Abstract
Successful infection by human immunodeficiency virus type 1 (HIV-1) requires the activation of target cells. Infection of quiescent peripheral CD4 lymphocytes by HIV-1 results in incomplete, labile, reverse transcripts. In the present study, we isolated highly purified quiescent T cells and utilized the CD3/CD28 activation pathways as well as cell cycle inhibitors to further define the role of costimulation and cell cycle progression in HIV-1 reverse transcription. Activation with αCD3 alone resulted in cell cycle progression into only G1a and incomplete HIV-1 reverse transcription. Costimulation through the CD28 receptor and transition into G1b was required to efficiently complete the reverse transcription process. These findings have relevance to immune activation in vivo, since lymphocytes rendered anergic by a single activation signal would be nonpermissive for productive infection with HIV-1. Importantly, these data also suggest that HIV vector-based genetic transduction strategies might be successful only in target cells that transition into the G1b phase of the cell cycle.
Retroviral replication is influenced greatly by the stage of the cell cycle at the time of infection (8, 44, 45, 49–52). Our previous studies have established that in contrast to stimulated lymphocytes, quiescent T cells become infected by human immunodeficiency virus (HIV) but fail to allow completion of viral reverse transcription (51, 52). In addition, the differences in the rates of reverse transcription in activated T lymphocytes and macrophages (38) may also exemplify the role that the cell cycle plays in the reverse transcription process.
Most circulating peripheral T lymphocytes and many in lymphoid tissues are in a G0 resting state. Diverse elements, including growth factors and antigen-mediated T-cell activation, are involved in the stimulation of these cells and may result in different states of cellular activation and cell cycle progression. Optimal T-cell activation requires signaling through the T-cell receptor (TCR) and additional stimulation that is provided through accessory molecules present on the surface of antigen-presenting cells (APC) (for reviews, see references 7, 24, and 27). The first signal, delivered through the antigen-specific TCR, results in the increased expression of certain lymphocyte activation molecules, such as the nuclear factor of activated T cells (NF-AT), and the activation of certain lymphokine genes and cellular oncogenes required for cell division, including interleukin-2 (IL-2) and c-myc, as well as a transition of the cell from the G0 to the G1 stage of the cell cycle (12). The second signal, which is provided by a costimulatory molecule(s) present on the surface of the APC, involves progression through late G1, S, and mitosis, and is controlled by IL-2–interleukin-2 receptor (IL-2R) interactions (26). In the absence of the second signal, the T cell enters an unresponsive state known as anergy, where it is incapable of producing IL-2 on subsequent stimulation (17, 40), and the cell cycle is arrested at the G1a/G1b transition point (22, 23). The G1a/G1b transition defines the point where cells which have entered the cell cycle are committed to progression to the S phase. The G1a phase of the cell cycle involves an increase in the levels of RNA and protein in preparation for entry into the G1b and S phases. Cells in the G1a phase contain higher levels of RNA than quiescent cells in the G0 phase of the cell cycle. The G1b phase is defined as the stage when RNA levels are equivalent to those seen in early S phase prior to DNA synthesis (13, 22). T cells in an unresponsive state may progress through the G1a phase but are prevented from achieving the critical threshold of RNA or protein required for progression into the G1b and S phases (22).
In the present study, we utilized physiological and immunologically relevant stimulatory signals to investigate the extent of T-cell activation and the phase of the cell cycle required for the completion of the HIV type 1 (HIV-1) reverse transcription process. We activated highly purified quiescent T lymphocytes with signals generated through the TCR alone or with the additional engagement of one of the major costimulatory molecules, CD28 (25, 26, 32). We employed cell cycle inhibitors and detailed cell cycle analysis to define early events in cell cycle progression. Our data indicate that stimulation through the TCR alone, which results in arrest of the cells in G1a, is insufficient to allow efficient completion of HIV-1 DNA synthesis. More specifically, costimulation and transition of the cell from the G1a phase of the cell cycle to the G1b phase are obligatory for the completion of the reverse transcription process.
MATERIALS AND METHODS
Isolation of cell populations.
Peripheral blood was obtained from healthy HIV-seronegative blood donors, and peripheral blood mononuclear cells were separated over a Ficoll-Hypaque gradient. Nonadherent (NA) cells were obtained after depleting macrophages by 3 h of adherence to plastic. The HLA-DR− population was depleted of monocytes and B cells by separation over a nylon wool column. For further purification of the DR− population, nylon wool-purified cells were incubated on ice with saturating amounts of the antibodies HLA-DR, CD19, and CD14 (Becton Dickinson, Mountain View, Calif.), washed extensively, resuspended in medium, and subjected to panning in goat anti-mouse antibody (GAM; Sigma, St. Louis, Mo.)-coated flasks, resulting in depletion of cells expressing major histocompatibility complex class II antigen, as well as B cells and macrophages, respectively. Purified cells were 99% pure, as assessed by flow cytometry.
Cell cultures and conditions.
Cells were cultured in RPMI 1640 supplemented with 10% human AB serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM glutamine. Cells were stimulated with 1 μg of anti-CD3 monoclonal antibody (MAb) per ml immobilized on GAM-coated plates alone to mimic TCR stimulation or were costimulated with anti-CD3 and soluble anti-CD28 (Pharmingen, San Diego, Calif.), each at a concentration of 1 μg/ml. Some cultures were treated with cell cycle inhibitors prior to their stimulation. The inhibitors used in these studies were n-butyrate and aphidicolin (Sigma) at 5 and 10 mM, respectively. Cellular proliferation was assessed by measuring DNA synthesis. At 3 days poststimulation, 105 cells were incubated for 4 h with [3H]thymidine (10 μCi/ml) and harvested onto glass fiber filters with a ClassicCell Harvester (Skatron Instruments, Lier, Norway) and thymidine incorporation was measured with a liquid scintillation counter.
