Abstract
Background
Pathogenic heterozygous mutations in the progranulin gene (GRN) are a key cause of frontotemporal dementia (FTD), leading to significantly reduced biofluid concentrations of the progranulin protein (PGRN). This has led to a number of ongoing therapeutic trials aiming to treat this form of FTD by increasing PGRN levels in mutation carriers. However, we currently lack a complete understanding of factors that affect PGRN levels and potential variation in measurement methods. Here, we aimed to address this gap in knowledge by systematically reviewing published literature on biofluid PGRN concentrations.
Methods
Published data including biofluid PGRN concentration, age, sex, diagnosis and GRN mutation were collected for 7071 individuals from 75 publications. The majority of analyses (72%) had focused on plasma PGRN concentrations, with many of these (56%) measured with a single assay type (Adipogen) and so the influence of mutation type, age at onset, sex, and diagnosis were investigated in this subset of the data.
Results
We established a plasma PGRN concentration cut-off between pathogenic mutation carriers and non-carriers of 74.8 ng/mL using the Adipogen assay based on 3301 individuals, with a CSF concentration cut-off of 3.43 ng/mL. Plasma PGRN concentration varied by GRN mutation type as well as by clinical diagnosis in those without a GRN mutation. Plasma PGRN concentration was significantly higher in women than men in GRN mutation carriers (p = 0.007) with a trend in non-carriers (p = 0.062), and there was a significant but weak positive correlation with age in both GRN mutation carriers and non-carriers. No significant association was seen with weight or with TMEM106B rs1990622 genotype. However, higher plasma PGRN levels were seen in those with the GRN rs5848 CC genotype in both GRN mutation carriers and non-carriers.
Conclusions
These results further support the usefulness of PGRN concentration for the identification of the large majority of pathogenic mutations in the GRN gene. Furthermore, these results highlight the importance of considering additional factors, such as mutation type, sex and age when interpreting PGRN concentrations. This will be particularly important as we enter the era of trials for progranulin-associated FTD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-024-01420-z.
Keywords: Frontotemporal dementia, Progranulin
Introduction
Heterozygous mutations in the progranulin gene (GRN) are a key cause of frontotemporal dementia (FTD) [2, 7]. These pathogenic GRN variants cause haploinsufficiency, resulting in a significantly decreased concentration of the progranulin protein (PGRN), a reduction which can be measured in biofluids [13, 15]. The discovery of this core pathophysiological process has led to the development of clinical trials that aim to increase the PGRN concentration in these mutation carriers by targeting key mechanisms involved in the biology of PGRN. However, to establish the efficacy of treatment, robust and effective measures of PGRN biofluid concentration are required. In turn, in order to correctly interpret results, a more comprehensive understanding of how PGRN concentrations change with disease and what factors influence such concentrations is required.
Despite multiple studies now being published on PGRN concentration, a number of questions remain. Firstly, what level signifies the presence of a pathogenic mutation? Although significantly reduced PGRN levels in mutation carriers occur compared with non-carriers, previous studies have shown some overlap between these groups, with debate on the best cut-off value to define pathogenicity: proposed plasma cut-offs have ranged from 61.5 ng/mL to 112.0 ng/mL [9, 14]. Secondly, what factors affect this variability in PGRN concentration? Little is known about the effect of pre-analytical or processing factors but existing literature indicates that different types of GRN mutation may cause lower or higher PGRN concentrations [22, 24, 28]. Moreover, previous studies have highlighted the influence of certain genetic polymorphisms, including TMEM106B rs1990622 and GRN rs5848, on PGRN concentration [10, 16, 25]. Finally, several studies have highlighted the influence of different clinical phenotypes and biological sex on PGRN concentration [1, 10, 30].
In this study, we aimed to explore these questions using a retrospective analysis of published data on biofluid PGRN concentration.
Methods and demographics
Data acquisition
We initially undertook a PubMed search for all publications reporting human biofluid PGRN concentrations in either disease or healthy controls (Supplementary Fig. 1). 154 publications were identified (up to a cut-off search date of January 2020): data was available online for 12 of these and for the further 142 publications, corresponding authors were contacted directly to enquire about the availability of data. As well as the specific PGRN concentration, the following data were requested: the assay that was used, whether a GRN mutation was present (and if so, which one), symptomatic vs presymptomatic status (if GRN mutation present), clinical diagnosis (e.g. behavioural variant frontotemporal dementia, Alzheimer’s disease etc. according to consensus diagnostic criteria), gender, age at sample, age at onset of disease, weight, and the genotype of two polymorphisms previously shown to potentially affect PGRN concentration: TMEM106B rs1990622 and GRN rs5848. Authors who did not initially respond were contacted one further time. Data that was collected, including the institute, country, assay type and sample type, alongside the number of data points provided, are listed in Supplementary Table 1.
Statistics
All statistical analysis was performed using GraphPad Prism (version 9.2.0), and data were tested for normality using the Shapiro Wilk test. Depending on normality, group differences were analysed using either a two tailed t-test or Mann Whitney test with p < 0.05 considered significant. Similarly, Pearson or Spearman correlations were calculated depending on normality. A ROC curve analysis was used for analysing clinical cut-off values with the maximum Youden’s index used to establish the best cut-off value.
Results
In total, data were shared from 75 publications, consisting of PGRN concentrations from 7071 individuals (Table 1, Supplementary Fig. 1). Of these, 56 used the Adipogen assay (Adipogen Inc., Seoul, Korea) and 19 used other assays (5 used A&G pharmaceutical (Columbia, MD), 1 used BioVendor (Brno, Czech Republic), 2 used Mediagnost (Reutlingen, Germany) and 11 used R&D systems (Minneapolis MN, USA)). We therefore decided to focus on the Adipogen assay data as there were limited data on each of the other individual assays. Of the 56 studies, 38 investigated plasma, 12 serum, and 12 CSF concentrations of PGRN (with some papers studying more than one sample type).
Table 1.
PGRN concentration data shared by assay and biofluid type. Total numbers are shown with available data on GRN mutation carriers shown in parentheses
| A&G | Adipogen | BioVendor | Mediagnost | R&D | Others | Total | |
|---|---|---|---|---|---|---|---|
| Total | 149 (7) | 5058 (564) | 56 (38) | 55 (0) | 1481 (6) | 272 (1) | 7071 (616) |
| Plasma | 0 (0) | 3301 (438) | 0 (0) | 0 (0) | 671 (0) | 147 (1) | 3986 (439) |
| Serum | 149 (7) | 758 (125) | 53 (35) | 49 (0) | 649 (6) | 0 (0) | 1658 (173) |
| CSF | 0 (0) | 1346 (19) | 32 (23) | 55 (0) | 0 (0) | 125 (0) | 1558 (42) |
PGRN concentrations in people with GRN variants
We initially examined concentrations in people with variants in the GRN gene. We considered different variants including those with nonsense, frameshift, or deletion mutations as well as splice site mutations. Missense variants were divided into mutations in the signal peptide and variants after the signal peptide (termed ‘other missense mutations’).
