Abstract
In human cells infected with herpes simplex virus (HSV), viral gene expression is initiated by the virion protein VP16. VP16 does not bind DNA directly but forms a multiprotein complex on the viral immediate-early gene promoters with two cellular proteins: the POU domain protein Oct-1 and host cell factor (HCF; also called C1, VCAF, and CFF). Despite its apparent role in stabilizing the VP16-induced transcription complex, the natural biological role of HCF is unclear. Only recently HCF has been implicated in control of the cell cycle. To determine the role of HCF in cells and answer why HSV has evolved an HCF-dependent mechanism for the initiation of the lytic cycle, we identified the first human ligand for HCF (R. Lu et al., Mol. Cell. Biol. 17:5117–5126, 1997). This protein, Luman, is a member of the CREB/ATF family of transcription factors that can activate transcription from promoters containing cyclic AMP response elements (CRE). Here we provide evidence that Luman and VP16 share two important structural features: an acidic activation domain and a common mechanism for binding HCF. We found that Luman, its homolog in Drosophila, dCREB-A (also known as BBF-2), and VP16 bind to HCF by a motif, (D/E)HXY(S/A), present in all three proteins. In addition, a mutation (P134S) in HCF that prevents VP16 binding also abolishes its binding to Luman and dCREB-A. We also show that while interaction with HCF is not required for the ability of Luman to activate transcription when tethered to the GAL4 promoter, it appears to be essential for Luman to activate transcription through CRE sites. These data suggest that the HCF-Luman interaction may represent a conserved mechanism for transcriptional regulation in metazoans, and HSV mimics this interaction with HCF to monitor the physiological state of the host cell.
In cells infected with herpes simplex virus (HSV), the transcription of HSV immediate-early (IE or α) genes is regulated by a virion protein, VP16 (also called Vmw65 or αTIF) (reviewed in references 23 and 33). Unlike most other transcription activators, VP16 does not bind to DNA directly but is recruited to IE gene promoters by its association with a cellular POU domain protein, Oct-1 (13, 19, 20, 24, 32). Another cellular protein, host cell factor (HCF; also called C1, VCAF, or CFF) is required to facilitate and stabilize the VP16–Oct-1 association (9, 11, 31, 42). Upon infection of permissive cells, VP16 first forms a complex with HCF. This association subsequently promotes its interaction with Oct-1, which is bound to the TAATGARAT motif (R is a purine), the cis-regulatory target of VP16 activation found in HSV IE promoters (2, 13, 20, 24, 26, 32).
Purified HCF consists of a family of polypeptides, originating from cleavage of a single large precursor. The resulting amino (N)- and carboxyl (C)-terminal polypeptides of HCF remain bound together by noncovalent linkages (12, 38, 40). Although the N-terminal portion of the molecule can bind VP16 on its own (14, 37), this association and subsequent interaction with DNA-bound Oct-1 is stabilized by the C-terminal portion of HCF (14).
HCF has largely been defined by its accessory role in VP16-activated transcription. Nonetheless the gene for HCF is conserved in species as diverse as humans (12, 38), mice (10), and nematodes (14), and an HCF-like activity in fruit flies has been reported (28). This suggests that HCF plays an important role in the biology of metazoans, and recently it has been implicated in the regulation of the cell cycle (5). The hamster cell line tsBN67 has a single point mutation in its gene for HCF, proline to serine at position 134 (P134S), which confers a temperature-sensitive phenotype on the protein. At the nonpermissive temperature, cell division in tsBN67 cells is arrested at the G0/G1-S decision point. Although at the nonpermissive temperature HCF stability and posttranslational processing are not affected by the mutation, VP16 is unable to interact with the mutant HCF in vitro, and transcription activation by VP16 is also disrupted in these cells (5, 37).
Recently, we (18) and others (4) identified a human basic domain-leucine zipper (bZIP) protein, Luman, that interacts with HCF both in vivo and in vitro. Luman is a transcription factor of the CREB/ATF gene family that can activate transcription from promoters containing cyclic AMP response elements (CREs) (18). We showed that Luman and VP16 compete with each other for the binding of HCF in vitro. In transfection assays, using a reporter gene with a promoter containing the GAL4 response element (the upstream activation sequence [UAS]), we also showed that VP16 inhibited transcription activation by a fusion protein of the GAL4 DNA-binding domain (DBD) and Luman (GAL-Luman). The results suggested that VP16 and Luman bind to HCF by similar mechanisms. Sequence analyses (18) revealed that, like VP16, Luman and its homologs in mice (LZIP [3]) and in fruit flies (dCREB-A; also called BBF-2 [1, 30]) have a terminal acidic domain and a possible HCF-binding motif. This sequence motif, (D/E)HXY(S/A) (where X stands for any amino acid), in VP16 has been implicated in HCF binding (7, 8, 15, 29, 41) and is also present in VP16 homologs in other members of Alphaherpesvirinae.
Here we provided evidence for the functional relevance of these structural similarities between VP16 and Luman. We show that Luman’s acidic amino terminus is a functional activation domain when fused to GAL4 DBD and that the (D/E)HXY(S/A) motif in Luman and dCREB-A is involved in their interaction with HCF. We also show that interaction with functional HCF appears to be essential for Luman to activate promoters through CRE sites. However, it is not required either for Luman recognition of the CRE in vitro or for its inherent ability to activate transcription when tethered to a GAL4 UAS. We propose that VP16 mimics Luman in its interaction with HCF and that HSV has evolved this mechanism to exploit an important cellular regulatory circuit.
