Abstract
Mouse adenovirus type 1 (MAV-1) mutants with deletions of conserved regions of early region 1A (E1A) or with point mutations that eliminate translation of E1A were used to determine the role of E1A in MAV-1 replication. MAV-1 E1A mutants expressing no E1A protein grew to titers comparable to wild-type MAV-1 titers on mouse fibroblasts (3T6 fibroblasts and fibroblasts derived from Rb+/+, Rb+/−, and Rb−/− transgenic embryos). To test the hypothesis that E1A could induce a quiescent cell to reenter the cell cycle, fibroblasts were serum starved to stop DNA replication and cellular replication and then infected with the E1A mutant and wild-type viruses. All grew to equivalent titers. Steady-state levels of MAV-1 early mRNAs (E1A, E1B, E2, E3, and E4) from 3T6 cells infected with wild-type or E1A mutant virus were examined by Northern analysis. Steady-state levels of mRNAs from the mutant-infected cells were comparable to or greater than the levels found in wild-type virus infections for most of the early regions and for two late genes. The E2 mRNA levels were slightly reduced in all mutant infections relative to wild-type infections. E1A mRNA was not detected from infections with the MAV-1 E1A null mutant, pmE109, or from infections with similar MAV-1 E1A null mutants, pmE112 and pmE113. The implications for the lack of a requirement of E1A in cell culture are discussed.
Early region 1A (E1A) produces a single transcript from the left end of the mouse adenovirus type 1 (MAV-1) genome (2). This transcript encodes a single 200-amino-acid (aa) protein that has an apparent molecular mass of approximately 30 kDa (2, 62). The MAV-1 E1A protein has three conserved regions found in most human adenovirus (hAd) 289-aa E1A proteins (49). MAV-1 E1A is approximately 40% similar to the hAd4, hAd5, and hAd7 289-aa E1A proteins within these three regions (3). Conserved region 2 (CR2) of MAV-1 E1A has the highest degree of similarity with other E1A proteins: it contains a pocket protein binding domain (DLXCXE) found not only in all hAd E1As but also in the transforming proteins of other DNA tumor viruses, such as the simian virus 40 large T antigen and the human papillomavirus E7 proteins (4, 13). This pocket protein binding domain is required by MAV-1 E1A to associate with mouse pRb and mouse p107 (62). CR1 also enhances binding of pRb and p107 to MAV-1 E1A but is not absolutely required.
hAds encode two major E1A proteins, a 243-aa and a 289-aa protein, which both contain CR1 and CR2. The 289-aa protein contains an additional internal 46-aa designated CR3. CR1 and CR2 are the regions of hAd E1A involved in its association with many cellular proteins, including the pRb oncoprotein and related members p107 and p130 (reviewed in references 42 and 48). Induction of cellular DNA synthesis and cell proliferation by either the 243- or the 289-aa E1A protein correlates with the ability of E1A proteins to bind to cell cycle regulators. E1A induces host cell DNA synthesis in mouse cells arrested in G0 by serum starvation (11) and in quiescent mouse 3T3 fibroblasts (65). Quinlan and Grodzicker showed that both cellular DNA synthesis and cell proliferation are induced by the 243-aa E1A in baby rat kidney epithelial cells (55). However, all of the above-mentioned experiments were done with cells from a nonpermissive host for hAds. In growth-arrested human lung fibroblasts, the natural target for some adenovirus infections in humans, the 243-aa E1A protein is required for maximal viral DNA replication, suggesting a biological function for this protein in the cells of the natural host (64).
Induction of cellular DNA synthesis and cell proliferation by the association of E1A with pRb and p107 can be explained by the subsequent dissociation of pRb and p107 from E2F, a transcription factor required for entry into S phase of the cell cycle (52). pRb and p107 are found in complexes with different E2F transcription factors. The association of E1A with pRb or p107, which occurs during an infection, results in an increase of free E2F transcription factors. E2F is then able to bind to and activate transcription from the promoters of many genes required for DNA replication and entry into S phase, such as the genes encoding thymidine kinase, dihydrofolate reductase, and DNA polymerase α (reviewed in references 38, 48, and 52). It is postulated that in quiescent cells, this virus-host protein interaction creates a more favorable environment for virus replication (71).
The leftmost portion (0 to 16.5 map units [m.u.]) of the MAV-1 genome, which contains both the E1A and E1B genes, as well as the polypeptide IX gene and part of the IVa2 gene, can transactivate a hAd E3 promoter in mouse cells (3). Even the segment from 0 to 6.0 m.u. (encompassing E1A and E1B) is capable of this transactivating activity. However, when the first 0.3 m.u. is removed, this region no longer transactivates the hAd E3 promoter. This may be due to inefficient transcription of E1A mRNA in the absence of critical upstream elements such as the CCAAT box (located at nucleotide [nt] 55; −54 with respect to the start of transcription). hAd E1A is both a transactivator of viral (E1B, E2, E3, E4, and major late promoter) (7, 30) and cellular (32, 66) promoters and a repressor of cellular enhanced transcription (27, 67, 70). Repression of cellular enhanced transcription requires the amino terminus and CR1 of hAd E1A. The transactivation function depends on the 289-aa E1A CR3, which has two domains. The amino-terminal region of CR3 contains a zinc finger motif required for the transactivation function of hAd E1A. The carboxy-terminal region of CR3 contains a domain required for association with various transcription factors such as ATF-2 (40, 41), E2F (35), E4F (56), TFIIIC (22), Oct-4 (59), p300 (16, 18, 72, 73), TATA box binding protein (TBP)-associated factor TAFII135 (46), and TBP (10, 23, 39). hAd E1A does not bind DNA directly (17) but is thought to act by bringing together sequence-specific transcription factors with the basal transcription machinery (e.g., TBP) in the vicinity of promoters. MAV-1 E1A CR3 has the amino-terminal zinc finger motif but does not have the second consensus sequence at the carboxy terminus (2).
