Abstract
The encapsidation signal of bovine leukemia virus (BLV) was previously shown by deletion analysis to be discontinuous and to extend into the 5′ end of the gag gene (L. Mansky et al., J. Virol. 69:3282–3289, 1995). The global minimum-energy optimal folding for the entire BLV RNA, including the previously mapped primary and secondary encapsidation signal regions, was analyzed. Two stable stem-loop structures (located just downstream of the gag start codon) were predicted within the primary signal region, and one stable stem-loop structure (in the gag gene) was predicted in the secondary signal region. Based on these predicted structures, we introduced a series of mutations into the primary and secondary encapsidation signals in order to explore the sequence and structural information contained within these regions. The replication efficiency and levels of cytoplasmic and virion RNA were analyzed for these mutants. Mutations that disrupted either or both of the predicted stem-loop structures of the primary signal reduced the replication efficiency by factors of 7 and 40, respectively; similar reductions in RNA encapsidation efficiency were observed. The mutant with both stem-loop structures disrupted had a phenotype similar to that of a mutant containing a deletion of the entire primary signal region. Mutations that disrupted the predicted stem-loop structure of the secondary signal led to similar reductions (factors of 4 to 6) in both the replication and RNA encapsidation efficiencies. The introduction of compensatory mutations into mutants from both the primary and secondary signal regions, which restored the predicted stem-loop structures, led to levels of replication and RNA encapsidation comparable to those of virus containing the wild-type encapsidation signal. Replacement of the BLV RNA region containing the primary and secondary encapsidation signals with a similar region from human T-cell leukemia virus (HTLV) type 1 or type 2 led to virus replication at three-quarters or one-fifth of the level of the parental virus, respectively. The results from both the compensatory mutants and BLV-HTLV chimeras indicate that the encapsidation sequences are recognized largely by their secondary or tertiary structures.
A retroviral vector contains all of the cis-acting elements necessary for retrovirus replication but is deficient in the production of some or all of the viral proteins necessary for replication and virus production. Detailed knowledge of the steps in retroviral replication have led to the successful use of retroviral vectors for gene transfer. An important cis-acting element for virus production is the encapsidation (packaging) signal (E or Ψ) (44).
The encapsidation signal of most retroviruses is located primarily in the 5′ untranslated region of the genome and is necessary for the packaging of two identical copies of retroviral RNA into virus particles. For simpler retroviruses such as spleen necrosis virus (SNV) and murine leukemia virus (MLV), the primary encapsidation signal is located between the major splice donor site and the gag start codon (2, 4, 17, 32, 33, 53). MLV has an extended encapsidation signal in the gag open reading frame, Ψ+, which increases viral RNA packaging and virus titer 10- to 200-fold (7, 38). The Rous sarcoma virus (RSV) primary encapsidation signal is located 5′ to the viral coding sequence and has the major splice donor site just downstream of the gag start codon (5, 6, 19, 26, 28, 54). Thus, unlike SNV and MLV, the RSV encapsidation signal is located not only on the unspliced viral RNA but also on the poorly encapsidated spliced env gene mRNA. The RSV encapsidation signal possibly extends into the gag region, including a region near the 3′ end of the genomic RNA (43, 49).
The encapsidation signals for human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (both are more complex retroviruses) include the viral sequences upstream of the major splice donor site, the leader region between the major splice donor site and the gag start codon, and may also extend into the 3′ end of the U5 region of the long terminal repeat and the 5′ end of the gag coding sequence (3, 10, 12, 20, 21, 30, 31, 41, 45). The primary site of RNA encapsidation for HIV-1 includes two stem-loop structures in the leader region between the major splice donor and the gag start codon (37). These data suggest a similar genomic location for the primary encapsidation signal regions of more complex and simpler retroviruses. However, additional regions were found to be important for encapsidation that may be unique to more complex retroviruses.
RNA secondary structures have been implicated as being important for the function of retroviral encapsidation signals (37, 55). Based on these observations, we reasoned that such RNA secondary structures in bovine leukemia virus (BLV) RNA may exist in the regions that we have mapped and may be essential for efficient RNA packaging and virus production. We have previously shown by deletion analysis that the BLV encapsidation signal is discontinuous (34). A primary signal, essential for RNA encapsidation, was mapped to a region starting in untranslated leader region (downstream of the primer binding site) to just downstream of the gag start codon (in the matrix domain). A secondary signal, which facilitates efficient RNA encapsidation, resides in a 132-nucleotide region toward the center of the gag gene (in the capsid domain).