MATERIALS AND METHODS
Materials.
All restriction endonucleases, modifying enzymes, oligonucleotide primers, Taq DNA polymerase, and other reagents were purchased from Canadian Life Technologies unless otherwise stated.
Plasmids and mutagenesis.
The construction of plasmids pMLuman (a mammalian expression vector for GAL4-Luman fusion protein), pGEXLuman (a bacterial expression vector for glutathione S-transferase [GST]–Luman fusion protein), and pcLuman (the coding sequence for Luman cloned in the expression vector pcDNA3; Invitrogen) has been described previously (18). Plasmids p-109C3 and p-68CRE, which have CRE-containing promoters linked to the coding sequences for chloramphenicol acetyltransferase (CAT) (27), were obtained from William Roesler, University of Saskatchewan. Plasmids pM1 and pM2, for constructing GAL4 fusion proteins, and pG5EC, a plasmid containing the CAT gene linked to five GAL4 UAS, were obtained from Ivan Sadowski, University of British Columbia.
The 3′-end deletion mutants of Luman were constructed by digestion and religation of pMLuman, except pMLuman1-315. The plasmid DNA of pMLuman was cut with EcoRI-NdeI, HindIII, HindIII-EcoRV, and HindIII-PstI individually, blunt ended with Klenow fragment (where needed), and subsequently religated. The resultant plasmids were pMLuman1-107, pMLuman1-119, pMLuman1-220, and pMLuman1-276, respectively. To generate plasmid pMLuman1-315, the stop codon TAG was introduced at amino acid 316 by oligonucleotide-directed mutagenesis.
Amino acid substitutions in Luman were generated by oligonucleotide-directed mutagenesis (22). These mutants were then cloned into the same expression vectors as Luman (18).
Plasmid p46CY, containing the coding sequence of the dCREB-A gene between HindIII-NotI sites, was kindly given by Sarah Smolik, Oregon Health Sciences University (30). To construct pc-dCREB-A, an oligonucleotide linker (5′-AAGCTTGTCCCATGGAATTCTACGC and 5′-GGCCGCGTAGAATTCCATGGGACA) was first inserted between the HindIII-NotI sites on pcDNA-Amp (Invitrogen) to generate vector pclinker, and then the EcoRI-NotI fragment from p46CY was cloned between the same restriction sites on pclinker. To construct pGEX-dCREB-A, the NcoI-SacI fragment from p46CY containing the dCREB-A gene was inserted into the pGEX-KG vector. Mutants of dCREB-A, constructed by oligonucleotide-directed mutagenesis, were cloned into the pGEX-KG and pclinker vectors by using the same strategy.
All of the HCF plasmids used in this study contain the functional version of HCF, HCF(NC) (14, 18). Plasmid SL2, an expression vector of GAL4 DBD and HCF(NC) fusion protein, was a gift from S. LaBoissière and P. O’Hare, Marie Curie Institute, Surrey, England. The P143S mutation of HCF was made by introducing a C-to-T transition by using a PCR strategy. The left or mutagenesis primer (5′-CAAAAACGGGCCCCCTtCGTGTCCTCGAC) and the right primer (5′-ACTCCCGGGGTGGTGGTAGGACC) had the restriction sites ApaI and SmaI, respectively (underlined). The 25-μl reaction mixture consisted of 50 ng of SL2 plasmid DNA, 0.2 mM each dATP, dTTP, dGTP, and dCTP, 0.2 μM primers, 1.75 mM MgCl2 and 1.25 U of Taq DNA polymerase. The PCR program began with one cycle of 2-min denaturation at 94°C, 2-min annealing at 58°C, and 2-min extension at 72°C, followed by 25 cycles of 45 s at 94°C, 30 s at 55°C, and 1 min at 72°C, ending with a final 8-min extension at 72°C. The PCR products were subjected to electrophoresis in a 1.6% agarose gel. The ∼200-bp DNA band was cut out, and DNA was eluted, purified by phenol-chloroform extraction, and precipitated in ethanol. The DNA fragment was then digested with ApaI and SmaI and was cloned between the same sites in SL2. The nucleotide sequences of all of the mutagenesis clones were confirmed by DNA sequencing.
Cell culture, transfections, and CAT assays.
The BHK-21-derived temperature-sensitive cell line tsBN67 was obtained from the RIKEN Gene Bank, Tsukuba, Japan. The growth conditions of COS7 cells and the method of transfection have been described previously (18, 21, 22). COS7 cells were transfected by using Lipofectamine (Canadian Life Technologies) and assayed for CAT expression by using a CAT enzyme-linked immunosorbent assay (ELISA) kit (Boehringer Mannheim) as instructed by the manufacturer. The BHK-21 and mutant tsBN67 cells were cultured at 37 and 33.5°C, respectively, in Dulbecco’s modified Eagle medium supplemented with 10% newborn calf serum. One day prior to transfection, BHK-21 and tsBN67 cells were seeded into six-well plates at a concentration of 2 × 105/well at the assay temperature (33.5 or 39.5°C). The cells were transfected with 12 μl of Lipofectamine and 2 μg of DNA (pUC19 was used to make up the total amount of DNA). CAT activity was measured 48 h posttransfection, using the CAT ELISA kit (Boehringer Mannheim). All transfection data were confirmed by at least three independent assays, and the results of representative assays are presented.