Plasma
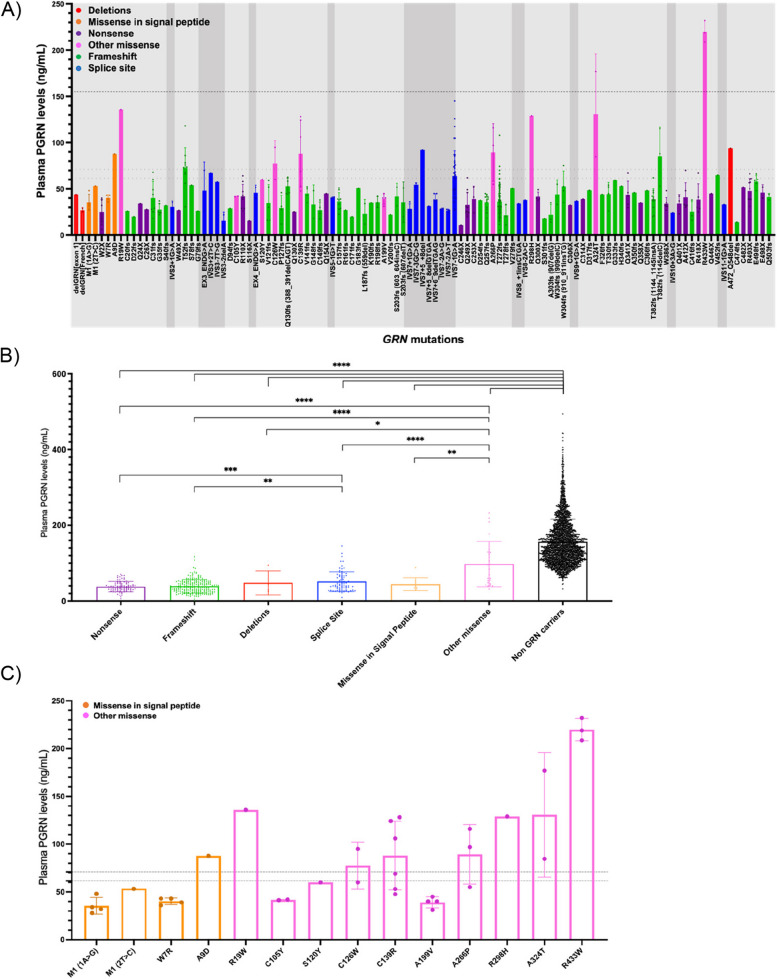
Data were available on plasma PGRN concentration using the Adipogen assay in 3301 individuals (Table 1). Concentrations were variable both between and within individual GRN variants (Fig. 1A). Grouping these variants by type of mutation (Fig. 1B), the other missense mutations had significantly higher plasma PGRN concentrations compared with all other mutation groups (versus missense in the signal peptide (p = 0.002), splice site (p < 0.0001), deletions (p = 0.042), frameshift (p < 0.0001) and nonsense mutations (p < 0.0001)). Significantly higher levels were also observed in the splice site group compared with both frameshift and nonsense mutations (p < 0.0001 and p = 0.001, respectively).
Fig. 1.
A Plasma PGRN concentrations across individual GRN variants. Light grey shading denotes exonic regions and darker grey shading intronic regions. Dotted lines denote previously published with cut-offs for pathogenicity 61.55 [14] and 71.00 [30] and average non-GRN plasma PGRN concentration (156.02 ng/mL). Different colours represent different types of variants. B Plasma PGRN concentrations by mutation type. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed Mann–Whitney Test. Small sample sizes in deletions (n = 4) and missense in signal peptide (n = 10). C Plasma PGRN concentrations in different GRN missense variants. Dotted lines denote cut-offs previously published of 61.55 [14] and 71.00 [30]. Error bars indicate standard error of the mean (SEM)
Focussing on missense mutations (Fig. 3), plasma PGRN levels were generally in the normal range for all mutations in the “other missense” group (i.e. those after the signal peptide). However, two mutations, C105Y and A199V, were exceptions to this, yielding low plasma PGRN levels (below both previously defined cut-offs of 61.5 ng/mL [14] and 71.0 ng/mL [30] in all measured cases. Interestingly, in some of the other missense mutations levels were variable, with concentrations both below and above the previously defined cut-offs (e.g. C139R, A266P).
Fig. 3.
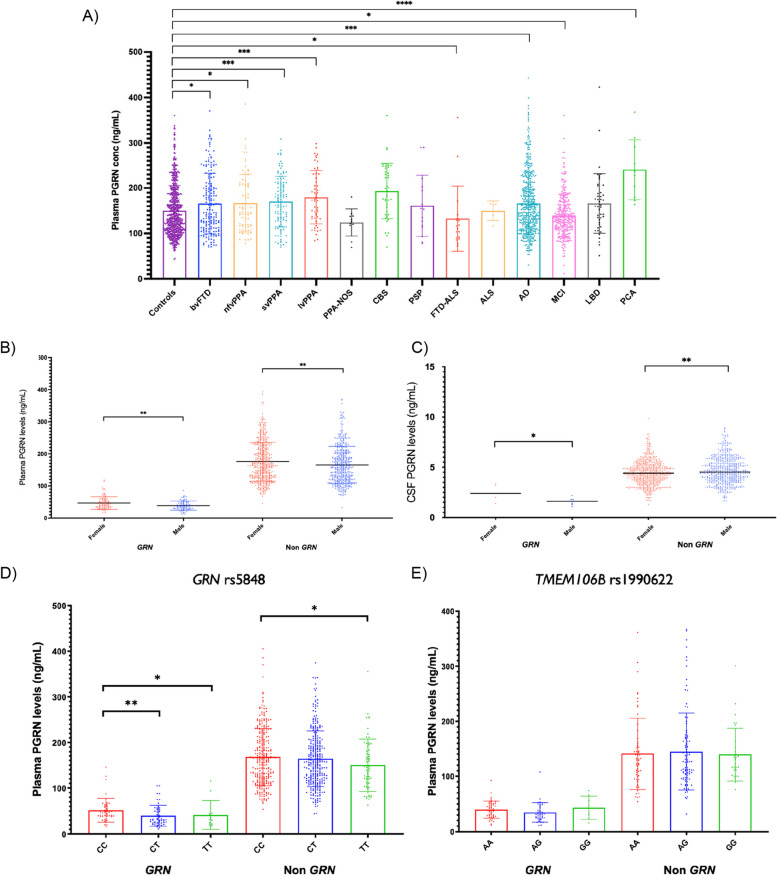
A Plasma PGRN concentrations across different clinical diagnoses. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed Mann–Whitney Test. Significant differences compared with controls are shown on the graph. Additionally, PGRN concentrations were significantly different for bvFTD v CBS (**), FTD-ALS (*), MCI (***) and PCA (**); lvPPA v PPA-NOS (**), FTD-ALS (***), MCI (****) and PCA (*); nfvPPA v CBS (*), FTD-ALS (***), MCI (**) and PCA (**); svPPA v PPA-NOS(**), CBS (*), FTD-ALS(**), MCI (****) and PCA (**); PPA-NOS v FTD-ALS (***), AD (*), LBD (*) and PCA (****); CBS v AD (**), LBD (*) and MCI (****); PSP v PCA (**); FTD-ALS v CBS (***), AD (**), LBD (**) and PCA (***); ALS v PCA (**); AD v MCI (****) and PCA (***); MCI v LBD (**) and PCA (****); LBD v PCA (**). (bvFTD: behavioural variant FTD, PPA: primary progressive aphasia, nfvPPA: nonfluent variant PPA, svPPA: semantic variant PPA, lvPPA: logopenic variant PPA, FTD-ALS: frontotemporal dementia—amyotrophic lateral sclerosis, ALS: amyotrophic lateral sclerosis, CBS: corticobasal syndrome, PSP: progressive supranuclear palsy, AD: Alzheimer’s disease, MCI: mild cognitive impairment, LBD: lewy body disease, PCA: posterior cortical atrophy). Small sample sizes in ALS (n = 