Analysis of the global minimum-energy optimal folding for the entire BLV RNA indicated stable stem-loop structures that overlapped the previously mapped primary and secondary encapsidation signal regions. Two stable stem-loop structures (located just downstream of the gag start codon) were predicted within the region containing the primary signal, and one stable stem-loop structure (in gag) was predicted as the secondary signal. Based on these predicted structures, we introduced a series of mutations into these stem-loop structures to test their effects on virus replication and RNA encapsidation. By analyzing the replication efficiencies and levels of cytoplasmic and virion RNA for these mutants, we found that mutations that disrupted either or both of the predicted stem-loop structures that overlapped the region containing the primary signal reduced the RNA encapsidation and replication efficiencies by factors of 7 and 40, respectively. The mutant with both stem-loops disrupted had a phenotype similar to that of a mutant containing a deletion of the entire primary signal region. This finding indicates that the two stem-loop structures are the primary signal. Mutations that disrupted the predicted stem-loop structure in the region containing the secondary signal led to similar reductions in both the replication and RNA encapsidation efficiencies (factors of 4 to 6), which indicates that this stem-loop structure is the secondary signal. Compensatory mutants in either the primary or secondary regions restored both the predicted stem-loop structures and the levels of replication and RNA encapsidation to that of the wild type. These results confirmed that these RNA structures comprise the encapsidation signal. When the BLV RNA region containing the primary and secondary encapsidation signals was replaced with a similar region from either human T-cell leukemia virus type 1 (HTLV-1) or type 2 (HTLV-2), replication occurred at a level either three-quarters or one-fifth respectively, of that of the parental virus.
Our results indicate that the primary BLV encapsidation signal includes two stem-loop structures located in the gag gene that are necessary for efficient RNA encapsidation. In addition, the secondary signal, which is also located in gag, is a stem-loop that is important for efficient replication and RNA encapsidation. The putative encapsidation signal regions of either HTLV-1 or HTLV-2 can lead to BLV replication, suggesting conservation of encapsidation signal function.
MATERIALS AND METHODS
Nomenclature.
Plasmid constructs are indicated by the nucleotide sequence coordinates of the first nucleotide used to create deletions. Plasmid constructs are indicated by the letter p (e.g., pRW 1) to distinguish them from viruses (e.g., RW 1). The nucleotide sequence coordinates used are relative to the 5′ end of the proviral DNA form of the complete BLV genome (46).
Construction of parental BLV vector and derivatives used for encapsidation signal region mutagenesis studies.
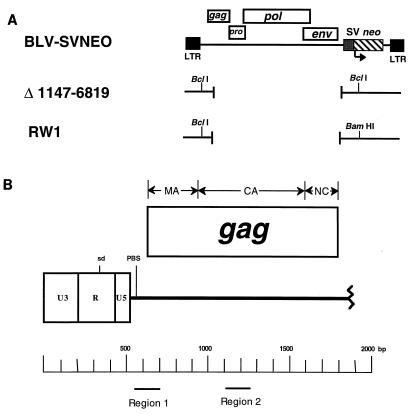
All BLV vectors used in this study were derivatives of pBLV-SVNEO (15). The parental BLV vector used in this study, pRW 1, was specifically derived from the BLV vector pΔ 1147-6819 (Fig. 1A) (34). To construct pRW 1, the BclI site in the region just upstream of the neo gene of pΔ 1147-6819 was mutated to BamHI by using a primary/combinatorial two-step PCR protocol (22). The introduced mutation was verified by DNA sequence analysis, and RW 1 was replicated in parallel with Δ 1147-6819 to ensure that the introduced mutation had no effect on the replication and RNA encapsidation efficiencies of the vector. All derivatives of pRW 1 made to test the structure-function relationship of the predicted RNA stem-loop structures located in the regions mapped as being part of the BLV encapsidation signal were made, using pRW 1 as the parent, by the primary/combinatorial two-step PCR protocol used to create the new BamHI site in pRW 1. The exception is the mutant p628/677, in which the 677 mutation was introduced into 628, and all compensatory mutants. Each mutant resulted in the creation of a SpeI site that could be used to help identify the introduction of the desired mutations. All mutants made were sequenced in the region bounded by the BclI and SalI sites to confirm the introduction of the desired mutations.
FIG. 1.