Expression and purification of GST fusion proteins, in vitro transcription and translation, and GST pull-down assays.
The GST fusion proteins were produced and purified by using glutathione-Sepharose beads (Pharmacia) from Escherichia coli BL21(DE3) (Novagen) (18). A rabbit reticulocyte in vitro transcription-translation system (TnT; Promega) was used to produce 35S-labeled proteins according to the manufacturer’s protocol. GST pull-down assays were performed as described previously (18). To ensure that each GST pull-down reaction contained the same amount of GST fusion protein, the concentration of each sample of glutathione beads with fusion proteins was adjusted so that when examined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), each reaction contained the same intensity of Coomassie blue-stained protein band.
EMSA.
Oligonucleotides representing binding sites for CRE (5′-AGCTGCC GGTGACGTCATCGCAT and 5′-CTAGATGCGATGACGTACCCGGC) were annealed, labeled with [α-32P]dCTP by using the Klenow fragment of E. coli DNA polymerase, and used as probes. In each electrophoretic mobility shift assay (EMSA) reaction (50 μl), approximately 1 ng of labeled DNA probe and 50 to 100 ng of recombinant GST-Luman and variant proteins were used, as measured by a protein assay kit (Bio-Rad) and SDS-PAGE. Details of the procedures are provided elsewhere (18, 21, 22).
RESULTS
Like VP16, Luman has a defined, acidic activation domain.
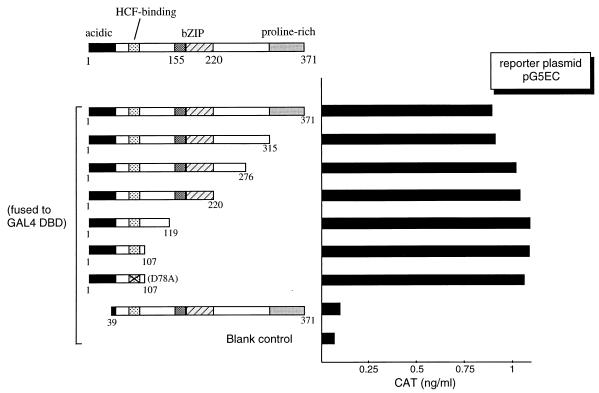
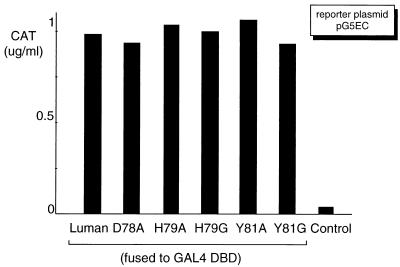
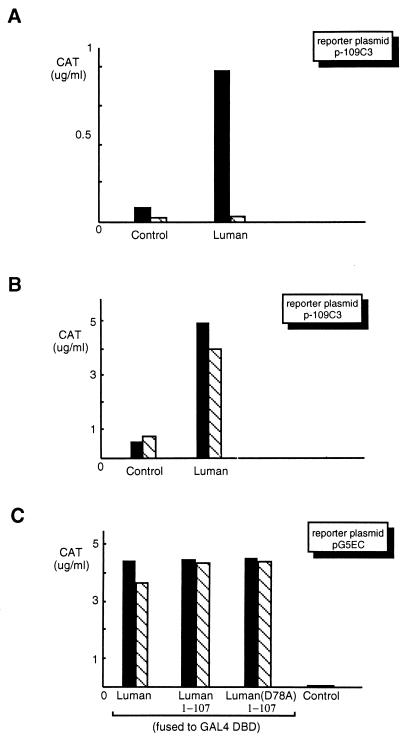
Previously, we identified a human bZIP protein, Luman, that interacts with HCF (18). Sequence analysis revealed that the N-terminal region of Luman is rich in negatively charged amino acids, suggesting the existence of a N-terminal acidic activation domain, as exemplified in VP16. To verify and map the N-terminal activation domain, we constructed a series of 3′ deletions (Fig. 1) to progressively remove the C-terminal proline-rich region, the segment between the proline-rich and the bZIP regions, the bZIP region, and the region between bZIP and the presumed HCF-binding motif. A clone bearing the first 107 amino acids of Luman with a mutation, D78A (an aspartic acid-to-alanine substitution at position 78), in the presumed HCF-binding motif (Fig. 2) was also included. (The same nomenclature is applied to other amino acid substitution mutations referred to in this paper.) These plasmids were cotransfected into COS7 cells with a reporter plasmid, pG5EC, that contains the coding sequences of CAT linked to five GAL4 UAS in its promoter region. The results (Fig. 1) showed that the first 107 amino acids in the N-terminal region were sufficient to activate transcription when fused to GAL4 DBD, and removal of the first 38 amino acids disrupted the activation. The D78A mutation, which abrogates HCF binding (see below), did not affect activation. The data suggest that the N-terminal acidic region of Luman is an activation domain and that transcription activation, at least in the context of a GAL4 UAS-containing promoter, does not require HCF.