Flow cytometry for surface markers.
To assess the purity of the populations, 5 × 105 cells were costained with MAbs (Becton Dickinson) against HLA-DR (major histocompatibility complex class II), CD25 (IL-2R), CD19 (B-cell marker), and CD14 (macrophage marker) cell surface markers. Anti-HLA-DR and anti-CD19 MAbs were conjugated directly to fluorescein isothiocyanate (FITC); anti-CD25 and anti-CD14 were conjugated directly to phycoerythrin (PE).
To determine the expression levels of activation markers, 5 × 105 cells were stained with MAbs against HLA-DR, CD38 (T-cell activation marker; may have role in cell adhesion and signal transduction), CD25, and CD69 (an early T-cell activation marker). For these stainings, anti-HLA-DR and anti-CD69 MAbs were conjugated directly to FITC; anti-CD38 and anti-CD25 were conjugated directly to PE.
To determine the expression levels of CD4 and the T-tropic HIV-1 coreceptor CXCR4 (19, 20), cells of the conditions indicated above were stained with peridinin chlorophyll protein-conjugated CD4 (Becton Dickinson) and CXCR4 MAbs, respectively. CXCR4 MAb 12G5 from J. Hoxie (19) was obtained through the AIDS Research and Reference Reagent Program, AIDS Program, National Institute of Allergy and Infectious Diseases. Cells stained for CXCR4 were counterstained with goat anti-mouse immunoglobulin G2a conjugated to PE (Pharmingen). The cells were then fixed in 2% paraformaldehyde. Data were accumulated on a FACStarplus flow cytometer and analyzed with the CellQuest program (Becton Dickinson).
Cell cycle analysis.
A total of 5 × 105 cells of each condition were stained for DNA and RNA content by using 7-amino-actinomycin D and pyronin Y (7AAD-PY) as previously described (47) with some modifications. Briefly, cells were suspended in a buffer containing 0.03% saponin (Sigma). Fifty microliters of 400 μM 7AAD (Calbiochem, La Jolla, Calif.) was added at a final concentration of 20 mM. The cells were incubated at room temperature for 30 min and cooled on ice for at least 5 min, and 3 μl of 1.7 mM PY (Polysciences, Warrington, Pa.) was added at a final concentration of 5 μM; the cells were then incubated for an additional 10 min on ice and analyzed. Data were accumulated on a FACStarplus flow cytometer and analyzed with the CellQuest program.
Virus and infections.
Stocks of the HIV-1 molecular clone NL4-3 (1) were obtained from 24-h harvests of supernatants from infected CEM cells. These supernatants generally contained 1 to 1.5 μg of viral p24 per ml as assessed by enzyme-linked immunosorbent assay (ELISA); consequently, when 1 ml of virus was used to infect 106 cells, the multiplicity of infection was approximately 0.2. To reduce the amount of contamination with viral DNA derived from cells lysed during culture, supernatants were filtered and treated with 2 μg of DNase (Worthington, Lakewood, N.J.) per ml for 30 min at room temperature in the presence of 0.01 M MgCl2. Infection was accomplished by incubating the cells for 1 to 2 h with virus in the presence of Polybrene (10 μg/ml). The cells were washed with medium three times to remove residual free virus and recultured under their respective conditions. Heat-inactivated virus controls were prepared by incubating the virus for 30 min at 60°C. Virus production by infected cells was analyzed by an ELISA specific for the p24 gag antigen (Coulter, Hialeah, Fla.).
Quantitative PCR.
Cells to be subjected to quantitative PCR were harvested and washed, and DNA was isolated by using the QIAamp blood kit (Qiagen, Chatsworth, Calif.). Quantitative PCR was performed on purified DNA samples by using primers specific for HIV-1 sequences as previously described (51). The primer pairs M667/AA55 (R/U5 region) and M667/M661 (LTR/gag region) were used to detect initiation and completion of the HIV-1 reverse transcription process, respectively. Primers specific for the human β-globin gene were utilized to determine the input of cellular DNA. One primer from each pair was end labeled with 32P as described previously (51). Following 25 cycles of PCR, samples were resolved on a 6% polyacrylamide gel, and quantitation was performed by comparison of values to a standard curve of known amounts of HIV-1 DNA, or cellular DNA from uninfected human peripheral blood mononuclear cells, by using an Ambis (San Diego, Calif.) radioanalytic imager.
RESULTS
Response of quiescent T cells to mitogens.