BLV vectors used in mutagenesis studies to identify the encapsidation signal within regions previously mapped by deletion analysis. (A) Vectors shown in proviral DNA form. Black boxes represent long terminal repeats (LTRs); black lines indicate viral sequence; rectangular boxes above the line indicate viral coding sequences, with the vertical locations of the boxes corresponding to the translational reading frame. Retroviral genes are indicated as gag, pro, pol, and env. The simian virus 40 (SV) promoter and the neo gene are indicated. (B) The 5′ end of the BLV genome, shown in proviral DNA form. The rectangular box at the left end of the line represents the 5′ BLV long terminal repeat, containing the U3 region, the R region, and the U5 region. The line indicates the viral sequence. The locations of the major splice donor site (sd) and the primer binding site (PBS) are indicated. The rectangular box above the lines indicates the BLV gag gene. The matrix (MA), capsid (CA), and nucleocapsid (NC) domains of gag are indicated above the rectangular box. The jagged line at the right end of the viral coding sequence indicates the end of the viral sequence shown in this diagram. The regions that overlap with the primary (region 1) and secondary (region 2) encapsidation signals are indicated at the bottom.
Chimeric BLV vectors containing the predicted encapsidation signal regions of HTLV-1 and HTLV-2 in place of the BLV encapsidation signal region were made by PCR amplifying the predicted encapsidation signal region from either HTLV-1 (nucleotides 798 to 1421 of the proviral DNA from pHTLV-1-CMVneo) (kindly provided by David Derse, National Cancer Institute, Frederick, Md.) (13, 47) or HTLV-2 (nucleotides 784 to 1410 of the proviral DNA from pH6neo) (11, 48), using primers containing BclI or SalI recognition sites. The PCR-amplified HTLV-1 and HTLV-2 DNAs were digested with BclI and SalI and inserted into pRW 1 digested with BclI and SalI. DNA sequencing was done to confirm the proper introduction of the HTLV-1 or HTLV-2 sequence in place of the BLV encapsidation signal region.
Cell lines, transfections, and cocultivations.
FLK-BLV cells were used to test virus production by use of BLV helper virus. FLK-BLV cells produce all of the BLV proteins necessary for virus production (52) and have been used previously for vector virus production. Madin-Darby bovine kidney (MDBK)-based BLV helper cells (34) were used to test RNA encapsidation efficiencies of selected mutants. All cells were grown in Temin-modified Eagle’s medium (51) supplemented with 10% fetal bovine serum (Sigma Chemical Co., St. Louis, Mo.). MDBK-based BLV helper cells and FLK-BLV cells were transfected by the dimethyl sulfoxide-Polybrene procedure as described previously (27).
FLK-BLV cells were transfected with each of the vectors tested. Two days posttransfection, cells were placed under G418 (900 μg/ml) selection. Approximately 100 G418-resistant colonies were pooled and used for cocultivation with fresh MDBK target cells. Infection of target cells was done by cocultivation of virus-producing cells with target cells as described by Mansky and Temin (35). Briefly, virus-producing cells (typically 2.5 × 105 in a 60-mm-diameter petri dish) were treated with mitomycin C (10 μg/ml), an inhibitor of host cell DNA synthesis, for 2 h at 37°C. The cells were then washed three times with fresh medium, and 2.5 × 105 FLK target cells were added. Two days after cocultivation, selective medium containing G418 was added. Control experiments were done with each cocultivation experiment to ensure that mitomycin C-treated, virus-producing cells did not proliferate and no longer adhered to the surfaces of culture dishes (35).
Determination of cytoplasmic and virion RNA levels of mutant vectors.
Selected derivatives of RW 1 were tested for RNA encapsidation efficiency. MDBK-based BLV helper cells were transfected with selected vectors from the replication experiments. Two days posttransfection, cells were placed under G418 selection (1 mg/ml). Approximately 100 G418-resistant colonies were pooled and used for cocultivation with fresh helper cells. Infection was done by cocultivation of virus-producing cells with fresh helper cells. Two days after cocultivation, selective medium containing G418 was added. Control experiments were done with each cocultivation experiment to ensure that mitomycin C-treated, virus-producing cells did not proliferate and no longer adhered to the surfaces of culture dishes (35).
Levels of vector viral RNA in infected helper cells and in virions produced from those cells were determined by RNA slot blot analysis as previously described (34). Cytoplasmic RNA was harvested from pools of G418-resistant clones that had been infected by cocultivation with mutants of RW 1. Virion RNA was purified from virions pelleted from cleared supernatant medium.