FIG. 1.
Mapping of the activation domain of Luman by deletion analysis. On the left is a schematic representation of the structure of the Luman protein, in which numbers indicate the positions of the amino acids. The X in the HCF-binding domain of D78A represents a point mutation (alanine replaces the aspartate residue at position 78). Features of the protein discussed in the text are labeled. Luman and its deletion mutants were fused to the GAL4 DBD. The same amount (0.5 μg) of each plasmid was introduced into COS7 cells along with the reporter plasmid, pG5EC (0.5 μg), which has five copies of GAL4 UAS in the promoter region linked to the cat gene. CAT activity was measured by ELISA 48 h posttransfection.
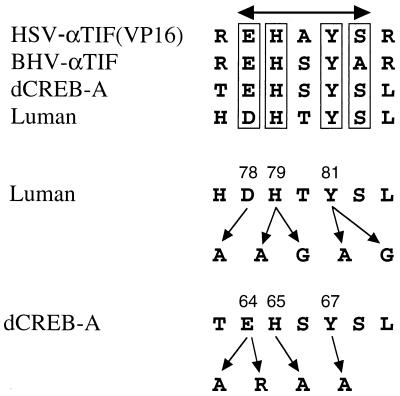
FIG. 2.
Putative HCF-binding domains and mutants. The amino acid sequence alignment shows that Luman and the Drosophila protein dCREB-A share, with the herpesvirus αTIF proteins, a motif that has been implicated in HCF binding in VP16 (HSV-αTIF). In this consensus (D/E)HXY(S/A) motif, we mutated amino acids D78, H79, and Y81 of Luman and amino acids E64, H65, and Y67 of dCREB-A. BHV, bovine herpesvirus.
Luman, along with dCREB-A, shares an HCF-binding motif with VP16.
Amino acid sequence alignment showed that Luman and the Drosophila protein dCREB-A (1, 30) share a motif with the αTIF proteins of alphaherpesviruses (18). This motif in VP16 (HSV αTIF) has been implicated in its association with HCF (7, 8, 15, 29, 41). In the consensus (D/E)HXY(S/A) motif (Fig. 2), we mutated amino acids D78, H79, and Y81 of Luman and amino acids E64, H65, and Y67 of dCREB-A to determine if these amino acids are involved in the HCF binding.
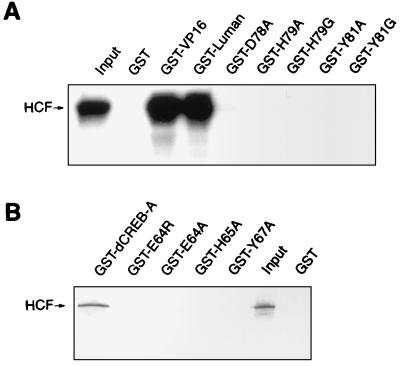
In the GST pull-down assays, the GST fusion proteins of Luman and dCREB-A were produced in bacteria and purified by using glutathione-Sepharose beads. The HCF protein was transcribed and translated in vitro in the presence of [35S]methionine. The GST and GST-VP16 fusion proteins were used as controls. Equivalent amounts of protein-beads were used for all samples, as estimated by SDS-PAGE. The result (Fig. 3) showed that the amino acid substitution mutations in the HCF-binding motifs of Luman or dCREB-A disrupted their association with HCF.
FIG. 3.
Mutants in the conserved residues of the putative HCF-binding motif of Luman and dCREB-A do not bind to HCF in vitro. All GST and GST fusion proteins were produced in E. coli BL21(DE3) and bound to glutathione-Sepharose beads. HCF was labeled with [35S]methionine by in vitro transcription and translation in the TnT system (Promega). Equivalent amounts of GST proteins attached to the beads were incubated with [35S]HCF for 45 min, washed, analyzed on an SDS–10% polyacrylamide gel, and visualized by autoradiography. The lane labeled Input represents 1/10 of the [35S]HCF incubation mixture. Panels A and B show the GST pull-down results for Luman and dCREB-A and their mutants, respectively.
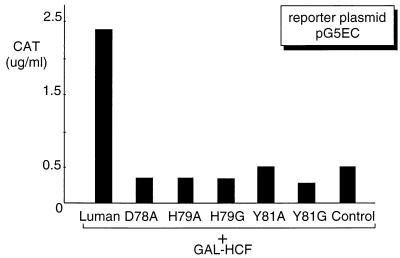
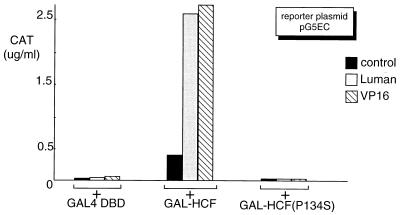
To test whether these mutants could also disrupt interaction with HCF in vivo, we used a mammalian two-hybrid system. We transfected COS7 cells with the expression plasmids for Luman or its mutants, GAL4 DBD-HCF fusion protein (GAL-HCF), as well as the reporter plasmid pG5EC. In this system, transcriptional activation by the Luman proteins is dependent on tethering to the GAL4 UAS-containing promoter in pG5EC via their interaction with GAL-HCF. The result (Fig. 4) showed that, in contrast to the wild-type Luman, the Luman mutants could not interact with HCF and therefore could not activate transcription on the GAL4 UAS promoter.
