Abstract
The GP3 protein of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus (PRRSV) was expressed in 293 cells by a recombinant human type 5 adenovirus carrying the open reading frame 3 gene. The protein exhibited a molecular mass of 42 kDa and comigrated with GP3 expressed in PRRSV-infected MARC-145 cells. Removal of N-linked glycans from GP3 resulted in a 27-kDa protein (P3), confirming its highly glycosylated nature. Pulse-chase experiments carried out either in the context of PRRSV infection or upon individual expression of GP3 in 293 cells showed that the protein remains completely endo-β-N-acetylglucosaminidase H-sensitive even after 4 h of synthesis. Thus, the transport of GP3 was restricted to the premedial Golgi compartment, presumably the endoplasmic reticulum (ER). However, a minor fraction of GP3 was found to be secreted in the culture medium as a soluble membrane-free form. This released protein (sGP3) was readily identified upon individual expression of GP3 in 293 cells as well as in the context of PRRSV infection, albeit at lower levels. The sGP3 migrated as a smear and displayed a molecular mass ranging from 43 to 53 kDa. The unglycosylated form of sGP3 comigrated with its intracellular deglycosylated counterpart, suggesting that the release from the cell of a subset of GP3 did not result from cleavage of a putative membrane-anchor sequence. Strikingly, unlike GP3, the sGP3 acquired Golgi-specific modifications of its carbohydrate side chains and folded into a disulfide-linked homodimer. Brefeldin A treatment completely abolished the release of sGP3, suggesting that the ER-to-Golgi compartment is an obligatory step in cellular secretion of sGP3. In contrast, 10 mM monensin did not prevent sGP3 release but inhibited the terminal glycosylation that confers on the protein its diffuse pattern. Since GP3 was found to be nonstructural in the case of the North American strain, secretion of a minor fraction of GP3 might be an explanation for its high degree of immunogenicity in infected pigs. Furthermore, this secreted protein might be relevant as a model for further studies on the cellular subcompartments involved in the sorting of proteins to the extracellular milieu.
Porcine reproductive and respiratory syndrome virus (PRRSV) belongs to the recently recognized Arteriviridae family within the genus Arterivirus, order Nidovirales, along with equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (5, 11). Mature PRRS virions are enveloped and contain an icosahedral capsid which encloses a linear plus-strand RNA genome of approximately 15 kb (41). The latter contains eight open reading frames (ORFs), designated (5′ to 3′) ORF1a, ORF1b, and ORF2 to ORF7. Aside from the genome-length mRNA, expression of the viral genome involves the synthesis of a 3′ coterminal nested set of six subgenomic mRNAs (8). The available evidence indicates that only the 5′-most ORF is translated from each one of these subgenomic mRNAs. According to sequence data, ORF1a and ORF1b are expected to produce functional products associated with virus replication (14).
At least five PRRSV structural proteins, designated GP2, GP4, GP5 (or E), M, and N, have been identified and found to be encoded by ORF2 and ORF4 to ORF7, respectively (38, 40, 42). While N (15 kDa) is the major protein of the capsid, GP2 (29 to 30 kDa), GP4 (28 to 31 kDa), GP5 (24 to 26 kDa), and M (18 to 19 kDa) are membrane associated. The latter two are present as disulfide-linked heterodimers and are the major components of the virus envelope (37). Among these structural proteins, only GP2, GP4, and GP5 are modified by N-linked glycans (37, 40, 42). With the exception of GP4, the other structural proteins (GP2, GP5, M, and N) have also been identified for EAV (12, 13) and LDV (18). Moreover, it has been shown that the envelope of Lelystad virus (LV), the European prototype of PRRSV, accomodates a fifth minor protein, of 45 to 50 kDa, encoded by ORF3 (42, 54). This finding was unexpected, since previous reports indicate that the ORF3-encoded protein is incorporated neither into LDV particles (19) nor into PRRS virions (27, 60). In the case of LDV, in vitro studies have shown that the ORF3 product is weakly associated with the membrane and tends to be soluble (19). Finally, by using a monospecific antiserum raised against Escherichia coli-expressed ORF3 product, we have recently confirmed the nonstructural nature of the GP3 of a reference Quebec strain of PRRSV (22).
The role of the ORF3 product in the life cycle of arteriviruses remains unknown. Therefore, full characterization of this viral protein is required. Apart from its nonstructural nature, recent reports indicate that the ORF3 product is highly antigenic (19, 22, 27), and in one experiment, this protein was shown to provide protection for piglets against PRRSV infection in the absence of a noticeable neutralizing-antibody response (49). These features have prompted us to undertake a detailed study aimed at characterizing this protein. In the present study, the intracellular processing of PRRSV ORF3 product was analyzed either in the context of PRRSV infection or upon individual expression of the protein in mammalian cells. Data demonstrated that a minor fraction of GP3 is secreted into cell culture fluids as a soluble protein (sGP3), whereas the bulk of the protein remains associated with the endoplasmic reticulum (ER). Unlike its intracellular counterpart, sGP3 appears as a smear due to extensive and complex Golgi-specific modifications of its carbohydrate side chains. By using protein transport inhibitors, the release of sGP3 from the cell has been found to occur by transit through the secretory pathway.
MATERIALS AND METHODS
Cells and viruses.
Cells of the MARC-145 line, a clone of MA-104 that is permissive to PRRSV, were kindly provided to us by J. Kwang (U.S. Meat Animal Research Center, Clay Center, Nebr.) and propagated as previously described (28). Adherent human 293 cells (24) were purchased from The American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 mg of streptomycin per ml.
The origin and propagation of the Quebec cytopathogenic IAF-Klop strain of PRRSV have been described previously (36). The human adenovirus (Ad) mutant Ad/CMVlacZ (1), which was used in cotransfection experiments, and the resulting Ad recombinants were prepared from infected 293 cell homogenates, and infectivity titers were determined by calculation of the 50% tissue culture infective doses (TCID50) (10).
Antisera.
Preparation of rabbit monospecific antisera (α7, α6, and α5) against the E. coli-expressed IAF-Klop PRRSV major structural proteins (N, M, and GP5, respectively) was detailed previously (37). Similarly, IAF-Klop PRRSV ORF3- and ORF4-encoded proteins were expressed in E. coli and monospecific antisera (α3 and α4) were produced (21, 23).
Generation of recombinant Ad type 5 expressing PRRSV GP3 protein.
The PRRSV GP3 coding sequence (ORF3) was amplified by PCR with the sense primer ETS3 (5′-GGATCCGGATCC GCCGCCGCC ATGGCTAATAGCCGTACA-3′) and the reverse primer ETR3 (5′-GGATCC GGATCC CTATCGCCGTGCGGCACT-3′). Both primers contain at their 5′ ends two recognition sites for BamHI, and in the case of ETS3, the ATG initiator codon is preceded by a triple GCC motif in order to provide an optimal Kozak consensus sequence for efficient translation (30). This pair of primers was designed from the sequence of the IAF-exp91 reference PRRSV strain (EMBL/GenBank accession no. L40898). The PCR product was inserted into the unique BamHI site of the Ad transfer vector pAdCMV5 (39) so that the GP3 coding sequence would be under the control of the constitutive human cytomegalovirus (CMV) immediate-early promoter and enhancer. In this cassette, which was derived from pAdBM5, expression of heterologous genes is also optimized by the presence of the Ad tripartite leader sequence and the Ad major late enhancer flanked by splice donor and acceptor sites. The recombinant plasmid was linearized at the unique ClaI site and rescued into the genome of Ad/CMVlacZ by homologous recombination in 293 cells, as described elsewhere (1). Upon cotransfection, virus plaques were isolated, amplified in 293 cells, and analyzed for expression of GP3 either by indirect immunofluorescence or by radioimmunoprecipitation assays. A recombinant Ad (AdCMV5/ORF3), which efficiently evoked the expression of GP3, was subjected to two consecutive rounds of plaque assays and used as virus stock in subsequent experiments.