Cytoplasmic and virion RNAs were denatured at 65°C for 15 min. Samples were twofold serially diluted in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and were blotted onto nitrocellulose paper (Schleicher & Schuell, Keene, N.H.) with a slot blot vacuum manifold (Millipore, Bedford, Mass.). The blots were then baked under vacuum at 80°C for 2 h. A DNA probe representing the U5 region of the 5′ end of the viral RNA, which is downstream from the splice donor site for the env mRNA, was used for hybridizations. Hybridization was visualized, and relative intensities were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Computer analysis of potential RNA secondary structures.
Calculation and analysis of minimal free energy and suboptimal RNA secondary structures of portions of the BLV, HTLV-1, or HTLV-2 RNA sequence were performed with the FOLDRNA (57) and MFOLD (23, 56) programs within the Genetics Computer Group (GCG) analysis package (version 8; GCG, Madison, Wis.), (16) using the Turner energy tables (18).
The global minimum free-energy structure for the complete genome was calculated with a standalone version of MFOLD implemented on a Silicon Graphics (Mountain View, Calif.) computer with 256 megabytes of memory and took more than 5 days to compute (47a). The RNA structures predicted by FOLDRNA and MFOLD in the GCG package were plotted with the GCG programs SQUIGGLES and PLOTFOLD, respectively. The complete genome was plotted with Michael Zuker’s NAVIEW graphics implementation (9). The STAR program (1) was used to analyze potential pseudoknot structures.
RESULTS
Global analysis of potential RNA secondary structures in the entire BLV RNA genome.
The entire 8,412-base genome of the BLV RNA (Fig. 2) was analyzed for global minimal and suboptimal free-energy structures in a single-step folding run (47a). The predicted secondary structure shown in Fig. 2 represents a global minimum free-energy optimal folding for BLV RNA. The BLV encapsidation signal region is located toward the 5′ end of the genome (see also Fig. 1B).
FIG. 2.
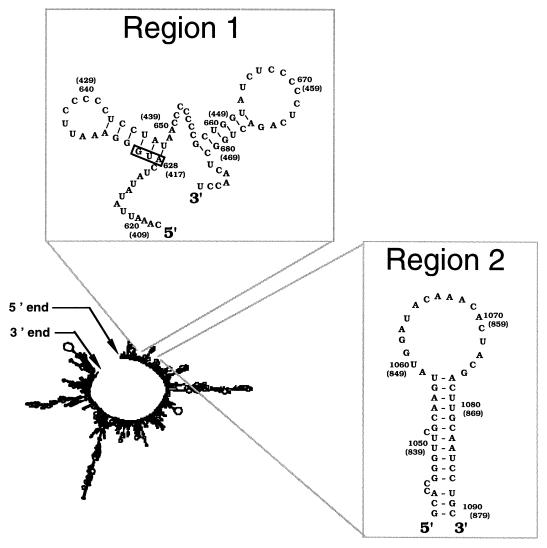
Global organization of the BLV genomic RNA. The predicted RNA secondary structure represents a global minimum-energy optimal folding. The BLV encapsidation signal region is located toward the 5′ end of the genome. The primary and secondary encapsidation signal regions are enlarged for easy visualization and to indicate their locations in the genomic RNA. The gag AUG is marked with a box. No very long range interactions that would bring the 5′ and 3′ ends closer together are observed.
The BLV RNA region containing the encapsidation signal region was also analyzed by folding of local regions (data not shown). The BLV RNA in the primary encapsidation signal region had two stable hairpin structures predicted. The first hairpin begins at the gag start codon. The predicted stable RNA structure in the secondary encapsidation region lies within the 132-nucleotide region (nucleotides 1015 to 1147) that was mapped by deletion analysis as being necessary for efficient virus production and RNA packaging. No pseudoknot foldings were predicted (data not shown) for the unusually large loop sequence with adjacent sequences by use of the STAR program (1).
Analysis of the global BLV RNA folding indicates that the structures predicted for the primary and secondary encapsidation signals (Fig. 2) are the same as those predicted in the local folding of the BLV RNA encapsidation signal region. That these structures are observed in the predicted secondary structure of the global BLV RNA genome folding shows that these segments can remain paired, according to the thermodynamics prediction, in spite of the many other potential partner choices elsewhere along the sequence. Since these structures are predicted in the context of the whole sequence, their biological relevance appears to be reinforced.