FIG. 4.
Luman mutants do not interact with HCF in vivo. Plasmids expressing wild-type Luman and its mutants (0.5 μg) were individually introduced in COS7 cells with pG5EC (0.5 μg) and a plasmid (0.5 μg) expressing GAL-HCF. Since transactivation by Luman depends on its tethering to GAL4 UAS through its interaction with HCF, CAT activity is an indicator of Luman-HCF interaction. The control represents the expression vector without an insert.
To prove that the loss of transactivation ability was not due to any changes that these mutations might have caused to the activation domain of Luman, the Luman mutants were fused to GAL4 DBD and tested for the ability to transactivate the GAL4 UAS promoter in pG5EC. The results (Fig. 5) showed that the activation domains of these Luman mutants were fully functional. The results also confirmed our observation [compare Luman1-107 and Luman(D78A)1-107 in Fig. 1] that mutation of the HCF-binding domain has little effect on the activation domain of Luman.
FIG. 5.
Mutations in the HCF-binding motif do not affect the activation domain of Luman. Luman and its mutants were fused to the GAL4 DBD to study the effects of the mutations on their ability to activate transcription when tethered to the GAL4 UAS. Equivalent amounts (0.5 μg) of each GAL-Luman fusion protein-expressing plasmid were cotransfected in COS7 cells with plasmid pG5EC (0.5 μg) as the reporter. The control represents the expression vector without an insert.
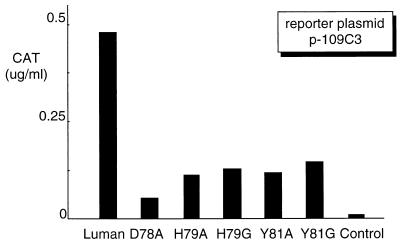
The HCF-binding mutants of Luman have reduced transcription activity on CRE-containing promoters, although they still retain the ability of binding to CRE in vitro. Our previous study showed that Luman binds to CRE and activates transcription from CRE-containing promoters (18). We therefore wanted to determine if Luman requires HCF for the activation of CRE-containing promoters in vivo. Plasmid p-109C3 (27), which has a CRE-containing promoter linked to the CAT gene, was used as the reporter plasmid to study the effect of mutations on the transcriptional activation by Luman. In the transfection assays, plasmids expressing Luman proteins were introduced into COS7 cells with p-109C3. Figure 6 shows that the Luman mutants that were unable to bind HCF were impaired in the ability to activate transcription from the CRE-containing promoter. The result was confirmed by using a different reporter plasmid, p-68CRE (data not shown).
FIG. 6.
Transcriptional activation by the Luman mutants on a CRE-containing promoter is reduced. Plasmids (0.5 μg each) expressing Luman and the mutants were introduced into COS7 cells with CAT reporter plasmid p-109C3 (0.5 μg), in which the promoter region has a CRE linked to the cat gene (27).
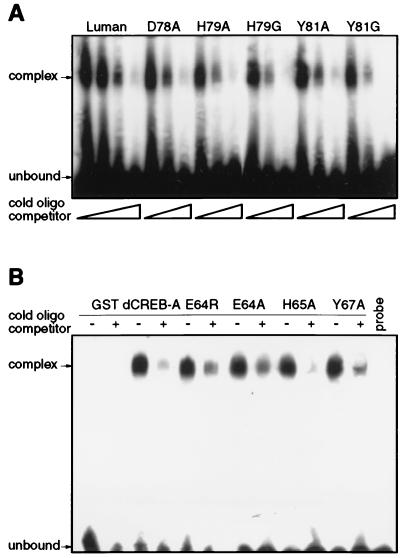
We had previously shown that Luman binds CRE in vitro in an HCF-independent manner (18). To eliminate the possibility that mutations in the HCF-binding domain indirectly affected their binding to CRE, we examined the purified mutant proteins in the HCF-independent CRE binding assay. The results (Fig. 7A) showed that all of the Luman mutants retained the ability of binding to CRE and that the binding could be competed by unlabeled CRE DNA probe. Similar EMSA results were also obtained for dCREB-A and its mutants (Fig. 7B).
FIG. 7.
Mutations in the putative HCF-binding domain of Luman and dCREB-A do not affect binding of CRE in vitro as detected by HCF-independent EMSA. A double-stranded oligonucleotide representing CRE was labeled with 32P, annealed, incubated with each of the purified Luman (A) or dCREB-A (B) GST fusion proteins, and analyzed on a 4% nondenaturing polyacrylamide gel. (A) For each GST-Luman fusion protein sample, 1 μl of unlabeled CRE oligonucleotide, at concentrations of 1 and 10 μM, was added to the incubation mix and used as a competitor. For wild-type Luman, an additional concentration of 0.1 mM was included. (B) In every other lane, 10 pmol of unlabeled CRE oligonucleotide was added to the GST–dCREB-A protein sample and used as a competitor. The last lane contains the labeled CRE probe alone.
Like VP16, Luman and dCREB-A do not interact with the HCF(P134S) mutant.