Virus infection and radiolabelling.
293 cells grown at 80% confluency were infected with AdCMV5/ORF3 at a multiplicity of infection (MOI) of 20 to 30 TCID50 per cell. After 1 h of adsorption at 37°C, cells were extensively washed with phosphate-buffered saline and the infection was allowed to continue for 17 h. Infection of MARC-145 cells with PRRSV was carried out essentially as previously described (36). Following a starvation period of 30 min in methionine-deprived DMEM, the infected cells were pulse-labelled with 100 μCi of [35S]methionine (specific activity, 1,120 Ci/mmol; Amersham Searle Co., Oakville, Ontario, Canada) for 15 or 30 min. Wherever indicated, labelled cells were chased in DMEM containing 5 mM l-methionine.
Radioimmunoprecipitation and SDS-PAGE.
Labelled cells were washed twice in cold phosphate-buffered saline and solubilized in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40 [NP-40], 0.1% sodium desoxycholate, 0.1% sodium dodecyl sulfate [SDS], 2 mM phenylmethylsulfonyl fluoride [PMSF], 1 mg of aprotinin/ml). Lysates of PRRSV-infected or mock-infected cells were clarified at 10,000 × g for 30 min, and 2 × 106-cpm aliquots were incubated overnight at 4°C with 5 μl of normal rabbit sera or specific antisera. Immune complexes were then collected by addition of protein A-Sepharose CL4B (Pharmacia) and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as previously described (37). For immunoprecipitation of extracellular proteins, culture fluids of radiolabelled cells were clarified by a centrifugation at 3,000 × g for 15 min at 4°C. Thereafter, protease inhibitors and lysis buffer were added, and the samples were processed for SDS-PAGE, as described above.
In vivo inhibition of N-linked glycosylation and protein transport.
For N-linked glycosylation and protein transport inhibition experiments, infected 293 cells were pretreated for 1 h either with 5 μg of tunicamycin (TUN) (Boehringer Mannheim)/ml, 5 μg of brefeldin A (BFA) (Sigma)/ml, or 10 μM monensin (MNS) (Sigma) in methionine-deprived DMEM prior to labelling with [35S]methionine in the presence of the aforementioned drugs. When labelling was carried out overnight in the presence of BFA, the latter was added freshly every 4 h, since it has been reported that the activity of BFA gradually decreases during incubation (7).
Carbohydrate analysis by glycosidase digestion.
Immunoprecipitated proteins bound to protein A-Sepharose CL4B beads were suspended in 200 μl of either endo-β-N-acetylglucosaminidase H (Endo H) buffer (50 mM sodium acetate [pH 6], 2 mM PMSF, 1 μg of aprotinin/ml), endoglycosidase F–N-glycosidase F (Glyco F) buffer (50 mM sodium acetate [pH 6.0], 10 mM EDTA, 1% [vol/vol] β-mercaptoethanol, 2 mM PMSF, 1 μg of aprotinin/ml), endo-β-galactosidase (Endo β) buffer (50 mM sodium acetate [pH 5.8], 200 μg of bovine serum albumin/ml, 2 mM PMSF, 1 μg of aprotinin/ml), or O-glycosidase buffer (Tris-maleate, [pH 6.8], 2 mM PMSF, 1 μg of aprotinin/ml) and treated overnight at 37°C with 4, 200, 3, and 4 mU of enzyme, respectively. Removal of sialic acid was performed by incubating immune complexes for 2 h at 37°C in a buffer containing 0.5% NP-40, 10 mM Tris (pH 7.4), 0.5 M NaCl, and 200 mU of neuraminidase per ml. All glycosidases were purchased from Boehringer Mannheim, Laval, Quebec, Canada. After the incubation period, immunoprecipitates were washed in 20 mM Tris-HCl, (pH 7.6), and analyzed by SDS-PAGE. Control samples were incubated under identical conditions but in the absence of enzyme. Alternatively, glycosidase treatments were performed on denatured proteins. Immunoprecipitated proteins were dissociated from the protein A-Sepharose CL4B beads by boiling the samples for 5 min in 20 μl of sample loading buffer containing 1% SDS, 10% glycerol, 5% β-mercaptoethanol, and 62 mM Tris (pH 6.8). The denatured proteins were suspended in 200 μl of the glycosidase buffer supplemented with 1% N-octylglucoside and treated with the corresponding glycosidase, as described above.
Treatment of extracellular GP3 protein with trypsin.
The culture fluid of [35S]methionine-labelled 293 cells expressing GP3 was clarified at 15,000 × g for 10 min, and the resulting supernatant was directly divided into five equal portions and processed as follows. Portion 1 was left untreated, portion 2 was incubated with trypsin (11,500 U/mg) at a final concentration of 300 μg/ml, portion 3 received trypsin plus 1% NP-40, portion 4 received trypsin plus soybean trypsin inhibitor (final concentration of 500 μg/ml), and portion 5 received only soybean trypsin inhibitor. All samples were incubated at room temperature for 30 min. Thereafter, trypsin inhibitor, PMSF, and aprotinin were added to all samples, which were then mixed with 5× lysis buffer and processed for immunoprecipitation as described above.
RESULTS
Expression of GP3 in 293 cells infected with a recombinant Ad carrying the ORF3.
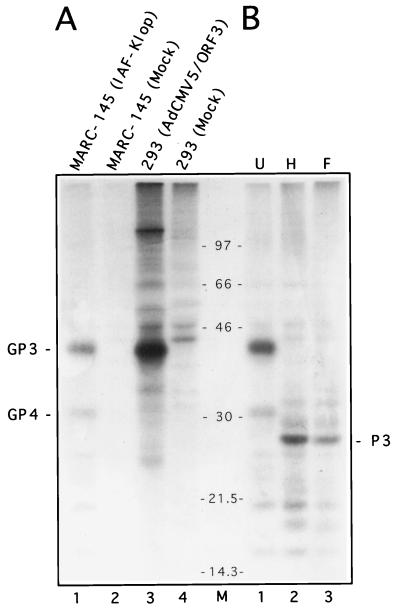
To firmly establish the nature of the PRRSV ORF3-encoded protein, a monospecific antiserum raised against the E. coli-expressed ORF3 product (α3) was developed as previously described (22, 37). From lysates of PRRSV-infected MARC-145 cells, α3 efficiently immunoprecipitated the GP3 protein, which migrated at the expected molecular size of 42 kDa (Fig. 1A, lane 1). To further characterize this protein, the cDNA sequence encoding the complete PRRSV GP3 was inserted into the E1A region of human type 5 Ad under the control of the CMV immediate-early promoter and enhancer, as described in Materials and Methods. AdCMV5/ORF3, a recombinant Ad carrying the PRRSV ORF3 gene, was identified by PCR, and the expression of GP3 in 293 cells was monitored by indirect immunofluorescence. Human 293 cells were then infected with plaque-purified AdCMV5/ORF3 (MOI of 30 TCID50/cell) and radiolabelled at 17 to 17.5 h postinfection (p.i.). Detergent extracts of infected or mock-infected cells were mixed with α3 antiserum and immune complexes were precipitated by protein A-Sepharose. A 42-kDa protein having a migration profile similar to that of the GP3 synthesized in PRRSV-infected cells was efficiently precipitated by α3 antiserum from AdCMV5/ORF3-infected 293 cells (Fig. 1A; compare lanes 1 and 3) but not by that from the mock-infected cells (Fig. 1A; compare lanes 3 and 4), confirming the identity of the PRRSV ORF3 product. It is noteworthy that a protein of approximately 31 kDa was substantially coprecipitated with GP3. This protein has been recently identified as the product of ORF4, referred to as GP4 (21). Coprecipitation of the latter protein still occurred when immunoprecipitates were extensively washed with lysis buffer containing SDS, and the amount of GP4 brought down by α3 antiserum did not increase when salts and SDS were omitted from the lysis buffer (data not shown). As previously reported by others, in studies with recombinant Ads expressing genes from other heterologous viruses (25, 39, 59), the 110-kDa hexon protein of Ad type 5 was consistently coprecipitated from lysates of AdCMV5/ORF3-infected cells (Fig. 1A, lane 3).