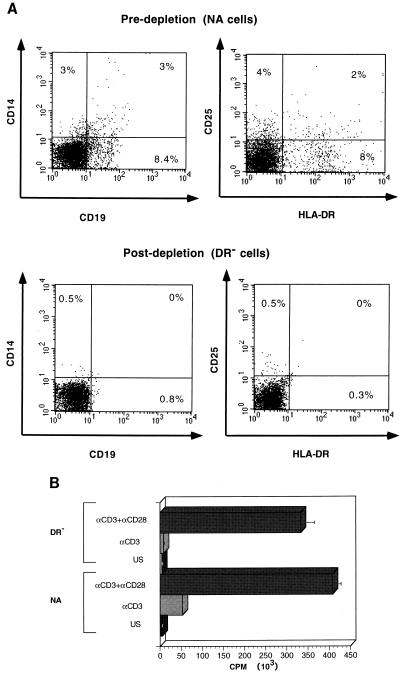
To study the role of costimulatory signals in HIV reverse transcription, we first established a highly purified population of quiescent T cells, free of HLA-DR+ lymphocytes, which could function as APC (Fig. 1A). In the present paper, this purified population is referred to as DR−, as opposed to the NA, macrophage-depleted population obtained by adherence to plastic alone (NA population). This NA population contains cells expressing HLA-DR that can act as APC, providing any necessary costimulatory signal. We initially characterized the DR− cells for their ability to respond to T-cell activation signals. As previously shown by others (11, 25), in contrast to the NA cells, the highly purified DR− population did not proliferate in response to immobilized anti-CD3 alone (Fig. 1B). However, following costimulation with anti-CD28, NA cells exhibited an 8-fold increase in proliferation and the DR− population exhibited a greater-than-30-fold enhancement of proliferation in comparison to cells stimulated with anti-CD3 alone (Fig. 1B). These results confirm that the DR− population is a highly purified population of quiescent T cells that requires two activation signals for optimal activation.
FIG. 1.
(A) Purification of lymphocyte populations. To assess the purity of the indicated populations, 5 × 105 unstimulated NA and DR− cells were costained with MAbs against HLA-DR, CD25, CD19, and CD14 cell markers as described in Materials and Methods. The DR− cell population is 99% pure in comparison to the NA population, which contains greater than 10% cells expressing HLA-DR, as well as macrophages and B cells. (B) Requirements of costimulation for proliferation of the DR− population. NA and DR− cells were cultured in medium alone (US), were stimulated with 1 μg of anti-CD3 MAb per ml immobilized on GAM-coated plates (αCD3), or were costimulated with anti-CD3 plus soluble anti-CD28 each at a concentration of 1 μg/ml (αCD3+αCD28) for 3 days. Cells were harvested in triplicate and assayed for thymidine incorporation as described in Materials and Methods. Results are the averages of triplicate wells. These results are representative of more than 10 experiments.
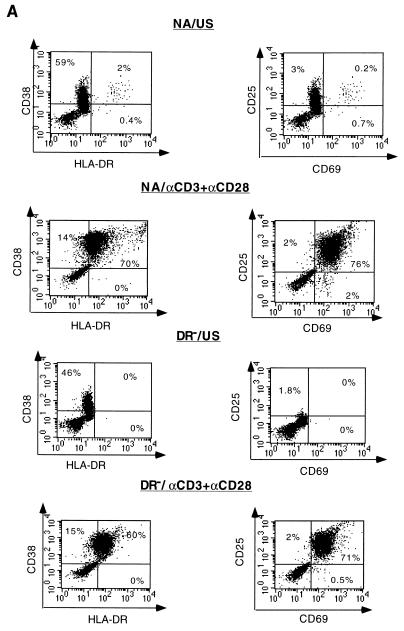
To further compare the responses of DR− and NA cells to the stimulatory signals, we correlated the expression levels of activation markers with progression through the cell cycle. Three days following costimulation with anti-CD3 and anti-CD28, the expression of all activation markers tested increased to similar levels in both populations (Fig. 2A). However when anti-CD3 stimulation alone was applied (Fig. 2B), expression of all activation markers was significantly induced only in NA cells. In contrast, only approximately 50% of DR− cells expressed the early activation marker CD69, expression of CD38 was slightly increased, and expression of other markers typically seen late in T-cell activation (HLA-DR and CD25) was not increased compared to expression levels in control (unstimulated) cells.
FIG. 2.
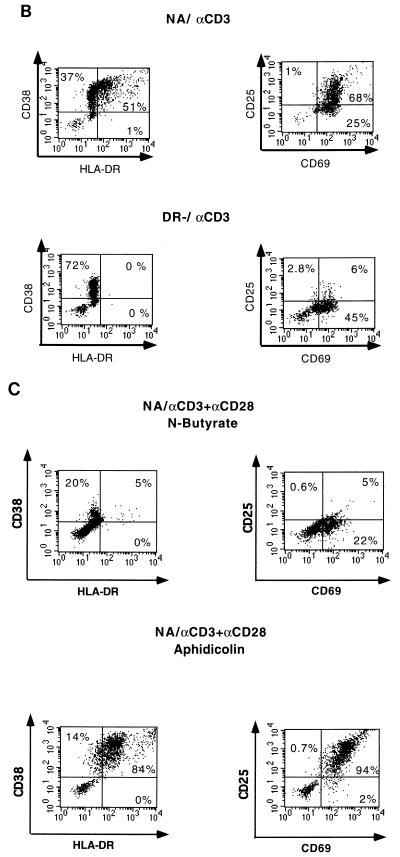
Expression levels of activation markers following stimulation. (A) NA and DR− unstimulated cells (US) and cells costimulated (αCD3+αCD28) for 3 days were stained with MAbs against the activation markers HLA-DR, CD38, CD69 and CD25 as described in Materials and Methods. The percentages of cells expressing the different activation markers are indicated in the corresponding quadrants. (B) Cells were stimulated with anti-CD3 alone for 3 days and stained as described for panel A. (C) NA cells were treated with the G1a/G1b cell cycle inhibitor n-butyrate or the G1b/S cell cycle inhibitor aphidicolin prior to costimulation with anti-CD3 plus anti-CD28 for 3 days and subsequently stained as described for panel A. The cells used for this experiment are the same as those used for the experiment illustrated in Fig. 1, and results are representative of more than 10 experiments.