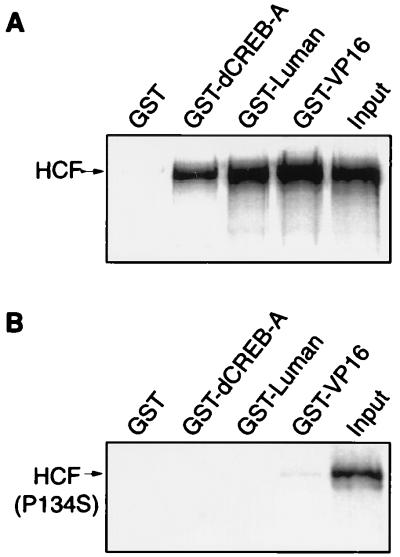
Recent studies (14, 37) suggest that VP16 targets the N-terminal region of HCF, which has six Kelch repeats. The P134S mutation, located in the third Kelch repeat of the HCF protein in tsBN67 cells, can cause cells to arrest at G0/G1. At the nonpermissive temperature, VP16 is unable to interact with the mutated HCF and cannot activate transcription in tsBN67 cells (5, 37). To further compare VP16 with Luman and dCREB-A in their modes of interaction with HCF, we also tested HCF(P134S) for its interaction with wild-type Luman and dCREB-A. Both the GST pull-down (Fig. 8) and mammalian two-hybrid (Fig. 9) assays, as described above, were performed with HCF(P134S) as well as wild-type HCF. The results of both assays clearly showed that, like VP16, Luman could not interact with the mutated form of HCF. The GST pull-down assay (Fig. 8B) and a separate mammalian two-hybrid assay on dCREB-A (data not shown) also indicated that dCREB-A could bind to wild-type HCF but not HCF(P134S).
FIG. 8.
A mutation in the VP16-binding site of HCF(P134S) prevents its association with VP16 as well as with Luman and dCREB-A in vitro. GST and GST fusion proteins of dCREB-A, Luman, and VP16 were incubated with an equivalent amount of 35S-labeled HCF (A) or HCF(P134S) (B) and analyzed on an SDS-polyacrylamide gel. Input (1/10) was run on the last lane of each gel.
FIG. 9.
A mutation in the VP16-binding site of HCF(P134S) prevents its association with VP16 as well as with Luman and dCREB-A in vivo. The parental expression vector pcDNA3 was used as a negative control, and VP16 was included as a reference. Along with the reporter plasmid pG5EC (0.5 μg), equivalent amounts (0.5 μg) of plasmids expressing Luman and VP16 or the empty expression vector pcDNA3 were introduced into COS7 cells with either the blank GAL fusion vector, a plasmid expressing GAL-HCF, or a plasmid expressing GAL-HCF(P134S).
A functional HCF is required for the ability of Luman to activate transcription from CRE-containing promoters but not from heterologous promoters.
To confirm that interaction with HCF is required for Luman to activate transcription through CRE, we used the tsBN67 cell line, in which the endogenous HCF(P134S) is nonfunctional at the nonpermissive temperature of 39.5°C. At either the permissive temperature of 33.5°C or the nonpermissive temperature, we transfected tsBN67 and the wild-type parental BHK-21 cells with plasmid pcLuman and the CRE-containing plasmid p-109C3 as a reporter. We found that in tsBN67 cells, Luman could activate transcription from the CRE-containing promoter only at the permissive temperature (Fig. 10A), while in BHK-21 cells, Luman could activate transcription at both temperatures (Fig. 10B). The result was also confirmed by using a different CRE-containing reporter plasmid, p-68CRE (data not shown). In contrast to these results, the GAL4 fusion proteins of full-length Luman, its amino-terminal portion containing the activation domain (Luman1-107), or the same segment with the D78A mutation, Luman(D78A)1-107, were all capable of activating transcription at both temperatures in the mutant tsBN67 cells (Fig. 10C) as well as in BHK-21 cells (data not shown).
FIG. 10.
Luman activates transcription from CRE-containing promoters in tsBN67 cells at the permissive (33.5°C) but not nonpermissive (39.5°C) temperature. (A and B) Plasmids expressing wild-type Luman (0.5 μg) and a blank vector (0.5 μg) were used to transfect tsBN67 (A) and parental BHK-21 (B) cells at both 33.5°C (▪) and 39.5°C (▧). (C) Plasmids (0.5 μg of each) expressing the GAL4 DBD fusion proteins of full-length Luman, the N-terminal portion containing the activation domain (Luman1-107) and the same segment with the mutation D78A were introduced into tsBN67 cells at both 33.5°C (▪) and 39.5°C (▧).
