Abstract
Nanosized bioprobes that can highlight diseased tissue can be powerful diagnostic tools. However, a major unmet need is a tool with adequate adhesive properties and contrast-to-dose ratio. To this end, this study demonstrates that targeted superparamagnetic nanoprobes engineered to present a worm-like shape and hydrophilic packaging enhance both adhesion efficiency to target substrates and magnetic resonance (MR) sensitivity. These nanoprobes were prepared by the controlled self-assembly of superparamagnetic iron oxide nanoparticles (SPIONs) into worm-like superstructures using glycogen-like amphiphilic hyperbranched polyglycerols functionalized with peptides capable of binding to defective vasculature. The resulting worm-like SPION clusters presented binding affinity to the target substrate 10-fold higher than that of spherical ones and T2 molar MR relaxivity 3.5-fold higher than that of conventional, single SPIONs. The design principles discovered for these nanoprobes should be applicable to a range of other diseases where improved diagnostics are needed.
Keywords: magnetic resonance imaging, nonspherical nanoparticle cluster, superparamagnetic iron oxide nanoparticle, hyperbranched polyglycerol, targeted imaging
Graphical Abstract
1. INTRODUCTION
Early diagnosis is a critical element in the complete and successful treatment of multiple acute, chronic, and malignant diseases. As such, a highly active but challenging area of research is focused on the development of smart beacons that can anchor exclusively to diseased tissue following systemic administration and provide high contrast bioimages.1–5 Toward this end, nonspherical nanoparticles have emerged as preferred carriers of bioimaging contrast agents because of their ability to adhere to target tissues with affinity greater than that of their spherical counterparts.6,7
A common method of controlling nanoparticle shape is to modulate the intermolecular packing parameter of self-assembling amphiphilic molecules.8–10 However, attaining an optimal packing parameter to achieve a desired, elongated morphology often requires a large hydrophobic component.11–15 As a consequence, the resulting nanoprobes have significantly limited functionality and contrast efficiency because the large hydrophobic layer surrounding the cargo substantially reduces the interaction between the nanoprobe and surrounding biological fluid. This is of particular importance when designing nanoprobes to carry contrast agents for magnetic resonance imaging (MRI), which rely mechanistically on the access of contrast agents to water molecules in the target tissue.16
We hypothesized that an MR-based nanoprobe with a highly elongated morphology, a hydrophilic surface coating, and peptides capable of binding diseased tissue would present superior tissue binding affinity without reducing relaxivity. We examined this hypothesis by changing the shape of clustered superparamagnetic iron oxide nanoparticles (SPIONs), a T2 MR contrast agent, from spherical to nonspherical using amphiphilic hyperbranched polyglycerols (HPGs) with a binary molecular weight distribution. Specifically, HPG with molecular weights of 3000 and 50 000 g/mol were mixed at different ratios to modulate the packing density of the SPIONs, in turn changing the curvature of the cluster. The HPG was specifically chosen to surround the SPION clusters because its unique glycogen-like hyperbranched architecture presents a larger number of hydroxyl groups in a small volume, thus maximizing the hydrophilicity of the polymers. The HPG further provided a platform to which oligopeptides containing the VHPKQHR sequence, termed VHPKQHR peptide, could be chemically coupled. This peptide bears homology to the very late antigen-4 (VLA-4) found on leukocytes and therefore binds with vascular cell adhesion molecule-1 (VCAM-1), which is overexpressed by inflamed endothelial cells.17
We assessed the ability of the bioactive, worm-like SPION nanoclusters to adhere to and serve as nanobeacons of vascular defects by examining the nanocluster morphology with transmission electron microscopy (TEM), measuring the MR relaxivity of phantom models of SPION clusters, and quantifying nanocluster binding kinetics in vitro using surface plasmon resonance (SPR) spectroscopy. Spherical SPION nanoclusters prepared with HPG with a single molecular weight were used as a control.
2. MATERIALS AND METHODS
2.1. Synthesis of Alkylated HPG.
All materials were purchased from Sigma-Aldrich unless otherwise noted. HPG was synthesized and alkylated according to the procedure previously described.18 Characterization of HPG and alkylated HPG is shown in Figures S1 and S2, respectively.
2.2. Conjugation of VCAM-1 Binding Peptides to HPG.
Alkylated HPG, HPG3k-g-C18(3) (107.3 mg, 1 equiv), was dried under high vacuum overnight 1 day before the reaction. Dried HPG was then dissolved in anhydrous DMF (20 mL), to which distilled triethylamine (467.5 μL, 312 equiv) was added. The reaction flask was placed in an ice bath before the addition of methacryloyl chloride to cool the reaction solution. Methacryloyl chloride (359 μL, 156 equiv) was then added dropwise, and the reaction continued to stir at room temperature for 24 h. The reaction of methacryloyl chloride with hydroxyl groups in HPG was monitored by 1H NMR of an aliquot of the reaction mixture. Then, the peptide solution in DMF (137.8 mg, 6 equiv, H2N-VHPKQHRGGSWGC-CONH2, Mimotopes, Victoria, Australia) was added to the reaction flask, and the reaction continued for another 24 h at 60 °C. The thiol group in cysteine reacted with the methacryloyl group on HPG via Michael addition. Afterward, the remaining methacryloyl groups on HPG were quenched with 3-mercaptopropane-1,2-diol. The reaction solution was then dialyzed for 2 days against an aqueous NaCl solution (10 g/L followed by 5 g/L) and then for 1 day against deionized water. The amount of peptide conjugated to HPG3k-g-C18(3) was determined by UV absorbance at 280 nm to be 0.457 mmol/g polymer.