To further define the extent of cellular activation, two cell cycle inhibitors, aphidicolin and n-acetyl butyric acid (n-butyrate) were used prior to costimulation of the NA population with anti-CD3 and anti-CD28. Aphidicolin, an inhibitor of cellular DNA polymerase alpha and lambda, blocks cell cycle progression between the G1b and S phases of the cell cycle without interfering with RNA and protein synthesis (42). This drug allows the expression of at least some early-replicative genes, such as Ha-ras, myc, and mos, in a mouse fibroblast cell line (41). The naturally occurring 4-carbon fatty acid n-butyrate has been shown to block cell cycle progression at the G1a phase of the cell cycle in different cell culture systems (14, 23, 28, 39). At least some of the effects of n-butyrate can be attributed to its ability to reversibly inhibit histone deacetylation and associated changes in chromatin structure (28). In T-cell clones, n-butyrate-induced arrest of cells in the G1a phase of the cell cycle was shown to require TCR occupation and has been used as a model for Th1 cell anergy (23).
NA cells treated with aphidicolin 30 min prior to costimulation with anti-CD3 and anti-CD28 expressed high levels of the activation markers CD69, CD25, and HLA-DR 3 days poststimulation (Fig. 2C). In contrast, cells treated with n-butyrate prior to costimulation expressed only the early surface activation marker CD69 (Fig. 2C). As shown in Fig. 2B, DR− cells stimulated with αCD3 alone exhibited staining profiles similar to that of n-butyrate-treated cells, suggesting that stimulation of DR− cells with αCD3 only allowed transition into G1a, without further progression through the cell cycle.
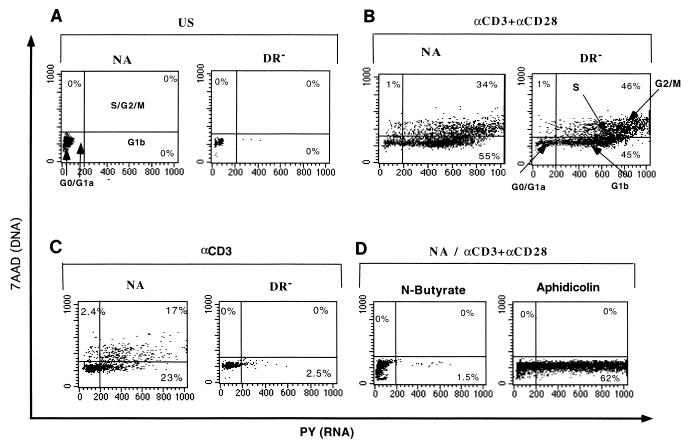
Cell cycle progression was determined for all cultures by utilizing a technique that permits simultaneous DNA and RNA quantitation by flow cytometry, i.e., staining with 7AAD and PY, respectively (47). Figure 3A and B show the cell cycle distribution of the unstimulated NA population, compared with the NA and the DR− populations costimulated with anti-CD3 and anti-CD28. While unstimulated cells remained in G0, the cells costimulated with both anti-CD3 and anti-CD28 progressed through the G1, S, G2, and M phases. No significant differences between the cell cycle progressions of costimulated NA and DR− populations were observed. NA cells activated with anti-CD3 alone also progressed through all stages of the cell cycle (Fig. 3C). In contrast, when the highly purified DR− cell population was activated with anti-CD3 alone, cell cycle progression was arrested in G1a. Figure 3D shows the expected G1a cell cycle arrest caused by n-butyrate treatment of the NA cells prior to costimulation and the G1b block of NA cells treated with aphidicolin prior to costimulation. The pattern of DR− cells stimulated with anti-CD3 alone is strikingly similar to that of NA cells treated with n-butyrate prior to stimulation with anti-CD3 and anti-CD28. Together, the results presented in Fig. 2 and 3 indicate that highly purified quiescent T cells stimulated through the TCR alone do not become fully activated and are blocked in the G1a cell cycle phase.
FIG. 3.
Cell cycle analysis of stimulated populations. The same cell populations as those illustrated in Fig. 2 were unstimulated (US) (A), costimulated with anti-CD3 and anti-CD28 for 3 days (B), stimulated with anti-CD3 alone (C), or treated with cell cycle inhibitor n-butyrate or aphidicolin prior to costimulation (D). A total of 5 × 105 cells for each condition were then stained for DNA and RNA levels for cell cycle analysis by using 7AAD and PY (47). The different cell cycle phases identified by this technique are indicated in panels A and B, and the percentage of cells in each stage of the cycle is indicated for each of the conditions.
Stimulatory signals required for productive infection.
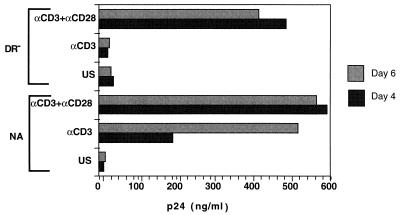
We next studied the ability of highly purified DR− quiescent T cells, activated through the TCR alone or costimulated with the additional signal provided through the CD28 costimulatory receptor, to support productive infection. Cells were infected with the CXCR4-tropic HIV-1 molecular clone NL4-3 at 3 days poststimulation, and culture supernatants were removed and assessed for viral p24 at 3 and 8 days following infection (Fig. 4). Costimulation with anti-CD3 and anti-CD28 rendered cells competent for productive infection. However, in contrast to NA cells, productive infection was not observed in DR− cells stimulated with anti-CD3 alone. Thus, productive HIV infection requires transition past the G1a block associated with stimulation of DR− cells with anti-CD3 alone.