DISCUSSION
Previously we identified a human cellular protein, Luman, through its interaction with HCF (18). Luman belongs to the CREB/ATF gene family of bZIP proteins. It can bind CRE in vitro and can activate transcription from promoters containing CRE in vivo. A contemporaneous study by Freiman and Herr (4) using the yeast two-hybrid system and immunoprecipitation assays also showed that VP16 and Luman (referred to as LZIP in that study) have a common mode of association with HCF. In the present study, we confirmed and extended these observations by showing the following. (i) Luman, like VP16, has a defined acidic activation domain. (ii) Luman and its Drosophila homolog dCREB-A interact with HCF similarly to VP16: Luman and dCREB-A share an HCF-binding motif, (D/E)HXY(S/A), present in VP16 and VP16 homologs in other alphaherpesviruses. Mutations of conserved residues in this motif drastically reduced the binding of Luman or dCREB-A to HCF both in vitro and in vivo, while these mutations had little effect on the activation potentials of these proteins. In addition, Luman, like VP16, did not interact with HCF with the P134S mutation, which in tsBN67 cells causes cell growth arrest at the G0/G1 stage of the cell cycle (5). (iii) The ability of Luman to activate CRE-containing promoters appears to require its association with HCF. The conservation of HCF in distantly related organisms (36, 39) and the presence of Luman and other HCF-binding bZIP proteins in species as diverse as humans and fruit flies suggests that the interaction between Luman and HCF may represent a mechanism for transcriptional regulation that has been retained during the evolution of metazoans.
In addition to Luman, we have recently isolated the cDNA for another human protein, named Zhangfei, that binds to HCF (unpublished data). Although the overall sequence homology between the Luman and Zhangfei proteins is not significant (∼30%), they share striking structural similarities: Zhangfei is also a bZIP protein with an acidic activation domain and an HCF-binding motif (DHDYAS). The mRNAs of both proteins are detected in a variety of adult and fetal tissues (reference 18 and unpublished data). The discovery of two HCF-binding proteins in human cells suggests that HCF is a key switch that coordinately regulates transcriptional activation by these and other ubiquitous HCF-binding proteins. In this role, we envisage HCF as an upstream trigger of a process like G0/G1-S transition. The response to HCF would be the immediate activation of one or more cascades regulated by these transcription activators. However, the withdrawal of HCF would not have a similar immediate effect. This might explain the long delay in achieving growth arrest after tsBN67 cells are elevated to the nonpermissive temperature (5).
Luman mutants that did not bind HCF also failed to activate transcription from CRE-containing promoters. These mutants nonetheless retained the ability to activate transcription when tethered to a heterologous promoter by the GAL4 DBD. In this respect, Luman is also similar to VP16, where inability to bind HCF compromises transcription from TAATGARAT-containing promoters but does not hinder activation by GAL4 DBD fusion proteins from GAL4 UAS-containing promoters (data not shown). For VP16, HCF is believed to stabilize the TAATGARAT–Oct-1–VP16 complex. It is likely that HCF also stabilizes the Luman-CRE association in vivo. However, our results show that at least in vitro, Luman does not require HCF to bind the CRE. This HCF-independent binding may be stabilized or enhanced by HCF, but we have not been able to obtain recombinant or native HCF of sufficiently high purity to test this hypothesis. The abundance of endogenous CRE-binding activity in cell lysates and the in vitro transcription-translation systems precludes the use of crude cell lysates or in vitro-synthesized HCF in this assay.
An alternative explanation for our observation is that the association with HCF does not change the DNA-binding ability of Luman. Instead, HCF may act by altering the conformation of the proteins with which it interacts. This hypothesis is supported by the observation that the basic DNA-binding domains of bZIP proteins are relatively independent of the adjoining protein structures and can readily bind to DNA by themselves after dimerization (25). In VP16, the conformational change resulting from HCF binding would allow it to form a stable complex with DNA-bound Oct-1 without effects on its activation domain. In Luman, the HCF-induced change might activate the transactivation domain of Luman on its cognate response element, CRE, or a similar motif. Many transcription factors have been found to possess masked activation domains, which can be unmasked by the binding of a factor, e.g., Leu3p (35) and YY1 (16), and/or by modification (e.g., phosphorylation; ATF-2 [17] and C/EBPβ [34]). It is puzzling that Luman mutants that do not bind HCF can nonetheless activate transcription when tethered to heterologous promoters by the GAL4 DBD. It is possible that fusion with the GAL4 DBD causes a conformational change in Luman that is equivalent to that caused by HCF. It has been shown that the GAL4 DBD when fused to transcription factors can alter their properties of transcription: fusion of VP16 to the GAL4 DBD allows the chimeric protein to activate from distal as well as proximal promoter sites, while without the GAL4 DBD, VP16 is restricted to activation from proximal binding sites (6).
We have also noticed that HCF itself can activate transcription. We observed low levels of transactivation activities in both mammalian cells (COS7 cells [control samples in Fig. 4 and 9]) and yeast cells (18). More interestingly, in the hamster fibroblast cell line, BHK-21, the GAL-HCF fusion protein was as potent as GAL-Luman in activating transcription from a GAL4 UAS-containing promoter (data not shown). Thus, in addition to the above hypotheses, HCF may be directly involved in the process of transcription activation, instead of merely stabilizing the transcription factor-DNA complex.
Wilson et al. (39) have suggested that the activation or synthesis of HCF might represent an important step in the commitment to exit G0/G1 phases of the cell cycle and that HSV uses HCF as a sensor of the physiological state of host cells to regulate IE gene expression. If this is indeed the case, HSV may have evolved a mechanism to tap into one of the more conserved and as yet little-understood triggers for the activation of genes needed for cell division.
