Abstract
Lysosome-targeted photodynamic therapy, which enhances reactive oxygen species (ROS)-responsive tumor cell death, has emerged as a promising strategy for cancer treatment. Herein, a uridine (dU)-modified Ru(II) complex (RdU) was synthesized by click chemistry. It was found that RdU exhibits impressive photo-induced inhibition against the growth of triple-negative breast cancer (TNBC) cells in normoxic and hypoxic microenvironments through ROS production. It was further revealed that RdU induces ferroptosis of MDA-MB-231 cells under light irradiation (650 nm, 300 mW/cm2). Additional experiments showed that RdU binds to lysosomal integral membrane protein 2 (LIMP-2), which was confirmed by the fact that RdU selectively localizes in the lysosomes of MDA-MB-231 cells and significantly augments the levels of LIMP-2. Molecular docking simulations and an isothermal titration calorimetry assay also showed that RdU has a high affinity to LIMP-2. Finally, in vivo studies in tumor-bearing (MDA-MB-231 cells) nude mice showed that RdU exerts promising photodynamic therapeutic effects on TNBC tumors. In summary, the uridine-modified Ru(II) complex has been developed as a potential LIMP-2 targeting agent for TNBC treatment through enhancing ROS production and promoting ferroptosis.
Keywords: PDT photosensitizer, uridine (dU)-modified Ru(II) complex, LIMP-2, lysosome-targeting distribution, ferroptosis
1. Introduction
Lysosome-targeted photodynamic therapy (PDT) has been developed as a promising strategy for cancer treatment. Significant progress has been made in both basic research and clinical applications of PDT in recent years.1 With distinct advantages over conventional surgery, radiotherapy, and chemotherapy, such as noninvasive treatment, spatiotemporal selectivity, and painless treatment, PDT has gradually developed into the fourth most commonly used minimally invasive treatment for tumors in clinical practice.2
Lysosomes play a critical role in various biological activities, including the degradation of proteins and nucleic acids, lipid transportation, energy metabolism, and remarkably, they respond to reactive oxygen species (ROS).3 Recently, it has been revealed that lysosomes play a crucial role in iron homeostasis. Lysosomal dysfunction may interfere with iron metabolism, resulting in the generation of lipid-based ROS that induces ferroptosis.4 Moreover, lysosomal membrane permeabilization and the consequent release of lysosomal material into the cytosol have been shown to trigger multiple forms of programmed cell death, including ferroptosis, pyroptosis, necroptosis, and autophagy.5−7 Rodriguez’s group reported that salinomycin and its derivative ionomycin specifically distribute in reproducible patterns when introduced in lysosomes of breast cancer stem cells. These ionophores effectively bind ferrous iron and reduce the amount of metabolically available iron in the cytosol. The load of lysosomal iron is further increased by increased ferritin breakdown, and the free ferrous/ferric ions appear to accelerate cellular lipid peroxidation and ROS production within the lysosomes. This was interpreted to indicate that lysosomes mediate a cell death pathway consistent with ferroptosis.8 Recently, it has been found that artemisinin and its derivative dihydroartemisinin induce lysosomal degradation of ferritin in a manner that is independent of autophagy. By increasing the level of cellular free iron, these drugs cause cells to become more sensitive to ferroptosis.9,10 These observations inspired us to consider the hypothesis that lysosome-enriched molecules may impact iron homeostasis to promote the accumulation of lipid peroxidation products, leading to ferroptosis in tumor cells.
Most clinically available photosensitizers are porphyrin derivatives like verteporfin, temporfin, and talaporfin,11−13 all of which have well-established photodynamic effects. In addition, other PDT photosensitizers with aromatic conjugated macroplanar molecular structures, including phenothiazines, phenolics, and metal phthalocyanines, have also received wide attention from researchers.14−16 However, due to their poor solubility and their low production of reactive oxygen toxins under conditions of tumor hypoxia, it is challenging to use these photosensitizers for tumor cell therapy. This difficulty limits their role in the clinic.17,18 Considering the heavy dependence of photosensitizers on oxygen, it is difficult to achieve high efficacy in the hypoxic microenvironment of tumors; therefore, developing effective, yet novel photosensitizers with low oxygen dependence is an important step forward toward improving the efficacy of PDT.
Ru(II) polypyridyl complexes possess the advantages of significant Stoke’s shift, strong spin coupling, efficient electron transfer, and rapid energy transfer.19 After light excitation, Ru(II) complexes boost the effectiveness of intersystem crossing (ISC), allowing molecules to reach the excited triplet state that favors ROS production through electron or energy transfer using oxygen or water molecules as grist for generating toxins that eventuate in the death of tumor cells under hypoxic conditions.20,21 In 2017, the first Ru-based photosensitizer, TLD1433, entered phase I clinical trials with effective PDT for non-muscle-invasive bladder cancer.22 Additional studies have shown that Ru(II) polypyridyl complexes are able to function as lysosome-enriched photodynamic photosensitizers that inhibit tumor cell growth.23−26 Peng et al. reported the synthesis of a red light-stimulated lysosome-targeted Ru(II) complex (Ru–I, [Ru(terpy)(Cl-7-IVQ)I]+), which exhibited significant therapeutic efficacy on breast cancer cells using radiation at 660 nm, both in vitro and in vivo.27 Chao et al. designed and synthesized a Ru(II) polypyridyl complex within cell targeting lysosomes that demonstrated good water solubility.28,29 This Ru(II) polypyridyl complex has good photostability and significant two-photon excitation efficiency, with high tumor kill rates. In addition, Mao et al. achieved targeted recognition of lysosomes by introducing nanocarbon-loaded polypyridine Ru(II) complexes. These effectively promote tumor cell death under light excitation.30 Collectively, these studies suggest that lysosome-targeting Ru(II) complexes will have promising applications as photosensitizers for clinical PDT treatments.
Previous studies on Ru(II) polypyridyl–lysosome complexes suggested that the presence of different heteroaromatic rings and long carbon chains substantially impacts target recognition, the valence electron structure, and energy transfer. Lysosomes have demonstrated good uptake of molecules containing aromatic heterocyclic structures such as purines, pyrimidines, and morpholines.31,32 At the optimal locations for incorporation within lysosomes, the pyrimidine ring covalently bonds through N terminal bridges of the ligand to the Ru(II) complex. In the present study, we explored a uridine (dU)-modified Ru(II) complex (RdU) that was synthesized by a click chemistry reaction, which targets lysosomal integral membrane protein 2 (LIMP-2) that localizes in the lysosomes of triple-negative breast cancer (TNBC) cells. The results indicate that the RdU complex strongly affects ROS production and remarkably inhibits TNBC cells under 650 nm light excitation. Exceptional ROS generation suggests that RdU may serve as a novel TNBC PDT photosensitizer. Through electron microscopy, proteomics, and other methods, we demonstrate that RdU complexes target LIMP-2 at lysosome localized sites to promote ROS production under photodynamic excitation. Interestingly, RdU promotes TNBC cell death even in the hypoxic tumor environment (Scheme 1). This study lays a foundation for studying lysosome-targeted Ru-pyridyl-dU photosensitizers and provides a new strategy for treating TNBC in the clinic.
Scheme 1. Illustration of RdU for the Photodynamic Therapy Mechanisms.
RdU targeted to the lysosomes and specifically recognizes LIMP-2 where it binds and undergoes photo-induced ROS-mediated cell death.
2. Results and Discussion
2.1. Synthesis and Physicochemical Property of RdU
RdU was synthesized by tethering the 2′-deoxyuridine (dU) group to the Ru(II) terminal cinnamyl group via click chemistry. The route of RdU synthesis is shown in Figure 1a. First, we prepared the intermediate product of [Ru(bpy)2dione](ClO4)2 using the precursor Ru(bpy)2Cl2 and 1,10-phenathrolinedione-5,6 as reactants. Second, the modified complex [Ru(bpy)2ASIP](ClO4)2 (RuA) with an azide group termination was synthesized from [Ru(bpy)2dione](ClO4)2 and 4-azidocinnamaldehyde. Finally, the complex [Ru(bpy)2PTdUIP](ClO4)2 (RdU) was obtained, with the dU targeting group conjugated to the terminal cinnamoyl group by linkage of the terminal alkynyl group to the azide group using click chemistry.33 RuA and RdU were structurally characterized by 1H NMR, 13C NMR, 1H–1H COSY, 1H–13C COSY, and mass spectrometry (Figures S1–S5). The purity of RdU is 96.8%, as confirmed by high-performance liquid chromatography (HPLC) as well as elemental analysis (Figure S6).
Figure 1.
RdU acts as a potential photosensitizer to promote ROS generation. (a) Synthesis of RdU by click chemistry. (b) Singlet oxygen production induced by RdU (10 μM) under increasing time of light irradiation through 1,3-diphenylisobenzofuran (DPBF) probe and the changes of hypochromicity compared with Ru(bpy)3Cl2 (10 μM). (c) The hydroxyl radical level induced by RdU (10 μM) under increasing time of light irradiation through aminophenyl fluorescein (APF) probe and the changes of intensity compared RdU with Ru(bpy)3Cl2. (d) The superoxide anion level induced by RdU (10 μM) under increasing time of light irradiation through the DHR123 probe and the changes of intensity compared RdU with Ru(bpy)3Cl2 (10 μM). (e) The energy, electronic configurations, and the associated frontier molecular orbitals for each state of RdU as determined by density functional theory (DFT) calculations. (f) The photochemical process (Part I) and the proposed mechanism for ROS generation (Part II) induced by RdU with light irradiation.