Replication and RNA encapsidation efficiencies of BLV encapsidation region 1 mutants.
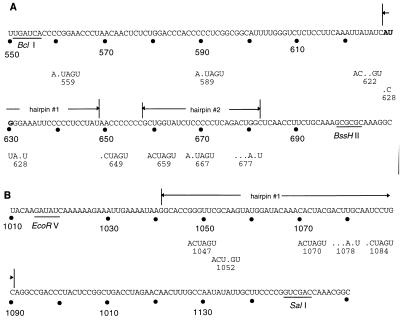
We made various mutations in RW 1 in order to determine the effects of these mutations on virus replication and RNA encapsidation. Figure 3A shows the locations of the mutations made in the primary encapsidation signal region (region 1). The mutations introduced into each vector resulted in the creation of a SpeI restriction site. Region 1 mutants spanned two predicted stem-loop structures in addition to sequences upstream of the hairpins. To test the ability of these vectors to replicate, we used a BLV helper virus as described in the Materials and Methods. FLK-BLV cells were stably transfected with each of the derivatives of pRW 1. Comparable numbers of G418-resistant colonies were observed for each mutant vector per microgram of transfected plasmid DNA (data not shown). Approximately 100 G418-resistant colonies were pooled for each mutant and used for cocultivation with MDBK target cells.
FIG. 3.
Locations of the BLV encapsidation signal region 1 (A) and region 2 (B) mutations. Numbering starts at the 5′ end of the proviral DNA. The locations of the recognition sites for the restriction enzymes BclI, BssHII, EcoRV, and SalI are underlined; the gag AUG is in boldface; regions that comprise the proposed RNA secondary structures important for RNA encapsidation are indicated by lines above the bases. The mutated bases in each mutant vector virus are shown below the wild-type BLV sequence; bases that are different from the wild-type bases are shown, and nonmutated wild-type bases are indicated by a dots. The names of the mutant vectors containing the mutations, as well as the coordinate of the first base of the newly created SpeI site, are indicated below each vector sequence. Nucleotide numbering is based on the complete BLV proviral DNA sequence (46).
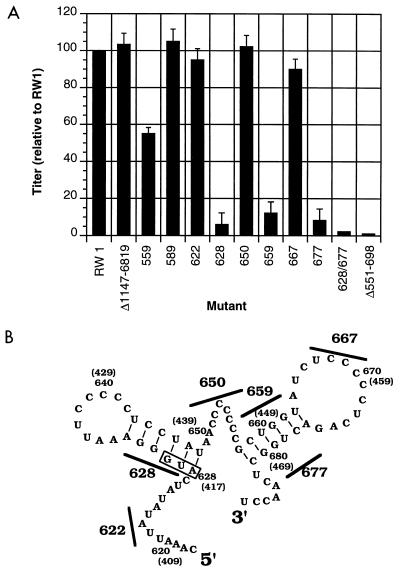
Figure 4 shows the results of vector virus production from FLK-BLV cells of the mutants containing mutations in region 1, a region defined by deletion analysis as overlapping with the primary encapsidation signal. The titers of vector virus mutants allow the mutants to be grouped into three classes. The titer for the first group, consisting of five mutants (628, 659, 677, 628/677, and Δ 551-698), was 7 to 40 lower than that of the parental vector. A second group, consisting of one mutant (559), had a titer lower than that of the parental RW 1 by a factor of 2. A third group (mutants 589, 622, 650, and 667) had vector virus titers that were similar to that of the parental vector. The vector Δ 1147-6819, from which RW 1 was derived, had a titer similar to that of RW 1. The titers of these vectors indicate that disruption of either stem (628, 659, and 677) reduces the replication efficiency by a factor of 7 and that disruption of both stems (628/677) results in a reduction of virus replication by a factor of 40. The replication of 628/677 was similar to the replication of a previously described mutant (34) containing a deletion of the primary signal region (i.e., Δ 551-698) (Fig. 4).
FIG. 4.