Using MAV-1 viruses with mutations in E1A, we studied the role of E1A during infection in mouse cells. The data show that E1A is not necessary for a productive infection in mouse 3T6 cells or embryonic fibroblasts. MAV-1 wild-type (wt) or E1A mutant viruses did not shut down host cell translation at late times in a productive infection.
MATERIALS AND METHODS
Cells and viruses.
Mouse 3T6 fibroblast cells and the E1A-expressing 37.1 cell line (62) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% heat-inactivated calf serum. For serum starvation, cells were grown in DMEM supplemented with 0.2% heat-inactivated calf serum. Viral mutants were derived from MAV-1 as described previously (62). pmE301 has a single point mutation in the first intron of E3 that eliminates the EcoRI site while maintaining the amino acid sequence of pVIII. This virus behaves like wt MAV-1 in all characteristics assayed to date (63) and is referred to as wt MAV-1 throughout this work. pmE109 has a single point mutation in the E1A initiator codon (ATG to TTG) (62). The E1A deletion mutants dlE102, dlE105, and dlE106 have all been described elsewhere (62). Briefly, they are as follows: dlE105 (CR1 deletion) lacks nt 383 to 514 (aa 36 to 77) (GenBank accession no. M22245) in the first exon of the E1A transcription unit, dlE102 (CR2 deletion) lacks nt 611 to 667 (aa 112 to 128), and dlE106 (CR3 deletion) lacks nt 682 to 742 (aa 134 to 153). pmE112 and pmE113 have point mutations changing the initiator methionine ATG codon to CAC and were constructed in a manner similar to that described for the other E1A mutants (62). Briefly, oligonucleotide-directed mutagenesis was performed with a single-stranded template containing the MAV-1 E1A sequence and a mixture of two oligonucleotides, MAVL281CAC3 (5′GCG ATT TTT CGA CTT TTG ACT CAC ACC CGC GGC TC 3′) and MAVL281CAC4 (5′GAC TCA CAC CCG CGG CTC CTA CGT 3′) (nucleotide changes from wt indicated in bold), differing only in the amount of sequence flanking the mutagenic site. The oligonucleotides also introduced an SstII site to aid in the screening of plaques for mutant virus. These nucleotide changes result in a methionine-to-histidine change and a serine-to-proline change (aa 1 and 2, respectively). The ATG mutation was transferred as a HindIII-BglII fragment to pMXD (3) which contains the rest of the E1A sequence. This new plasmid, pMXDcacc, was digested with HindIII and XbaI, treated with EcoRI methylase, and cotransfected into 37.1 cells along with pmE301 viral DNA that had been digested with EcoRI and then end filled with Klenow polymerase as described previously (62). Mutant virus was purified by three rounds of plaque purification and confirmed by sequencing. All virus stocks were grown and titers were determined on 37.1 cells. Twenty-four hours before infection, 37.1 cells were treated with 10−5 M dexamethasone to induce E1A expression.
Primary mouse embryo fibroblasts were prepared from E12.5 embryos derived from a pregnant Rb+/− female that had been mated with a Rb+/− male. Genotypes of the individual embryos were determined by PCR amplification using Rb-specific primers (26). Cells were passaged once prior to infection.
Metabolic labeling.
For both the immunoprecipitation and protein analysis experiments, 3T6 cells grown on 60-mm-diameter plates were infected with viruses at a multiplicity of infection (MOI) of 5 PFU/cell. After 1 h of virus adsorption at 37°C, cells were incubated with 1× DMEM containing 2% heat-inactivated calf serum. At the specified times postinfection (p.i.), the cells were washed three times with phosphate-buffered saline and labeled for 3 h with 3 ml of medium lacking methionine and containing 30 μCi of Tran 35S-label (ICN) per ml and 2% dialyzed fetal calf serum. Cells were then scraped in their medium, pelleted, and washed twice in cold phosphate-buffered saline by low-speed centrifugation. Cells (1.2 × 106) were resuspended in 50 μl of 1× sodium dodecyl sulfate (SDS) sample buffer (37) and boiled for 5 min, and then 15-μl aliquots of the samples were resolved by polyacrylamide gel electrophoresis (PAGE) on an SDS-polyacrylamide gel. Protein bands were visualized with a PhosphorImager and ImageQuant software (Molecular Dynamics). Anti-MAV-1 E1A antiserum was used for immunoprecipitation of MAV-1 E1A from the infected cell lysates (62).
Viral growth curves.
Cells were infected at an MOI of 5 and harvested at various times p.i. by scraping in their medium. The cell suspension was subjected to three cycles of freezing and thawing before the cell debris was centrifuged out of the supernatant. Tenfold serial dilutions of supernatants taken from infected cells at the indicated times were plated in triplicate on 3T6 or 37.1 cells, and plaque development was monitored.
Northern blot analysis.
3T6 cells grown on 150-mm-diameter plates were infected with viruses at an MOI of 1, 5, or 10. At 1 h p.i., the inoculum was replaced with 1× DMEM supplemented with 2% heat-inactivated calf serum. At 22 h p.i., cells were harvested and total cytoplasmic RNA was isolated from mock- or virus-infected cells by Nonidet P-40 lysis and phenol-chloroform extraction as described previously (7). RNA samples (20 μg) were resolved by electrophoresis through denaturing formaldehyde–1.2% agarose gels and transferred to nitrocellulose. Blots were hybridized and washed as described previously (2). Probes were random-primer-labeled DNAs as follows. Plasmid pMHF-2B contains MAV-1 DNA sequence from nt 1 to 360 in the E1A region and was derived from pMHF-2 (3) by digestion with BamHI and religation. pHSP23 contains MAV-1 DNA sequence from nt 1430 to 1647 of the E1B coding region and was constructed by ligation of the smallest HindIII-SstI fragment of pZ136 (2) into pATH23 (34). pH61 contains MAV-1 E2A DNA sequence from 64 to 66.3 m.u. inserted into pBluescriptIISK+ and was constructed by ExoIII deletion of a BamHI (64 m.u.)-HindIII (77 m.u.) fragment cloned in pBluescript IISK+. The right end of the fragment in pH61 at 66.3 m.u. corresponds to nt 715 in GenBank accession no. U23770. pZU14 is a class 1 E3 cDNA clone (5) that hybridizes to E3, fiber, and pVIII messages. pZ483 is an E4 cDNA clone with MAV-1 sequence from 88.3 to 96 m.u. This cDNA corresponds to those cDNAs which can encode a protein consisting of the a/b open reading frame (36). To control for sample loading efficiency, a 28S rRNA-specific oligonucleotide (5′AACGATCAGAGTAGTGGTATTTCACC3′) was labeled with [γ-32P]ATP and used as a probe. Relative amounts of each message were quantitated by PhosphorImager analysis of blots. Before reprobing, blots were stripped by incubating twice for 10 min in boiling water.