2.2. ROS Generation Mechanism under Normoxia and Hypoxia
Effective photosensitizers for PDT should strongly enhance the production of ROS, especially type I ROS, but this remains a significant challenge. Therefore, we comprehensively evaluated the ability of light-induced Ru(II) complexes to enhance the production of different types of ROS. The specific indicator 1,3-diphenylisobenzofuran (DPBF) reacts promptly with singlet oxygen (1O2) to form an endoperoxide, which is then transformed to produce 1,2-dibenzoylbenzene.34 The generation of 1O2 was detected by measuring the decrease in optical absorbance of DPBF at 410 nm. As shown in Figure 1b, under 650 nm light-emitting diode (LED) light irradiation (300 mW/cm2), RdU (10 μM) strongly diminishes the characteristic DPBF absorption as the irradiation time is extended. The characteristic peak at 410 nm decreased to 60.4% after ∼80 s of exposure, indicating efficient 1O2 generation. The quantum yield for 1O2 generation is comparable to that of [Ru(bpy)3]Cl2 (a prominent 1O2 inducer),35 as exhibited by the pronounced hypochromic change of 72.8% at 3 min exposure (Figure S7). While the absorbance of the blank control (DPBF minus RdU) is essentially unchanged under 650 nm light irradiation, the absorbance by DPBF in the presence of RdU is significantly reduced, showing that RdU effectively generates 1O2 after light irradiation.
To determine other ROS that are generated, we monitored the production of hydroxyl radicals (•OH) and superoxide radicals (•O2–) using two radical indicators, aminophenyl fluorescein (APF) and dihydroxylamine 123 (DHR123), respectively.36,37Figure 1c shows that after 11 min of exposure to 650 nm light, RdU increased the fluorescence intensity of APF almost 3.4-fold, whereas [Ru(bpy)3]Cl2 increased the fluorescence intensity of APF by only 1.3-fold. Additionally, when DHR123 was used as a superoxide anion indicator, RdU produced a significantly faster increase in DHR123 fluorescence than did [Ru(bpy)3]Cl2. After 210 s under 650 nm light irradiation, RdU and [Ru(bpy)3]Cl2 enhanced the fluorescence intensity of DHR123 5.6-fold and 1.7-fold, respectively. The overall capacity to enhance ROS generation is summarized in Figure 1d. As previously mentioned, RdU is a highly effective photosensitizer with good quantum yields at 650 nm that is capable of generating a variety of ROS, especially type I and type II, as well as oxygen-independent free radicals under light irradiation. It should be emphasized that light-exposed RdU results in the production of a remarkable amount of •O2– in a brief period, indicating that it may qualify as a potent type I PDT agent.
To gain greater insight into the mechanism by which RdU enhances the generation of type I and II ROS, relevant frontier molecular orbitals were estimated in both optimized ground and excited states, with the energy difference being determined by density functional theory (DFT) calculations.38,39 In RdU, the lowest transition in the singlet manifold was attributed to the highest occupied molecular orbital (HOMO) → the lowest unoccupied molecular orbital (LUMO) with an energy gap of 1.04 eV. The HOMO comprises a significant electron density due to a lone dipyridyl pair (Figure 1e). The calculated elements of the spin–orbit matrix are based on the single-group excited states and listed in Tables S1 and S2. It is evident that the lowest-lying S1 state has a ππ* character. In striking contrast, the higher singlet excitation S2 state is attributed to a HOMO → LUMO + 2 transition with an energy gap of 1.06 eV that exhibits a πdx2–y2 configuration.40 The results support that the observed visible absorption is due to the S0 → S2 πdx2–y2 transition with sizable molar extinction coefficients.
The lowest triplet state T1 is attributed to a HOMO → LUMO + 1 transition in a πdxy configuration with an energy gap of 0.63 eV and the higher triplet state T2 that is attributed to a HOMO → LUMO + 2 transition in a πdx2–y2 configuration with an energy gap of 0.89 eV. The transition between the photosensitizer’s ground state (S0) and the lowest excited singlet state (S1) is typically used to achieve excitation.41 The triplet state of the sensitizer is generated via an ISC transition (T1). This excited state responds via an electron transfer or energy transfer process, producing a free radical (type I) or singlet oxygen (type II) (Figure 1f). It should be emphasized that for RdU, the HOMO is distributed on the nitrogen heteroaromatic ring moiety (two dipyridyl units and a phenanthroline unit) coordinated with the Ru atom. However, the LUMO is assigned to an imidazole-styrene-triazole moiety, which is characterized by a clear separation.20 Indeed, a more extensively separated HOMO–LUMO distribution will result in a lower ΔEST value. For RdU, the ΔEST band gap was calculated to be 0.1631 eV, which shows a more facile ISC process and a much higher ROS production efficiency.
2.3. Phototoxicity Studies
The results of long-term (72 h) UV characteristic absorption peaks and mass spectrometric detection demonstrate that the structures of the compounds dissolved in the physiological solution are stable (Figure S8). Furthermore, the photocytotoxic effects of RdU were studied in the dark and under light irradiation (650 nm, 300 mW/cm2). When treated with RdU (5 μM) at light exposure for 10 min, MDA-MB-231 cells shrink and produce a clear vacuole near the cell membrane (Figure 2a). After longer periods of irradiation, cellular morphology changes, noticeably and progressively leading to cell death.20,38 In addition, two breast cancer cell lines (MDA-MB-231 and MCF-7) and a normal breast cell line (MCF-10A) were cultured under normoxic (21% O2) and hypoxic conditions (0.1% O2). The cells were then treated with different concentrations of RdU (0, 1.56, 3.13, 6.25, 12.5, 25, 50, and 100 μM) for 24 h, and the cells were washed once with phosphate buffered saline (PBS) and continued to be cultured for another 48 h. During 72 h culture, the cells were irradiated at 650 nm for 5 min at 24 and 48 h points (Figure S9).
Figure 2.
RdU acts as a type I photosensitizer to promote ROS generation to overcome tumor hypoxia. (a) Cellular morphology and the distribution of RdU (5 μM) over time in MDA-MB-231 cells with fluorescence excited by 650 nm light irradiation. (b) Survival rate of MDA-MB-231 cells treated with different concentrations of RdU (0, 1.5625, 3.125, 6.25, 12, 25, 50, and 100 μM) with and without 650 nm light irradiation for 10 min under normoxic and hypoxic conditions. The total ROS generation by RdU with and without light irradiation under normoxic (c) and hypoxic (d) conditions. (e) Total ROS level analysis was measured by 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) assay.
RdU also demonstrated comparable inhibitory effects against MDA-MB-231 cells cultured under normoxic and hypoxic conditions. IC50 values were 36.41 ± 3.97 and 32.06 ± 5.87 μM, respectively (Table 1) in the dark. However, RdU exhibited weak inhibitory effects on MCF-7 and MCF-10A cells under normoxic conditions in the dark with IC50 values of 54.29 ± 8.31 and 73.34 ± 6.43 μM, respectively. In the absence of irradiation and under hypoxic conditions, the IC50 values for MCF-7 and MCF-10A cells were greater than 100 μM. Under light exposure and normal oxygen, RdU exhibited impressive phototoxic effects on MDA-MB-231 cells (IC50 = 1.09 ± 0.22 μM, photocytotoxicity index, PI = 33.4). Under irradiation and hypoxic conditions, RdU exhibited even stronger toxicity (IC50 = 0.36 ± 1.61 μM, PI = 89.1) (Figure 2b and Table 1). In addition, RdU produced somewhat less effective phototoxicity in cancerous MCF-7 cells and evidenced low phototoxicity in normal MCF-10A breast cells under normoxic conditions (IC50/MCF-7 = 8.16 ± 1.73 μM and IC50/MCF-10A = 36.57 ± 4.17 μM, with PIMCF-7 = 3.39 and PIMCF-10A = 2.01, respectively). Under hypoxic conditions with light activation, RdU remained effectively phototoxic to MCF-7 cells (Figure S10). These results show that RdU exerts strong phototoxic effects on TNBC MDA-MB-231 cells under both normoxic and hypoxic conditions but somewhat lower toxicity on cancerous MCF-7 cells under the same conditions.
Table 1. Cytotoxicity of Ru(II) Complex RdU toward Varied Human Breast Cancer Cells and Normal Breast Cells.
| inhibitory
activity (IC50, μM) |
||||||
|---|---|---|---|---|---|---|
| MDA-MB-231 | MCF-7 | MCF-10A | ||||
| condition | normoxia | hypoxia | normoxia | hypoxia | normoxia | hypoxia |
| dark | 36.41 ± 3.97 | 32.06 ± 5.87 | 54.29 ± 8.31 | >100 | 73.34 ± 6.43 | >100 |
| lighta | 1.09 ± 0.22 | 0.36 ± 1.61 | 8.16 ± 1.73 | 36.03 ± 5.26 | 36.57 ± 4.17 | >100 |
| PIb | 33.4 | 89.1 | 3.39 | 2.01 | ||
Irradiated at 650 nm by LED light (180 J/cm2).
Photocytotoxicity index: PI = IC50(Dark)/IC50(Light).