FIG. 4.
Virus production in response to stimulation. DR− and NA cells were infected with 1.5 μg of strain NL4-3 prior to culture. Supernatants from the DR− and NA cell populations were assayed for HIV-1 p24 antigen production by ELISA on days 3 and 8 postinfection. Results are representative of four different experiments. US, unstimulated; αCD3+αCD28, costimulated with anti-CD3 and anti-CD28; αCD3, stimulated with anti-CD3 alone.
Mechanism of block to productive infection in cells in G1a.
The discovery of various coreceptors which act in concert with CD4 to allow virus entry into target cells has increased our understanding of entry events in HIV infection (3, 15, 18, 20). Recent studies have shown down-regulation of the mRNA of the CCR5 coreceptor, but not the CXCR4 coreceptor, following T-cell costimulation with anti-CD3 and anti-CD28 (6). To determine whether expression levels of HIV-1 coreceptors might have an effect on viral entry in our system, cells stimulated under various conditions were analyzed by flow cytometry for cell surface expression of the coreceptor CXCR4 (19), which allows entry of the NL4-3 strain used in these studies. As shown in Table 1, the majority of CD4+ cells continue to express CXCR4 at surface densities comparable to those of unstimulated or costimulated cells. These data suggest that the block of productive infection in cells activated with anti-CD3 alone in our system is not likely due to differences in receptor and coreceptor expression.
TABLE 1.
Expression of CD4 and CXCR4 on the surface of DR− cellsa
| DR− cell stimulation conditionb | % of CD4+ cells expressing CXCR4 | Mean fluorescence of CXCR4 on CD4+ cells |
|---|---|---|
| US | 97 | 97 |
| Anti-CD3 | 70 | 90 |
| Anti-CD3 + anti-CD28 | 92 | 89 |
DR− cells subjected to the indicated stimulation conditions were costained with peridinin chlorophyll protein-conjugated anti-CD4 and PE-conjugated anti-CXCR4 MAbs. Data were accumulated and analyzed as described in Materials and Methods. Data are representative of three experiments.
US, unstimulated; anti-CD3, TCR stimulation alone (with anti-CD3); anti-CD3 + anti-CD28, costimulation.
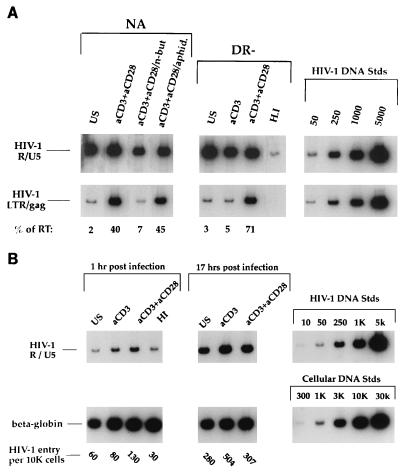
Our previous studies had identified a block to reverse transcription in quiescent cells (51, 52). To determine the mechanism responsible for lack of productive infection in quiescent cells stimulated with anti-CD3 alone, we utilized quantitative PCR employing oligonucleotide primers specific for DNA structures present at various stages of the reverse transcription process (51, 52). These primers differentially detect the first region of the viral genome synthesized (R/U5 region of the long terminal repeat), versus regions present only following complete or nearly complete reverse transcription (LTR/gag junction). Figure 5A shows that the levels of initiation of reverse transcription were comparable for all tested cultures. This result confirms that entry of HIV into quiescent cells is not impaired. As was expected from previous studies, neither unstimulated NA cells nor unstimulated DR− cells support synthesis of full-length viral DNA, whereas costimulated cells quite efficiently complete viral DNA synthesis. The level of full-length viral DNA produced in NA cells pretreated with aphidicolin prior to costimulation is comparable to that of cells costimulated with anti-CD3 and anti-CD28. In contrast, NA cells treated with n-butyrate prior to costimulation, and consequently arrested in G1a, do not support full-length viral DNA synthesis. The highly purified quiescent DR− population, activated with anti-CD3 alone, also was incapable of efficiently producing full-length viral DNA. Taken together with data presented in Fig. 3, these results demonstrate that while cell DNA synthesis is not required, a G1a cell cycle block renders T cells less permissive for completion of HIV-1 reverse transcription and that progression to G1b is required to allow complete synthesis of viral DNA.
FIG. 5.
(A) HIV-1 reverse transcription following various stimulation treatments. Cells under the conditions described in the legends to Fig. 2 and 3 were infected with the CXCR4-tropic HIV-1 molecular clone NL4-3. At 17 h postinfection, DNA was harvested and subjected to a quantitative PCR. The primer pairs M667/AA55 (R/U5) and M667/M661 (LTR/gag) were used to detect initiation and completion of the HIV-1 reverse transcription process, respectively, in equivalent amounts of infected cell DNA. Copy numbers were determined by comparison of samples under each condition to the appropriate standard curve by utilizing radioanalytic image analysis. Percentages of initiated reverse transcripts that completed the reverse transcription process (% of RT) were determined by utilizing the following formula: %RT = (completed DNA copies/ initiated DNA copies) × 100; these values are indicated for each of the conditions. Data are representative of seven experiments and are derived from the same cells as those represented in Fig. 2 and 3. (B) HIV DNA is due to de novo reverse transcription. Highly purified DR− cells were left unstimulated, were stimulated with anti-CD3 alone, or were costimulated with anti-CD3 and anti-CD28 for 3 days. Cells were infected as described in the legend to Fig. 5A and harvested and subjected to quantitative PCR at 1 and 17 h postinfection. The number of HIV-1 entry sequences per 104 cells was calculated based on total cell genomes assessed, as determined by quantitative PCR for human β-globin DNA sequences, and is indicated for each of the conditions. Standards for both HIV and cell DNAs that were amplified in parallel are shown on the right (K, thousand). US, unstimulated; aCD3, stimulated with anti-CD3 alone; aCD3+aCD28, costimulated with anti-CD3 and anti-CD28.