RESULTS
E1A protein production from MAV-1 E1A mutant viruses.
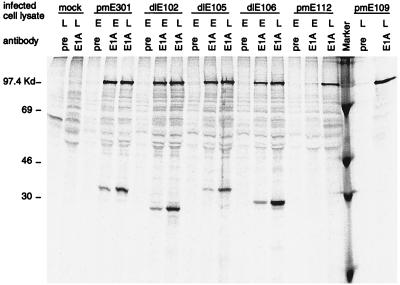
Mouse 3T6 cells were infected with either wt or E1A mutant virus at an MOI of 5 PFU/cell and radioactively labeled with [35S]methionine and [35S]cysteine for 4 h prior to harvesting at 20 h (early) or 42 h (late) p.i. The ability of each of the mutants to produce E1A protein was tested by immunoprecipitating E1A from these infected lysates with anti-MAV-1 E1A antiserum. Wild-type (pmE301)-infected cells produced an E1A protein that migrated slightly slower than the 30-kDa marker (Fig. 1). This protein was precipitated at both early and late times p.i. The CR2 deletion (dlE102) and CR3 deletion (dlE106) mutants produced E1A proteins that migrated faster than the wt E1A protein, as expected for deletions of 19 and 20 aa, respectively. The CR1 deletion (dlE105) mutant, however, produced an E1A protein that migrated anomalously slowly for a deletion of 39 aa. This was reported previously for this mutant (62) as well as a hAd E1A CR1 deletion mutant (16) and is probably due to the amino acid sequence in this region. The two initiator codon mutants, pmE112 and pmE109, produced no detectable E1A protein at either early or late times p.i. All of the mutants that produced an E1A protein did so at levels comparable to those for wt MAV-1.
FIG. 1.
Expression of E1A protein in wt- and mutant-infected mouse 3T6 cells. Mouse 3T6 cells were infected with either wt virus (pmE301) or the indicated mutant virus at an MOI of 5, or they were mock infected. [35S]methionine- and [35S]cysteine-labeled infected cell lysates were harvested at 22 h (early [E]) and 40 h (late [L]) p.i. and immunoprecipitated with an anti-MAV-1 E1A antibody (E1A) or preimmune serum (pre). Immunoprecipitated products were electrophoresed on an SDS–12% polyacrylamide gel followed by phosphorimager analysis. Marker designates 14C-labeled protein standards, whose sizes are indicated on the left.
E1A is not required for efficient viral replication in dividing or serum-starved mouse 3T6 cells or pRb− transgenic embryo fibroblasts.
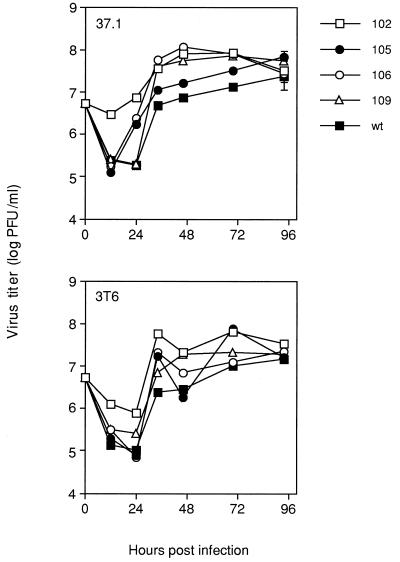
hAd E1A deletion mutants replicate to much lower levels than wt in cell culture (7, 30). This has been attributed to the role of E1A in transactivating other adenovirus early proteins that are required for efficient virus replication. The ability of MAV-1 E1A mutants to grow in mouse fibroblasts was compared to that of wt virus to determine the importance of MAV-1 E1A in viral replication. Virus titers were determined on both 3T6 and 37.1 cells (37.1 is a 3T6 cell line expressing MAV-1 E1A under the control of an inducible promoter [62]) (Fig. 2). Wild-type virus titers were approximately 2 × 107 PFU/ml at 96 h p.i. on 3T6 cells. The titers of the mutants were never more than 1 log different from the wt titer at 96 h p.i. The same result was found for 37.1 cells. Therefore, deletion of CR1, CR2, or CR3 of E1A did not have a dramatic affect on the ability of MAV-1 to replicate in mouse 3T6 cells. Most surprisingly, the initiator codon mutant, pmE109, from which no detectable E1A protein was produced, grew to wt titers in 3T6 and 37.1 cells.
FIG. 2.
Growth curve of wt and mutant viruses on mouse fibroblasts. Mouse 3T6 or 37.1 cells were infected with the indicated viruses at an MOI of 5. Virus growth was monitored through 96 h p.i. by determining titers on 37.1 cells. Two assays of the titers at the 96-h time point were done, and the standard error bars determined from multiple plaque assay plates are indicated. The standard errors for the stocks grown on 3T6 cells are not visible because they are so small, ranging from 2.0 × 106 to 1.4 × 107.
The ability of E1A mutant viruses to grow in 3T6 cells may be explained by the fact that these are established cell lines that are continually dividing. We reasoned that if the virus requires E1A to induce DNA replication during an infection, then these continually dividing cells would not stringently test such a requirement. Therefore, we serum starved 3T6 cells and then infected them at an MOI of 5 with wt or E1A mutant virus. Serum starvation for 3 days resulted in a significant decrease in both cellular replication and DNA synthesis as assayed by cell counts and [3H]thymidine incorporation (data not shown). Serum starvation had no effect on the ability of any of the viruses to grow in 3T6 cells (Fig. 3). There was never more than 1 log difference in virus titer between mutant and wt viruses.