All of these discoveries indicate that the introduction of triazole coupling of dU is an effective strategy to enhance ROS production, which may not only promote 1O2 formation but also significantly boost the production of free radicals. RdU may be a potential type I photosensitizer which is not oxygen-dependent and exhibits promising therapeutic effects in the hypoxic tumor microenvironment. To confirm the ability of RdU to enhance ROS production under hypoxic conditions, MDA-MB-231 cells were incubated with different ROS probes (2′,7′-dichlorodihydrofluorescein diacetate [DCFH-DA] for total ROS, APF for •OH, and DHR123 for •O2–), then exposed to light irradiation. Cellular fluorescence for each of these probes was monitored by flow cytometry. Following the addition of RdU plus light irradiation, the total ROS level increased by 26.49 and 28.22% under normoxic and hypoxic conditions, respectively (Figure 2c–e). In addition, a minor increase in •OH was observed under normoxic and hypoxic conditions (Figure S11A–C). Surprisingly, after RdU incubation plus light irradiation, the level of •O2– generation perceptibly increased, as did the total ROS level, under both normoxic and hypoxic conditions (Figure S10D–F). A stronger fluorescence signal was seen immediately following light irradiation, demonstrating the ability of RdU to enhance •OH and •O2– production under hypoxic conditions in a cellular environment. The above results indicate that after photoexcitation, lysosome-targeted RdU enhances ROS generation, thereby inhibiting the growth of MDA-MB-231 cells in a hypoxic microenvironment.
2.4. Ferroptosis Induced by RdU under Light Irradiation
We further studied the nature of cell death stimulated by RdU via flow cytometry. After treatment with RdU (5 μM) combined with light irradiation, almost all cells shrink in size, indicating that photoexcited RdU engenders death in tumor cells (Figure 3a). However, few changes in the cell cycle distribution of MDA-MB-231, MCF-7, and MCF-10A cells were observed after treatment either with or without RdU or light irradiation (Figures 3b, S12, and S13). Furthermore, after treatment with RdU or light irradiation, the apoptosis rate is less than 10%. However, after incubation with RdU (5 μM) plus light (650 nm, 300 mW/cm2) irradiation for 10 min (5 min at the point of 24 and 48 h each time), the percentage of apoptotic cells increased dramatically to 23.58% of MDA-MB-231 cells (Figures 3c, S12, and S13). However, JC-1 assays of the mitochondrial membrane potential revealed no significant change in MDA-MB-231 cells (Figure S14). The cytotoxicity of RdU against MDA-MB-231 cells reveals an IC50 value of 1.09 ± 0.22 μM, yet the percentage of apoptotic cells after treatment with 5 μM RdU is surprisingly much lower than 50%. These findings suggest that photoexcited RdU induces alternative cell death mechanisms in MDA-MB-231 cells other than apoptosis or cell cycle arrest.
Figure 3.
RdU induces MDA-MB-231 cell death through promoting ferroptosis. (a) Cellular morphology of MDA-MB-231 cells cultured with different treatments of control, light (10 min), RdU (5 μM), and RdU (5 μM) with light (10 min). Cell cycle distribution (b) and apoptosis (c) of MDA-MB-231 cells after different treatments of the control, light (10 min), RdU (5 μM), and RdU (5 μM) with light (10 min). (d) Ultramicrostructure of MDA-MB-231 cells after different treatments of control, light, RdU (5 μM), and RdU (5 μM) with light (10 min) as observed by biological transmission electron microscope (Bio-TEM). The distribution and the variation of the ferroptosis marker protein GPX4 (e) and SLC7A11 (f) in MDA-MB-231 cells after different treatments of control, light (10 min), RdU (5 μM), and RdU (5 μM) with light (10 min).
To determine the cell death pathway triggered by photoexcited RdU, the changes in cell morphology and microstructure were closely observed by using a biological transmission electron microscope (Bio-TEM). Absent any treatment, TEM images of MDA-MB-231 cells reveal ideal lysosome structures, an intact cytoplasm, and intact outer and inner mitochondrial membranes (white arrows in Figure 3d). After treatment with 5 μM RdU or light irradiation, there was no significant change in the lysosomal and mitochondrial structure. However, upon treatment with 5 μM RdU plus light irradiation, several vacuoles appeared in the cytoplasm and some lysosomes visibly disintegrated.42 Mitochondrial staining showed conspicuous disruption; bilayer membranes disappeared, and mitochondrial cristae degraded. This suggested that photoexcited RdU-induced cell death might occur through ferroptosis.
Numerous studies have demonstrated that GPX4 and SLC7A11, two key transcription factors involved in ferroptosis, tightly regulate the ferroptosis pathway brought about by lipid peroxidation.43,44 We assessed the alterations in GPX4 and SLC7A11 expression levels and monitored by immunofluorescence its distribution in MDA-MB-231 cells treated with RdU plus light irradiation. As shown in Figure 3e, GPX4 protein was distributed throughout the MDA-MB-231 cells without any treatment, but it is somewhat more enriched in the nucleus. Additionally, alone RdU treatment or light irradiation resulted in little change in GPX4 expression. However, following RdU with light irradiation, the intensity of GPX4 in the cells significantly decreased. Moreover, in untreated MDA-MB-231 cells, the SLC7A11 protein was primarily dispersed in the cytoplasm with some foci near the nucleus. Furthermore, no changes were observed after treatment with RdU or light irradiation alone. However, after treatment with both RdU and light, the amount of SLC7A11 in the cytoplasm substantially dropped and the number of foci surrounding the nucleus increased (Figure 3f). The overall analysis of GPX4 and SLC7A11 fluorescence intensity revealed that photoexcited RdU decreased GPX4 and SLC7A11 expression. These findings show that RdU causes death in the MDA-MB-231 cell potentially by inducing ferroptosis. This hypothesis needs further study to confirm.
2.5. RdU Targeting Localized in Lysosome through Selectively Recognized LIMP-2
These promising early findings have motivated researchers to focus on identifying the underlying biological mechanism(s) of action. Utilizing the luminescence properties of RdU, its subcellular localization was studied by laser scanning confocal microscopy. We compared its luminescence distribution pattern with fluorescence patterns showing localization from organelle-specific dyes. It is worth noting that the deep red fluorescent foci of RdU are primarily found in the cytoplasm, exhibiting marked differences from the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI) and the mitochondrial markers (Figure 4a). And it is found that red fluorescent foci of RdU were partially matched with endoplasmic reticulum in three-dimensional (3D) tomoscan imaging (Figure 4b). Notably, an overlap ratio of about 100% was obtained between the red foci of RdU and the lysosome probe Lyso-Blue, demonstrating that the two labels colocalize. Red fluorescence also fills the entire lysosome in three-dimensional tomoscan imaging. The staining patterns of RdU and LysoTracker were closely matched (Figure 4c). These findings demonstrate that RdU exhibits a perfect subcellular lysosome-specific localization23,25 and inhibits the growth of breast tumor cells under light irradiation.
Figure 4.
Lysosome-targeted localization of RdU in MDA-MB-231 cells. (a) Comparison of luminescence distribution pattern of RdU (5 μM) with fluorescence patterns showing the localization from organelle-specific dyes in MDA-MB-231 cells for 6 h, including nucleus dyed in blue by DAPI, mitochondria highlighted in green by MitoTracker, endoplasmic reticulum stained in green by ER-tracker and lysosome lightened by LysoTracker. The 3D scan for endoplasmic reticulum (b) and lysosome (c) stained MDA-MB-231 cells with RdU.
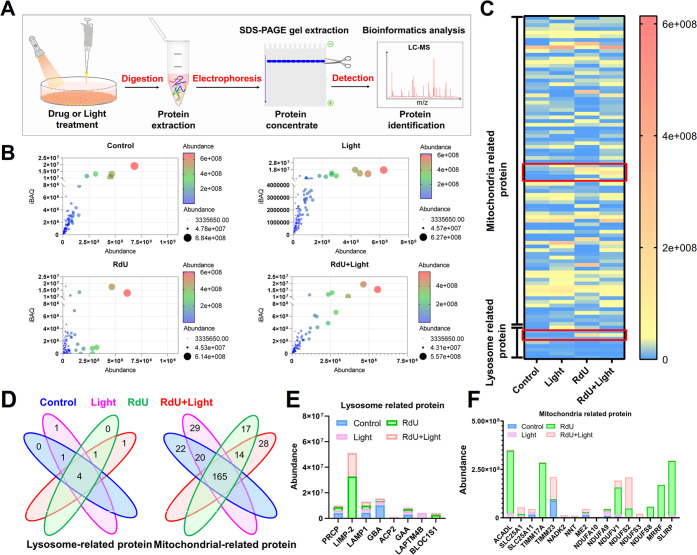
To explore why RdU locates in the lysosomes, we identified the specific target species by proteomics identification technology (Figure 5a). More than 2000 proteins were identified in each of the four sample groups (control, light alone, RdU, RdU + light) that were extracted from mixed protein gels. Data quality and protein abundance met the analysis criteria in all four treatment groups (Figure 5b). Additionally, because RdU not only targets lysosomes but also potentially enhances ROS production, changes in these biological responses have implications for mitochondrial metabolic processes. The differential expression of lysosome- and mitochondrial-related proteins was analyzed in depth by liquid chromatography-mass spectrometry (LC-MS) from cutouts from gels containing cell protein homogenates. Interestingly, the abundance of the lysosomal protein LIMP-2 was frequently elevated in the RdU and RdU plus light groups, but hardly changed in the control and light-only groups (Figure 5c). Eight lysosomal proteins were identified. Two of these eight proteins were specifically expressed in the groups RdU and RdU plus light exposure (Figure 5d,e). But many mitochondrial proteins were also identified in the RdU and RdU plus light groups (Figure 5d,f).