2.3. Formation of SPION Nanoclusters.
Hydrophobic SPIONs stabilized with oleic acid were suspended in chloroform at 10 mg/mL.19 To form nanoclusters, SPIONs were sonicated with varying ratios of alkylated HPG50k to HPG3k dissolved in water. Removal of the chloroform was then accomplished by rotary evaporation, and the resultant clusters were purified from unclustered materials by centrifugation. For the assembly of biofunctionalized SPION clusters, peptide-conjugated HPG3k-g-C18 was incorporated with either HPG50k-g-C18 or unmodified HPG3k-g-C18 at a 3:1 molar ratio to prepare worm-like and spherical clusters, respectively.
2.4. Microscopic Analysis of Nanoclusters.
HPG-SPION nanoclusters were imaged with a JEOL 2100 transmission electron microscope (JEOL 2100 cryo TEM). The accelerating voltage was set at 200 kV, and specimens were air-dried on a holey carbon-coated copper grid. Quantitative analysis was conducted with ImageJ software. Approximately 50 clusters were analyzed per condition. Additionally, to support the stability of worm-like clusters in serum, clusters were incubated at 37 °C in a solution of phosphate buffered saline (PBS) containing 10% bovine serum for 30 min, followed by analysis by TEM.
2.5. Determination of Kinetic Binding Parameters for Adhesion of SPION Nanoclusters onto a Target Substrate.
The targeted binding of peptide-conjugated nanoclusters was measured with a Biacore 3000 (GE Healthcare, United States). A gold sensor chip was modified with 11-mercaptoundecanoic acid to present reactive carboxylic acid groups on the chip surface. The carboxyl groups were then activated by flowing 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) over the chip, followed by a recombinant human VCAM-1 (R&D systems) to attach the protein to the gold surface. SPION nanoclusters functionalized with VCAM-1 binding peptides and unmodified SPION nanoclusters at an iron concentration of 420 μM in PBS were then flowed over the surface at a rate of 5 μL/min to measure association and dissociation rates with the VCAM-1 coated surface. For control experiments, free peptides at a concentration of 6 mg/mL were flowed over the surface for 20 min at a rate of 2 μL/min prior to introduction of nanoclusters to saturate the binding sites. Binding parameters were determined from a 1:1 Langmuir binding model20 with BIAevaluation software version 4.1.
2.6. MR Relaxivity Characterization.
A spin echo pulse sequence was used on a 3 T Siemens Magnetom Trio scanner (Siemens AG, Erlangen, Germany) to produce phantom MR images of SPION clusters suspended in water. For MR imaging, the echo time (TE) ranged from 12 to 490 ms, while the repetition time (TR) was kept at 1200 ms for all images. Signal intensities were measured with ImageJ, and the T2 relaxation was determined by least-squares curve fitting of signal as a function of TE.18 When the samples are imaged with longer echo times, the signal intensity for a fully relaxed image was obtained, thus enabling a more accurate fit to determine T2. After imaging, phantoms were digested in nitric acid, and the iron content was measured with inductively coupled plasma optical emission spectroscopy (ICP-OES, PerkinElmer Optima 2000 DV, Norwalk, CT). Relaxivity was then determined by linear regression of the relaxation rate (1/T2) versus iron content.
2.7. Analysis of Saturation Magnetization.
SPION magnetization was examined with a magnetometer (MPMS, Quantum Design) as previously described.18 The saturation magnetization was determined to be 71 emu/g, which yielded a calculated maximum relaxivity for the SPIONs of 742 mM−1 s−1.
3. RESULTS AND DISCUSSION
3.1. Fabrication of SPION Clusters.
As the first step, HPGs with molecular weights of 3000 and 50 000 g/mol, referred to as HPG3k and HPG50k, respectively, were prepared by varying the ratio of glycidol to an anionic ring opening initiator. The resultant HPGs were alkylated with octadecyl chains by reaction with bromooctadecane. The HPG3k was found to have an average of three octadecyl groups per macromolecule, designated HPG3k-g-C18(3), whereas its HPG50k counterpart had 15 chains, termed HPG50k-g-C18(15). Separately, oleic acid-coated SPIONs were fabricated via the high-temperature thermal decomposition of iron oleate. These SPIONs had an average core diameter of 5 nm, as confirmed by images captured with TEM.21
Clusters of SPIONs were fabricated by emulsifying SPIONs dispersed in chloroform with HPG dissolved in deionized water (Figure 1a). The clusters had a spherical morphology when HPGs with unary molecular weight were used to form the SPION clusters (Figures 1a and b). Interestingly, however, when the aqueous phase of the emulsion consisted of a binary mixture of the HPG50k-g-C18(15) and HPG3k-g-C18(3), there was a notable change in morphology (Figures 1a and b). Specifically, as , defined in eq 1, increased, the clusters became elongated.
Figure 1.