To rule out the possibility that the differences in the levels of PCR products in Fig. 5A were due to the presence of partial reverse transcripts incorporated into virions attached to the cell surface (4, 33, 48, 53) rather than actual infection, we compared the extent of HIV-1 DNA synthesis at 1 h postinfection with that at 17 h postinfection. Highly purified DR− cells were left unstimulated, were stimulated with anti-CD3 alone, or were costimulated with anti-CD3 and anti-CD28 prior to infection. All cells were rinsed thoroughly at 1 h postinfection to remove unbound virions. Thus, any increase seen at 17 h versus 1 h in the amount of HIV DNA detectable by the R/U5 primers must be due to de novo reverse transcription. As shown in Fig. 5B, similar to what we previously reported (52), levels of specific HIV-1 proviral DNA (determined following subtraction of background signal from that of heat-inactivated control) were three- to ninefold higher in cells harvested at 17 h than in cells harvested at 1 h postinfection, thus confirming that these are newly synthesized DNA products, as opposed to virion-associated reverse transcripts.
DISCUSSION
In the present work, we studied the extent of T-cell activation that is required for completion of HIV-1 reverse transcription. Since the activation of the highly purified DR− population through the TCR alone is not sufficient for cell division, the system we studied allowed us to further define the role of activation signal pathways and cell cycle progression in HIV-1 infection and to shed light on the mechanisms involved in HIV-1 replication. Using this system, we showed that both TCR and costimulatory signals are needed to allow efficient full reverse transcription and productive infection of target cells. Cell division per se is not required for reverse transcription, since cells that progress only as far as the G1b phase allow complete reverse transcription to occur. These results are in agreement with previously obtained data from our laboratory and others (30, 45, 50) indicating that cellular activation but not cellular DNA synthesis is required for efficient reverse transcription by HIV-1. Here we extend these findings and further demonstrate that highly purified quiescent T cells, free of contaminating APC, that are stimulated through the TCR alone progress to G1a only and are less permissive for full viral reverse transcription. This has particular relevance to activation of the immune response in that it suggests that lymphocytes in vivo which have received only a single activation signal (i.e., they are rendered anergic) will be refractory to productive infection.
Some recent studies looking at the effect of activation pathways on HIV-1 infection reported successful infection in T cells activated through the TCR alone (29, 43). These experiments were conducted by using cell populations that were not depleted of HLA-DR-bearing cells which could provide the costimulatory signal. One of these studies (29) also reported that costimulation of CD4 cells with anti-CD3 and anti-CD28 can render these cells nonpermissive for HIV-1 reverse transcription and productive infection. The macrophage-tropic Ba-L strain of HIV-1 was used in that study. However, it was subsequently shown (6) that CD28 costimulation of T lymphocytes down-regulates mRNA for the coreceptor CCR5 but not that for CXCR4; thus, the antiviral effect described by Levine et al. (29) was operating at the level of viral entry. In our experiments, we used the CXCR4-tropic HIV-1 molecular clone NL4-3 and demonstrated (Table 1) that coreceptor expression remained high.
Our system was designed to isolate the two activation pathways and to study their particular role in cellular activation that pertains to productive HIV-1 infection of T cells. Our results differ from those of a recent study by Sun et al. (46) that reported complete reverse transcription in infected resting T lymphocytes after activation with anti-CD3 alone. It is not clear why different results were obtained; however, our use of the cell cycle inhibitor n-butyrate confirms the block of reverse transcription found for cells arrested in G1a. Our results showing a lack of productive infection in cells stimulated with anti-CD3 alone, however, are in agreement with those of Sun et al. (46).
The mechanism(s) involved in the reverse transcription block remains undefined, although some suggest that low levels of deoxyribonucleotides may contribute to this phenomenon (34, 35). We cannot exclude this possibility, and additional studies will be needed to further elucidate the role of nucleotide pools in our system. Nonetheless, our findings identify a specific cell cycle stage required for the optimal completion of the reverse transcription process in T lymphocytes and thus provide us with a more precise window of opportunity to be studied and possibly inhibited. In preliminary studies (data not shown), we have found that cultured macrophages reside primarily in the G1b phase of the cell cycle. Their infectibility by HIV is consistent with our finding that G1b is permissive for HIV reverse transcription in T cells. It will be of interest to determine what factors allow productive infection of neurons by HIV vectors (37), since these cells are terminally differentiated and not progressing through the cell cycle. Recent studies have identified some of the intracellular signaling pathways that are blocked in T cells activated through the TCR alone. There is a normal induction and translocation of the dephosphorylated cytoplasmic form of NF-AT to the nucleus, as well as its normal binding to the DNA at the IL-2 NF-AT response element. However, in cells stimulated with anti-CD3 alone, tyrosine phosphorylation is defective in certain substrates such as p34, p38, and p74 (9). In particular, activation of the mitogen-activated protein kinases ERK-1 and -2 and JNK-1 and -2 is impaired (16, 31). The GTP-binding protein p21Ras also remains unactivated (21). This block in Ras could result in the inability to induce and activate the transcription factors c-Fos and JunB, thus preventing IL-2 gene transcription in T cells stimulated with anti-CD3 alone (36). These numerous differences between cells in G1a and G1b may also include the absence of a cellular cofactor needed to augment reverse transcription in suboptimally activated cells.