Figure 5.
Proteins potentially recognized by RdU as determined by proteomics. (a) Proteins that are upregulated after RdU treatment, as determined by LC-MS. (b) Data quality and protein abundance. (c) Heat map showing the abundance of mitochondrial and lysosome-related proteins in MDA-MB-231 cells in different treatment groups (control, light, RdU, and RdU with light). (d) Comparisons of mitochondrial-related proteins and lysosomal-related proteins in the control, light, RdU, and RdU with light groups. (e) Abundance analysis of eight lysosome-related proteins. (f) Top 15 differentially regulated mitochondrial-related proteins resulting from different treatments.
These findings suggest that RdU may initially and preferentially target lysosomes by attaching to the lysosomal membrane protein LIMP-2. We discovered, however, that photoexcited RdU significantly also increases the levels of mitochondrial-associated proteins, such as SLC25A1 (a mitochondrial citrate carrier), SLC25A11 (a mitochondrial oxoglutarate carrier), TIMM23 (an inner mitochondrial membrane translocase), NDUFS2 (a core subunit of mitochondrial complex I), and NDUFV1 (a mitochondrial NADH dehydrogenase). Notably, SLC25A1, TIMM23, and NDUFV1 play a role in mitochondrial ROS generation;45−47 NDUFS2 is essential for acute oxygen-sensing in a hypoxic environment;48 and SLC25A11 may control mitochondrial GSH levels that antagonize ferroptosis (Figure 5f).49 Our findings are consistent with the view that photoexcited RdU might target lysosomes, thereby enhancing ROS generation, and activate the mitochondrial-mediated ferroptosis pathway.
Based on the above results, the binding of RdU with LIMP-2 was studied in greater depth. Two 3D structures of the human LIMP-2 protein are available in the PDB database at pH 4.8 and 7.5. Considering the acidic environment within lysosomes, we selected the configuration at pH 4.8 to perform docking simulations of the interaction of RdU with LIMP-2. The results show that the “head” group (the Ru-coordinated pyridine segment) of RdU is small enough to be inserted into a hydrophobic pocket (Figure 6a) that consists of amino acid residues Phe273, Met356, His357, and Pro358. Our model allows binding through weak noncovalent interactions, including hydrophobic interactions, van der Waals forces, and π–π stacking (Domain III of LIMP-2).50,51 Interestingly, the “tail” group (the triazole-coupled uridine part) of RdU closely interacts with the surface of amino acid residues Val86, Gly87, and Asp127 (Figure 6b) through numerous noncovalent interactions (Domain I of LIMP-2), including hydrophobic interactions, van der Waals forces, and π–π stacking. In particular, two oxygen atoms of the terminal hydroxyl in uridine could potentially form three hydrogen bonds with Gly87 (Figure 6b).52 Moreover, the binding energy of this proposed configuration is calculated to be −12.72 kcal/mol, and the inhibition constant Ki is 475.44 pM. In addition, RdU exhibited high affinity with a binding energy of −9.42 kcal/mol and an inhibition constant Ki of 125.21 nM for the configuration of LIMP-2 evaluated at pH 7.5 (Figure S15). These results indicate that RdU has the potential to bind to LIMP-2 with a strong affinity.
Figure 6.
Interaction of RdU with human LIMP-2 protein in vitro. (a) Binding site and mode of interaction of RdU with LIMP-2 as determined by molecular docking analysis. (b) Residues of LIMP-2 and the chemical groups of RdU that are involved in intermolecular interactions and identification of the related binding groups. (c) Electronic absorption spectrum and (d) fluorescence emission spectrum of RdU with increasing concentrations of LIMP-2. (e) Isothermal titration calorimetry (ITC) titration of RdU (100 μM) binding with human LIMP-2 (amino acids 27–432, 10 μM) in Tris–HCl buffer (pH 7.4) at 298 K. (f) Distribution and changes of LIMP-2 in MDA-MB-231 cells of different treatment groups (control, light, RdU, and RdU with light).
We then used the electronic absorption and fluorescence emission spectra to confirm the binding properties of RdU with LIMP-2. The absorption of 10 μM RdU was followed at 287 nm as LIMP-2 was titrated into the solution (Figure 6c). When LIMP-2 reached a concentration of 1.667 μM, the mixture showed a hypochromicity of 18.5% as well as a red shift of 3 nm from 287 to 290 nm. No further changes in the absorption intensity were seen at LIMP-2 concentrations greater than 1.667 μM, indicating that the interaction between RdU and LIMP-2 had saturated. Interestingly, the fluorescence emission intensity of RdU at 585 nm also increased rapidly upon addition of LIMP-2, then remained steady when the LIMP-2 concentration exceeded 2.333 μM (Figure 6d). The above results demonstrate that RdU binds to LIMP-2 with high affinity.
To further understand the thermodynamics and kinetic changes of the interaction between RdU and LIMP-2, we carried out isothermal titration calorimetry (ITC) experiments. Figure 6e shows the ITC curves, and Table S3 shows the results of the thermodynamic analyses. The measured binding enthalpy values, ΔH, provided by ITC studies enable the calculation of binding entropy (ΔG = ΔH – TΔS). Plots of heat exchange versus molar ratio were produced by titrating RdU (100 μM) into LIMP-2 (10 μM) solution. The titration plot strongly supports the idea that RdU binds to LIMP-2 in a two-site manner and saturates at a 2:1 (RdU/ LIMP-2) stoichiometry. The binding constants calculated for RdU are [K1] = 5.69 × 106 M–1 and [K2] = 1.02 × 103 M–1. For the first site of binding, ΔH1 = −3.801 kJ/mol and TΔS1 = 34.764 kJ/mol, indicating that it is a slightly exothermic process with a significant entropy increase and large binding equilibrium constants, both of which render the process exothermic. Hence, the first type of binding shows a synergistic enthalpy–entropy-driven gain, with the driving force being mainly electrostatic.
For the second site of binding, ΔH2 = 200 kJ/mol and TΔS2 = 217.112 kJ/mol, respectively, indicating that these processes are associated with heat absorption and a significant entropy increase. The binding equilibrium constant is smaller, probably because the hydrophobic part of RdU is deeply embedded in the hydrophobic part of the protein. RdU displaces all or part of the solvent water in the deeper hydrophobic cavity to enter the solution and transform into free water molecules, thereby increasing entropy. Therefore, site #2 is an entropy-driven binding process in which the hydrophobic effect dominates. Amazingly, these interactions are far stronger than that of LIMP-2 with β-glucocerebrosidase (an LIMP-2 binding protein),51 indicating that both types of binding processes proceed spontaneously and that the molecular structures of the resulting products are relatively stable.
To follow up on the above cellular assays, we subsequently examined the distribution of RdU and LIMP-2 in the cytoplasm of cancer cells to see if their intracellular localization overlapped. As shown in Figure 6f, MDA-MD-231 cells in the presence of RdU present a partial overlap of RdU (red fluorescence) with prominent LIMP-2 foci (green fluorescence) in the cytoplasm. The number of LIMP-2 foci increases after combined RdU and light irradiation treatment.53 Taken altogether, these results suggest that lysosomes are the likely targets of RdU dye at two dye-specific binding sites in LIMP-2 membrane proteins.
2.6. In Vivo Biodistribution and Phototherapeutic Studies
Encouraged by these promising results, the effect of PDT with RdU was investigated in vivo by constructing a TNBC transplant tumor model of MDA-MB-231 cells in nude mice (Figure 7a).54 When mice were injected intravenously with RdU (10 mg/kg), the fluorescence intensity in the tumor region increased significantly with time, indicating that RdU was distributed to the tumor site through the blood circulation (Figure 7b). Comparison of fluorescence intensity showed that the concentration of RdU reached a maximum at 2 h after tail vein injection, suggesting that RdU is either slowly metabolized and/or excreted by the body. Fluorescence images of the internal organs and tumors removed 24 h after administration of RdU, indicated that RdU primarily accumulated in the liver and tumor, while low concentration was detected in the brain. This fluorescence measurement is consistent with the Ru localization results obtained by inductively coupled plasma (ICP) mass spectroscopy, which showed that the concentration of Ru atoms was high in the tumor and liver tissues and low in other organs (Figure 7c). This result suggests that RdU efficiently accumulates in tumors, slowly metabolizes, and is removed through the liver. Eighteen days after the initial drug or light irradiation treatment, significant differences in tumor size were discovered between the different treatment groups. Specifically, in the RdU plus light irradiation group, the tumor volume was significantly smaller than in the other groups (Figure 7d,e).
Figure 7.
In vivo PDT activity of RdU. (a) Protocol of the in vivo experiment that evaluates the therapeutic efficacy of PDT with RdU. (b) Biodistribution of RdU (10 mg/kg) in MDA-MB-231 solid tumor implanted in mice axilla. Biodistribution measurements were measured up to 24 h after administration of RdU in the tail vein and 9 days after the tumor was xenographed. The fluorescence intensity of organs (heart, liver, spleen, lung, kidney, and tumor) after mice were sacrificed is shown. (c) The content of RdU in different organs as determined by ICP analysis of the Ru element. (d) Photographs of tumors removed from the sacrificed mice on day 26. Tumor weight (e), relative tumor volume (f), and body weight (g) in tumor-bearing mice of four groups.