FIG. 1.
Mammalian expression of the PRRSV GP3 protein and analysis of its glycosylated nature. (A) Human 293 cells (107) were infected by the recombinant Ad AdCMV5/ORF3 carrying the coding sequence of the PRRSV IAF-Klop strain ORF3 gene and pulse-labelled for 30 min with [35S]methionine at 17 to 17.5 h p.i. Solubilized labelled proteins were subjected to radioimmunoprecipitation with α3 antiserum (anti-ORF3 product) and analyzed by SDS–12% PAGE. As a negative control, mock-infected 293 cells were processed similarly. For comparison, IAF-Klop-infected or mock-infected MARC-145 cells (107) labelled for 30 min at 24 h p.i. were processed in parallel. The α3 monospecific antiserum immunoprecipitated a 42-kDa protein from both PRRSV-infected (lane 1) and AdCMV5/ORF3-infected (lane 3) cells. In the latter case, coprecipitation of the Ad hexon protein (110 kDa) was also observed. (B) α3 immunoprecipitates from IAF-Klop-infected MARC-145 cells (subjected to 30 min of labelling) were either treated with Endo H (lane 2) or Glyco F (lane 3) or left untreated (lane 1) and were then analyzed by SDS–12% PAGE. In lane M, the positions of [14C]Rainbow Marker standards (Amersham) are indicated (in kilodaltons). P3 refers to the unglycosylated precursor of GP3.
Glycosylation of GP3 synthesized in PRRSV-infected MARC-145 cells.
To analyze the nature of the carbohydrate content of GP3, α3 immunoprecipitates from PRRSV-infected cells were subjected to treatment with Glyco F or Endo H. Under the conditions used, GP3 was sensitive to Glyco F, an enzyme mixture which hydrolyzes N-glycans of the high-mannose and complex type. The resulting nonglycosylated product (P3) displayed a molecular mass of approximately 27 kDa (Fig. 1B, lane F), which was consistent with that predicted from the amino acid sequence of the ORF3 product after removal of the predicted N-terminal signal sequence (38, 58). However, it remains to be determined whether the cleavage of the putative N-terminal signal sequence has occurred. A similar electrophoretic profile was obtained after treatment with Endo H (Fig. 1B, lane H), an enzyme which cleaves high-mannose and hybrid oligosaccharides which are added in the ER compartment but not complex-type oligosaccharides which occur in the Golgi complex. No resistance, not even partial, could be observed with Endo H. Taking into account that cells were labelled for a period of 30 min, this indicates that in PRRSV-infected cells, GP3 protein carbohydrates might be processed slowly to the complex type, a finding that has been previously reported for PRRSV GP5 (37) as well as for GP2 and GP4 (40, 43).
Intracellular stability and transport of GP3 protein expressed in the context of PRRSV infection.
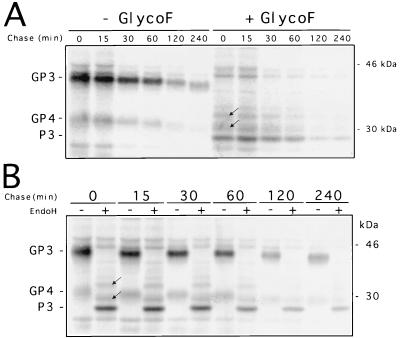
Pulse-chase experiments were performed to examine in greater detail the processing of the GP3 of the IAF-Klop strain of PRRSV. At 24 h p.i., PRRSV-infected MARC-145 cells were pulse-labelled with [35S]methionine for 15 min (time zero) and processing of intracellular GP3 was monitored during a chase period that varied from 15 to 240 min. Cell lysates were immunoprecipitated with α3 antiserum and subjected or not to treatment with either Glyco F or Endo H. As shown in Fig. 2A, GP3 was readily detected within the pulse interval and migrated at the expected size. A gradual decrease in the mobility of the protein became apparent after a 2-h chase period, which was most likely due to the sequential removal of glycan moieties, as the mobility of the Glyco F-treated products remained unchanged during the entire chase. Also during the chase period, a marked decrease in the GP3 amount occurred, presumably as a result of proteolysis. It is worth noting that in comparison with the three PRRSV major structural proteins (N, M, and GP5), the breakdown of the GP3 protein was much more considerable (data not shown). As mentioned above, GP4 was consistently coprecipitated with GP3.
FIG. 2.
Intracellular stability (A) and transport (B) of GP3 protein expressed during IAF-Klop infection of MARC-145 cells. Infected cells were pulse-labelled with [35S]methionine for 15 min and chased in the presence of 5 mM nonradioactive methionine for the indicated periods. Cell lysates were immunoprecipitated with α3 antiserum and were then subjected to treatment with Glyco F or Endo H or were left untreated. Numbers on the right indicate the positions and sizes of marker proteins. The arrows show the two protein bands resulting from incomplete deglycosylation. Since they were also generated by Glyco F digestion, they must not be considered partially resistant to Endo H.
Strikingly, neither complete nor partial resistance to Endo H was observed during the pulse and the whole chase period (Fig. 2B), suggesting that during PRRSV infection, the transport of GP3 was restricted to the premedial Golgi compartment. Two minor bands (estimated Mrs, 29 and 32 kDa) migrating slightly slower than the nonglycosylated GP3 were observed after Endo H treatment (Fig. 2). The possibility that these two additional precipitated bands correspond to cellular proteins recognized by the α3 antiserum was ruled out since they were not recovered from lysates of mock-infected cells. Since they were also generated upon Glyco F treatment, they were instead considered the results of incomplete deglycosylation. In the case of LV, the reference European PRRSV strain, anti-GP3 monoclonal antibodies also immunoprecipitated at least four proteins with apparent molecular masses ranging from 28 to 44 kDa from Sf21 cells infected with baculovirus-ORF3 recombinants (54). It was demonstrated that these protein bands correspond to the GP3 that was N-glycosylated at different degrees (54).
Processing of GP3 individually expressed in 293 cells by a recombinant Ad and identification of an extracellular form.