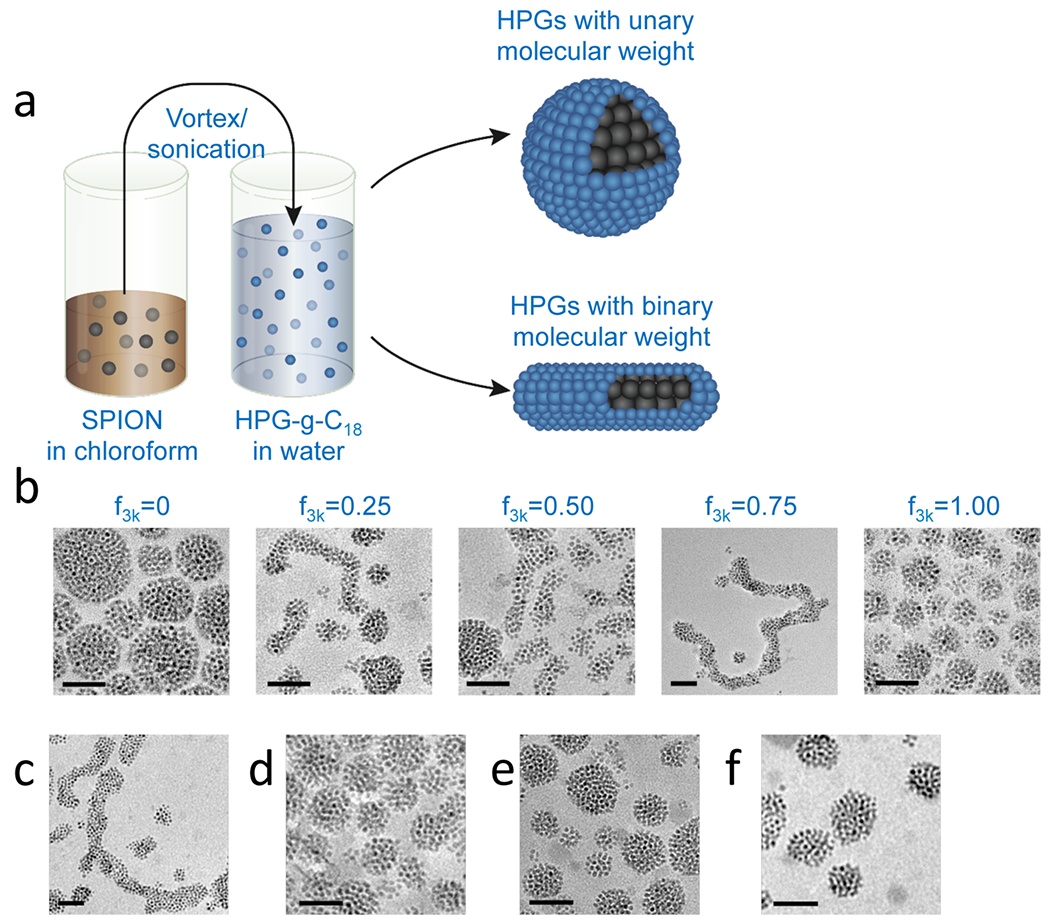
Morphological control of SPION clusters. (a) Schematic describing the emulsification process to fabricate SPION clusters with controlled shapes. The oleic acid-capped SPIONs suspended in chloroform were emulsified with HPG-g-C18 of controlled molecular weight dissolved in water. (b) TEM images showing the sphere-to-worm transition of SPION nanoclusters controlled by . As HPGs with MWs of 3000 and 50 000 g/mol were incorporated into water for the emulsification, the resultant SPION clusters became elongated with maximal length occurring at a 3:1 HPG3k-g-C18(3):HPG50k-g-C18(15) ratio. (c) A 3:1 ratio of low:high molecular weight HPG still resulted in worm-like clusters when both had an average of two C18 chains per molecule. (d) Alkylated HPG3k was unable to produce worm-like clusters by varying only the degree of alkylation. Similarly, using an intermediately sized HPG, HPG8k-g-C18(2), resulted only in spheres when mixed with alkylated (e) HPG3k and (f) HPG50k. All scale bars represent 50 nm.
| (1) |
Here, and are defined as the molar amount of HPG3k-g-C18(3) and HPG50k-g-C18(15), respectively. Notably, at a critical value of 0.75, the SPION clusters were maximally extended, forming worm-like structures (Figure 1b). These worm-like clusters had an average aspect ratio of 10. More than 50% of the SPIONs took the form of worm-like cluster under these conditions, whereas at other values, such as 0.25 and 0.50, only approximately 15% of the SPION clusters presented an elongated shape, with an average aspect ratio of 4 (Figure 1b). The distribution of aspect ratios in these samples can be found in Figure S3.
It is proposed that the morphological changes of the SPION clusters arise from the molecular weight mismatch of the HPGs, independent of the degree of alkylation. For example, the mixture of HPG50k and HPG3k, both of which are conjugated with two C18 chains per molecule, still resulted in assemblies with a worm-like appearance when was 0.75 (Figure 1c). In contrast, mixing HPG3k with two different degrees of alkylation, namely HPG3k-g-C18(3) and HPG3k-g-C18(5), did not result in a worm-like cluster (Figure 1d). It was also demonstrated that the difference in the molecular weight of the HPGs is a key factor in forming the worm-like SPION clusters. Alkylated but intermediately sized HPGs with a molecular weight of 8000 g/mol, HPG8k-g-C18(2), were not able to form worm-like SPION clusters when mixed with either HPG3k-g-C18(3) or HPG50k-g-C18(15), which may be due to a small difference in size (Figures 1e and f). Aqueous suspensions of these nanoclusters remained stable for over a month at room temperature. Addition of these nanoclusters into phosphate buffered saline (PBS) supplemented with 10% serum at 37 °C did not result in any morphological changes or aggregation (Figure S4).
Additionally, the SPIONs themselves served to promote the formation of worm-like structures.22 When was 0.75, the HPG3k-g-C18(5) and HPG50k-g-C18(15) created a self-assembled structure by themselves without SPIONs. However, the vast majority of the polymeric structures produced were spherical (Figure S5).
We postulate that formation of the worm-like SPION cluster results from the packing behavior of HPG50k and HPG3k by analogy to classical arguments for high-curvature spherical vs lower-curvature worm-like micelles. Coarse-grained simulations depicted in the Supporting Information highlight geometric differences between the alkylated HPG50k and HPG3k, in particular the presence of voids in the expanded HPG50k unit (Figure 2a). We suggest that HPGs self-assemble in a “wedge” geometry on the spherical SPION, in which the HPG unit can be highly stretched to accommodate the size disparity compared to the hydrophobic alkyl chains. Such molecular stretching would drive an increase in the curvature and form the spherical SPION cluster (Figure 2b). In contrast, when the binary mixture of HPG3k and HPG50k with an intermediate (e.g., 0.75) induces the SPION nanocluster, it is likely that HPG3k readily fills the interstitial volume between the HPG50k molecules (Figure 2c). HPG3k spaces out the large HPG50k molecules, thereby decreasing the HPG-induced curvature and forming worm-like SPION clusters at intermediate .
Figure 2.