1), PCA (n = 10), PPA-NOS (n = 11), FTD-ALS (n = 18), PSP (n = 16). B Plasma and C CSF PGRN concentrations in females and males in both GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). * P < 0.05, ** P < 0.01, two-tailed Mann–Whitney Test. D Plasma PGRN concentrations in those with different GRN rs5848 and E TMEM106b rs1990622 polymorphisms in both GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). * P < 0.05, ** P < 0.01, two-tailed Mann–Whitney Test. Error bars indicate standard error of the mean (SEM)
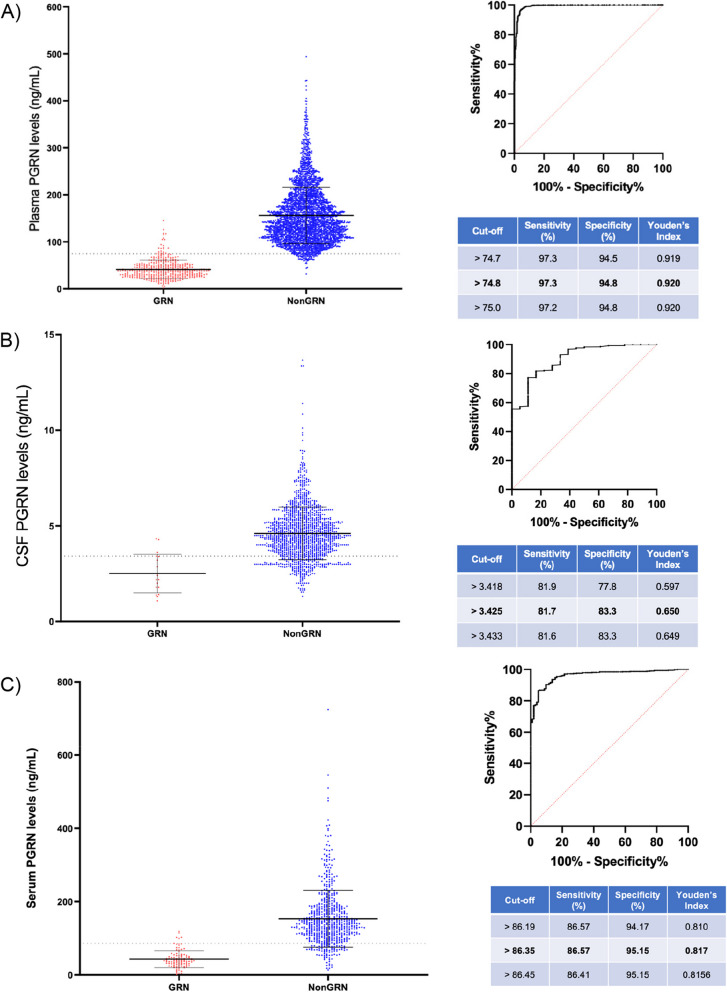
Using this large dataset of plasma PGRN concentrations, a cut-off for GRN mutation pathogenicity was established as 74.8 ng/mL with a Youden’s index of 0.92 (sensitivity 97.3; specificity 94.8), based on 3265 individuals (401 GRN mutation carriers (excluding other missense mutations) and 2864 non-carriers (both healthy and disease controls)) (Fig. 2A).
Fig. 2.
A PGRN plasma concentrations in GRN mutation carriers (GRN) and non-mutation carriers (Non-GRN). Cut-off determined using the optimal Youden’s index. B PGRN CSF concentrations in GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). Cut-off determined using the optimal Youden’s index. C Serum PGRN concentrations in both GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). Cut-off determined using the optimal Youden’s index. Error bars indicate standard error of the mean (SEM)
Serum
Less data were available for serum concentrations using the Adipogen assay, with measures available for 758 individuals. A cut-off for GRN mutation pathogenicity was established as 86.3 ng/mL with a Youden’s index of 0.82, based on 125 GRN mutation carriers and 633 non-carriers (Fig. 2C). Serum levels across all cases (mutation carriers and non-carriers) showed a trend for correlation with plasma levels, r = 0.67, p = 0.0696 (Supplementary Fig. 3).
CSF (Fig. 2B)
CSF PGRN concentrations using the Adipogen assay were available in 1346 individuals. A cut-off for GRN mutation pathogenicity was established as 3.43 ng/mL with a Youden’s index of 0.65, based on 19 GRN mutation carriers and 1327 non-carriers (Fig. 2B). CSF levels correlated with plasma levels, r = 0.33, p < 0.001 but only showed a trend to correlation with serum concentrations, r = 0.15, p = 0.0780 (Supplementary Fig. 3).
PGRN concentrations by clinical phenotype
Neurodegenerative disorders
In the GRN mutation carrier group, no differences were observed in plasma PGRN concentrations between patients with a diagnosis of behavioural variant FTD (bvFTD) and those with a primary progressive aphasia (PPA) (Supplementary Fig. 4).
In those without GRN mutations, plasma PGRN concentrations were significantly higher than controls in multiple groups (Fig. 3A), including not only those with FTD syndromes, but also those with typical and atypical forms of Alzheimer’s disease (including posterior cortical atrophy).
PGRN concentrations were also available for a number of non-neurodegenerative conditions, with lower plasma PGRN levels seen in bipolar disorder compared with controls (p = 0.018), but not in diabetes (p = 0.543) (Supplementary Fig. 5A). Serum PGRN levels were found to be lower in Gaucher’s disease compared with controls (p < 0.001) but concentrations in both osteoarthritis and rheumatoid arthritis groups were higher (both p < 0.001) (Supplementary Fig. 5B).