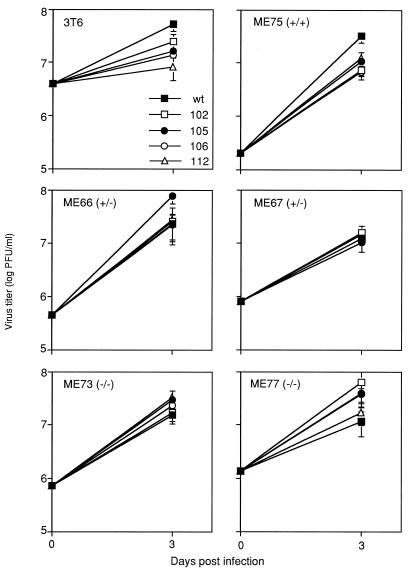
FIG. 3.
Virus titers on serum-starved mouse cell lines. Monolayers of mouse 3T6 cells or secondary cells derived from embryos of Rb+/+, Rb+/−, or Rb−/− knockout mice were serum starved for 3 days prior to infection. Virus titers were determined on day 3 p.i. for wt and mutant MAV-1. The cell type is given in the upper left corner of each panel. Two independent Rb+/− cell strains and two independent Rb−/− cell strains were tested.
MAV-1 E1A interacts with mouse pRb and effectively inactivates it (62). Therefore, we compared the ability of the E1A mutants and wt virus to grow on cells derived from embryos of Rb transgenic mice (26). Cells with Rb+/+, Rb+/−, and Rb−/− genotypes were infected with wt and E1A mutant viruses, and there was no difference in the ability of any of the mutants to grow in comparison to wt virus (data not shown). In another experiment, these cells were serum starved 3 days prior to infection with the viruses, and the E1A mutants were able to grow to titers comparable to wt virus titers (Fig. 3).
E1A is not required for the transactivation of other MAV-1 early promoters during an infection of mouse 3T6 cells.
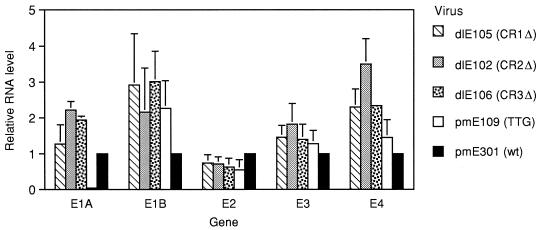
One of the first functions attributed to hAd E1A was its ability to transactivate other early adenovirus promoters, such as E1B, E2, and E3 (7, 29). Genome sequences from the left end of the MAV-1 genome transactivate a heterologous E3 promoter from hAd5 (3), suggesting that MAV-1 E1A may also have the ability to transactivate MAV-1 early promoters. Using E1A viral mutants, we examined the ability of wt and mutant E1A proteins to transactivate homologous MAV-1 promoters. Northern analysis was done on 3T6 cells infected with wt or E1A mutant virus to determine the effect of E1A mutations on viral early gene mRNA levels. Levels of viral mRNA isolated at early times p.i. for early genes E1A, E1B, E2, E3, and E4 were assayed and quantified by using gene-specific probes as described in Materials and Methods (Fig. 4). Relative mRNA levels were compared to the levels found in wt-infected cells, which were defined as 1 for each message. The most dramatic difference in mRNA levels compared to wt virus was found in an infection with the initiator codon mutant, pmE109, where there was no detectable E1A mRNA produced. The pmE109 mutation is a single-nucleotide point mutation that changes the initiator codon from ATG to TTG. Results from both Western analysis (62) and immunoprecipitation (Fig. 1) with anti-E1A antiserum have shown that no detectable E1A protein is produced from pmE109. Additional Northern analysis was done with two independent isolates of a different initiator codon mutant (pmE112 and pmE113; ATG→CAC [data not shown]). Similar to the results with pmE109, little or no E1A mRNA was detected at early or late times p.i. with pmE112 and pmE113, while other early mRNA (E2, E3, and E4) levels were comparable to the levels made in the other initiator mutant (pmE109) and wt virus.
FIG. 4.
Northern analysis of MAV-1 early mRNAs from wt- or mutant-infected 3T6 cells. Total cytoplasmic RNA was isolated at 20 h p.i. from mouse 3T6 cells infected with wt virus and each of the indicated E1A mutant viruses at an MOI of 5. Steady-state levels of E1A, E1B, E2, E3, and E4 mRNAs were determined by using gene-specific probes and were normalized to 1 for wt levels. The results represent the means of three independent experiments.
E1A message levels from the CR1, CR2, and CR3 deletion mutants were comparable to or greater than the levels in wt-infected cells. In fact, the E1A mRNA levels were significantly higher than wt levels in the CR2 and CR3 deletion mutant infections as determined by Student’s t test (t values of 8.93 and 14.33, respectively; P < 0.001). Every E1A mutant infection had steady-state E1B and E3 mRNA levels comparable to or greater than those in wt-infected cells. The E2 message levels were slightly reduced in all mutants relative to the E2 message levels in a wt infection. Steady-state levels of E4 mRNA were consistently higher in CR1, CR2, and CR3 deletion mutant infections in comparison to wt infections (t values of 4.3, 6.2, and 30.7; P < 0.05, 0.05, and 0.001, respectively), but they were comparable to wt levels in the TTG mutant infections. Two late-specific mRNAs (pVIII and fiber) were also examined in cells infected with each of the mutants, and the mRNA levels were comparable to the levels made during a wt infection (data not shown).
MAV-1 does not exhibit host translation shutoff at late times in infection.