We recorded the tumor volume and body weight daily. As shown in Figure 7f, during the first 10 days of photodynamic treatment, no significant difference in tumor volume was observed. However, from the 11th day forward, tumor volume in the RdU plus light irradiation group was much smaller than that in other groups. All mice behaved normally, showing no signs of distress. Their body weight remained largely stable, indicating that RdU is biocompatible (Figure 7g).
Following treatment, all major organs (i.e., heart, liver, spleen, lung, kidney, and brain) and tumor tissue were examined histologically using hematoxylin–eosin (H&E) staining. Although no pathological changes or damage was observed in any organs (Figure 8a), significant damage was noted in the tumor tissue, including deepened nuclear staining, reduced cellular tight junctions, disrupted cellular arrangement, and the formation of numerous vacuoles (Figure 8b).55 To further clarify the efficacy underlying the effect of RdU in vivo, the status of cell death within the tumor tissue was analyzed by the terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) staining assay. A faint green fluorescence signal was observed when tumors were treated with RdU or light alone, indicating no significant tumor cell death. A significantly stronger signal was observed after combined treatment with RdU plus light, indicating effective treatment in vivo (Figure 8c,e). However, it remained unclear whether these cells died due to ferroptosis or another process. Therefore, we completed additional immunofluorescence assays to investigate the expression and distribution of ferroptosis marker protein SLC7A11 in the tumor tissues of mice from different treatment groups. The results showed that the expression of SLC7A11 significantly diminished in the tumor tissues of mice treated with RdU plus light irradiation (Figure 8d,f). This further supports the notion that photoexcited RdU induces ferroptosis and promotes ROS production in tumor cells after targeting lysosomes. Overall, this study shows that RdU offers good treatment outcomes against hypoxic TNBC in vivo with low toxicity in the absence of photodynamic stimulation.
Figure 8.
Toxicity and efficacy of RdU in vivo. (a) H&E staining of the heart, liver, spleen, lung, kidney, and brain from the mouse used in the toxicity analysis. The control group (PBS), light groups (650 nm LED light exposed for 5 min every 2 days), RdU groups (10 mg/kg for every 2 days), and RdU + light groups (10 mg/kg drug on even days and 650 nm LED light exposed for 5 min on odd days), n = 4 per group, magnification (40×). H&E staining (b), TUNEL apoptosis assay (c), and SLC7A11 expression analysis (d) in tumor tissue. (e) The intensity of TUNEL and (f) SLC7A11 changed with different treatments.
3. Discussion
Initially, we designed the introduction of uridine into the Ru(II) complex with the expectation that it would target the insertion of DNA into rapidly proliferating tumor cells, thereby inducing DNA damage to inhibit tumor cell proliferation. However, in-depth studies revealed that the covalent linkage of uridine in RdU can target recognition of the lysosomal protein LIMP-2, effectively promoting lysosomal labeling of tumor cells and improving the efficiency of lysosomal targeting of PDT. Moreover, targeting lysosomes to interfere with their metabolic function has emerged as an effective strategy for tumor suppression.56−58 Accumulating evidence reveals that increasing the size and capacity of lysosomes promotes cellular metabolism in tumor cells and reduces the stability of lysosomal membranes, making tumor cells more accessible to abundant nutrients.59,60 LIMP-2, also known as scavenger receptor class B member 2 (SCARB2),50 is one of the most abundant proteins distributed on the surface of the lysosomal membrane, and is primarily responsible for delivering lipids to the lysosomes, including cholesterol, low-density lipoprotein, and glucoencephalosidase.51,61 Recent studies also indicate that the variants of LIMP-2 are closely related to multiple diseases, such as Parkinson’s, Gaucher, and cancer.62,63 The distribution of pathological forms, associated with LIMP-2 lesions, occurs in the gastrointestinal tract, lungs, and brain in humans. A dramatic decline in glucoencephalosidase activity in LIMP-2-deficient brain tissue results in lipid storage, abnormal autophagic/lysosomal function, and α-synuclein buildup.64,65 Hence, there exists a considerable track record of previous studies involving small molecules that explicitly bind to LIMP-2 and have been developed to target lysosomes to suppress tumor progression.66
Lysosomes are the primary organelles that respond to various stimuli by eliciting a ROS cascade. Their size and number increase abnormally in tumor cells,57 facilitating the degradation of proteins, nucleic acids, polysaccharides, and other biological substances to provide energy and building blocks for the unlimited proliferation of tumor cells.67 These findings have encouraged researchers to investigate the feasibility of using small molecule ligands as potential anticancer agents to target lysosomes and inhibit tumor cell growth. Lysosome-targeted PDT may be a practical approach to suppress metastatic and recurrent TNBC. Based on this view, lysosome-targeted Ru(II) polypyridyl complexes were designed and synthesized to investigate their efficacy as PDT photosensitizers.
4. Conclusions
In this study, we synthesized an uridine-modified polypyridine Ru(II) complex RdU by linking sections using a click chemistry reaction. We demonstrated that the covalent linkage of uridine in RdU is able to target and recognize the lysosomal protein LIMP-2, effectively promotes the tagging of lysosomes in tumor cells, and enhances the efficiency of lysosomal targeting PDT. Under physiological conditions, the photosensitizer RdU demonstrates effective photoexcitation properties and enhances the production of different types of ROS, especially superoxide anion, under light irradiation. This suggests that RdU may function as a type I PDT photosensitizer. In addition, in vitro and in vivo experiments revealed that RdU exhibits an effective tumor suppression action in TNBC under light irradiation. These combined results argue that RdU inhibits the growth of TNBC cells by binding specifically to LIMP-2 and promoting the generation of copious amounts of ROS. It appears capable of generating toxin radicals even in the hypoxic tumor environment where type II photodynamic excitation is severely limited. In summary, lysosome-targeted dU-modified Ru(II) complex (RdU) was fashioned in this study by a simple synthetic route. It exhibited high photosensitivity, and good prospects for clinical applications. There are ongoing efforts to evaluate the efficacy of these small molecule assemblies as toxins and probes for clinical imaging and therapy. We expect that the successful development of these drugs will provide promising treatment options for refractory hypoxic TNBCs.
5. Experimental Section
5.1. Chemicals
Unless otherwise specified, all chemicals and solvents were purchased commercially and utilized without additional purification. A click chemistry reaction was employed to create the polypyridyl Ru(II) complexes with triazole rings using an Anton Paar Monowave 300 microwave reactor. Electron spray ionization mass spectra (ESI-MS) were acquired from dimethyl sulfoxide (DMSO) solutions on an Agilent 1100 ESI-MS system that was operated at room temperature. The 1H NMR, 13C NMR, 1H–1H COSY, and 13C–1H COSY spectra were captured in a dimethyl-d6 sulfoxide (DMSO-d6) solution using a Bruker Avance III 500 spectrometer operating at ambient temperature. The purity of the synthesized Ru(II) complex RdU was verified to be at least 95% using HPLC (Agilent 1290 Infinity II) with methanol/acetonitrile (3:7) as solvent.
5.2. Synthesis and Characterization
5.2.1. Synthesis of [Ru(bpy)2ASIP](ClO4)2 (RuA)
A mixture of HAc (10 mL) with [Ru(bpy)2dione]Cl2 (125 mg, 0.2 mmol), (2E)-3-(4-azidophenyl)acrolein (51.9 mg, 0.3 mmol), and NH4Ac (662.2 mg, 8 mmol) was placed in a quartz tube and heated in a microwave cavity to 130 °C for 30 min to protect it from the effect of nitrogen. After the reaction ended, the mixture was cooled to room temperature, 20 mL of distilled water was added, and the pH was adjusted to 7.2 using concentrated ammonia. A significant amount of precipitate was formed, filtered, and dried to produce a red crude product. After the product was purified by chromatography with anhydrous acetonitrile as the mobile phase and neutral alumina of 200–300 mesh as the stationary phase, a red solid powder was recovered with a yield of 52%. ESI-MS (in CH3CN, m/z): 388.0 ([M – 2ClO4–]2+). 1H NMR (600 MHz, DMSO-d6) yielded: δ 9.02 (dd, J = 17.0, 8.0 Hz, 2H), 8.87 (ddd, J = 23.0, 8.2, 4.5 Hz, 5H), 8.27–8.18 (m, 3H), 8.12 (ddd, J = 7.9, 6.4, 2.6 Hz, 3H), 8.09–8.02 (m, 4H), 7.97–7.89 (m, 3H), 7.89–7.74 (m, 5H), 7.67–7.56 (m, 6H), 7.40–7.32 (m, 3H), 7.21 (t, J = 8.8 Hz, 1H). 13C NMR (151 MHz, DMSO-d6): δ 172.50 (s), 157.25 (s), 157.03 (s), 151.92 (s), 138.36 (s), 128.30 (s), 126.81 (s), 124.89 (s), 49.07 (s), 40.47 (s), 40.23 (s), 40.13 (s), 39.99 (s), 39.85 (s), 39.71 (s), 39.57 (s).