PGRN concentration differences by sex
Plasma PGRN concentrations were significantly higher in women compared to men in GRN mutation carriers (mean (standard deviation) 46.6 (19.6) ng/mL vs (38.9 (14.2) ng/mL, p = 0.007), as well as in non-carriers (175.7 (60.7) ng/mL vs (165.7 (58.1) ng/mL p = 0.006, Fig. 3B). CSF PGRN levels were also significantly higher in women (2.5 (0.7) ng/mL) compared to men (1.6 (0.4) ng/mL) in the GRN mutation carrier group (p = 0.037), but not in the non-carrier group, where the opposite result was seen (men 4.7 (1.3) ng/mL, women 4.5 (1.2) ng/mL, p = 0.003) (Fig. 3C).
PGRN concentration with increasing age
Plasma PGRN concentrations showed a weak but significant positive correlation with age at sampling (r = 0.09, p < 0.0001) in non-carriers with a similar correlation seen in GRN mutation carriers (r = 0.15, p = 0.0031) (Supplementary Fig. 6).
There was no significant correlation of plasma PGRN concentration with age at symptom onset in GRN mutation carriers. However, there was a significant positive correlation in an FTD cohort without mutations, r = 0.18, p < 0.0001. In contrast, there was a significant negative correlation in those with Alzheimer’s disease (AD), r = -0.24, p < 0.0001 i.e. lower concentrations associated with older age at onset (Supplementary Fig. 7).
PGRN concentration by weight
No significant correlations were seen between plasma PGRN levels and weight in either GRN mutation carriers (r = -0.08, p = 0.7354) or in those without mutations (r = -0.07, p = 0.0751) (Supplementary Fig. 8).
PGRN concentration in relation to the GRN rs5848 and TMEM106b rs1990622 polymorphisms
Significantly higher plasma PGRN levels were seen in those with the GRN rs5848 CC genotype compared to those with both the CT and TT genotypes in GRN mutation carriers (p = 0.003 and p = 0.027 respectively; note that for heterozygous individuals these analyses did not take into account which rs5848-allele produced PGRN. Significantly higher concentrations were also seen in non-mutation carriers with the CC genotype compared with those with TT (p = 0.041) (Fig. 3D).
We found no significant differences between concentrations in those with the AA, AG or GG TMEM106B rs1990622 genotypes, either in GRN mutation carriers or non-carriers (Fig. 3E).
Discussion
This study has highlighted a number of important factors that influence biofluid PGRN concentrations across a large cohort of individuals including type of GRN variant, clinical phenotype, age at sampling and at symptom onset, sex and the GRN rs5848 polymorphism.
Firstly, we highlighted the variability of plasma PGRN levels across GRN mutation types, most notably showing levels for missense mutations after the signal peptide to be significantly higher than the other mutation groups, suggesting that the majority of these mutations may not be pathogenic but more likely either risk factors for FTD or just benign polymorphisms. This is supported by earlier work indicating missense mutations yield PGRN levels higher than other GRN mutation carriers [9]. However, this was not true for all mutations, as two mutations in this group, C105Y and A199V led to pathogenic levels of plasma PGRN. Previous functional studies of these mutations have shown defects in RNA splicing, PGRN secretion and proteolytic processing likely to cause a pathological effect (The A199V is in fact two nucleotides away from the splice site and thus is technically a splice-site mutation) [22, 20, 24]. This supports the suggestion that these two missense mutations are pathogenic. Furthermore, as suspected, the missense mutations in the signal peptide generally yielded low, likely pathogenic, levels of plasma PGRN which is corroborated by existing reports of disrupted protein interactions and low PGRN levels in these mutations [28, 29]. These findings provide a key insight into the potential pathogenicity of certain GRN mutations and may help identify suspected pathogenic mutations in the future. However, what these studies do not tell us about is whether missense mutations cause abnormal functioning progranulin in the presence of normal PGRN concentrations in biofluids, and further work is required to understand the complex downstream pathways that might lead to neurodegeneration in these cases.
Secondly, we established a novel cut-off value for GRN mutation pathogenicity at a plasma PGRN level of 74.8 ng/mL based on data from 3265 individuals (excluding individuals with ‘other missense mutations’). This newly calculated cut-off is very close to one initially reported in the literature (74.4 ng/ml) [13]. Other previously established cut-offs were generally lower than this value, at 61.5 ng/mL and 71.0 ng/mL [14, 30] although an early study reported a much higher cut-off of 112.0 ng/mL [9]. This variation could be due to the inclusion criteria, with different populations, smaller sample sizes in some studies, and in some cases a different set of mutation types. There may also have been pre-analytical or analytical differences between centres which we are unaware of. Lastly, the control group of ‘non-carriers’ may have varied between studies. The cut-off generated here is based on the largest sample size reported to our knowledge, incorporating different mutation types and a spread of populations. However, as seen with previous studies, overlap in plasma PGRN levels between mutation carriers and non-carriers is still observed (i.e. there is no absolute cut-off), likely reflecting the influence of other biological factors on these levels such as hormonal or metabolic disorders.
We also identified cut-offs for CSF and serum PGRN levels in this cohort, which could prove beneficial for a more complete picture of how PGRN levels are analysed. Notably however, the lack of tight correlations between different fluid PGRN measures found here could suggest differential PGRN expression throughout the body. This is supported by prior findings of significantly lower CSF PGRN levels compared with serum levels in some GRN mutation carriers and other reports proposing differentially derived PGRN in CSF and blood [8, 26, 34].
Although GRN mutation carriers are associated with lower PGRN concentrations, we found that across multiple neurodegenerative diseases PGRN levels were generally higher than a control population e.g. in AD and non-GRN-FTD, as reported in a previous multicentre Italian study [14]. Numerous reviews have highlighted PGRN’s key role in neurodegeneration [5, 19, 27], although other studies have not reported differences in plasma PGRN levels in AD despite a reported increase in PGRN mRNA [6, 15]. Similarly, previous research has suggested that people with FTD without GRN mutations have similar levels to controls [15, 30]. Further work is needed to better understand the role of increased PGRN in these disorders.
We also analysed whether PGRN differences were observed in non-neurodegenerative diseases, finding that certain conditions, such as bipolar disorder and Gaucher disease, were linked to lower PGRN concentrations whereas others, such as arthritis, had higher biofluid PGRN concentration. This mirrors findings reported previously for these conditions, emphasising the multitude of processes PGRN is involved in and the need to consider comorbidities when interpreting levels [1, 3, 11, 18, 21]. For example, PGRN concentrations may be more challenging to interpret in the differential diagnosis of people with FTD compared with bipolar disorder [12, 21].