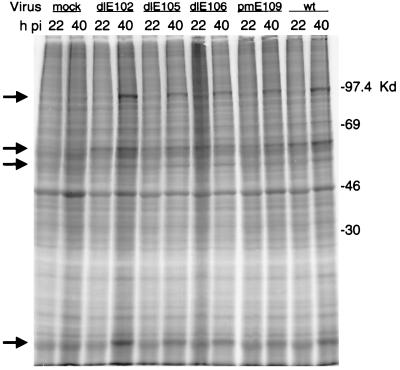
To determine whether E1A had any detectable effects on late viral protein synthesis or host cell shutoff, we examined protein expression at late times in mouse 3T6 cells infected with either wt or mutant virus. Cells were labeled and harvested at 22 and 40 h p.i., and cell lysates were analyzed by SDS-PAGE. When the mock-infected cell lysates were compared to the wt-infected cell lysate, no reduction in host cell-specific proteins was evident at either time p.i. (Fig. 5). There was also no detectable decrease in host protein expression between 22 and 40 h p.i. in the wt- or mutant-infected cells. Levels of predominant late infection-specific proteins were similar in wt- and mutant-infected cells.
FIG. 5.
Viral and host protein production at early (22 h p.i.) and late (40 h p.i.) times of infection of 3T6 cells with wt or mutant virus. [35S]methionine- and [35S]cysteine-labeled cellular extracts were harvested from mock-, wt-, or mutant-infected cells at 22 or 40 h p.i. as indicated and analyzed by SDS-PAGE (10% polyacrylamide gel). The migration of protein size markers is indicated on the right. Several virus-infected cell-specific proteins are indicated by the arrows at the left.
DISCUSSION
Using MAV-1 mutants with deletions in the E1A region, we have shown that E1A is not required for a productive infection in mouse 3T6 cells. An E1A null mutant in which the initiator codon ATG was changed to TTG produced no E1A protein yet grew to titers no more than 1 log different from wt titers when initial MOIs were 5 (Fig. 2). Other E1A mutants, with deletions of each of the three conserved regions, also grew to wt titers. The ability of E1A mutants to achieve the same titers as wt may be because the infections at an MOI of 5 are effectively high-multiplicity infections. High-multiplicity infection with a hAd E1A mutant results in early gene expression (MOI of 200 [53]) and the production of viral progeny (MOI of 800 [60]). This is probably due to low-level transcription occurring from the large number of mutant DNA templates. These same mutants replicate to significantly lower titers than wt virus on most cell lines tested (7, 30). Because transcription occurs from hAd early genes in the absence of E1A, E1A is not essential for transactivation of these genes, although it does enhance transcription. Imperiale et al. suggested that the growth rate of a cell is important for early gene expression (25), based on measurements of E2 expression during an infection with hAd dl312 at high multiplicities in various cell lines. Only human tumor cell lines or an undifferentiated stem cell line supported early viral gene expression in the absence of E1A (25). If the requirement for E1A is to induce the cell to reenter the cell cycle and begin DNA synthesis and cell proliferation, then it is possible that in growing, dividing cells in culture, the E1A protein is not required. Therefore, we serum starved the 3T6 cells to significantly slow down growth and DNA synthesis to determine if the rate of cell growth was a factor during an MAV-1 infection. E1A mutants grew to the same titers as wt virus in serum-starved cells (Fig. 3). Even in growth-arrested, serum-starved 3T6 cells, E1A was not a requirement for efficient virus replication. The same results were obtained from cells derived from Rb transgenic embryos. Mutant viruses grew to titers comparable to wt on cells derived from Rb+/+, Rb+/−, and Rb−/− embryos, with or without serum (Fig. 3). One possible explanation for this is that these cell lines may have an E1A-like activity that enables the virus to replicate in the absence of a virally encoded E1A protein.
Early gene expression was examined by Northern analysis of 3T6 cells infected with each of the mutants. The only early gene expression dramatically affected by E1A mutation was that of the E1A mRNA itself in the E1A null mutants. These mutants have a mutation that changes the initiator codon from ATG to TTG (pmE109) or to CAC (pmE112 and pmE113), leaving the rest of the gene intact. There are several possible explanations for the absence of detectable E1A mRNA during infection with these initiator mutants. MAV-1 E1A may be autoregulatory, such that in the absence of E1A protein, E1A mRNA is transcribed poorly if at all. If this is so, then such an autoregulatory function cannot be assigned to any one of the three MAV-1 E1A conserved regions, since the deletion mutants each produce equivalent or greater levels of E1A mRNAs compared to wt. Another possibility is that due to the presence of premature termination codons in the E1A coding region, the E1A mRNA becomes extremely unstable and is rapidly degraded. Such instability of mRNA in the absence of translation of eukaryotic proteins has been reported (reviewed in references 20 and 43).
Except for the E1A expression in E1A null mutants noted above, most of the early gene regions assayed had steady-state levels of mRNA in mutant-infected 3T6 cells comparable to or greater than those found in wt-infected cells. The steady-state levels of E4 mRNAs were significantly higher in the CR1, CR2, and CR3 deletion mutants compared to wt. E1A mRNA levels in the CR2 and CR3 mutants were also significantly higher than the wt E1A mRNA levels. One possible explanation for this result is that deletion of large regions (19, 20, and 39 aa) of E1A causes a significant change in the conformation of the protein. This could lead to an increase in transcription directly or indirectly by causing a nonspecific or fortuitous interaction (e.g., with E4- and E1A-specific transcription factors or enhancer binding proteins) that does not normally occur with the full-length E1A protein. Alternatively, wt MAV-1 E1A may act as a repressor of enhanced transcription from the E4 and E1A promoters. A conformational change in E1A could cause inactivation of a transcriptional repressor, resulting in an increase of transcription. hAd E1A has been shown to repress transcription from viral (8, 69, 70) and cellular enhancers (21, 27, 67, 68). In the presence of MAV-1 E1A, enhanced transcription of certain MAV-1 early genes, including E1A itself, could be repressed, but with an E1A mutant this repression is relieved. Potential enhancer sequences have not been identified for MAV-1 early genes, and it is possible that enhancers that are repressed by MAV-1 E1A exist in the genome of MAV-1.