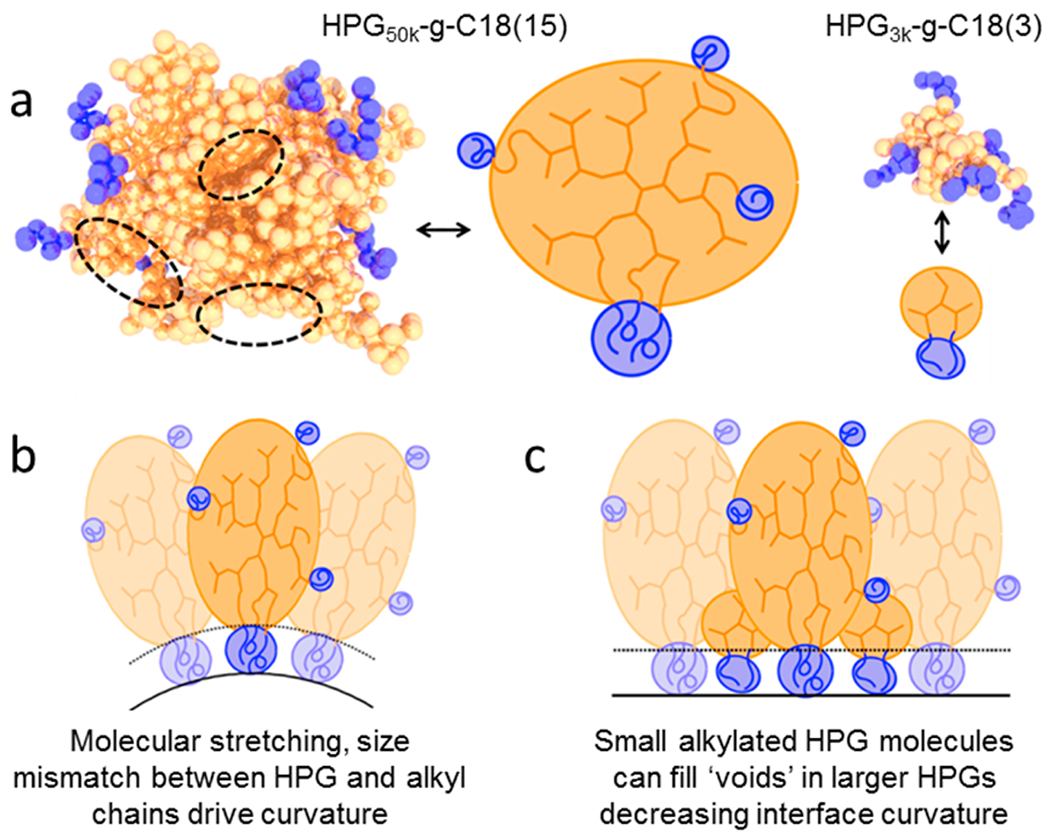
Suggested mechanism of worm-like SPION cluster formation. (a) Simulated images demonstrating the structure of alkylated HPG50k and HPG3k along with simplified representations. We note the presence of voids in the hyperbranched conformation (dotted, black circles). (b) HPG50k and alkyl components drive curvature, leading to spherical SPION clusters. (c) Introduction of smaller HPG3k molecules will fill voids in HPG50k molecules, decreasing curvature and allowing for worm-like SPION clusters.
3.2. MR Relaxivities of SPION Clusters.
According to MR images of phantom models prepared by dispersing SPIONs or SPION clusters in water, both spherical and worm-like clusters created substantial negative contrast at 0.07 mM Fe (Figure 3a). In contrast, the single SPIONs (FeREX, BioPAL Inc.) failed to generate such negative contrast even at a higher concentration of 0.14 mM Fe (Figure 3a). Measurement of T2 molar relaxivity from the phantom images yielded a value of 675 mM−1 s−1 for the worm-like clusters, nearly reaching the theoretical maximum value, 742 mM−1 s−1. The theoretical maximum value was calculated with saturation magnetization measured with MPMS.
Figure 3.
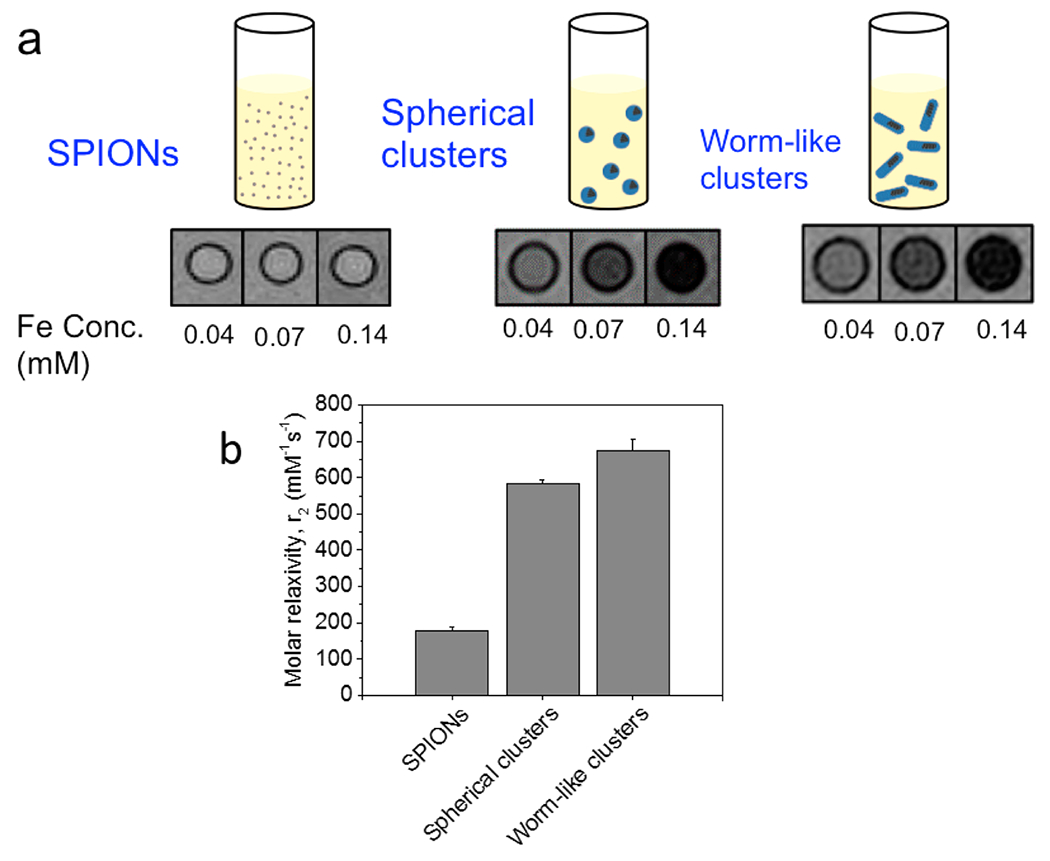
Analysis of MR relaxivity of SPION clusters. (a) MR images of the phantom model prepared with SPIONs, spherical clusters, and worm-like clusters dispersed in water. (b) MR relaxivity of SPION clusters. The T2 molar relaxivity of worm-like clusters was slightly higher than that of spherical clusters; however, resultant differences in contrast capability at given Fe concentrations were minimal. Error bars represent standard deviation of the fit parameter.