Sex differences have been previously reported in FTD [17] including PGRN levels across different biofluids. Nicholson and colleagues investigated the differential regulation between CSF and plasma PGRN, suggesting that this is linked to biological sex, with plasma PGRN levels higher in women and CSF PGRN levels higher in men [26]. This is consistent with our findings on sex differences in this cohort as well as other prior studies, with female plasma PGRN levels higher than males [30]. We also observed higher CSF PGRN levels in men in the non-carrier group, supporting the hypothesis of Nicholson et al. but notably found the opposite pattern in GRN mutation carriers, who yielded higher CSF PGRN levels in females. This potentially indicates a GRN mutation specific sex difference in PGRN activity in the central nervous system. Certainly, sex differences appear to be important in normal PGRN function, with only female GRN mutation mice developing bone formation defects compared with wildtype, and GRN expression linked to oestrogen activity during brain development [32, 33]. Collectively, these findings reveal the complexity of PGRN activity both peripherally and centrally and highlight the effect of biological sex on the function of this protein, something which should be considered for upcoming therapeutic trials.
A previous multicentre Italian study reported that the lowest plasma PGRN levels in GRN mutation carriers were associated with an anticipation of disease onset of about 9 years [14], although a further study did not find any correlation with either age at onset or age at sample [25]. In the present study, we found a significant positive correlation between age at symptom onset and plasma PGRN level in people with FTD without GRN mutations, where an earlier onset is seen in those with lower PGRN levels. However, we also saw a significant positive correlation between age at sampling and PGRN level in these non-carriers, which could be influencing our results, indicating a general increase in PGRN level with age. The reason for this increase is unclear although may be due to inflammatory changes with increasing age. In contrast, when analysing correlations in people with AD, we saw a significant negative correlation between plasma PGRN level and age at onset, indicating that higher levels of PGRN are linked to earlier ages of onset. Interestingly, Suarez-Calvet and colleagues report an increase in CSF PGRN level as the disease course progresses supporting the idea that PGRN plays a role in the pathogenicity of this condition [31], although the exact relationship between AD and PGRN requires further study.
We additionally found that the GRN rs5848 polymorphism influences plasma PGRN levels in both GRN mutation carriers and non-carriers, with the lowest levels in those with the TT genotype. This finding was previously reported by Hsiung and colleagues [16], who reported reduced serum PGRN levels for this genotype and speculated that this was related to miR-659 dependent translational inhibition. This was also highlighted in work by Nicholson and colleagues [26] who demonstrated a significant association between rs5848 and CSF PGRN concentration. Additionally, it has been previously reported that rs5848 significantly affects GRN mRNA levels both centrally and peripherally and has been linked to both AD and Parkinson’s disease [4, 23]. Together, these findings suggest rs5848 is a key influencer of PGRN levels and could also help understand the role of PGRN in other neurodegenerative diseases. Interestingly, the TMEM106b rs1990622 polymorphism was not associated with differences in plasma PGRN levels in this large cohort. This finding suggests that the influence of this well-known risk factor for GRN-associated FTD is unlikely to be modulated through PGRN levels, as speculated previously [10]. However, more research is required to understand this fully.
Lastly, it is important to note the limitations of this study. One reason for variation in PGRN levels between studies included here is that despite testing the same fluid type with the same assay, the tests are performed by different researchers in different laboratories with different assay batches. Secondly, although the numbers of cases with available concentrations to study was large, once stratified, numbers for individual comparisons were often much smaller. Thirdly, for many of the conditions studied (including AD), we did not have access to robust disease severity measures. Finally, it should also be noted that in the absence of a certified reference material and a value assigned for PGRN concentration by a certified reference method, different assays are not standardized to each other; hence, laboratory- and assay-specific validation of the cut-points reported here remains important.
In summary, this large and diverse cohort of PGRN levels has allowed us to firstly refine the plasma PGRN cut-off level to predict GRN mutations, secondly, confirm the differential impact of the mutation type on blood PGRN levels, thirdly, highlight the high variability across missense mutations in GRN, and lastly, highlight the numerous factors which influence PGRN biofluid levels both GRN mutation carriers and non-carriers (i.e. clinical diagnosis, sex, age, GRN rs5848 genotype). These factors should be considered when utilising this marker, generating a more personalised approach to treatment. These results also reflect the need for the identification of additional factors which affect PGRN biofluids levels and thus hopefully modulate disease onset and/or progression, providing a more comprehensive picture of this disease as we continue into the era of therapeutic trials.
Supplementary Information
Additional file 1: Supplementary Figure 1. Data collection flowchart. Supplementary Figure 2. Serum PGRN concentrations in both GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). Cut-off determined using the optimal Youden’s index. Supplementary Figure 3. A) Correlation between serum and plasma PGRN levels in this cohort (p= 0.0696). B) Correlation between CSF and serum PGRN levels in this cohort (p = 0.0780). C) Correlation between CSF and plasma in this cohort (p<0.0001). Supplementary Figure 4. No significant difference in plasma PGRN levels between bvFTD (behavioural variant FTD) and PPA (primary progressive aphasia) GRN mutation carriers.Supplementary Figure 5. A) Correlation between plasma PGRN levels and age at sampling in GRN mutation carriers. B) Correlation between plasma PGRN levels and age at sampling in non GRN mutation carriers. ** P < 0.01,**** P < 0.0001, Spearman correlation. Supplementary Figure 6. A) Correlation between plasma PGRN levels and age of onset in GRN mutation carriers. B) Correlation between plasma PGRN levels and age of onset in non-GRN mutation carriers. C) Correlation between plasma PGRN levels and age of onset in people with AD. **** P < 0.0001, Spearman correlation. Supplementary Figure 7. A) Correlation between plasma PGRN levels and weight in GRN mutation carriers. B) Correlation between plasma PGRN levels and weight in non GRN mutation carriers. Supplementary Figure 8. A) Differences in plasma PGRN levels measured with the Adipogen assay between clinical diagnoses in this cohort. B) Differences in serum PGRN levels measured with the Adipogen assay between clinical diagnoses in this cohort. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed Mann-Whitney Test. Supplementary Table 1. List of institutions, countries, assay types and sample types included in this dataset alongside the number of data point provided.
Acknowledgements
We thank the research participants for their contribution to the study.