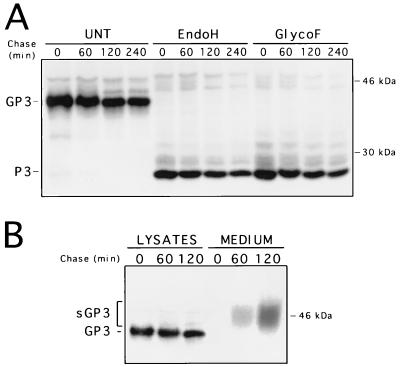
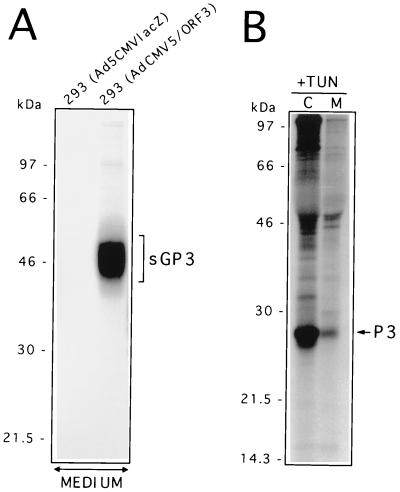
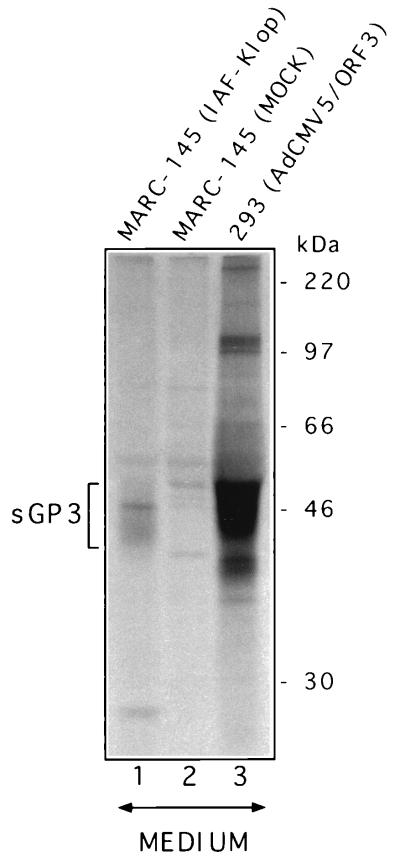
The results described above indicate that GP3 transport might be restricted to the premedial Golgi compartment, most probably in the ER, as the carbohydrate moieties of the protein remain Endo H sensitive even 4 h after synthesis. Since GP3 was not found to be packaged into the IAF-Klop virus particle (22), the possibility that GP3 might be retained in that compartment (presumably through its interaction with other viral proteins) was investigated. The finding that GP4 coprecipitates with GP3 has reinforced such an assumption. A pulse-chase experiment was therefore conducted on 293 cells expressing GP3 following their infection by the recombinant AdCMV5/ORF3 virus. As shown in Fig. 3A, the GP3 was efficiently expressed at levels consistently higher than that observed with PRRSV-infected MARC-145 cells. This is due to the higher MOI used with the recombinant Ad that could not be attained with the IAF-Klop strain of PRRSV, as well as to the optimized expression cassette that drives the synthesis of GP3 from the recombinant Ad (39). As in the case of PRRSV-infected MARC-145 cells, no resistance to Endo H could be detected at any time following GP3 synthesis when the protein was individually expressed. The Endo H profile was identical to that obtained with Glyco F treatment. Thus, the processing of GP3 expressed in the absence of any other viral product appeared similar to that observed in the context of PRRSV infection. Consequently, the ability of GP3 to restrict its own transport to the premedial Golgi compartment appears to be an intrinsic property. Strikingly, when the supernatant of AdCMV5/ORF3-infected 293 cells was analyzed by radioimmunoprecipitation with α3 antiserum, a protein band which migrated as a smear of approximately 43 to 53 kDa became apparent after a 1 h chase period and could be readily observed after 2 h of chase (Fig. 3B). Since no other proteins were brought down in the α3 immunoprecipitates from the supernatant of AdCMV5/ORF3-infected 293 cells, we hypothesized that this protein band, although having a different migration pattern than GP3, may represent a fraction of GP3 (sGP3) that was efficiently transported and has undergone other modifications leading to its secretion in the culture medium. Comparison of the intensities of the protein bands obtained from the supernatant fluids or lysates of Ad-infected cells indicated that approximately 20% of the GP3 was secreted from the cells. The fact that sGP3 occurred only in the culture medium of 293 cells expressing GP3 and was completely absent from that of cells infected with the nonrecombinant parental Ad (Fig. 4A) also strongly suggested that sGP3 was indeed another form of GP3. This assumption was definitely confirmed by demonstrating that in the presence of TUN, the nonglycosylated product of this released form comigrated with P3, the nonglycosylated cell-associated form of GP3 (Fig. 4B). Finally, the finding that sGP3, displayed a different mobility pattern than GP3 (Fig. 3B) ruled out the possibility that it may have resulted from Ad-induced cell lysis and indicated that it was an active secreting process instead. Indeed, at the time when the pulse-chase experiment was performed (16 to 17 h p.i.), trypan blue staining of the Ad-infected cell monolayers revealed not more than 1 to 2% cell death, a level which was similar to that obtained with mock-infected cells (data not shown).
FIG. 3.
Processing and location of PRRSV GP3 protein individually expressed in 293 cells with the recombinant AdCMV5/ORF3 virus. (A) Adherent 293 cells at 80% confluency (107) were infected with AdCMV5/ORF3 at an MOI of 20 to 30 TCID50/cell. At 24 h p.i., cells were pulse-labelled with [35]methionine for 30 min (time zero), chased for the indicated times, and processed for radioimmunoprecipitation with α3 antiserum. Aliquots of immunoprecipitates were treated with Endo H or Glyco F or were left untreated (UNT) and were then analyzed by SDS–12% PAGE. The data indicated that Ad-expressed GP3 was much more stable than PRRSV-expressed GP3. (B) Cells infected with the recombinant AdCMV5/ORF3 virus were pulse-labelled for 30 min and chased for the indicated times as described for panel A. Expression of GP3 was monitored by radioimmunoprecipitation in either the cell lysates or the culture medium. Numbers on the right indicate the positions and sizes of 14C-radiolabelled marker proteins.
FIG. 4.
Identification of an extracellular form of PRRSV GP3 protein. (A) 293 cells (107) infected with the nonrecombinant parental adenovirus (Ad5CMVlacZ) or infected with the recombinant adenovirus expressing GP3 (AdCMV5/ORF3) were labelled for 1 h and then chased for 6 h. The culture medium was clarified at 3,000 × g, and 5× lysis buffer was added prior to radioimmunoprecipitation with α3 antiserum. The immunoprecipitates were then analyzed by SDS–12% PAGE. (B) 293 cells infected with AdCMV5/ORF3 were starved in DMEM lacking methionine and containing 5 μg of TUN/ml and then labelled and chased as described for panel A in the presence of the same drug. After the chase, the clarified culture medium (M) and the cell lysates (C) were processed for radioimmunoprecipitation with α3 antiserum and SDS–12% PAGE. Positions and sizes of 14C-radiolabelled marker proteins are on the left.
sGP3 is also generated in the context of PRRSV infection.
During our preliminary experiments using α3 antiserum, a very faint diffuse band was constantly precipitated from the whole culture medium of PRRSV-infected MARC-145 cells (data not shown). In order to rule out the possibility that the latter band corresponds to a cellular contaminant that nonspecifically reacts with α3 antiserum rather than sGP3, as identified in the case of infection with AdCMV5/ORF3 virus, the following experiments were conducted. Confluent MARC-145 cells growing in 150-cm2 flasks (3 × 107 cells) were infected with IAF-Klop at an MOI of 10 TCID50/cell. After 12 h p.i., the infected cells were labelled with 1.5 mCi of [35S]methionine for an additional 12 h. At that time, cytopathic effects and cell death were observed in less than 1 to 2% of the cells. The whole medium was centrifuged for 2 h at 100,000 × g to pellet the virus and the supernatant was subjected to immunoprecipitation with α3 antiserum. As a negative control, a mock-infected culture of MARC-145 cells was similarly processed. As a positive control, 293 cells (3 × 107 cells) were infected with AdCMV5/ORF3 virus (MOI of 20 to 30) and labelled as described above. A protein migrating as a diffuse band, with the same mobility as sGP3 precipitated from the supernatant fluid of AdCMV5/ORF3-infected 293 cells, could be recovered in smaller amounts from the supernatant fluid of PRRSV-infected but not from that of mock-infected MARC-145 cells (Fig. 5). A contaminant cellular protein of <30 kDa occasionally coprecipitated from the supernatant fluid of PRRSV-infected cultures. Since the latter could not be recovered from the supernatant fluid of AdCMV5/ORF3-infected cultures, it was not considered to represent the nonglycosylated form of GP3. Therefore, data showed that during PRRSV infection of MARC-145 cells, a fraction of GP3 (about 10 to 20% of the amount of GP3 immunoprecipitated from PRRSV-infected cell lysates) was able to reach the extracellular compartment in a fashion which seems to be similar to that which occurs during individual expression of the protein. By radioimmunoprecipitation, neither GP3 nor its released form, sGP3, could be identified in the viral pellet (even though highly concentrated virus stocks were used [22]), which was in agreement with GP3 being released in the culture medium as a non-virion-associated form.