5.2.2. Synthesis of [Ru(bpy)2PTdUIP](ClO4)2 (RdU)
First, [Ru(bpy)2ASIP](ClO4)2 (155.4 mg, 0.2 mmol), 2′-deoxy-5-ethynyluridine (E832486, MACKLIN, China) (75.7 mg, 0.3 mmol), 1 mol % CuSO4·5H2O, and 1 mol % sodium ascorbate were combined in a 30 mL microwave reaction tube. Subsequently, 18 mL of DMSO and 2 mL of distilled water were added as solvents, and the mixture was passed through nitrogen for 10 min. Then, 50 mL of distilled water was added, followed by incubation at 65 °C for 25 min. The precipitate was filtered, dried, and passed through a drying column containing sodium perchlorate. On the column, the fluid was dried repeatedly and eluted with a modest amount of acetonitrile. The final bright red powder was produced with a yield of 17.4%. ESI-MS (in CH3CN, m/z): 514.9 ([M – 2ClO4–]2+), 1027.6 ([M – ClO4–]+). 1H NMR (500 MHz, DMSO-d6) yielded: δ 9.03 (dd, J = 22.9, 8.5 Hz, 4H), 8.93 (d, J = 10.4 Hz, 2H), 8.84 (dd, J = 18.6, 8.0 Hz, 4H), 8.68 (d, J = 6.3 Hz, 2H), 8.57 (d, J = 8.6 Hz, 4H), 8.20 (dt, J = 9.5, 1.5 Hz, 4H), 8.13–8.06 (m, 2H), 8.06–8.00 (m, 1H), 7.96 (d, J = 8.7 Hz, 4H), 7.94–7.84 (m, 2H), 7.65–7.50 (m, 1H), 7.37–7.30 (m, 1H), 6.24 (m, 1H), 5.07 (d, J = 11.2 Hz, 2H), 4.30 (s, 2H). 13C NMR (126 MHz, DMSO-d6): δ 171.50 (s), 161.12 (s), 156.67 (s), 151.61 (s), 149.60 (s), 144.93 (s), 140.21 (s), 137.85 (s), 136.93 (s), 136.30 (s), 136.55 (s), 132.08 (s), 130.41 (s), 128.53 (s), 127.82 (s), 126.20 (s), 124.41 (s), 120.49 (s), 119.93 (s), 119.45 (s), 104.67 (s), 87.49 (s), 84.92 (s), 70.63 (s), 61.38 (s), 43.02 (s), 22.50 (s), 18.91 (s), 13.43 (s).
5.3. Theoretical Calculations
The molecular structure of RdU was graphically modeled and energetically optimized using the ADF 2019.104 suite program (Computer Code: ADF 2019.199:80:00:0b:aa:1d:ce) with the GGA:BP86 level of theory and the Mopac method. Frontier molecular orbitals and optical excitation energies were derived by using the time-dependent DFT approach. To describe the nature of the first excited singlet and triplet states for those with complicated transition compositions, natural transition orbitals were examined.
5.4. In Vivo Antitumor Activity Evaluation
All animal experiments were carried out in accordance with the Guangdong Pharmaceutical University Experimental Animal Center’s ethical guidelines. All BALB/c nude mice (female, 5 weeks old, and weighing 18–23 (g)) were purchased from Changzhou Cavens Laboratory Animals Co., Ltd. (Changzhou, China, animal license No. 202242314). All animals were kept in a specific pathogen-free environment with unrestricted access to food and water at a temperature of 25 °C and a relative humidity of 50%. MDA-MB-231 cells (1 × 107 cells/mouse) in 200 μL of buffer (PBS/Matrigel = 3:1) were injected into the right axilla of mice. The tumor volume in mice grew to 100 mm3 after 9 days. The mice were randomly divided into four groups (n = 4 per group): the control group (PBS), the light group (exposed to 650 nm LED light for 5 min every 2 days), the RdU minus light group (10 mg/kg every 2 days), and the RdU plus light group (10 mg/kg drug on odd days and 650 nm 300 mW/cm2 LED light for 5 min on even days). The negative control group consisted of five mice that did not receive an injection of MDA-MB-231 cells. Every day, body weight was recorded, and tumor dimensions (length, width, and height) were determined using a vernier caliper. After the mice were anesthetized with isoflurane on the final day, the tumors and organs were harvested.
5.5. Statistical Analysis
All quantitative data were expressed as mean ± standard deviation (SD) and were performed at least three times. One-way analysis of variance (ANOVA) and the unpaired Student’s t test were used to determine whether there was statistical significance between the control group and the experimental group using GraphPad Prism 9.0. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 were considered statistically significant.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 81572926), the National Natural Science Foundation of China for Young Scholars (grant no. 81703349), the China Postdoctoral Science Foundation (grant no. 2017M610576), the Provincial Major Scientific Research Projects at Universities of Guangdong Province (grant no. 2014KZDXM053), and the Science and Technology Project of Guangdong Province (grant no. 2014A020212312). The authors thank LetPub (www.letpub.com) for linguistic assistance and presubmission expert review.
Data Availability Statement
All data supporting the conclusions in this paper are presented in the paper and/or the Supporting Information. The original data sets are available from the corresponding author upon reasonable request.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.3c00808.
Synthetic details for the complexes, materials, and structural characterization data; analytical and 1H NMR, 13C NMR, 1H–1H COSY, and mass spectra of complexes; and inhibitory effect of RdU against MCF-7 cells and MCF-10A cells (PDF)
Author Contributions
CRediT: Qiong Wu conceptualization, funding acquisition, methodology, writing-original draft; Chanling Yuan formal analysis, methodology, visualization; Jiacheng Wang methodology, visualization; Guohu Li methodology, visualization; Chunguang Zhu methodology, validation; Li Li methodology, validation; Zongtao Wang investigation, methodology; Qingshuang Lv investigation, methodology; Wenjie Mei funding acquisition, investigation, supervision, writing-review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Dolmans D. E.; Fukumura D.; Jain R. K. Photodynamic therapy for cancer. Nat. Rev. Cancer 2003, 3 (5), 380–387. 10.1038/nrc1071. [DOI] [PubMed] [Google Scholar]
- Vickerman B. M.; Zywot E. M.; Tarrant T. K.; Lawrence D. S. Taking phototherapeutics from concept to clinical launch. Nat. Rev. Chem. 2021, 5 (11), 816–834. 10.1038/s41570-021-00326-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Cheng X.; Yu L.; Yang J.; Calvo R.; Patnaik S.; Hu X.; Gao Q.; Yang M.; Lawas M.; Delling M.; Marugan J.; Ferrer M.; Xu H. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 2016, 7, 12109 10.1038/ncomms12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J.; Lemberg K. M.; Lamprecht M. R.; Skouta R.; Zaitsev E. M.; Gleason C. E.; Patel D. N.; Bauer A. J.; Cantley A. M.; Yang W. S.; Morrison B. 3rd; Stockwell B. R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012, 149 (5), 1060–1072. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C.; Fraldi A.; Medina D. L.; Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell. Biol. 2013, 14 (5), 283–296. 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim W.-Y.; Mizushima N. Lysosome biology in autophagy. Cell Discovery 2020, 6, 6 10.1038/s41421-020-0141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavčič N.; Pegan K.; Turk B. Lysosomes in programmed cell death pathways: from initiators to amplifiers. Biol. Chem. 2017, 398 (3), 289–301. 10.1515/hsz-2016-0252. [DOI] [PubMed] [Google Scholar]
- Mai T. T.; Hamai A.; Hienzsch A.; Caneque T.; Muller S.; Wicinski J.; Cabaud O.; Leroy C.; David A.; Acevedo V.; Ryo A.; Ginestier C.; Birnbaum D.; Charafe-Jauffret E.; Codogno P.; Mehrpour M.; Rodriguez R. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017, 9 (10), 1025–1033. 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. Q.; Benthani F. A.; Wu J.; Liang D.; Bian Z. X.; Jiang X. Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ. 2020, 27 (1), 242–254. 10.1038/s41418-019-0352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.; Zhang Z.; Chen L.; Zhou Y.; Zou P.; Feng C.; Wang L.; Liang G. Dihydroartemisinin (DHA) induces ferroptosis and causes cell cycle arrest in head and neck carcinoma cells. Cancer Lett. 2016, 381 (1), 165–175. 10.1016/j.canlet.2016.07.033. [DOI] [PubMed] [Google Scholar]
- Lim T. H.; Lai T. Y. Y.; Takahashi K.; Wong T. Y.; Chen L. J.; Ruamviboonsuk P.; Tan C. S.; Lee W. K.; Cheung C. M. G.; Ngah N. F.; Patalauskaite R.; Margaron P.; Koh A.; Group E. I. S. Comparison of ranibizumab with or without verteporfin photodynamic therapy for polypoidal choroidal vasculopathy: the EVEREST II randomized clinical trial. JAMA Ophthalmol. 2020, 138 (9), 935–942. 10.1001/jamaophthalmol.2020.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe A.; Senge M. O. The photosensitizer temoporfin (mTHPC) - chemical, pre-clinical and clinical developments in the last decade. Photochem. Photobiol. 2023, 99 (2), 356–419. 10.1111/php.13730. [DOI] [PubMed] [Google Scholar]