The T2 molar relaxivity of worm-like SPION clusters was slightly higher than that of spherical clusters prepared solely with the HPG3k-g-C18(5). One explanation is that HPG50k significantly increased the hydrophilicity of the cluster surface, noting that HPG50k is more hydrophilic than HPG3k because of the larger number of hydroxyl groups in a given volume. In contrast, the spherical cluster prepared by HPG3k may present zones unoccupied by HPG because of the excluded volume effect. Another explanation could be due to size effects, as aggregate size can play a major role in relaxivity enhancement. Previous work examined the effects of HPG molecular weight and cluster size on relaxivity.18 Furthermore, anisotropy has been implicated in affecting molar relaxivity in other systems and could cause the worm-like clusters to have better contrast capability.23–26 Spherical and worm-like clusters both presented relaxivities that were substantially greater than that of the single SPIONs, which yielded a relaxivity less than 200 mM−1 s−1.
Furthermore, because our approach of tuning the morphology of the nanoclusters relies on a size mismatch of the HPG units rather than hydrophobicity, we maintained a high degree of hydrophilicity. The surface hydrophilicity is important in the design of any MRI contrast agent. In addition, the clustered nature of the SPIONs that make up the worm-like structure may prove advantageous over linearly aligned SPIONs,27–29 as thicker clusters could allow relaxivity to be maximized within the static dephasing regime (SDR).30,31 As a result of the hydrophilicity coupled with the high degree of clustering, the relaxivity of the clusters was higher than previous reports of elongated nanocarriers of SPIONs.15,32,33 Additional relaxivity improvement may be possible by inducing clustering of SPIONs of a larger diameter. Whereas 5 nm is the diameter typical of commercial biomedical SPIONs such as FeREX, a core diameter of 10 nm, for example, would likely provide higher saturation magnetization and therefore could result in higher relaxivity.34
3.3. Targeting Efficiency of Peptide-Modified SPION Clusters.
We next modified SPION clusters with targeting ligands by conjugating VHPKQHR peptides to HPG so as to examine the ability of the HPG-SPION cluster to accumulate at simulated defective tissue sites of interest. The peptide was conjugated to an acrylate-modified HPG3k-g-C18(3) via the terminal cysteine residue (Figure 4a). The peptide-conjugated HPG was subsequently used to induce SPION clustering. The presence of the peptide did not interfere with the morphology of the clusters (Figure 4b).
Figure 4.
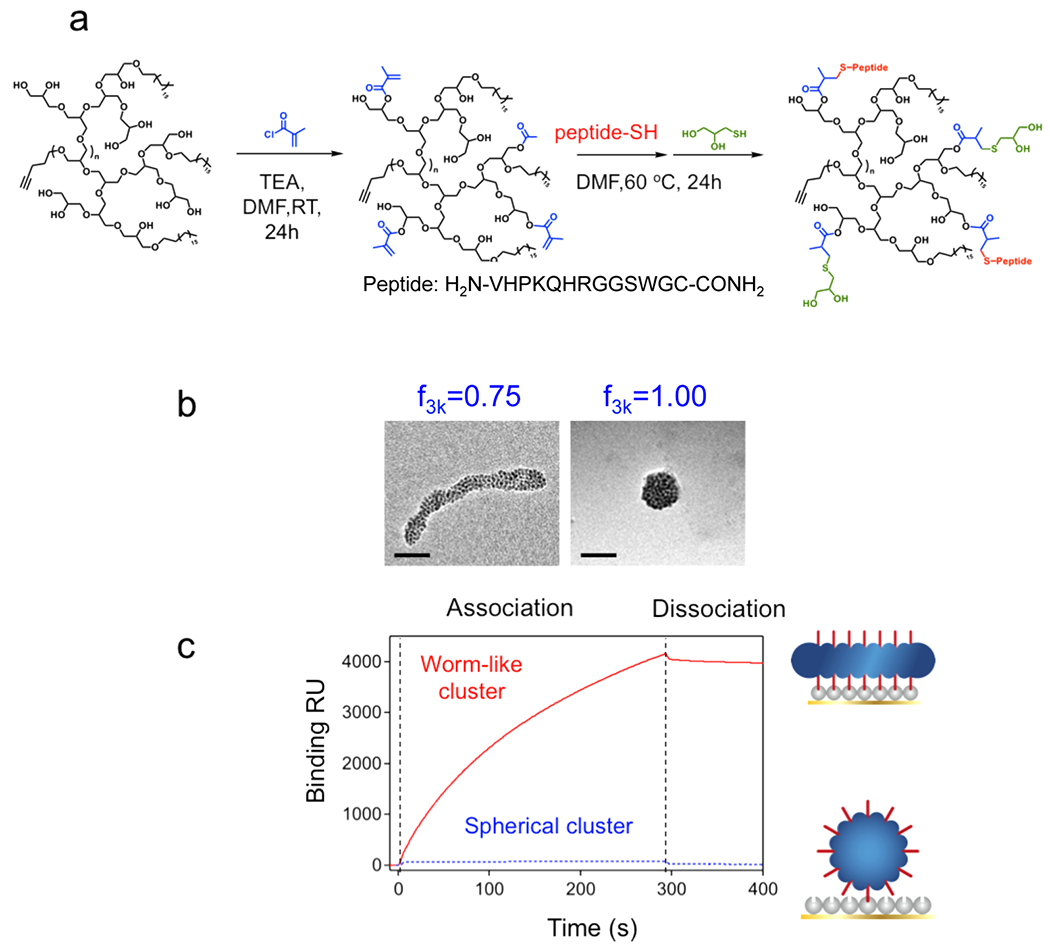
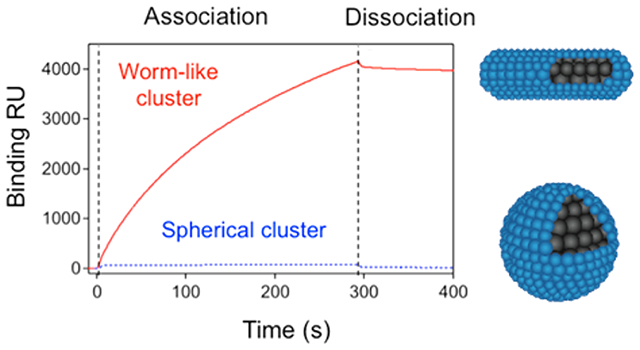
Shape-controlled adhesion properties of the SPION clusters. (a) Conjugation of the VCAM-1 targeting peptide VHPKQHR to the alkylated HPG backbone. (b) Addition of the VHPKQHR peptide did not interfere with cluster formation, as SPIONs formed a worm-like cluster at of 0.75, while they formed a spherical cluster at of 1. Scale bars represent 50 nm. (c) SPR kinetic binding analysis of targeted worm-like (red, solid curve) and spherical (blue, dotted curve) nanoclusters during association and dissociation with VCAM-1 receptors.
The kinetics of binding between SPION clusters and inflammatory sites was examined by measuring association and dissociation rates with VCAM-coated substrates using SPR spectroscopy. VCAM-1 receptors were immobilized on an SPR chip, and SPION clusters were introduced into the flow to examine their binding properties. The binding resonance unit (RU) for the worm-like clusters, indicative of molecular mass bound to the SPR chip, was dramatically higher than that of the spherical clusters at the same iron dose (Figure 4c). When the binding parameters are taken into account, the association rate constant, , was 1 order of magnitude higher per iron dose for worm-like SPION clusters than that of spherical ones (Table 1). Additionally, the dissociation rate constant, , was 2 orders of magnitude lower. As a result, the overall binding constant, a ratio of to , was over 3 orders of magnitude greater for the worm-like clusters (Table 1).