There are at least two different models that can explain why E1A is not needed for transactivation of these early viral genes in 3T6 cells. It has been proposed that there exists in some cell lines an inhibitor of viral early gene transcription that hAd E1A is capable of inactivating, allowing transcription to occur. This hypothesis came from the observation that inhibition of protein synthesis prior to and during infection with hAd E1A mutants resulted in the partial activation of viral gene expression (33, 53). It is possible that mouse 3T6 cells and mouse embryonic fibroblasts lack such a cellular factor that inhibits adenovirus early transcription in the absence of E1A. It may be that endothelial cells and lymphoid cells, the targets of MAV-1 during an infection in the mouse (31), do contain this inhibitor. This could explain the significant decrease in the pathogenicity of MAV-1 mutants in mice and the decrease in the amount of virus found in the organs of mice infected with low doses of mutant virus in comparison to the levels of wt virus (61). It has also been proposed by Imperiale et al. that certain cell lines may express a cellular E1A-like activity, allowing the virus to express early regions even in the absence of E1A (25). Mouse 3T6 cells and primary embryo fibroblasts may contain a cellular MAV-1 E1A-like activity that could enable the E1A null mutants to replicate as efficiently as wt virus.
It was surprising that MAV-1 E1A mutants, unlike those of hAds, did not reduce steady-state levels of early mRNAs, since in hAds E1A is involved in transactivation of the early viral genes. The fact that the left end of the MAV-1 genome is capable of transactivating a heterologous promoter in mouse 3T6 cells (3) yet is not necessary for transactivation of its own promoters during an infection (Fig. 4) may be due to differences in promoter sequence and/or transcription factor requirements. It may also be due to differences in effective levels of E1A proteins and target promoters in infection compared to transfection. The MAV-1 sequences that the hAd5 E3 promoter requires for transactivation in transfected mouse cells are evidently different from those required by the MAV-1 early promoters in an infection.
In addition to the interactions of E1A with host proteins that both regulate viral and cellular promoters and inactivate certain cellular proteins, adenoviruses manipulate their environment by causing a shutdown in host cell translation at late times in infection so that viral mRNAs are preferentially expressed (reviewed in references 1, 74, and 75). We investigated the ability of mutant and wt MAV-1 to synthesize late viral proteins and to inhibit host translation during infection. No effect on viral late protein synthesis was seen, nor was a decrease in the production of cellular proteins observed at either early or late times in infection (Fig. 5). There are currently two known ways that adenovirus blocks cellular translation. The first to be described is the ability of the virus-associated (VA) RNAs to inactivate protein kinase R (PKR) in the vicinity of viral mRNA (reviewed in reference 45). PKR is activated in the presence of double-stranded RNA or interferon, both of which are abundant during a hAd infection. Activated PKR phosphorylates eIF-2α, an initiation factor required for translation initiation, thereby inactivating it. VA RNAs, however, have the ability to inactivate PKR by preventing phosphorylation of eIF-2 (44, 54). VA RNAs copurify with viral mRNAs, and it has been proposed that this allows translation to occur preferentially in the vicinity of viral mRNA but not cellular mRNA (57, 58). The entire MAV-1 genome sequence has recently been completed, and no VA RNA equivalent was found by sequence analysis or biochemical assays (47). This lack of VA RNA in MAV-1 may contribute to the inability of this virus to shut down host cell translation.
A second mechanism used by adenoviruses to regulate translation during an infection is inactivation of the cap binding protein complex (eIF-4F). At late times in infection, eIF-4F, composed of eIF-4A, eIF-4E, and p220, becomes inactivated due to hypophosphorylation of eIF-4E (24). eIF-4F is required for initiation of translation from capped mRNAs. All late hAd mRNAs have an ∼200-nt 5′ noncoding region, the tripartite leader, which allows them to be translated in a cap-independent manner (6, 14, 15, 28). It is currently unknown what hAd factor is responsible for the inactivation of eIF-4F, although it is believed to be a late-specific factor since host cell translation shutoff occurs only during the late phase of infection. In mouse 3T6 cells, MAV-1 does not shut off translation of host mRNAs. MAV-1 does not appear to need preferential translation of viral mRNAs for efficient virus production. Alternatively, MAV-1 may require continued synthesis of some host protein for efficient replication and therefore does not shut off any translation in order to produce such a host protein. Two cellular proteins known to augment hAd DNA replication are nuclear factors I and III (9, 12, 19, 50, 51).
Our studies revealed a significant lack of requirement for E1A during MAV-1 infection of mouse 3T6 cells. Virus titers were not significantly reduced and expression of the other early viral regions was not decreased in the absence of E1A. E1A is a highly conserved gene among the adenoviruses, and adenovirus genomes are compact. This makes it highly unlikely that MAV-1 encodes an unnecessary E1A gene. The primary targets for MAV-1 in the mouse are endothelial and lymphoid cells (31). Thus, the ability of E1A mutants to efficiently grow and express the early viral genes at wt levels in 3T6 fibroblasts may not adequately reflect what actually occurs during an infection with these mutants in mice. 3T6 cells may have an activity that replaces the need for E1A in cell culture, whereas endothelial cells or macrophages may not have this activity. E1A null mutants are significantly less pathogenic in mice than the wt virus, supporting the hypothesis that E1A is an essential gene in the animal (61). There is a requirement for 100,000 times more mutant virus than wt virus in mice to produce the same pathology (61). This may be due to inefficient transcription of the early genes in the absence of E1A in the organs of the mouse. Viral mRNA levels have not been examined in mice infected with these mutant viruses. Steady-state levels of early gene mRNAs may be significantly reduced in the organs of mice infected with these mutants. The decreased virulence of MAV-1 E1A mutants in mice may also be due to requirements for virus-host protein interactions. E1A may also help the virus escape the immune response. Future studies of these E1A mutant viruses in cell culture and mice will help elucidate the role of MAV-1 E1A in viral pathogenesis.