- Yano T.; Minamide T.; Takashima K.; Nakajo K.; Kadota T.; Yoda Y. Clinical practice of photodynamic therapy using talaporfin sodium for esophageal cancer. J. Clin. Med. 2021, 10 (13), 2785. 10.3390/jcm10132785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins W. K.; Santos N. F.; Rocha C. S.; Bacellar I. O. L.; Tsubone T. M.; Viotto A. C.; Matsukuma A. Y.; Abrantes A. B. P.; Siani P.; Dias L. G.; Baptista M. S. Parallel damage in mitochondria and lysosomes is an efficient way to photoinduce cell death. Autophagy 2019, 15 (2), 259–279. 10.1080/15548627.2018.1515609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W.; Zhang Y.; Liu M.; Zhang H.; Zhang J.; Li C. Universal scaffold for an activatable photosensitizer with completely inhibited photosensitivity. Angew. Chem., Int. Ed. 2019, 58 (46), 16601–16609. 10.1002/anie.201907510. [DOI] [PubMed] [Google Scholar]
- Rak J.; Kabesova M.; Benes J.; Pouckova P.; Vetvicka D. Advances in liposome-encapsulated phthalocyanines for photodynamic therapy. Life 2023, 13 (2), 305. 10.3390/life13020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming L.; Cheng K.; Chen Y.; Yang R.; Chen D. Enhancement of tumor lethality of ROS in photodynamic therapy. Cancer Med. 2021, 10 (1), 257–268. 10.1002/cam4.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.; Huang B.; Nawaz M. H.; Zhang W. Recent advances of multi-dimensional porphyrin-based functional materials in photodynamic therapy. Coord. Chem. Rev. 2020, 420, 213410 10.1016/j.ccr.2020.213410. [DOI] [Google Scholar]
- Heinemann F.; Karges J.; Gasser G. Critical overview of the Use of Ru(II) polypyridyl complexes as photosensitizers in one-photon and two-photon photodynamic therapy. Acc. Chem. Res. 2017, 50 (11), 2727–2736. 10.1021/acs.accounts.7b00180. [DOI] [PubMed] [Google Scholar]
- Roque Iii J. A.; Cole H. D.; Barrett P. C.; Lifshits L. M.; Hodges R. O.; Kim S.; Deep G.; Frances-Monerris A.; Alberto M. E.; Cameron C. G.; McFarland S. A. Intraligand excited states turn a ruthenium oligothiophene complex into a light-triggered ubertoxin with anticancer effects in extreme hypoxia. J. Am. Chem. Soc. 2022, 144 (18), 8317–8336. 10.1021/jacs.2c02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemin E.; Ong Y. C.; Thomas C. M.; Gasser G. Polymer encapsulation of ruthenium complexes for biological and medicinal applications. Nat. Rev. Chem. 2019, 3 (4), 261–282. 10.1038/s41570-019-0088-0. [DOI] [Google Scholar]
- Monro S.; Colon K. L.; Yin H.; Roque J. 3rd; Konda P.; Gujar S.; Thummel R. P.; Lilge L.; Cameron C. G.; McFarland S. A. Transition metal complexes and photodynamic therapy from a tumor-centered approach: challenges, opportunities, and highlights from the development of TLD1433. Chem. Rev. 2019, 119 (2), 797–828. 10.1021/acs.chemrev.8b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N. L.; Liao J. X.; Huang M. Y.; Zhang Y. Q.; Chen J. X.; Zhang Z. W.; Yang Z. X.; Long X. E.; Wu X. T.; Sun J. Lysosome-targeted ruthenium(II) complexes induce both apoptosis and autophagy in HeLa cells. J. Inorg. Biochem. 2022, 229, 111729 10.1016/j.jinorgbio.2022.111729. [DOI] [PubMed] [Google Scholar]
- Chen J.; Tao Q.; Wu J.; Wang M.; Su Z.; Qian Y.; Yu T.; Wang Y.; Xue X.; Liu H. K. A lysosome-targeted ruthenium(II) polypyridyl complex as photodynamic anticancer agent. J. Inorg. Biochem. 2020, 210, 111132 10.1016/j.jinorgbio.2020.111132. [DOI] [PubMed] [Google Scholar]
- Liu B.; Gao Y.; Jabed M. A.; Kilina S.; Liu G.; Sun W. Lysosome targeting bis-terpyridine ruthenium(II) complexes: photophysical properties and in vitro photodynamic therapy. ACS Appl. Bio Mater. 2020, 3 (9), 6025–6038. 10.1021/acsabm.0c00647. [DOI] [PubMed] [Google Scholar]
- Paul S.; Pathak S.; Sahoo S.; Maji R. C.; Bhattacharyya U.; Nandi D.; Chakravarty A. R. Bichromophoric ruthenium(II) bis-terpyridine-BODIPY based photosensitizers for cellular imaging and photodynamic therapy. Dalton Trans. 2022, 51 (27), 10392–10405. 10.1039/D2DT01137A. [DOI] [PubMed] [Google Scholar]
- He G.; Xu N.; Ge H.; Lu Y.; Wang R.; Wang H.; Du J.; Fan J.; Sun W.; Peng X. Red-light-responsive Ru complex photosensitizer for lysosome localization photodynamic therapy. ACS Appl. Mater. Interfaces 2021, 13 (17), 19572–19580. 10.1021/acsami.0c22551. [DOI] [PubMed] [Google Scholar]
- Huang H.; Yu B.; Zhang P.; Huang J.; Chen Y.; Gasser G.; Ji L.; Chao H. Highly charged ruthenium(II) polypyridyl complexes as lysosome-localized photosensitizers for two-photon photodynamic therapy. Angew. Chem., Int. Ed. 2015, 54 (47), 14049–14052. 10.1002/anie.201507800. [DOI] [PubMed] [Google Scholar]
- Qiao L.; Liu J.; Han Y.; Wei F.; Liao X.; Zhang C.; Xie L.; Ji L.; Chao H. Rational design of a lysosome-targeting and near-infrared absorbing Ru(ii)-BODIPY conjugate for photodynamic therapy. Chem. Commun. 2021, 57 (14), 1790–1793. 10.1039/D0CC06926D. [DOI] [PubMed] [Google Scholar]
- Zhang D. Y.; Zheng Y.; Zhang H.; He L.; Tan C. P.; Sun J. H.; Zhang W.; Peng X.; Zhan Q.; Ji L. N.; Mao Z. W. Ruthenium complex-modified carbon nanodots for lysosome-targeted one- and two-photon imaging and photodynamic therapy. Nanoscale 2017, 9 (47), 18966–18976. 10.1039/C7NR05349E. [DOI] [PubMed] [Google Scholar]
- Li X.; Rydzewski N.; Hider A.; Zhang X.; Yang J.; Wang W.; Gao Q.; Cheng X.; Xu H. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 2016, 18 (4), 404–417. 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano O.; Kazetani K.; Funata M.; Fukuda Y.; Matsui J.; Iwata H. Vacuolin-1 inhibits autophagy by impairing lysosomal maturation via PIKfyve inhibition. FEBS Lett. 2016, 590 (11), 1576–1585. 10.1002/1873-3468.12195. [DOI] [PubMed] [Google Scholar]
- White J. D.; Osborn M. F.; Moghaddam A. D.; Guzman L. E.; Haley M. M.; DeRose V. J. Picazoplatin, an azide-containing platinum(II) derivative for target analysis by click chemistry. J. Am. Chem. Soc. 2013, 135 (32), 11680–11683. 10.1021/ja402453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- żamojć K.; Zdrowowicz M.; Rudnicki-Velasquez P. B.; Krzyminski K.; Zaborowski B.; Niedzialkowski P.; Jacewicz D.; Chmurzynski L. The development of 1,3-diphenylisobenzofuran as a highly selective probe for the detection and quantitative determination of hydrogen peroxide. Free Radical Res. 2017, 51 (1), 38–46. 10.1080/10715762.2016.1262541. [DOI] [PubMed] [Google Scholar]
- Liang C.; Xie J.; Luo S.; Huang C.; Zhang Q.; Huang H.; Zhang P. A highly potent ruthenium(II)-sonosensitizer and sonocatalyst for in vivo sonotherapy. Nat. Commun. 2021, 12 (1), 5001 10.1038/s41467-021-25303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn C. A.; Simon S. R.; Schoonen M. A. Comparison of fluorescence-based techniques for the quantification of particle-induced hydroxyl radicals. Part. Fibre Toxicol. 2008, 5, 2. 10.1186/1743-8977-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L.; Jiang Y.; Xia D.; Wei Z.; Spinney R.; Dionysiou D. D.; Minakata D.; Xiao R.; Xie H. B.; Chai L. Mechanistic understanding of superoxide radical-mediated degradation of perfluorocarboxylic acids. Environ. Sci. Technol. 2022, 56 (1), 624–633. 10.1021/acs.est.1c06356. [DOI] [PubMed] [Google Scholar]
- Cole H. D.; Roque J. A. 3rd; Shi G.; Lifshits L. M.; Ramasamy E.; Barrett P. C.; Hodges R. O.; Cameron C. G.; McFarland S. A. Anticancer agent with inexplicable potency in extreme hypoxia: characterizing a light-triggered ruthenium ubertoxin. J. Am. Chem. Soc. 2022, 144 (22), 9543–9547. 10.1021/jacs.1c09010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Wu S.; Zhang L.; Xu S.; Dai C.; Gan S.; Xie G.; Feng G.; Tang B. Z. Cationization to boost both type I and type II ROS generation for photodynamic therapy. Biomaterials 2022, 280, 121255 10.1016/j.biomaterials.2021.121255. [DOI] [PubMed] [Google Scholar]
- Wei F.; Kuang S.; Rees T. W.; Liao X.; Liu J.; Luo D.; Wang J.; Zhang X.; Ji L.; Chao H. Ruthenium(II) complexes coordinated to graphitic carbon nitride: Oxygen self-sufficient photosensitizers which produce multiple ROS for photodynamic therapy in hypoxia. Biomaterials 2021, 276, 121064 10.1016/j.biomaterials.2021.121064. [DOI] [PubMed] [Google Scholar]