FIG. 5.
The extracellular form of GP3 (sGP3) is also generated during PRRSV infection of MARC-145 cells. Mock-infected or IAF-Klop-infected MARC-145 cells (3 × 107 cells infected at a MOI of 10) were radiolabelled for 12 h. The culture medium was then clarified at 100,000 × g for 2 h in order to pellet the virus and was used for immunoprecipitation with α3 antiserum. For comparison, 293 cells (3 × 107) were similarly infected with AdCMV5/ORF3 virus (MOI of 20 to 30) and the culture medium was used for radioimmunoprecipitation and SDS-PAGE analysis. Numbers on the right indicate the positions and sizes of 14C-radiolabelled marker proteins.
sGP3 is released from the cell as a membrane-free form.
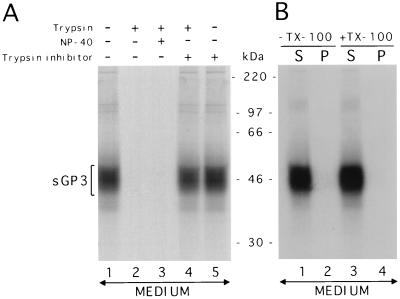
The membrane topography of sGP3 was investigated by assessing either its sensitivity to a dose of 350 U of trypsin/ml or its ability to be pelleted in association with membranes of microorganelles or small cytoplasmic vesicles. The results obtained are consistent with sGP3 being released as a membrane-free form (Fig. 6). Although we could not demonstrate that trypsin-protected fragments of <6 kDa were generated, it appeared that the major part of the protein was not protected by a lipid bilayer if the enzymatic digestion was done in either the presence or absence of the detergent NP-40 (Fig. 6A). However, the protein was completely protected from digestion with trypsin in the presence of soybean trypsin inhibitor. Furthermore, when the clarified culture medium of AdCMV5/ORF3-infected 293 cells was subjected to a centrifugation of 1 to 2 h at 200,000 × g, which is sufficient to pellet very small cytoplasmic vesicles (6), no sGP3 could be recovered in the pellet fraction with or without prior treatment with 1% Triton X-100 (Fig. 6B). Taken together, the above results indicated that the released form of GP3 is soluble.
FIG. 6.
The sGP3 of the IAF-Klop strain of PRRSV is released from the cell as a membrane-free form. (A) Sensitivity of sGP3 to trypsin digestion in the presence or absence of detergent. Prior to radioimmunoprecipitation of sGP3, the culture medium of 293 cells infected with AdCMV5/ORF3 was supplemented with trypsin, NP-40, and/or trypsin inhibitor as indicated in Materials and Methods. The culture medium was allowed to stand for 30 min at 25°C and was then analyzed by radioimmunoprecipitation. (B) In a second experiment, Triton X-100 was added to the culture medium, which was next subjected to centrifugation at 200,000 × g for 1 to 2 h. The resulting pellet (P) and supernatant (S) fractions were assessed by radioimmunoprecipitation for the presence of sGP3. In both experiments, protein bands between 5 and 30 kDa were not revealed (data not shown). Positions and sizes of 14C-radiolabelled marker proteins are indicated between the two gels.
The carbohydrate side chains of sGP3 acquire Golgi-specific modifications.
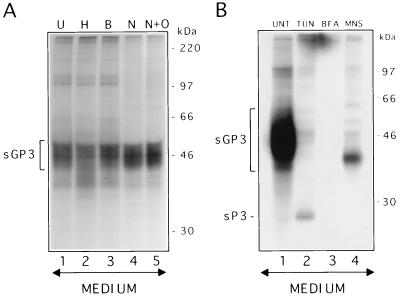
The fact that sGP3 exhibited a smear pattern with a molecular mass greater than that of the intracellular GP3 suggested that this soluble form may have been subjected to further modifications of its carbohydrate side chains. To determine whether the shift in mobility and the smear pattern of sGP3 were indeed the results of complex-type glycosylation, α3 immunoprecipitates from the supernatant of AdCMV5/ORF3-infected 293 cells were subjected to several glycosidases that cleave carbohydrates of a different nature. The first indication of a complex-type glycosylation of sGP3 came from treatment with Endo H. Indeed, unlike its intracellular counterpart, sGP3 was resistant to digestion by Endo H, suggesting that it had at least reached the medial Golgi compartment. However, digestion with Endo H resulted in a more diffuse pattern of sGP3 than in the untreated sample (Fig. 7A; compare lanes H and U). This result was obtained in two independent experiments and suggests that some carbohydrate side chains may still contain N-linked glycans of the high-mannose or hybrid oligosaccharides. Endo β did not affect sGP3, indicating that the smear structure was not due to the presence of poly-N-acetyllactosamine residues. Finally, this released form of GP3 was not modified by sialic acids or type O glycosylation as judged by the resistance of sGP3 to treatment by neuraminidase or neuraminidase plus O-glycosidase.
FIG. 7.
Glycosylation and effect of protein transport inhibitors in the release of sGP3. (A) sGP3 was immunoprecipitated from the medium of AdCMV5/ORF3-infected and [35S]methionine-labelled 293 cells with α3 antiserum. The immunoprecipitates were left untreated (U) or were treated with either Endo H (H), Endo β (B), neuraminidase (N), and neuraminidase plus O-glycosidase (N+O). (B) 293 cells were infected with AdCMV5/ORF3 virus, in the absence of any drug (UNT) or in the presence of either TUN, BFA, or MNS. The culture medium was then processed for radioimmunoprecipitation with α3 antiserum. Positions and sizes of 14C-radiolabelled marker proteins are on the right. sP3 refers to the nonglycosylated released form of GP3.
The release of sGP3 from the cell occurs by transit through the secretory pathway.
Involvement of the secretory pathway in the transit of the GP3 subset that is destined to be exported from the cell was first suspected after the demonstration that sGP3 was essentially Endo H resistant. Thus, to better characterize the sorting process leading to the shedding of a GP3 fraction in the culture fluids, the influence of protein transport inhibitors such as TUN, BFA, and MNS was studied. TUN is known to prevent modification of proteins by N-linked glycans at very early stages of their synthesis, which may affect their transport. The fungicide BFA inhibits the transport of proteins from the ER to the Golgi apparatus (29, 33, 44), whereas the monovalent ionophore MNS alters protein translocation through the Golgi network, most likely by blocking the transport at the medial regions of this organelle (15, 46, 48, 53). Data obtained indicate that inhibition of N-linked glycosylation strongly reduces the rate of sGP3 release without completely inhibiting this process. Indeed, a small fraction of the nonglycosylated GP3 (sP3) could be recovered in cells expressing GP3 in the presence of TUN (Fig. 7B). A complete inhibition of sGP3 release was observed in the presence of BFA (Fig. 7B). It is worth noting that throughout the course of BFA treatment, GP3 was efficiently synthesized and remained Endo H sensitive (data not shown). MNS at 10 mM was found to interfere with sGP3 terminal glycosylation but not with its release from the cell. In the presence of this drug, sGP3 displayed a migration pattern similar to that of the intracellular GP3 (Fig. 7B). This released form was Endo H sensitive (data not shown). Taken together, the results indicate that the intracellular itinerary leading to the secretion of a subset of GP3 involves the vesicular traffic, and that the ER-to-Golgi compartment step is a crucial one for the protein to be released from the cell. Terminal glycosylation of sGP3, which results in its diffuse pattern, most likely takes place in a very distal compartment between the medial Golgi and the cell surface. Such a late transport destination appeared not to affect the export of sGP3 from the cell.