Table 1.
Kinetic Binding Values for the Association and Dissociation between Nanoclusters and VCAM-Immobilized Substrate as Determined by SPR Spectroscopya
| morphology of targeted cluster | association rate constant (M−1 s−1) | dissociation rate constant (s−1) | binding constant (M−1) |
|---|---|---|---|
| spherical | 1.0 | 1.3 × 10−3 | 7.7 × 102 |
| worm-like | 14.7 | 1.5 × 10−5 | 1.0 × 106 |
Values are given per molar concentration of iron.
In addition to morphology, the importance of the targeting peptide was evaluated in control experiments using SPION nanoclusters either free of targeting peptides or peptide-conjugated clusters flowed over VCAM presaturated with peptide (Figure S6). Without targeting peptide, the binding was significantly reduced compared to binding between VCAM-1 and the peptide-conjugated worm-like HPG-SPIONs. Similarly, by occupying the binding sites of VCAM-1 prior to administration of the nanoclusters, the VHPKQHR peptide-conjugated SPION clusters minimally adhered to the substrate. The advantage of the targeted worm-like clusters over targeted spheres is further highlighted by considering the number of peptides per iron dose. According to thermogravimetric analysis (TGA), the molar amount of peptides per iron for spherical and worm-like clusters was approximately 1:36 and 1:112, respectively, as described in the Supporting Information. In this way, despite fewer peptides per iron dose, the worm-like clusters were able to bind significantly more iron to the simulated target site in SPR experiments.
The transport mechanism presented herein mimics the way in which several strains of bacteria and parasites have developed rod-shaped or filamentous morphologies to maximize contact surface area with the endothelium of their host under shear forces of blood flow.35 Others have directly used such structures as templates for assembling nanoparticles to increase the number of particles immobilized per binding event.36 Similarly, our approach was able to bring a large number of SPION clusters to a model target site compared with that using spherical SPION clusters. Additionally, the elongated morphology may allow for extended circulation in vivo compared to spherical structures,26,37 which could also promote accumulation at sites of inflammation.
4. CONCLUSION
Taken together, this study demonstrated SPION clusters with worm-like morphology and HPG-induced hydrophilic surfaces present superior MR relaxivity compared to that of a single SPION, and a targetability higher than that of spherical SPION clusters. In future studies, the worm-like structures may be purified from their spherical counterparts to yield a uniform population with enhanced targetability. Additionally, by understanding the specific parameters that result in the wormlike clusters, it may be possible to fabricate populations that are more monomodal. In this way, efforts to study this mechanism of self-assembly will be critical in further improving these targeted clusters. Hence, this study also has the potential to substantially enhance imaging-based diagnostic capabilities. We also believe that the assembly strategy presented here could be readily extended to other nanoparticle types and is therefore broadly applicable to colloidal systems.