- Lee Y. L.; Chou Y. T.; Su B. K.; Wu C. C.; Wang C. H.; Chang K. H.; Ho J. A.; Chou P. T. Comprehensive thione-derived perylene diimides and their bio-conjugation for simultaneous imaging, tracking, and targeted photodynamic therapy. J. Am. Chem. Soc. 2022, 144 (37), 17249–17260. 10.1021/jacs.2c07967. [DOI] [PubMed] [Google Scholar]
- Lai Y.; Lu N.; Luo S.; Wang H.; Zhang P. A photoactivated sorafenib-ruthenium(II) prodrug for resistant hepatocellular carcinoma therapy through ferroptosis and purine metabolism disruption. J. Med. Chem. 2022, 65 (19), 13041–13051. 10.1021/acs.jmedchem.2c00880. [DOI] [PubMed] [Google Scholar]
- Koppula P.; Zhuang L.; Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12 (8), 599–620. 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S.; SriRamaratnam R.; Welsch M. E.; Shimada K.; Skouta R.; Viswanathan V. S.; Cheah J. H.; Clemons P. A.; Shamji A. F.; Clish C. B.; Brown L. M.; Girotti A. W.; Cornish V. W.; Schreiber S. L.; Stockwell B. R. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156 (1–2), 317–331. 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez H. R.; Gadre S. M.; Tan M.; Graham G. T.; Mosaoa R.; Ongkeko M. S.; Kim K. A.; Riggins R. B.; Parasido E.; Petrini I.; Pacini S.; Cheema A.; Varghese R.; Ressom H. W.; Zhang Y.; Albanese C.; Uren A.; Paige M.; Giaccone G.; Avantaggiati M. L. The mitochondrial citrate carrier, SLC25A1, drives stemness and therapy resistance in non-small cell lung cancer. Cell Death Differ. 2018, 25 (7), 1239–1258. 10.1038/s41418-018-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S.; Ruan M.; Li Y.; Yang J.; Bai S.; Richter C.; Schwalbe H.; Xie C.; Shen B.; Wang J. Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide. Cell Res. 2021, 31 (7), 821–824. 10.1038/s41422-020-00452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin G.; Margiotta D.; Kasahara A.; Bassoy E. Y.; Walch M.; Thiery J.; Lieberman J.; Martinvalet D. Granzyme B-induced mitochondrial ROS are required for apoptosis. Cell Death Differ. 2015, 22 (5), 862–874. 10.1038/cdd.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Snary K. J.; Wu D.; Potus F.; Sykes E. A.; Mewburn J. D.; Charles R. L.; Eaton P.; Sultanian R. A.; Archer S. L. Ndufs2, a Core subunit of mitochondrial complex I, is essential for acute oxygen-sensing and hypoxic pulmonary vasoconstriction. Circ. Res. 2019, 124 (12), 1727–1746. 10.1161/CIRCRESAHA.118.314284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta N.; Qu C.; Wu H.; Zhang D.; Sun T.; Li Y.; Wang J.; Wang X.; Tang T.; Chen Q.; Liu L. Mitochondrial outer membrane protein FUNDC2 promotes ferroptosis and contributes to doxorubicin-induced cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (36), e2117396119 10.1073/pnas.2117396119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Ren J.; Padilla-Parra S.; Fry E. E.; Stuart D. I. Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor. Nat. Commun. 2014, 5, 4321 10.1038/ncomms5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neculai D.; Schwake M.; Ravichandran M.; Zunke F.; Collins R. F.; Peters J.; Neculai M.; Plumb J.; Loppnau P.; Pizarro J. C.; Seitova A.; Trimble W. S.; Saftig P.; Grinstein S.; Dhe-Paganon S. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 2013, 504 (7478), 172–176. 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- Conrad K. S.; Cheng T. W.; Ysselstein D.; Heybrock S.; Hoth L. R.; Chrunyk B. A.; Am Ende C. W.; Krainc D.; Schwake M.; Saftig P.; Liu S.; Qiu X.; Ehlers M. D. Lysosomal integral membrane protein-2 as a phospholipid receptor revealed by biophysical and cellular studies. Nat. Commun. 2017, 8 (1), 1908 10.1038/s41467-017-02044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.; Zhang J.; Zhang X.; Wang Y.; Yu H.; Yin X.; Li J.; Du P.; Plumas J.; Chaperot L.; Chen J.; Su L.; Liu Y.; Zhang L. SCARB2/LIMP-2 regulates IFN production of plasmacytoid dendritic cells by mediating endosomal translocation of TLR9 and nuclear translocation of IRF7. J. Immunol. 2015, 194 (10), 4737–4749. 10.4049/jimmunol.1402312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.; Zhou Y.; Liu Y.; Jiang X.; Zeng W.; Gong Z.; Zheng G.; Sun D.; Dai Z. Asymmetric, amphiphilic RGD conjugated phthalocyanine for targeted photodynamic therapy of triple negative breast cancer. Signal Transduction Targeted Ther. 2022, 7 (1), 64 10.1038/s41392-022-00906-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia Hernandez R.; Lee M. C.; Yadav A. K.; Chan J. Repurposing cyanine photoinstability to develop near-infrared light-activatable nanogels for in vivo cargo delivery. J. Am. Chem. Soc. 2022, 144 (39), 18101–18108. 10.1021/jacs.2c08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S.; Amaravadi R. K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016, 1371 (1), 45–54. 10.1111/nyas.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam S. R.; Wang F.; Muller S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discovery 2019, 18 (12), 923–948. 10.1038/s41573-019-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M.; Luo X.; Wu K.; He X. Targeting lysosomes in human disease: from basic research to clinical applications. Signal Transduction Targeted Ther. 2021, 6 (1), 379 10.1038/s41392-021-00778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.; Wang X. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 2021, 220 (6), e202102001 10.1083/jcb.202102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.; Yang Z. Y.; Wang D.; Yang X. Y.; Wang J.; Li L.; Wen Q.; Gao L.; Bian X. W.; Yu S. C. The role of lysosomes in cancer development and progression. Cell Biosci. 2020, 10 (1), 131 10.1186/s13578-020-00489-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heybrock S.; Kanerva K.; Meng Y.; Ing C.; Liang A.; Xiong Z. J.; Weng X.; Ah Kim Y.; Collins R.; Trimble W.; Pomes R.; Prive G. G.; Annaert W.; Schwake M.; Heeren J.; Lullmann-Rauch R.; Grinstein S.; Ikonen E.; Saftig P.; Neculai D. Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) is involved in lysosomal cholesterol export. Nat. Commun. 2019, 10 (1), 3521 10.1038/s41467-019-11425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y.; Pei J.; Wang Y.; Chen Q.; Sun M.; Kang L.; Zhang X.; Zhang L.; Gao X.; Lin Z. The deficiency of SCARB2/LIMP-2 impairs metabolism via disrupted mTORC1-dependent mitochondrial OXPHOS. Int. J. Mol. Sci. 2022, 23 (15), 8634 10.3390/ijms23158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcalay R. N.; Levy O. A.; Wolf P.; Oliva P.; Zhang X. K.; Waters C. H.; Fahn S.; Kang U.; Liong C.; Ford B.; Mazzoni P.; Kuo S.; Johnson A.; Xiong L.; Rouleau G. A.; Chung W.; Marder K. S.; Gan-Or Z. SCARB2 variants and glucocerebrosidase activity in Parkinson’s disease. npj Parkinson’s Dis. 2016, 2, 16004 10.1038/npjparkd.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothaug M.; Zunke F.; Mazzulli J. R.; Schweizer M.; Altmeppen H.; Lullmann-Rauch R.; Kallemeijn W. W.; Gaspar P.; Aerts J. M.; Glatzel M.; Saftig P.; Krainc D.; Schwake M.; Blanz J. LIMP-2 expression is critical for beta-glucocerebrosidase activity and alpha-synuclein clearance. Proc. Natl. Acad. Sci. U.S.A. 2014, 111 (43), 15573–15578. 10.1073/pnas.1405700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X. Y.; Guo L.; Huang D. Y.; Chang X. L.; Qiu Q. C. Distribution of EV71 receptors SCARB2 and PSGL-1 in human tissues. Virus Res. 2014, 190, 40–52. 10.1016/j.virusres.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Pei J.; Wang G.; Feng L.; Zhang J.; Jiang T.; Sun Q.; Ouyang L. Targeting lysosomal degradation pathways: new strategies and techniques for drug discovery. J. Med. Chem. 2021, 64 (7), 3493–3507. 10.1021/acs.jmedchem.0c01689. [DOI] [PubMed] [Google Scholar]
- Gros F.; Muller S. The role of lysosomes in metabolic and autoimmune diseases. Nat. Rev. Nephrol. 2023, 19 (6), 366–383. 10.1038/s41581-023-00692-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the conclusions in this paper are presented in the paper and/or the Supporting Information. The original data sets are available from the corresponding author upon reasonable request.