Authors’ contributions
Imogen Swift and Jonathan Rohrer analysed and interpreted the data and wrote the initial draft of the manuscript. All authors (Rosa Rademakers, NiCole Finch, Matt Baker, Roberta Ghidoni, Luisa Benussi, Giuliano Binetti, Giacomina Rossi, Matthis Synofzik, Carlo Wilke, David Mengel, Caroline Graff, Leonel T. Takada, Raquel Sánchez-Valle, Anna Antonell, Daniela Galimberti, Chiara Fenoglio, Maria Serpente, Marina Arcaro, Stefanie Schreiber, Stefan Vielhaber, Philipp Arndt, Isabel Santana, Maria Rosario Almeida, Fermín Moreno, Myriam Barandiaran, Alazne Gabilondo, Johannes Stubert, Estrella Gómez-Tortosa, Pablo Agüero, M. José Sainz, Tomohito Gohda, Maki Murakoshi, Nozomu Kamei, Sarah Kittel-Schneider, Botne Sando, Gøril Rolfseng Grøntvedt, Camilla Lauridsen, Amanda Heslegrave, Carolin Heller, Emily Abel, Alba Gómez-Núñez, Roger Puey, Andrea Arighi, Enmanuela Rotondo, Lize C. Jiskoot, Lieke HH Meeter, João Durães, Marisa Lima, Miguel Tábuas-Pereira, João Lemos, Bradley Boeve, Ronald C. Petersen, Dennis W. Dickson, Neill R. Graff-Radford, Isabelle LeBer, Leila Sellami, Foudil Lamari, Fabienne Clot, Barbara Borroni, Valentina Cantoni, Jasmine Rivolta, Ignacio Illán-Gala, Lucie Andres-Cerezo, Philip Damme, Jordi Clarimon, Petra Steinacker, Emily Feneberg, Markus Otto, Emma L. Ende, John C. Swieten, Harro Seelaar, Henrik Zetterberg, Aitana Sogorb-Esteve) read and approved the final manuscript.
Funding
The Dementia Research Centre is supported by Alzheimer's Research UK, Alzheimer's Society, Brain Research UK, and The Wolfson Foundation. This work was supported by the NIHR UCL/H Biomedical Research Centre, the Leonard Wolfson Experimental Neurology Centre (LWENC) Clinical Research Facility, and the UK Dementia Research Institute, which receives its funding from UK DRI Ltd, funded by the UK Medical Research Council, Alzheimer's Society and Alzheimer's Research UK. ASE is funded by Race Against Dementia fellowship, supported by Alzheimer’s Research UK (ARUK-RADF2021A-003). ASE is also supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK. IJS is supported by the Alzheimer’s Association. HZ is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2022–01018 and #2019–02397), the European Union’s Horizon Europe research and innovation programme under grant agreement No 101053962, Swedish State Support for Clinical Research (#ALFGBG-71320), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809–2016862), the AD Strategic Fund and the Alzheimer's Association (#ADSF-21–831376-C, #ADSF-21–831381-C, and #ADSF-21–831377-C), the Bluefield Project, the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2022-0270), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE), the European Union Joint Programme – Neurodegenerative Disease Research (JPND2021-00694), the National Institute for Health and Care Research University College London Hospitals Biomedical Research Centre, and the UK Dementia Research Institute at UCL (UKDRI-1003). JDR has received funding from an MRC Clinician Scientist Fellowship (MR/M008525/1) and an NIHR Rare Disease Translational Research Collaboration (BRC149/NS/MH); his work is also supported by the MRC UK GENFI grant (MR/M023664/1), the Bluefield Project and the JPND GENFI-PROX grant (2019–02248). The IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy and the Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico are is supported by the Italian Ministry of Health (Ricerca Corrente). MO was supported by the EU Joint Programme-Neurodegenerative Diseases networks Genfi-Prox (01ED2008A) and bPride (01ED2001) the German Federal Ministry of Education and Research (FTLDc 01GI1007A, Moodmarker 01EW200), the EU (MIRIADE 860197, FAIR-PARK II 633190), the German Research Foundation/DFG (SFB1279), the foundation of the state Baden-Württemberg (D.3830), Boehringer Ingelheim Ulm University BioCenter (D.5009) and the Thierry Latran Foundation. MS is supported by the Italian Ministry of Health, grant GR-2019–12369100. This work was supported by JPND grant “GENFI-prox” (by DLR/BMBF to M.S, joint with JR, MO, and BB), and the Clinician Scientist programme "PRECISE.net" funded by the Else Kröner-Fresenius-Stiftung (to CW, DM and MS). LTT was funded by FAPESP grant number 2013/017584. RP was funded by NIH grant P30 AG062677.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The London Queen Square Ethics committee approved the study. The study complies with the Declaration of Helsinki.
Consent for publication
All participants provided written informed consent at enrolment including consent to publication.
Competing interests
HZ has served at scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, Alzinova, ALZPath, Annexon, Apellis, Artery Therapeutics, AZTherapies, Cognito Therapeutics, CogRx, Denali, Eisai, Nervgen, Novo Nordisk, Optoceutics, Passage Bio, Pinteon Therapeutics, Prothena, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave, has given lectures in symposia sponsored by Alzecure, Biogen, Cellectricon, Fujirebio, Lilly, and Roche, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). JDR has served on medical advisory boards and consultancy for Alector, Arkuda Therapeutics, Wave Life Sciences, and Prevail Therapeutics. Consultancy for UCB, AC Immune, Astex Pharmaceuticals, Biogen, Takeda and Eisai. SKS has received author’s and advisory honoraria from Takeda/Shire and Medice Arzneimittel Pütter GmbH in the last 3 years. AL participated in advisory boards from Biogen, Eisai, Fujirebio-Europe, Grifols, Novartis, Roche, Otsuka Pharmaceutical, Nutricia, Zambón, y NovoNordisk. AL declares a filed patent application (WO2019175379 A1 Markers of synaptopathy in neurodegenerative disease). DA participated in advisory boards from Fujirebio-Europe, Roche Diagnostics, Grifols S.A. and Lilly, and received speaker honoraria from Fujirebio-Europe, Roche Diagnostics, Nutricia, Krka Farmacéutica S.L., Zambon S.A.U. and Esteve Pharmaceuticals S.A. DA declares a filed patent application (WO2019175379 A1 Markers of synaptopathy in neurodegenerative disease). MS has received consultancy honoraria from Ionis, UCB, Prevail, Orphazyme, Servier, Reata, GenOrph, AviadoBio, Biohaven, Zevra,and Lilly, all unrelated to the present manuscript. IIG participated in advisory boards from UCB Pharma S.A., and received speaker honoraria from Nutricia, Kern Pharma S., Krka Farmacéutica S.L., Zambon S.A.U. and Esteve Pharmaceuticals S.A.