ACKNOWLEDGMENTS
This work was funded by an operating grant to V.M. from the Natural Sciences and Engineering Research Council of Canada. R.L. is a postdoctoral fellow of the Saskatchewan Health Services Utilization and Research Commission.
REFERENCES
- 1.Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. . (Erratum, 7:719, 1993.) [DOI] [PubMed] [Google Scholar]
- 2.apRhys C M, Ciufo D M, O’Neill E A, Kelly T J, Hayward G S. Overlapping octamer and TAATGARAT motifs in the VF65-response elements in herpes simplex virus immediate-early promoters represent independent binding sites for cellular nuclear factor III. J Virol. 1989;63:2798–2812. doi: 10.1128/jvi.63.6.2798-2812.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burbelo P D, Gabriel G C, Kibbey M C, Yamada Y, Kleinman H K, Weeks B S. LZIP-1 and LZIP-2: two novel members of the bZIP family. Gene. 1994;139:241–245. doi: 10.1016/0378-1119(94)90763-3. [DOI] [PubMed] [Google Scholar]
- 4.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3137. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 6.Hagmann M, Georgiev O, Schaffner W. The VP16 paradox: herpes simplex virus VP16 contains a long-range activation domain but within the natural multiprotein complex activates only from promoter-proximal positions. J Virol. 1997;71:5952–5962. doi: 10.1128/jvi.71.8.5952-5962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haigh A, Greaves R, O’Hare P. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature. 1990;344:257–259. doi: 10.1038/344257a0. [DOI] [PubMed] [Google Scholar]
- 8.Hayes S, O’Hare P. Mapping of a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J Virol. 1993;67:852–862. doi: 10.1128/jvi.67.2.852-862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katan M, Haigh A, Verrijzer C P, van der Vliet P C, O’Hare P. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 1990;18:6871–6880. doi: 10.1093/nar/18.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristie T M. The mouse homologue of the human transcription factor C1 (host cell factor). Conservation of forms and function. J Biol Chem. 1997;272:26749–26755. doi: 10.1074/jbc.272.42.26749. [DOI] [PubMed] [Google Scholar]
- 11.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein–DNA complexes with the HSV alpha TIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 13.Kristie T M, Roizman B. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc Natl Acad Sci USA. 1987;84:71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaBoissière S, Walker S, O’Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai J S, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J S, See R H, Galvin K M, Wang J, Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsden H S, Campbell M E, Haarr L, Frame M C, Parris D S, Murphy M, Hope R G, Muller M T, Preston C M. The 65,000-Mr DNA-binding and virion trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKnight J L, Kristie T M, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra V, Walker S, Hayes S, O’Hare P. The bovine herpesvirus alpha gene trans-inducing factor activates transcription by mechanisms different from those of its herpes simplex virus type 1 counterpart VP16. J Virol. 1995;69:5209–5216. doi: 10.1128/jvi.69.9.5209-5216.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misra V, Walter S, Yang P, Hayes S, O’Hare P. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 24.O’Hare P, Goding C R. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52:435–445. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- 25.O’Neil K T, Hoess R H, DeGrado W F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 26.Preston C M, Frame M C, Campbell M E. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell. 1988;52:425–434. doi: 10.1016/s0092-8674(88)80035-7. [DOI] [PubMed] [Google Scholar]
- 27.Roesler W J, Simard J, Graham J G, McFie P J. Characterization of the liver specific component of the cAMP response unit in the phosphoenolpyruvate carboxykinase (GTP) gene promoter. J Biol Chem. 1994;269:14276–14283. [PubMed] [Google Scholar]
- 28.Rose R E, Gallaher N M, Andrew D J, Goodman R H, Smolik S M. The CRE-binding protein dCREB-A is required for Drosophila embryonic development. Genetics. 1997;146:595–606. doi: 10.1093/genetics/146.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmen K A, Newell A, Robinson M, Mills J S, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smolik S M, Rose R E, Goodman R H. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stern S, Herr W. The herpes simplex virus trans-activator VP16 recognizes the Oct-1 homeo domain: evidence for a homeo domain recognition subdomain. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 32.Stern S, Tanaka M, Herr W. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson C, McKnight S. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 34.Twamley Stein G, Kowenz Leutz E, Ansieau S, Leutz A. Regulation of C/EBP beta/NF-M activity by kinase oncogenes. Curr Top Microbiol Immunol. 1996;211:129–136. [PubMed] [Google Scholar]
- 35.Wang D, Hu Y, Zheng F, Zhou K, Kohlhaw G B. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3p. J Biol Chem. 1997;272:19383–19392. doi: 10.1074/jbc.272.31.19383. [DOI] [PubMed] [Google Scholar]
- 36.Werstuck G H, Capone J P. An unusual cellular factor potentiates protein-DNA complex assembly between Oct-1 and Vmw65. J Biol Chem. 1993;268:1272–1278. [PubMed] [Google Scholar]
- 37.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 39.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 40.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 41.Wu T J, Monokian G, Mark D F, Wobbe C R. Transcriptional activation by herpes simplex virus type 1 VP16 in vitro and its inhibition by oligopeptides. Mol Cell Biol. 1994;14:3484–3493. doi: 10.1128/mcb.14.5.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao P, Capone J P. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol Cell Biol. 1990;10:4974–4977. doi: 10.1128/mcb.10.9.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]