Evidence that sGP3 folds into a disulfide homodimer.
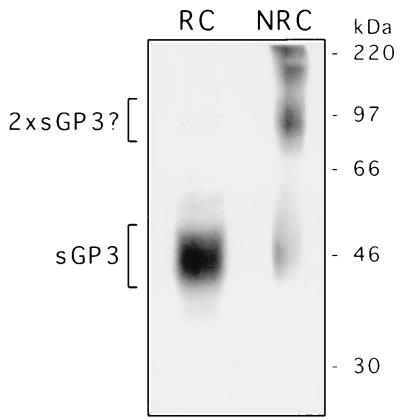
Although the PRRSV GP3 protein of the IAF-Klop strain of PRRSV was found to diverge significantly from its European homolog (from the LV strain) in terms of amino acid sequence, several cysteine residues are conserved between the two strains (38). Therefore, the participation of such conserved residues in the establishment of intra- or intermolecular disulfide bonds could be possible. For this purpose, the GP3 migration pattern from N-ethylmaleimide-treated and [35S]methionine pulse-labelled 293 cells was analyzed under nonreducing conditions. No evidence of the formation of disulfide-linked oligomers of GP3 was found (data not shown). The only modification was that GP3 migrated slightly slower than its authentic molecular mass, a finding which could be attributed to the addition of the alkylating agent N-ethylmaleimide. Strikingly, analysis of the released form of GP3 under nonreducing conditions yielded a predominant protein band of approximately 97 kDa. Given its molecular mass, this protein was considered to be a disulfide-linked homodimer of sGP3 (Fig. 8). This result was reproduced in two independent experiments. The small amount of sGP3 monomers observed under nonreducing conditions might have been generated during our experimental procedure. It is important to note that, like sGP3, the sGP3 dimers, which presumably are formed in the ER, could not be detected in cell lysates but were revealed only in the supernatant of 293 cells expressing GP3 after a lag time similar to that required for the appearance of the monomeric sGP3 (data not shown). Collectively, these results indicate that GP3 in its monomeric state was selectively retained in the ER, as demonstrated by its complete and permanent sensitivity to Endo H, whereas the transport-competent fraction, which acquired resistance to Endo H and was released into the culture fluids, appeared to fold into a disulfide-linked dimer.
FIG. 8.
The sGP3 of the IAF-Klop strain of PRRSV occurs in a dimeric form. The culture medium of AdCMV5/ORF3-infected and labelled 293 cells was subjected to radioimmunoprecipitation with α3 antiserum and analyses by SDS–12% PAGE under reducing (RC) or nonreducing (NRC) conditions. On the right, the positions and sizes of 14C-radiolabelled marker proteins are shown.
DISCUSSION
In the present study, the fate of the PRRSV GP3 (ORF3 product) of a reference North American strain of PRRSV (IAF-Klop strain) was analyzed either in the context of PRRSV infection or by individual expression with recombinant Ad. Data obtained are consistent with the existence of two maturation pathways regardless of the presence or absence of the other viral components.
It has been demonstrated that the bulk of the GP3 remained associated with the premedial Golgi compartment, presumably in the ER, as no resistance to Endo H was observed at any time after the protein synthesis. However, the viral protein was shown to be highly glycosylated, suggesting that most of the potential N-linked glycosylation sites were exposed to the lumen of the ER. Although double staining assays were not performed, the fluorescence pattern displayed by GP3 in 293 cells that have been infected with AdCMV5/ORF3, a recombinant Ad expressing the PRRSV ORF3 gene under the control of the CMV promoter, was quite similar to that observed following incubation of the cells with a rabbit antiserum directed against calnexin (data not shown). Since the latter is an ER resident membrane protein, the data provide strong evidence that PRRSV GP3 was localized in the ER. Moreover, in comparison to the other PRRSV proteins that are incorporated into virions, GP3 seems to be rapidly subjected to degradation. This is a characteristic reminiscent of proteins targeted to the ER membrane, presumably because higher concentrations of proteases are found in that subcellular compartment (32, 55). Data obtained also suggested that the association of GP3 with the ER does not occur through interactions with other viral products, since no resistance to Endo H could be observed upon individual expression of GP3.
The possibility that GP3 might be intrinsically retained within the ER has led us to examine the primary sequence for the presence of any potential ER-targeting sequence. We have been unable to identify any obvious ER-targeting motifs like the tetrapeptide KDEL-COOH (45), the dilysine ER-targeting signal KKXX-COOH (20, 26), the consensus sequence YXXLXXR-COOH (34) (where X is any amino acid), or the double arginine motif NH2-RR (52). The absence of a putative ER retention signal correlates with the ability of a GP3 fraction to be transport competent and finally secreted from the cell. Such a finding raises the question of whether the putative ER retention signal, if one exists, needs to be proteolytically removed in order to allow the protein to be transport competent. Should this be the case, a difference in the molecular mass between the nonglycosylated released fraction and the nonglycosylated ER-associated form would be observed. Data showed that both forms of GP3 comigrated, and hence, the proteolytic removal of a putative ER targeting sequence may not account for retention of the bulk of GP3 in the ER. A previous study conducted in vitro with the ORF3 product of LDV allowed its authors to conclude that the protein might be weakly associated with membranes through its uncleaved signal peptide (19). If we assume that it is also the case for PRRSV GP3, then the above finding precludes the possibility that the uncleaved signal sequence may act as an ER retention peptide. In fact, several proteins with uncleaved signal sequences were shown to remain fully transport competent (57).
Aside from the absence of any recognizable ER retention signal or a putative cleavable membrane anchor sequence, secretion of GP3 from the cell is in fair agreement with amino acid sequence analysis and in vitro studies with LDV. Indeed, sequence analysis of the ORF3 did not reveal any potential sufficiently hydrophobic domain to act as a membrane anchor sequence. Furthermore, the amino terminus of GP3 was not found to contain charged residues which might influence the membrane topography of the protein (3). These molecular predictions are similar to those previously reported for the ORF3 product of LDV. The latter was also shown in vitro to be soluble, to associate weakly with membranes, and most probably not to be incorporated into virions (19). The similarity in the fates of the ORF3 products of both LDV and the Quebec reference strain of PRRSV contrasts with previous findings that LV GP3 is packaged into virus particles and hence can be described as structural (54). Most likely, critical residues in the GP3 primary sequence might determine the difference in its fate.