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arrant AE, Roth JR, Boyle NR, Kashyap SN, Hoffmann MQ, Murchison CF, Ramos EM, Nana AL, Spina S, Grinberg LT, Miller BL, Seeley WW, Roberson ED. Impaired β-glucocerebrosidase activity and processing in frontotemporal dementia due to progranulin mutations. Acta Neuropathol Commun. 2019;7(1):218. doi: 10.1186/s40478-019-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 3.Cerezo LA, Kuklová M, Hulejová H, Vernerová Z, Kaspříková N, Veigl D, Pavelka K, Vencovský J, Šenolt L. Progranulin Is associated with disease activity in patients with rheumatoid arthritis. Mediators Inflamm. 2015;2015:740357. doi: 10.1155/2015/740357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang K-H, Chen C-M, Chen Y-C, Hsiao Y-C, Huang C-C, Kuo H-C, Hsu H-C, Lee-Chen G-J, Wu Y-R. Association between GRN rs5848 polymorphism and Parkinson’s disease in Taiwanese population. PLoS ONE. 2013;8(1):e54448. doi: 10.1371/journal.pone.0054448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitramuthu BP, Bennett HPJ, Bateman A. Progranulin: a new avenue towards the understanding and treatment of neurodegenerative disease. Brain. 2017;140(12):3081–3104. doi: 10.1093/brain/awx198. [DOI] [PubMed] [Google Scholar]
- 6.Cooper YA, Nachun D, Dokuru D, Yang Z, Karydas AM, Serrero G, Yue B, Alzheimer's Disease Neuroimaging Initiative. Boxer AL, Miller BL, Coppola G. Progranulin levels in blood in Alzheimer’s disease and mild cognitive impairment Annals of clinical and translational neurology. Ann Clin Transl Neurol. 2018;5(5):616–629. doi: 10.1002/acn3.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin J-J, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 8.Feneberg E, Steinacker P, Volk AE, Weishaupt JH, Wollmer MA, Boxer A, Tumani H, Ludolph AC, Otto M. Progranulin as a candidate biomarker for therapeutic trial in patients with ALS and FTLD. J Neural Transm. 2016;123(3):289–296. doi: 10.1007/s00702-015-1486-1. [DOI] [PubMed] [Google Scholar]
- 9.Finch N, Baker M, Crook R, Swanson K, Kuntz K, Surtees R, Bisceglio G, Rovelet-Lecrux A, Boeve B, Petersen RC, Dickson DW, Younkin SG, Deramecourt V, Crook J, Graff-Radford NR, Rademakers R. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132(Pt 3):583–91. doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, Crook J, Finger E, Hantanpaa KJ, Karydas AM, Sengdy P, Gonzalez J, Seeley WW, Johnson N, Beach TG, Mesulam M, Forloni G, Kertesz A, Knopman DS, Uitti R, White CL, Caselli R, Lippa C, Bigio EH, Wszolek ZK, Binetti G, Mackenzie IR, Miller BL, Boeve BF, Younkin SG, Dickson DW, Petersen RC, Graff-Radford NR, Geschwind DH, Rademakers R. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76(5):467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouad NA, Nassr MH, Fathi HM, Zaki OM, Negm AA, Senara SH. Potential value of serum progranulin as an activity biomarker in rheumatoid arthritis patients: relation to musculoskeletal ultrasonographic evaluation. The Egyptian Rheumatologist. 2019;41(2):93–97. doi: 10.1016/j.ejr.2018.07.004. [DOI] [Google Scholar]
- 12.Galimberti D, Dell'Osso B, Fenoglio C, Villa C, Cortini F, Serpente M, Kittel-Schneider S, Weigl J, Neuner M, Volkert J, Leonhard C, Olmes DG, Kopf J, Cantoni C, Ridolfi E, Palazzo C, Ghezzi L, Bresolin N, Altamura AC, Scarpini E, Reif A. Progranulin gene variability and plasma levels in bipolar disorder and schizophrenia. PLoS ONE. 2012;7(4):e32164. doi: 10.1371/journal.pone.0032164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghidoni R, Benussi L, Glionna M, Franzoni M, Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71(16):1235–1239. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- 14.Ghidoni R, Stoppani E, Rossi G, Piccoli E, Albertini V, Paterlini A, Glionna M, Pegoiani E, Agnati LF, Fenoglio C, Scarpini E, Galimberti D, Morbin M, Tagliavini F, Binetti G, Benussi L. Optimal plasma progranulin cutoff value for predicting null progranulin mutations in neurodegenerative diseases: a multicenter Italian study. Neurodegener Dis. 2012;9(3):121–127. doi: 10.1159/000333132. [DOI] [PubMed] [Google Scholar]
- 15.Goossens J, Bjerke M, Van Mossevelde S, Van den Bossche T, Goeman J, De Vil B, Sieben A, Martin J-J, Cras P, De Deyn PP, Van Broeckhoven C, van der Zee J, Engelborghs S. Diagnostic value of cerebrospinal fluid tau, neurofilament, and progranulin in definite frontotemporal lobar degeneration. Alzheimer’s Res Ther. 2018;10(1):31. doi: 10.1186/s13195-018-0364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsiung G-YR, Fok A, Feldman HH, Rademakers R, Mackenzie IRA. rs5848 polymorphism and serum progranulin level. J Neurol Sci. 2011;300(1–2):28–32. doi: 10.1016/j.jns.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Illán-Gala I, Casaletto KB, Borrego-Écija S, Arenaza-Urquijo EM, Wolf A, Cobigo Y, Goh SYM, Staffaroni AM, Alcolea D, Fortea J, Blesa R, Clarimon J, Iulita MF, Brugulat-Serrat A, Lladó A, Grinberg LT, Possin K, Rankin KP, Kramer JH, Rabinovici GD, Boxer A, Seeley WW, Sturm VE, Gorno-Tempini ML, Miller BL, Sánchez-Valle R, Perry DC, Lleó A, Rosen HJ. Sex differences in the behavioral variant of frontotemporal dementia: A new window to executive and behavioral reserve. Alzheimers Dement. 2021;17(8):1329–1341. doi: 10.1002/alz.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jian J, Zhao S, Tian Q-Y, Liu H, Zhao Y, Chen W-C, Grunig G, Torres PA, Wang BC, Zeng B, Pastores G, Tang W, Sun Y, Grabowski GA, Kong MX, Wang G, Chen Y, Liang F, Overkleeft HS, Saunders-Pullman R, Chan GL, Liu C-J. Association between progranulin and gaucher disease. EBioMedicine. 2016;11:127–137. doi: 10.1016/j.ebiom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao AW, McKay A, Singh PP, Brunet A, Huang EJ. Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci. 2017;18(6):325–333. doi: 10.1038/nrn.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karch CM, Ezerskiy L, Redaelli V, Giovagnoli AR, Tiraboschi P, Pelliccioni G, Pelliccioni P, Kapetis D, D’Amato I, Piccoli E, Ferretti MG, Tagliavini F, Rossi G. Missense mutations in progranulin gene associated with frontotemporal lobar degeneration: study of pathogenetic features. Neurobiol Aging. 2016;38:215.e1–215.e12. doi: 10.1016/j.neurobiolaging.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kittel-Schneider S, Weigl J, Volkert J, Geßner A, Schmidt B, Hempel S, Kiel T, Olmes DG, Bartl J, Weber H, Kopf J, Reif A. Further evidence for plasma progranulin as a biomarker in bipolar disorder. J Affect Disord. 2014;157:87–91. doi: 10.1016/j.jad.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Kleinberger G, Capell A, Brouwers N, Fellerer K, Sleegers K, Cruts M, Van Broeckhoven C, Haass C. Reduced secretion and altered proteolytic processing caused by missense mutations in progranulin. Neurobiol Aging. 2016;39:220.e17–26. doi: 10.1016/j.neurobiolaging.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Lee M-J, Chen T-F, Cheng T-W, Chiu M-J. rs5848 variant of progranulin gene is a risk of Alzheimer’s disease in the Taiwanese population. Neurodegener Dis. 2011;8(4):216–220. doi: 10.1159/000322538. [DOI] [PubMed] [Google Scholar]