Supplementary Material
ACKNOWLEDGMENTS
The authors are thankful for MRI assistance provided by R. Larsen and B. Odintsov at the Beckman Institute for Advanced Science and Technology. The authors also thank A. Ferguson in the Department of Materials Science and Engineering at the University of Illinois for insightful discussion. Electron microscopy was performed at the Frederick Seitz Materials Research Laboratory Central Facilities at the University of Illinois. Mass spectrometry was performed at the Mass Spectrometry Laboratory, School of Chemical Sciences, University of Illinois. Funding was provided by the National Institutes of Health (Grant 1R01 HL109192 to H.J.K., S.C.Z., and S.M., Grant 1R01 HL098967 to S.M., and Chemistry-Biology Interface Training Grant 5T32-GM070421 to C.E.S.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsami.6b10891.
Simulation of HPG structure, chemical characterization of HPG, TEM evaluation of worm-like structures, and additional SPR results (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Choi HS; Liu W; Liu F; Nasr K; Misra P; Bawendi MG; Frangioni JV Design Considerations for Tumour-Targeted Nanoparticles. Nat. Nanotechnol 2010, 5, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Byrne JD; Betancourt T; Brannon-Peppas L Active Targeting Schemes for Nanoparticle Systems in Cancer Therapeutics. Adv. Drug Delivery Rev 2008, 60, 1615–1626. [DOI] [PubMed] [Google Scholar]
- (3).Yun Y; Cho YW; Park K Nanoparticles for Oral Delivery: Targeted Nanoparticles with Peptidic Ligands for Oral Protein Delivery. Adv. Drug Delivery Rev 2013, 65, 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gu F; Zhang L; Teply BA; Mann N; Wang A; Radovic-Moreno AF; Langer R; Farokhzad OC Precise Engineering of Targeted Nanoparticles by Using Self-Assembled Biointegrated Block Copolymers. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Davis ME; Zuckerman JE; Choi CHJ; Seligson D; Tolcher A; Alabi CA; Yen Y; Heidel JD; Ribas A Evidence of RNAi in Humans from Systemically Administered siRNA via Targeted Nanoparticles. Nature 2010, 464, 1067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lai M-H; Jeong JH; DeVolder RJ; Brockman C; Schroeder C; Kong H Ellipsoidal Polyaspartamide Polymersomes with Enhanced Cell-Targeting Ability. Adv. Funct. Mater 2012, 22, 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chen J; Clay NE; Park N.-h.; Kong H Non-Spherical Particles for Targeted Drug Delivery. Chem. Eng. Sci 2015, 125, 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Blanazs A; Armes SP; Ryan AJ Self-Assembled Block Copolymer Aggregates: From Micelles to Vesicles and Their Biological Applications. Macromol. Rapid Commun 2009, 30, 267–277. [DOI] [PubMed] [Google Scholar]
- (9).Cheng H; Yuan X; Sun X; Li K; Zhou Y; Yan D Effect of Degree of Branching on the Self-Assembly of Amphiphilic Hyper-branched Multiarm Copolymers. Macromolecules 2010, 43, 1143–1147. [Google Scholar]
- (10).Chhatre A; Duttagupta S; Thaokar R; Mehra A Mechanism of Nanorod Formation by Wormlike Micelle-Assisted Assembly of Nanospheres. Langmuir 2015, 31, 10524–10531. [DOI] [PubMed] [Google Scholar]
- (11).Zhang L; Eisenberg A Multiple Morphologies of “Crew-Cut” Aggregates of Polystyrene-B-Poly(Acrylic Acid) Block Copolymers. Science 1995, 268, 1728–1731. [DOI] [PubMed] [Google Scholar]
- (12).Rajagopal K; Mahmud A; Christian DA; Pajerowski JD; Brown AEX; Loverde SM; Discher DE Curvature-Coupled Hydration of Semicrystalline Polymer Amphiphiles Yields Flexible Worm Micelles but Favors Rigid Vesicles: Polycaprolactone-Based Block Copolymers. Macromolecules 2010, 43, 9736–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Won Y-Y; Davis HT; Bates FS Giant Wormlike Rubber Micelles. Science 1999, 283, 960–963. [DOI] [PubMed] [Google Scholar]
- (14).Wang L; Neoh KG; Kang ET; Shuter B Multifunctional Polyglycerol-Grafted Fe(3)O(4)@Sio(2) Nanoparticles for Targeting Ovarian Cancer Cells. Biomaterials 2011, 32, 2166–73. [DOI] [PubMed] [Google Scholar]
- (15).Yang X; Grailer JJ; Rowland IJ; Javadi A; Hurley SA; Steeber DA; Gong S Multifunctional SPIO/DOX-Loaded Wormlike Polymer Vesicles for Cancer Therapy and MR Imaging. Biomaterials 2010, 31, 9065–73. [DOI] [PubMed] [Google Scholar]
- (16).Smith CE; Shkumatov A; Withers SG; Yang B; Glockner JF; Misra S; Roy EJ; Wong C-H; Zimmerman SC; Kong H A Polymeric Fastener Can Easily Functionalize Liposome Surfaces with Gadolinium for Enhanced Magnetic Resonance Imaging. ACS Nano 2013, 7, 9599–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nahrendorf M; Jaffer FA; Kelly KA; Sosnovik DE; Aikawa E; Libby P; Weissleder R Noninvasive Vascular Cell Adhesion Molecule-1 Imaging Identifies Inflammatory Activation of Cells in Atherosclerosis. Circulation 2006, 114, 1504–1511. [DOI] [PubMed] [Google Scholar]