The fact that only a fraction of PRRSV GP3 was shown to undergo efficient processing and transport within the secretory pathway leading to its release into the culture medium suggests that this fraction was properly folded in a manner competent for secretion. Thus, the most plausible explanation for the retention of the bulk of GP3 within the ER might be its inability to undergo efficient folding and its subjection to quality control, a mechanism which prevents misfolded, incorrectly assembled, or unassembled proteins from exiting the ER (for a review, see reference 16). Most often, proteins that do not pass the ER-associated quality control system aggregate, and they are frequently rapidly degraded. The observation that the Endo H-sensitive intracellular pool of GP3 displayed a relatively rapid turnover is consistent with the above explanation. Another intriguing finding which might be associated with the inability of GP3 to undergo efficient intracellular transport is that dimerization of the protein was shown to occur only for a small subset of the protein which is transport competent. Thus, it is tempting to speculate that dimerization of GP3 is a prerequisite for its efficient transport. This observation is not without precedent and was previously documented for several viral proteins. For instance, the ORF2 product of EAV (Gs protein) is retained in the ER in its monomeric state, whereas dimers become Endo H resistant and are selectively incorporated into virus particles (13). Heterotrimer formation between the authentic membrane-integrated G protein of vesicular stomatitis virus and the smaller soluble Gs fraction is required for secretion of the latter into the growth medium (51). By using monomer- and oligomer-specific antibodies to influenza virus hemagglutinin, it was shown that monomers are restricted to the ER, whereas assembled molecules are able to reach distal points in the transport pathway (9). Furthermore, a fraction of the hemagglutinin of measles virus expressed from a vaccinia virus recombinant was found to be released into the culture medium as a dimer (35). These are but a few examples emphasizing the importance of the oligomeric assembly in the efficient transport of viral glycoproteins, which may be regarded as a likely explanation for the retention of the PRRSV GP3 in the ER. In this respect, it would be of interest to look for a possible association of GP3 with molecular chaperones such as immunoglobulin heavy-chain-binding protein and calnexin, which are known to bind to improperly assembled proteins trapped as large covalent or noncovalent aggregates in the ER (2, 31, 47).
Data also indicate that only a very small fraction of GP3 was able to exit the ER and transit through the secretory pathway to its final destination, the extracellular medium. This conclusion stems from the following observations: (i) sGP3 could not be precipitated from the cell lysates, (ii) it appears in the culture medium after a lag time of at least 1 h, and (iii) the amount of this extracellular secreted form increased as a function of time. As mentioned above, these features of sGP3 also characterize the dimeric form of sGP3. It is actually unclear why only a small fraction of GP3 became transport competent. Is it for functional purposes that the bulk of GP3 was restricted to the ER? Or is it a regulation mechanism that allowed the secretion of only a small fraction of GP3 that is sufficient to fulfill a yet-unidentified role? In fact, secretion of a subset of GP3 in the culture medium contrasts with the tendency of PRRSV to localize and assemble its glycoproteins in intracytoplasmic membranes, where budding appears to occur (50). The propensity of PRRSV glycoproteins to localize to the premedial Golgi compartment now becomes more evident (37, 40, 43). Indeed, unlike GP3, the GP2, GP4, and GP5 proteins displayed a very slow processing of their carbohydrate moieties to the complex type. Thus, as with some coronaviruses, bunyaviruses, and hepadnaviruses (4, 5, 7, 56), intracytoplasmic assembly and budding of PRRSV might be viewed as a strategy to evade host immune detection and hence, secretion of a subset of GP3 would not be in line with such a mechanism. At present, the release of sGP3 may explain why the proteins, though nonstructural, elicits a strong antibody response in infected animals, at least in the case of the North American IAF-Klop strain (22). Such a finding was also reported for the GP3 counterpart in LDV (19). The role of anti-GP3 antibodies in the evolution of the PRRSV and LDV infections remains to be defined.
The coding sequence of GP3 is the most variable among PRRSV strains (17, 38), the major amino acid variations being at the amino terminus. Since this region carries signals that influence the destiny of most proteins, it is conceivable that the difference in the fate of GP3 in North American and European strains may lie in this portion of the protein. Further studies involving the construction of GP3 chimeras and the generation of mutants may certainly help us to learn more about the biogenesis and the role of this protein. However, one can speculate that since GP3 is not packaged in the case of the North American IAF-Klop PRRSV strain, this protein may not be necessary for the initiation of the virus infection.
ACKNOWLEDGMENTS
We thank Nicole Sawyer, Louise Wilson, and François Bouthillier for excellent technical assistance. Particular thanks go to Julie Dionne for the preparation of the figures and to Maria Koutromanis for comments on the manuscript.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC Strategic Grants STR0167446 and STP0202083). P. Gonin is a recipient of a postdoctoral grant from the Institut Armand-Frappier, and C. A. Gagnon is a recipient of a fellowship from the Medical Research Council of Canada.
Footnotes
This is NRC publication 41419.
REFERENCES
- 1.Ascadi G, Jani A, Massie B, Simoneau M, Holland P, Blaschuk K, Karpati G. A differential efficiency of adenovirus-mediated in vivo gene transfer into skeletal muscle cells of different maturity. Hum Mol Genet. 1994;3:578–584. doi: 10.1093/hmg/3.4.579. [DOI] [PubMed] [Google Scholar]
- 2.Bole D G, Hendershot L M, Kearney J F. Post-translational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino J S, Cosson P, Shah N, Klausner R D. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 6.Chambers J J A, Rickwood D. Fractionation of subcellular organelles by differential centrifugation. In: Rickwood D, editor. Centrifugation: a practical approach. Washington, D.C: Information Retrieval Ltd.; 1978. pp. 33–47. [Google Scholar]
- 7.Chen S-Y, Matsuoka Y, Compans R W. Assembly and polarized release of Punta Toro virus and effects of brefeldin A. J Virol. 1991;65:1427–1439. doi: 10.1128/jvi.65.3.1427-1439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conzelmann K K, Visser N, Van Woensel P, Thiel H J. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329–339. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland C S, Zimmer K-P, Wagner K R, Healey G A, Mellman I, Helenius A. Folding, trimerization, and transport are sequential events in the biogenesis of influenza hemagglutinin. Cell. 1988;53:197–209. doi: 10.1016/0092-8674(88)90381-9. [DOI] [PubMed] [Google Scholar]
- 10.Dea S, Bilodeau R, Athanassious R, Sauvageau R, Martineau G P. Swine reproductive and respiratory syndrome in Quebec: isolation of an enveloped virus serologically-related to Lelystad virus. Can Vet J. 1992;33:801–808. [PMC free article] [PubMed] [Google Scholar]
- 11.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries A A F, Raamsman M J B, van Dijk H A, Horzinek M C, Rottier P J M. The small envelope glycoprotein (Gs) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J Virol. 1995;69:3441–3448. doi: 10.1128/jvi.69.6.3441-3448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewar R L, Vasudevachari M B, Natarajan V, Salzman N P. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J Virol. 1989;63:2452–2456. doi: 10.1128/jvi.63.6.2452-2456.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doms R W, Lamb R A, Rose J K, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 17.Drew T W, Lowings J P, Yapp F. Variation in open reading frames 3, 4, and 7 among porcine reproductive and respiratory syndrome virus isolates in the UK. Vet Microbiol. 1997;55:209–221. doi: 10.1016/s0378-1135(96)01328-4. [DOI] [PubMed] [Google Scholar]
- 18.Faaberg K S, Plagemann P G W. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology. 1995;212:512–525. doi: 10.1006/viro.1995.1509. [DOI] [PubMed] [Google Scholar]