Our studies have important implications for pathogenesis since they might help to identify a potential cellular factor(s) involved in HIV replication. In addition, specific inhibitors of cell activation might be useful in decreasing the efficiency of reverse transcription in infected individuals. While it is unlikely that HIV-positive individuals will benefit from immunosuppressive therapy, this approach might be appropriate for immediate therapy after known virus exposure, such as that via needle stick injury or perinatal transmission. The inhibition of required cellular activities will likely not be overcome by the development of resistant mutants, which makes this area of research relevant even in the era of improved therapeutic treatment of viral functions. It is interesting that in vivo full-length HIV DNA is found in resting cells (5, 10). Our results suggest that this would be mediated by initial activation and infection of the cells and subsequent return to a resting state following removal of the activating stimulus. This is supported by the observation that these sequences are found only in CD45RO+ memory T cells (10).
Our results also have important relevance in the gene therapy arena, as the ability of HIV-based retroviral vectors to enter nondividing cells has generated much interest (2, 37). Our data suggest that target cells in the G0 or G1a phase of the cell cycle (as is presumed for hematopoietic stem cells) might be refractory for reverse transcription of these HIV-based vectors. Thus, additional studies regarding appropriate activation of these hematopoietic precursor cells are still required to achieve optimal transduction efficiencies with these vectors.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI 33259, the University of California—Los Angeles CFAR, and an AMGEN Fellowship from the UCLA AIDS Institute (to Y.D.K.).
We thank Ingrid Schmid for her help in establishing the flow cytometric technique for the cell cycle analysis and Beth D. Jamieson and Livia Pedroza-Martins for helpful suggestions on manuscript preparation.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkina R K, Walton R M, Chen M L, Li Q-X, Planelles V, Chen I S Y. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Arts E J, Mak J, Kleiman L, Wainberg M A. DNA found in human immunodeficiency virus type 1 particles may not be required for infectivity. J Gen Virol. 1994;75:1605–1613. doi: 10.1099/0022-1317-75-7-1605. [DOI] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–427. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll R G, Riley J L, Levine B L, Feng Y, Kaushal S, Ritchey D W, Bernstein W, Weislow O S, Brown C R, Berger E A, June C H, St. Louis D C. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science. 1997;276:273–276. doi: 10.1126/science.276.5310.273. [DOI] [PubMed] [Google Scholar]
- 7.Chambers C A, Allison J P. Co-stimulation in T cell responses. Curr Opin Immunol. 1997;9:396–404. doi: 10.1016/s0952-7915(97)80087-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen I S Y, Temin H M. Establishment of infection by spleen necrosis virus: inhibition in stationary cells and the role of secondary infection. J Virol. 1982;41:183–191. doi: 10.1128/jvi.41.1.183-191.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho E A, Riley M P, Sillman A L, Quill H. Altered protein tyrosine phosphorylation in anergic Th1 cells. J Immunol. 1993;151:20–28. [PubMed] [Google Scholar]
- 10.Chun T, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 11.Costello R, Cerdan C, Pavon C, Brailly H, Hurpin C, Mawas C, Olive D. The CD2 and CD28 adhesion molecules induce long-term autocrine proliferation of CD4+ T cells. Eur J Immunol. 1993;23:608–613. doi: 10.1002/eji.1830230304. [DOI] [PubMed] [Google Scholar]
- 12.Crabtree G R. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 13.Darzynkiewicz Z, Sharpless T, Staiano-Coico L, Melamed M R. Subcompartments of the G1 phase of cell cycle detected by flow cytometry. Proc Natl Acad Sci USA. 1980;77:6696–6699. doi: 10.1073/pnas.77.11.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darzynkiewicz Z, Traganos F, Xue S B, Melamed M R. Effect of n-butyrate on cell cycle progression and in situ chromatin structure of L1210 cells. Exp Cell Res. 1981;136:279–293. doi: 10.1016/0014-4827(81)90006-9. [DOI] [PubMed] [Google Scholar]
- 15.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 16.DeSilva D R, Feeser W S, Tancula E J, Scherle P A. Anergic T cells are defective in both jun NH2-terminal kinase and mitogen-activated protein kinase signaling pathways. J Exp Med. 1996;183:2017–2023. doi: 10.1084/jem.183.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeSilva D R, Urdahl K B, Jenkins M K. Clonal anergy is induced in vitro by T cell receptor occupancy in the absence of proliferation. J Immunol. 1991;147:3261–3267. [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 21.Fields P E, Gajewski T F, Fitch F W. Blocked Ras activation in anergic CD4+ T cells. Science. 1996;271:1276–1278. doi: 10.1126/science.271.5253.1276. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert K M, Ernst D N, Hobbs M V, Weigle W O. Effects of tolerance induction on early cell cycle progression by Th1 clones. Cell Immunol. 1992;141:362–372. doi: 10.1016/0008-8749(92)90155-i. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert K M, Weigle W O. Th1 cell anergy and blockade in G1a phase of the cell cycle. J Immunol. 1993;151:1245–1254. [PubMed] [Google Scholar]
