/NEJMsb1609216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