- 19.Faaberg K S, Plagemann P G W. ORF3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology. 1997;227:245–251. doi: 10.1006/viro.1996.8310. [DOI] [PubMed] [Google Scholar]
- 20.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonin P, Gagnon C A, Massie B, Dea S. Abstracts of the 47th Annual Meeting of the Canadian Society of Microbiologists 1997. Ottawa, Ontario, Canada: Canadian Society of Microbiologists; 1997. Procaryotic and eucaryotic expression of ORFs 2 to 4 of porcine reproductive and respiratory syndrome virus, abstr. VM5; p. 86. [Google Scholar]
- 22.Gonin, P., H. Mardassi, C. A. Gagnon, B. Massie, and S. Dea. The ORF3 gene of the Quebec IAF-Klop strain of porcine reproductive and respiratory syndrome virus encodes a nonstructural and antigenic glycoprotein. Arch. Virol., in press. [DOI] [PMC free article] [PubMed]
- 23.Gonin, P., B. Pirzadeh, C. A. Gagnon, and S. Dea. Seroneutralization of porcine reproductive and respiratory syndrome virus correlates with antibody response to the GP5 major envelope glycoprotein. J. Vet. Diagn. Invest., in press. [DOI] [PubMed]
- 24.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 25.Hanke T, Graham F L, Rosenthal K L, Johnson D C. Identification of an immunodominant cytotoxic T-lymphocyte recognition site in glycoprotein B of herpes simplex virus by using recombinant adenovirus vectors and synthetic peptides. J Virol. 1991;65:1177–1186. doi: 10.1128/jvi.65.3.1177-1186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson M R, Nilsson T, Peterson P A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. Cell. 1990;56:801–813. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz J B, Shafer A L, Eernisse K A, Landgraf J G, Nelson E A. Antigenic differences between European and American isolates of porcine reproductive and respiratory syndrome virus (PRRSV) are encoded by the carboxyterminal portion of viral open reading frame 3. Vet Microbiol. 1995;44:65–76. doi: 10.1016/0378-1135(94)00113-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H S, Kwang J, Yoon I J, Joo H S, Frey M L. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogenous subpopulation of MA-104 cell line. Arch Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 29.Klausner R D, Donaldson J G, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 31.Kozutsumi Y, Segal M, Normington K, Gething M-J, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signal the induction of glucose-regulated proteins. Nature (London) 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 32.Krishna N K, Weldon R A, Jr, Willis J W. Transport and processing of the Rous sarcoma virus Gag protein in the endoplasmic reticulum. J Virol. 1996;70:1570–1579. doi: 10.1128/jvi.70.3.1570-1579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin-A. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallabiabarrena A, Jimenez M A, Rico M, Alarcon B. A tyrosine-containing motif mediates ER retention of CD3-epsilon and adopts a helix-turn structure. EMBO J. 1995;14:2257–2268. doi: 10.1002/j.1460-2075.1995.tb07220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malvoisin E, Wild F. Characterization of a secreted form of measles virus haemagglutinin expressed from a vaccinia virus recombinant. J Gen Virol. 1994;75:3603–3609. doi: 10.1099/0022-1317-75-12-3603. [DOI] [PubMed] [Google Scholar]
- 36.Mardassi H, Athanassious R, Mounir S, Dea S. Porcine reproductive and respiratory syndrome virus: morphological, biochemical and serological characteristics of Quebec isolates associated to acute and chronic outbreaks of PRRS. Can J Vet Res. 1994;58:55–64. [PMC free article] [PubMed] [Google Scholar]
- 37.Mardassi H, Massie B, Dea S. Intracellular synthesis, processing, and transport of porcine reproductive and respiratory syndrome virus proteins encoded by ORFs 5, 6, and 7. Virology. 1996;221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- 38.Mardassi H, Mounir S, Dea S. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Québec reference strain. Arch Virol. 1995;140:1405–1418. doi: 10.1007/BF01322667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massie B, Couture F, Lamoureux L, Mosser D D, Guilbault C, Jolicoeur P, Bélanger F, Langelier Y. Inducible overexpression of a toxic protein by an adenovirus vector with a tetracycline-regulatable expression cassette. J Virol. 1998;72:2289–2296. doi: 10.1128/jvi.72.3.2289-2296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meulenberg J J M, den Besten A P. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology. 1996;225:44–51. doi: 10.1006/viro.1996.0573. [DOI] [PubMed] [Google Scholar]
- 41.Meulenberg J J M, Hulst M M, de Meijer E J, Moonen P L J M, den Besten A, de Kluyver E P, Wensvoort G, Moormann R J M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS) is related to LDV and EAV. Virology. 1993;192:62–74. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meulenberg J J M, Petersen-den Besten A, De Kluyver E P, Moormann R J, Schaaper W M, Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meulenberg J J M, van Nieuwstadt A P, van Essen-Zandbergen A, Langeveld J P M. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J Virol. 1997;71:6061–6067. doi: 10.1128/jvi.71.8.6061-6067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller S G, Carnell L, Moore H H. Post-Golgi membrane traffic: brefeldin A inhibits export from distal Golgi compartments to the cell surface but not recycling. J Cell Biol. 1992;118:267–283. doi: 10.1083/jcb.118.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munro S, Pelham H R B. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1997;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 46.Oda K, Ikehara Y. Monensin inhibits the conversion of proalbumin to serum albumin in cultured rat hepatocytes. Biochem Biophys Res Commun. 1982;105:766–772. doi: 10.1016/0006-291x(82)91500-5. [DOI] [PubMed] [Google Scholar]
- 47.Ou W-J, Cameron P H, Thomas D Y, Bergeron J J M. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature (London) 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- 48.Pesonen M, Kaariainen L. Incomplete complex oligosaccharides in Semliki Forest virus envelope proteins arrested within the cell in the presence of monensin. J Mol Biol. 1982;158:213–230. doi: 10.1016/0022-2836(82)90430-2. [DOI] [PubMed] [Google Scholar]
- 49.Plana-Duran J, Climent I, Sarraseca J, Urniza A, Cortes E, Vela C, Casal J I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes. 1997;14:19–29. doi: 10.1023/a:1007931322271. [DOI] [PubMed] [Google Scholar]
- 50.Pol J, Wagenaar F. Morphogenesis of Lelystad virus in porcine lung alveolar macrophages. Am Assoc Swine Pract Newsl. 1992;4:29. [Google Scholar]
- 51.Schmidt C, Grünberg J, Kruppa J. Formation of heterotrimers between the membrane-integrated and the soluble glycoproteins of vesicular stomatitis virus leads to their intracellular cotransport. J Virol. 1992;66:2792–2797. doi: 10.1128/jvi.66.5.2792-2797.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schutze M-P, Peterson P A, Jackson M R. A N-terminal double arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tartakoff A M. Perturbation of vesicular traffic with the carboxylic ionophore monensin. Cell. 1983;32:1026–1028. doi: 10.1016/0092-8674(83)90286-6. [DOI] [PubMed] [Google Scholar]
- 54.van Nieuwstadt A P, Meulenberg J J M, van Essen-Zandbergen A, Peterson-den Besten A, Bende R J, Moormann R J M, Wensvoort G. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J Virol. 1996;70:4767–4772. doi: 10.1128/jvi.70.7.4767-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 56.Vennema H, Heijnen L, Zijderveld A, Horzinek M C, Spaan W J M. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verner K, Schatz G. Protein translocation across membranes. Science. 1988;241:1307–1313. doi: 10.1126/science.2842866. [DOI] [PubMed] [Google Scholar]
- 58.Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wesseling J G, Godeke G-J, Schijns V E C J, Prevec L, Graham F L, Horzinek M C, Rottier P J M. Mouse hepatitis virus spike and nucleocapsid proteins expressed by adenovirus vectors protect mice against a lethal infection. J Gen Virol. 1993;74:2061–2069. doi: 10.1099/0022-1317-74-10-2061. [DOI] [PubMed] [Google Scholar]
- 60.Wieczorek-Krohmer M, Weiland F, Conzelmann K, Kohl D, Visser N, van Woensel P, Thiel H-J, Weiland E. Porcine reproductive and respiratory syndrome virus (PRRSV): monoclonal antibodies detect common epitopes on two viral proteins of European and U.S. isolates. Vet Microbiol. 1996;51:257–266. doi: 10.1016/0378-1135(96)00047-8. [DOI] [PubMed] [Google Scholar]