- (18).Smith CE; Ernenwein D; Shkumatov A; Clay NE; Lee JY; Melhem M; Misra S; Zimmerman SC; Kong H Hydrophilic Packaging of Iron Oxide Nanoclusters for Highly Sensitive Imaging. Biomaterials 2015, 69, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kim BH; Lee N; Kim H; An K; Park YI; Choi Y; Shin K; Lee Y; Kwon SG; Na HB; Park JG; Ahn TY; Kim YW; Moon WK; Choi SH; Hyeon T Large-Scale Synthesis of Uniform and Extremely Small-Sized Iron Oxide Nanoparticles for High-Resolution T-1 Magnetic Resonance Imaging Contrast Agents. J. Am. Chem. Soc 2011, 133, 12624–12631. [DOI] [PubMed] [Google Scholar]
- (20).Kortt AA; Nice E; Gruen LC Analysis of the Binding of the Fab Fragment of Monoclonal Antibody Nc10 to Influenza Virus N9 Neuraminidase from Tern and Whale Using the Biacore Biosensor: Effect of Immobilization Level and Flow Rate on Kinetic Analysis. Anal. Biochem 1999, 273, 133–41. [DOI] [PubMed] [Google Scholar]
- (21).Clay N; Baek K; Shkumatov A; Lai M-H; Smith CE; Rich M; Kong H Flow-Mediated Stem Cell Labeling with Superparamagnetic Iron Oxide Nanoparticle Clusters. ACS Appl Mater. Interfaces 2013, 5, 10266–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhu J; Hayward RC Spontaneous Generation of Amphiphilic Block Copolymer Micelles with Multiple Morphologies through Interfacial Instabilities. J. Am. Chem. Soc 2008, 130, 7496–7502. [DOI] [PubMed] [Google Scholar]
- (23).Zhao Z; Zhou Z; Bao J; Wang Z; Hu J; Chi X; Ni K; Wang R; Chen X; Chen Z; Gao J Octapod Iron Oxide Nanoparticles as High-Performance T2 Contrast Agents for Magnetic Resonance Imaging. Nat. Commun 2013, 4, 2266. [DOI] [PubMed] [Google Scholar]
- (24).Zhou Z; Zhao Z; Zhang H; Wang Z; Chen X; Wang R; Chen Z; Gao J Interplay between Longitudinal and Transverse Contrasts in Fe3O4 Nanoplates with (111) Exposed Surfaces. ACS Nano 2014, 8, 7976–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wyss PP; Lamichhane S; Rauber M; Thomann R; Kramer KW; Shastri VP Tripod USPIONs with High Aspect Ratio Show Enhanced T2 Relaxation and Cytocompatibility. Nanomedicine (London U. K.) 2016, 11, 1017–1030. [DOI] [PubMed] [Google Scholar]
- (26).Park J-H; von Maltzahn G; Zhang L; Derfus AM; Simberg D; Harris TJ; Ruoslahti E; Bhatia SN; Sailor MJ Systematic Surface Engineering of Magnetic Nanoworms for in Vivo Tumor Targeting. Small 2009, 5, 694–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gossuin Y; Disch S; Vuong QL; Gillis P; Hermann RP; Park J-H; Sailor MJ NMR Relaxation and Magnetic Properties of Superparamagnetic Nanoworms. Contrast Media Mol. Imaging 2010, 5, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Park J-H; von Maltzahn G; Xu MJ; Fogal V; Kotamraju VR; Ruoslahti E; Bhatia SN; Sailor MJ Cooperative Nanomaterial System to Sensitize, Target, and Treat Tumors. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lo JH; von Maltzahn G; Douglass J; Park J-H; Sailor MJ; Ruoslahti E; Bhatia SN Nanoparticle Amplification via Photothermal Unveiling of Cryptic Collagen Binding Sites. J. Mater. Chem. B 2013, 1, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yablonskiy DA; Haacke EM Theory of NMR Signal Behavior in Magnetically Inhomogeneous Tissues: The Static Dephasing Regime. Magn. Reson. Med 1994, 32, 749–763. [DOI] [PubMed] [Google Scholar]
- (31).Pöselt E; Kloust H; Tromsdorf U; Janschel M; Hahn C; Maßlo C; Weller H Relaxivity Optimization of a PEGylated Iron-Oxide-Based Negative Magnetic Resonance Contrast Agent for T2-Weighted Spin–Echo Imaging. ACS Nano 2012, 6, 1619–1624. [DOI] [PubMed] [Google Scholar]
- (32).Park J-H; von Maltzahn G; Zhang L; Schwartz MP; Ruoslahti E; Bhatia SN; Sailor MJ Magnetic Iron Oxide Nanoworms for Tumor Targeting and Imaging. Adv. Mater 2008, 20, 1630–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang G; Inturi S; Serkova NJ; Merkulov S; McCrae K; Russek SE; Banda NK; Simberg D High-Relaxivity Superparamagnetic Iron Oxide Nanoworms with Decreased Immune Recognition and Long-Circulating Properties. ACS Nano 2014, 8, 12437–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Jun Y.-w.; Huh Y-M; Choi J.-s.; Lee J-H; Song H-T; Kim K-Y; Yoon S; Kim K-S; Shin J-S; Suh J-S; Cheon J. Nanoscale Size Effect of Magnetic Nanocrystals and Their Utilization for Cancer Diagnosis via Magnetic Resonance Imaging. J. Am. Chem. Soc 2005, 127, 5732–5733. [DOI] [PubMed] [Google Scholar]
- (35).Young KD The Selective Value of Bacterial Shape. Microbiol. Mol. Biol. Rev 2006, 70, 660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ghosh D; Lee Y; Thomas S; Kohli AG; Yun DS; Belcher AM; Kelly KA M13-Templated Magnetic Nanoparticles for Targeted in Vivo Imaging of Prostate Cancer. Nat. Nanotechnol 2012, 7, 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Geng Y; Dalhaimer P; Cai S; Tsai R; Tewari M; Minko T; Discher DE Shape Effects of Filaments Versus Spherical Particles in Flow and Drug Delivery. Nat. Nanotechnol 2007, 2, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