ACKNOWLEDGMENTS
We thank Andy High for technical assistance with the Northern analysis of the pmE112 and pmE113 mutants. We also thank Tyler Jacks and Lee Remington for providing Rb+/− mice and for advice in their use in preparation of primary cells. We thank Lois Miller, Bob Ivarie, and the Spindler lab members for comments on the manuscript, and we thank Gwen Hirsch and Liz LaRue for technical assistance throughout this work.
This work was supported by NIH grant AI23762 and American Cancer Society grant VM-176 to K.R.S. and by NIH predoctoral traineeship GM 07103 to K.S. K.R.S. is the recipient of an NIH Research Career Development Award.
REFERENCES
- 1.Babich A, Feldman C T, Nevins J R, Darnell J E, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol Cell Biol. 1983;3:1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball A O, Beard C W, Redick S D, Spindler K R. Genome organization of mouse adenovirus type 1 early region 1: a novel transcription map. Virology. 1989;170:523–536. doi: 10.1016/0042-6822(89)90444-3. [DOI] [PubMed] [Google Scholar]
- 3.Ball A O, Williams M E, Spindler K R. Identification of mouse adenovirus type 1 early region 1: DNA sequence and a conserved transactivating function. J Virol. 1988;62:3947–3957. doi: 10.1128/jvi.62.11.3947-3957.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa M S, Edmonds C, Fisher C, Schiller J T, Lowy D R, Vousden K H. The region of the HPV E7 oncoprotein homologous to adenovirus E1A and SV40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. EMBO J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beard C W, Ball A O, Wooley E H, Spindler K R. Transcription mapping of mouse adenovirus type 1 early region 3. Virology. 1990;175:81–90. doi: 10.1016/0042-6822(90)90188-w. [DOI] [PubMed] [Google Scholar]
- 6.Berget S M, Moore C, Sharp P A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berk A J, Lee F, Harrison T, Williams J, Sharp P A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979;17:935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- 8.Borelli E, Hen R, Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transcription. Nature. 1984;312:608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- 9.Bosher J, Robinson E C, Hay R T. Interactions between the adenovirus type 2 DNA polymerase and the DNA binding domain of nuclear factor 1. New Biol. 1990;2:1083–1090. [PubMed] [Google Scholar]
- 10.Boyer T G, Berk A J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993;7:1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- 11.Braithwaite A W, Cheetham B F, Li P, Parish C R, Waldron-Stevens L K, Bellett A J D. Adenovirus-induced alterations of the cell growth cycle: a requirement for expression of E1A but not of E1B. J Virol. 1983;45:192–199. doi: 10.1128/jvi.45.1.192-199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Mermod N, Horwitz M S. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor-I mediate formation of the DNA replication preinitiation complex. J Biol Chem. 1990;265:18634–18642. [PubMed] [Google Scholar]
- 13.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large T antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 14.Dolph P J, Huang J, Schneider R J. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64:2669–2677. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolph P J, Racaniello V, Villamarin A, Palladino F, Schneider R J. The adenovirus tripartite leader eliminates the requirement for cap binding protein complex during translation initiation. J Virol. 1988;62:2059–2066. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egan C, Jelsma T N, Howe J A, Bayley S T, Ferguson B, Branton P E. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson B, Krippl B, Andrisani O, Jones N, Westphal H, Rosenberg M. E1A 13S and 12S mRNA products made in Escherichia coli both functions as nucleus-localized transcription activators but do not directly bind DNA. Mol Cell Biol. 1985;5:2653–2661. doi: 10.1128/mcb.5.10.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Whyte P, Franza B R J, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfield L, Hearing P. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J Virol. 1993;67:3931–3939. doi: 10.1128/jvi.67.7.3931-3939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hearing P, Shenk T. Sequence-independent autoregulation of the adenovirus type 5 E1A transcription unit. Mol Cell Biol. 1985;5:3214–3221. doi: 10.1128/mcb.5.11.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hen R, Borelli E, Chambon P. Repression of the immunoglobulin heavy chain enhancer by the adenovirus-2 E1A products. Science. 1985;230:1391–1394. doi: 10.1126/science.2999984. [DOI] [PubMed] [Google Scholar]
- 22.Hoeffler W K, Kovelman R, Roeder R G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 23.Horikoshi N, Maguire K, Kralli A, Maldonado E, Reinberg D, Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 1991;88:5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Schneider R J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- 25.Imperiale M J, Kao H-T, Feldman L T, Nevins J R, Strickland S. Common control of the heat shock gene and early adenovirus genes: evidence for a cellular E1A-like activity. Mol Cell Biol. 1984;4:867–874. doi: 10.1128/mcb.4.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 27.Janaswami P M, Kalvakolanu D V R, Zhang Y, Sen G C. Transcriptional repression of interleukin-6 gene by adenoviral E1A proteins. J Biol Chem. 1992;267:24886–24891. [PubMed] [Google Scholar]
- 28.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones N, Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 30.Jones N C, Shenk T. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajon A E, Brown C C, Spindler K R. Distribution of mouse adenovirus type 1 in intraperitoneally and intranasally infected adult outbred mice. J Virol. 1998;72:1219–1223. doi: 10.1128/jvi.72.2.1219-1223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao H-T, Nevins J R. Transcriptional activation and subsequent control of the human heat shock gene during adenovirus infection. Mol Cell Biol. 1983;3:2058–2065. doi: 10.1128/mcb.3.11.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katze M G, Persson H, Philipson L. Control of adenovirus early gene expression: posttranscriptional control mediated by both viral and cellular gene products. Mol Cell Biol. 1981;1:807–813. doi: 10.1128/mcb.1.9.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koerner T J, Hill J E, Myers A M, Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE-fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 35.Kovesdi I, Reichel R, Nevins J. Identification of a cellular transcription factor involved in E1A transactivation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 36.Kring S C, Ball A O, Spindler K R. Transcription mapping of mouse adenovirus type 1 early region 4. Virology. 1992;190:248–255. doi: 10.1016/0042-6822(92)91211-c. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.La Thangue N B. DP and E1F proteins: components of a heterodimeric transcription factor implicated in cell cycle control. Curr Opin Cell Biol. 1994;6:443–450. doi: 10.1016/0955-0674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 39.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 40.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Green M R. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- 42.Ludlow J W, Skuse G R. Viral oncoprotein binding to pRB, p107, p130, and p300. Virus Res. 1995;35:113–121. doi: 10.1016/0168-1702(94)00094-s. [DOI] [PubMed] [Google Scholar]