- 24.Janeway C A, Jr, Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins M K, Taylor P S, Norton S D, Urdahl K B. CD28 delivers a costimulatory signal involved in antigen-specific IL-2 production by human T cells. J Immunol. 1991;147:2461–2466. [PubMed] [Google Scholar]
- 26.June C H, Ledbetter J A, Linsley P S, Thompson C B. Role of the CD28 receptor in T-cell activation. Immunol Today. 1990;11:211–216. doi: 10.1016/0167-5699(90)90085-n. [DOI] [PubMed] [Google Scholar]
- 27.Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 28.Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 29.Levine B L, Mosca J D, Riley J L, Carroll R G, Vahey M T, Jagodzinski L L, Wagner K F, Mayers D L, Burke D S, Weislow O S, St. Louis D C, June C H. Antiviral effect and ex vivo CD4+ T cell proliferation in HIV-positive patients as a result of CD28 costimulation. Science. 1996;272:1939–1943. doi: 10.1126/science.272.5270.1939. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Simm M, Potash M J, Volsky D J. Human immunodeficiency virus type 1 DNA synthesis, integration, and efficient viral replication in growth-arrested T cells. J Virol. 1993;67:3969–3977. doi: 10.1128/jvi.67.7.3969-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Whaley C D, Mondino A, Mueller D L. Blocked signal transduction to the ERK and JNK protein kinases in anergic CD4+ T cells. Science. 1996;271:1272–1276. doi: 10.1126/science.271.5253.1272. [DOI] [PubMed] [Google Scholar]
- 32.Linsley P S, Brady W, Grosmaire L, Aruffo A, Damle N K, Ledbetter J A. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J Exp Med. 1991;173:721–730. doi: 10.1084/jem.173.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lori F, di Marzo Veronese F, De Vico A L, Lusso P, Reitz M S, Jr, Gallo R C. Viral DNA carried by human immunodeficiency virus type 1 virions. J Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lori F, Malykh A, Cara A, Sun D, Weinstein J N, Lisziewicz J, Gallo R C. Hydroxyurea as an inhibitor of human immunodeficiency virus-type 1 replication. Science. 1994;266:801–805. doi: 10.1126/science.7973634. [DOI] [PubMed] [Google Scholar]
- 35.Meyerhans A, Vartanian J-P, Hultgren C, Plikat U, Karlsson A, Wang L, Eriksson S, Wain-Hobson S. Restriction and enhancement of human immunodeficiency virus type 1 replication by modulation of intracellular deoxynucleoside triphosphate pools. J Virol. 1994;68:535–540. doi: 10.1128/jvi.68.1.535-540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller D L, Jenkins M K. Molecular mechanisms underlying functional T-cell unresponsiveness. Curr Opin Immunol. 1995;7:375–381. doi: 10.1016/0952-7915(95)80113-8. [DOI] [PubMed] [Google Scholar]
- 37.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien W A, Namazi A, Kalhor H, Mao S-H, Zack J A, Chen I S Y. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad K N. Butyric acid: a small fatty acid with diverse biological functions. Life Sci. 1980;27:1351–1358. doi: 10.1016/0024-3205(80)90397-5. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz R H. A cell culture model for T lymphocyte clonal anergy. Science. 1990;248:1349–1356. doi: 10.1126/science.2113314. [DOI] [PubMed] [Google Scholar]
- 41.Sorscher D H, Cordeiro-Stone M. Gene replication in the presence of aphidicolin. Biochemistry. 1991;30:1086–1090. doi: 10.1021/bi00218a030. [DOI] [PubMed] [Google Scholar]
- 42.Spadari S, Sala F, Pedrali-Noy G. Aphidicolin and eukaryotic DNA synthesis. Adv Exp Med Biol. 1984;179:169–181. doi: 10.1007/978-1-4684-8730-5_17. [DOI] [PubMed] [Google Scholar]
- 43.Spina C A, Prince H E, Richman D D. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Invest. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson M, Brichacek B, Heinzinger N, Swindells S, Pirruccello S, Janoff E, Emerman M. Molecular basis of cell cycle dependent HIV-1 replication. Implications for control of virus burden. Adv Exp Med Biol. 1995;374:33–45. doi: 10.1007/978-1-4615-1995-9_4. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Pinchuk L M, Agy M B, Clark E A. Nuclear import of HIV-1 DNA in resting CD4+ T cells requires a cyclosporin A-sensitive pathway. J Immunol. 1997;158:512–517. [PubMed] [Google Scholar]
- 47.Toba K, Winton E F, Koike T, Shibata A. Simultaneous three-color analysis of the surface phenotype and DNA-RNA quantitation using 7-amino-actinomycin D and pyronin Y. J Immunol Methods. 1995;182:193–207. doi: 10.1016/0022-1759(95)00050-k. [DOI] [PubMed] [Google Scholar]
- 48.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varmus H E, Padgett T, Heasley S, Simon G, Bishop J M. Cellular functions are required for the synthesis and integration of avian sarcoma virus-specific DNA. Cell. 1977;11:307–319. doi: 10.1016/0092-8674(77)90047-2. [DOI] [PubMed] [Google Scholar]
- 50.Zack J A. The role of the cell cycle in HIV-1 infection. Adv Exp Med Biol. 1995;374:27–31. doi: 10.1007/978-1-4615-1995-9_3. [DOI] [PubMed] [Google Scholar]
- 51.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 52.Zack J A, Haislip A M, Krogstad P, Chen I S Y. Incompletely reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Bagasra O, Niikura M, Poiesz B J, Pomerantz R J. Intravirion reverse transcripts in the peripheral blood plasma of human immunodeficiency virus type 1-infected individuals. J Virol. 1994;68:7591–7597. doi: 10.1128/jvi.68.11.7591-7597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]