- 24.Luzzi S, Colleoni L, Corbetta P, Baldinelli S, Fiori C, Girelli F, Silvestrini M, Caroppo P, Giaccone G, Tagliavini F, Rossi G. Missense mutation in GRN gene affecting RNA splicing and plasma progranulin level in a family affected by frontotemporal lobar degeneration. Neurobiol Aging. 2017;54:214.e1–214.e6. doi: 10.1016/j.neurobiolaging.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Meeter LHH, Patzke H, Loewen G, Dopper EGP, Pijnenburg YAL, van Minkelen R, van Swieten JC. Progranulin levels in plasma and cerebrospinal fluid in granulin mutation carriers. Dementia and geriatric cognitive disorders extra. 2016;6(2):330–340. doi: 10.1159/000447738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson AM, Finch NA, Thomas CS, Wojtas A, Rutherford NJ, Mielke MM, Roberts RO, Boeve BF, Knopman DS, Petersen RC, Rademakers R. Progranulin protein levels are differently regulated in plasma and CSF. Neurology. 2014;82(21):1871–1878. doi: 10.1212/WNL.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37(7):388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Pinarbasi ES, Karamyshev AL, Tikhonova EB, Wu I-H, Hudson H, Thomas PJ. Pathogenic signal sequence mutations in progranulin disrupt SRP interactions required for mRNA stability. Cell Rep. 2018;23(10):2844–2851. doi: 10.1016/j.celrep.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saracino D, Sellami L, Clot F, Camuzat A, Lamari F, Rucheton B, Benyounes I, Roué-Jagot C, Lagarde J, Sarazin M, Jornea L, Forlani S, LeGuern E, Dubois B, Brice A, Le Ber I. The missense p.Trp7Arg mutation in GRN gene leads to progranulin haploinsufficiency. Neurobiol Aging. 2020;85:154.e9–154.e11. doi: 10.1016/j.neurobiolaging.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Sellami, L., Rucheton, B., Ben Younes, I., Camuzat, A., Saracino, D., Rinaldi, D., Epelbaum, S., Azuar, C., Levy, R., Auriacombe, S., Hannequin, D., Pariente, J., Barbier, M., Boutoleau-Bretonnière, C., Couratier, P., Pasquier, F., Deramecourt, V., Sauvée, M., Sarazin, M., Lagarde, J., Roué-Jagot, C., Forlani, S., Jornea, L., David, I., French Research Network on FTLD/FTLD-ALS, PREVDEMALS and Predict-PGRN Groups, LeGuern, E., Dubois, B., Brice, A., Clot, F., Lamari, F. and Le Ber, I Plasma progranulin levels for frontotemporal dementia in clinical practice: a 10-year French experience. Neurobiol Aging. 2020;91:167.e1–167.e9. doi: 10.1016/j.neurobiolaging.2020.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Suárez-Calvet M, Capell A, Araque Caballero MÁ, Morenas-Rodríguez E, Fellerer K, Franzmeier N, Kleinberger G, Eren E, Deming Y, Piccio L, Karch CM, Cruchaga C, Paumier K, Bateman RJ, Fagan AM, Morris JC, Levin J, Danek A, Jucker M, Masters CL, Rossor MN, Ringman JM, Shaw LM, Trojanowski JQ, Weiner M, Ewers M, Haass C, Dominantly Inherited Alzheimer Network; Alzheimer's Disease Neuroimaging Initiative CSF progranulin increases in the course of Alzheimer’s disease and is associated with sTREM2, neurodegeneration and cognitive decline. EMBO Mol Med. 2018;10(12):e9712. doi: 10.15252/emmm.201809712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Lee H-C, Kayasuga Y, Chiba S, Nedachi T, Matsuwaki T, Yamanouchi K, Nishihara M. Roles of progranulin in sexual differentiation of the developing brain and adult neurogenesis. J Reprod Dev. 2009;55(4):351–355. doi: 10.1262/jrd.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Roth T, Nakamura MC, Nissenson RA. Female-specific role of progranulin to suppress bone formation. Endocrinology. 2019;160(9):2024–2037. doi: 10.1210/en.2018-00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilke C, Gillardon F, Deuschle C, Hobert MA, Jansen IE, Metzger FG, Heutink P, Gasser T, Maetzler W, Blauwendraat C, Synofzik M. cerebrospinal fluid progranulin, but not serum progranulin, is reduced in GRN-negative frontotemporal dementia. Neurodegener Dis. 2017;17(2–3):83–88. doi: 10.1159/000448896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. Data collection flowchart. Supplementary Figure 2. Serum PGRN concentrations in both GRN mutation carriers (GRN) and non-mutation carriers (non-GRN). Cut-off determined using the optimal Youden’s index. Supplementary Figure 3. A) Correlation between serum and plasma PGRN levels in this cohort (p= 0.0696). B) Correlation between CSF and serum PGRN levels in this cohort (p = 0.0780). C) Correlation between CSF and plasma in this cohort (p<0.0001). Supplementary Figure 4. No significant difference in plasma PGRN levels between bvFTD (behavioural variant FTD) and PPA (primary progressive aphasia) GRN mutation carriers.Supplementary Figure 5. A) Correlation between plasma PGRN levels and age at sampling in GRN mutation carriers. B) Correlation between plasma PGRN levels and age at sampling in non GRN mutation carriers. ** P < 0.01,**** P < 0.0001, Spearman correlation. Supplementary Figure 6. A) Correlation between plasma PGRN levels and age of onset in GRN mutation carriers. B) Correlation between plasma PGRN levels and age of onset in non-GRN mutation carriers. C) Correlation between plasma PGRN levels and age of onset in people with AD. **** P < 0.0001, Spearman correlation. Supplementary Figure 7. A) Correlation between plasma PGRN levels and weight in GRN mutation carriers. B) Correlation between plasma PGRN levels and weight in non GRN mutation carriers. Supplementary Figure 8. A) Differences in plasma PGRN levels measured with the Adipogen assay between clinical diagnoses in this cohort. B) Differences in serum PGRN levels measured with the Adipogen assay between clinical diagnoses in this cohort. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001, two-tailed Mann-Whitney Test. Supplementary Table 1. List of institutions, countries, assay types and sample types included in this dataset alongside the number of data point provided.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.