- 43.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 44.Mathews M B. Binding of adenovirus VA RNA to mRNA: a possible role in splicing. Nature. 1980;285:575–577. doi: 10.1038/285575a0. [DOI] [PubMed] [Google Scholar]
- 45.Mathews M B, Shenk T. Adenovirus virus-associated RNA and translational control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazzarelli J M, Mengus G, Davidson I, Ricciardi R P. The transactivation domain of adenovirus E1A interacts with the C terminus of human TAFII135. J Virol. 1997;71:7978–7983. doi: 10.1128/jvi.71.10.7978-7983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meissner J D, Hirsch G N, LaRue E A, Fulcher R A, Spindler K R. Completion of the DNA sequence of mouse adenovirus type 1: sequence of E2B, L1, and L2 (18-51 map units) Virus Res. 1997;51:53–64. doi: 10.1016/s0168-1702(97)00079-8. [DOI] [PubMed] [Google Scholar]
- 48.Moran E. Mammalian cell growth controls reflected through protein interactions with the adenovirus E1A gene products. Semin Virol. 1994;5:327–340. [Google Scholar]
- 49.Moran E, Mathews M B. Multiple functional domains in the adenovirus E1A gene. Cell. 1987;48:177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- 50.Mul Y M, van der Vliet P C. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 1992;11:751–760. doi: 10.1002/j.1460-2075.1992.tb05108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mul Y M, Verrijzer C P, van der Vliet P C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 53.Nevins J R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981;26:213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- 54.O’Malley R P, Mariano T M, Siekierka J, Mathews M B. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell. 1986;44:391–400. doi: 10.1016/0092-8674(86)90460-5. [DOI] [PubMed] [Google Scholar]
- 55.Quinlan M P, Grodzicker T. Adenovirus E1A 12S protein induces DNA synthesis and proliferation in primary epithelial cells in both the presence and absence of serum. J Virol. 1987;61:673–682. doi: 10.1128/jvi.61.3.673-682.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raychaudhuri P, Rooney R, Nevins J R. Identification of an E1A-inducible cellular factor that interacts with regulatory sequences within the adenovirus E4 promoter. EMBO J. 1987;6:4073–4081. doi: 10.1002/j.1460-2075.1987.tb02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reichel P A, Merrick W C, Siekierka J, Mathews M B. Adenovirus VA RNA1 regulates the activity of a protein synthesis initiator factor. Nature. 1985;313:196–200. doi: 10.1038/313196a0. [DOI] [PubMed] [Google Scholar]
- 58.Schneider R J, Safer B, Munemitsu S M, Samuel C E, Shenk T. Adenovirus VAI RNA prevents phosphorylation of the eukaryotic initiation factor 2 alpha subunit subsequent to infection. Proc Natl Acad Sci USA. 1985;82:4321–4324. doi: 10.1073/pnas.82.13.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schöler H R, Ciesolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 60.Shenk T, Jones N, Colby W, Fowlkes D. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harbor Symp Quant Biol. 1979;44:367–375. doi: 10.1101/sqb.1980.044.01.041. [DOI] [PubMed] [Google Scholar]
- 61.Smith K, Brown C C, Spindler K R. The role of mouse adenovirus type 1 early region 1A in acute and persistent infections in mice. J Virol. 1998;72:5699–5706. doi: 10.1128/jvi.72.7.5699-5706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith K, Ying B, Ball A O, Beard C W, Spindler K R. Interaction of mouse adenovirus type 1 early region 1A protein with cellular proteins pRb and p107. Virology. 1996;224:184–197. doi: 10.1006/viro.1996.0520. [DOI] [PubMed] [Google Scholar]
- 63.Spindler, K. R. Unpublished observations.
- 64.Spindler K R, Eng C Y, Berk A J. An adenovirus early region 1A protein is required for maximal viral DNA replication in growth-arrested human cells. J Virol. 1985;53:742–750. doi: 10.1128/jvi.53.3.742-750.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stabel S, Argos P, Philipson L. The release of growth arrest by microinjection of adenovirus E1A DNA. EMBO J. 1985;4:2329–2336. doi: 10.1002/j.1460-2075.1985.tb03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein R, Ziff E B. HeLa cell β-tubulin gene transcription is stimulated by adenovirus 5 in parallel with viral early genes by an E1a-dependent mechanism. Mol Cell Biol. 1984;4:2792–2801. doi: 10.1128/mcb.4.12.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stein R W, Ziff E B. Repression of insulin gene expression by adenovirus type 5 E1A proteins. Mol Cell Biol. 1987;7:1164–1170. doi: 10.1128/mcb.7.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Velcich A, Ziff E. Adenovirus E1A proteins repress transcription from the SV40 early promoter. Cell. 1985;40:705–716. doi: 10.1016/0092-8674(85)90219-3. [DOI] [PubMed] [Google Scholar]
- 70.Wang H-G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 72.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 73.Yee S P, Branton P E. Detection of cellular proteins associated with human adenovirus type 5 early region 1A polypeptides. Virology. 1985;147:142–153. doi: 10.1016/0042-6822(85)90234-x. [DOI] [PubMed] [Google Scholar]
- 74.Yoder S S, Robberson B L, Leys E J, Hook A G, Al-Ubaidi M, Yeung C Y, Kellems R E, Berget S M. Control of cellular gene expression during adenovirus infection: induction and shut-off of dihydrofolate reductase gene expression by adenovirus type 2. Mol Cell Biol. 1983;3:819–828. doi: 10.1128/mcb.3.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Schneider R. Adenovirus inhibition of cellular protein synthesis and the specific translation of late viral mRNAs. Semin Virol. 1993;4:229–236. [Google Scholar]