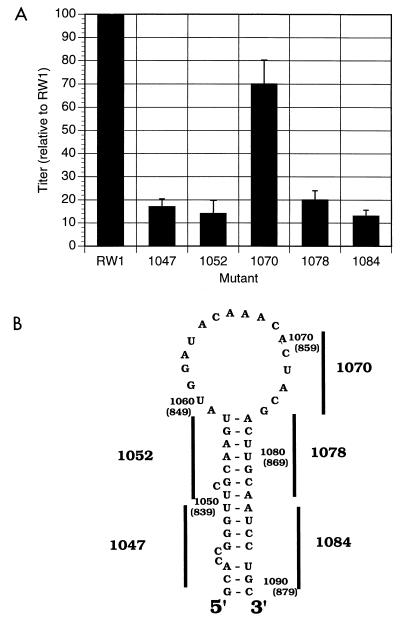
Replication and relative titers of vectors with mutations in the primary encapsidation signal. (A) Relative titers (normalized to that of RW 1) from three independent experiments, presented as the average with the standard deviation indicated. The average absolute titer of RW 1 from these three experiments was 4 × 102 CFU/5 × 104 FLK target cells. (B) Predicted stable RNA structures in region 1 that represent the primary encapsidation signal and general locations of the mutations made in each mutant (black bars with numbers) to disrupt the RNA stem structure. Detailed locations of mutations made are shown in Fig. 3. Numbers indicate coordinates relative to the proviral DNA or the genomic RNA (numbers in parentheses).
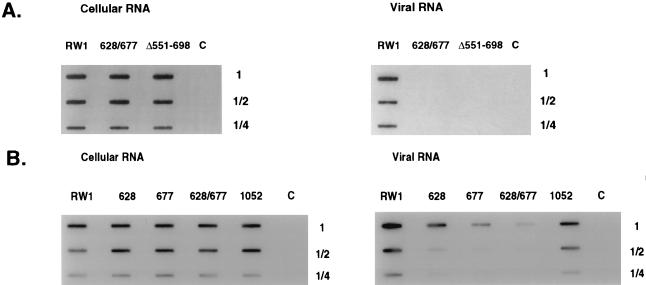
The RNA encapsidation efficiency of selected region 1 mutants (i.e., 628, 677, and 628/677) was tested (Fig. 5). As a measure of the sensitivity of the assay, the expression of viral RNA in cells and the amount encapsidated into virus particles were compared between the mutant 628/677 and a previously described mutant (Δ 551-698) (34) that was derived from the BLV vector Δ 1147-6819 (Fig. 5A). For each of these mutants, the expression of viral RNA in cells was similar to that for RW 1. However, viral RNA from virus particles was poorly detected for both 628/677 and Δ 551-698, indicating that these two mutants lead to a minimal 40-fold reduction in RNA encapsidation. Expression of viral RNA in cells for each of the mutants 628 and 677 was similar to that for pRW 1 (Fig. 5B). In contrast, the amount of viral RNA detected in virus particles of each mutant tested was drastically lower than that of RW 1 (Fig. 5B). Viral RNA detected for 628 and 677 was sevenfold lower than that of RW 1. Viral RNA detected for 628/677 was 40-fold lower than that of RW 1. Titers of vectors from helper cells were similar to that observed from FLK-BLV cells (data not shown). These data indicate that the measured reductions in replication efficiency of these vectors correlates to reductions in RNA encapsidation efficiency. These results support the conclusion that mutation of either or both of the two stems disrupts the primary encapsidation signal.
FIG. 5.
Analysis of relative cytoplasmic and virion RNA levels. (A) Sensitivity of BLV RNA detection. (B) Measurement of RNA levels from selected derivatives of RW 1. For cytoplasmic RNA analysis, RNA from five petri dishes (100-mm diameter) of infected or uninfected cells was twofold serially diluted and blotted onto nitrocellulose paper. For virion RNA analysis, RNA from the equivalent of the supernatant medium from 25 petri dishes (100-mm diameter) (250 ml, total) was twofold serially diluted and blotted. The probe used was a randomly primed probe in the U5 region of the BLV long terminal repeat. Lanes C, viral RNA from uninfected cells.
Replication and RNA encapsidation efficiencies of BLV encapsidation region 2 mutants.
Figure 6 shows the results of vector virus production from FLK-BLV cells of the mutants containing mutations in region 2, the previously defined region that overlaps with the secondary encapsidation signal. Figure 3B shows the locations of the mutations made in the secondary encapsidation signal region. The mutations introduced into each mutant resulted in the creation of a SpeI restriction site. Mutants in region 2 spanned one predicted stable stem-loop structure.
FIG. 6.
Replication of vectors with mutations in the secondary encapsidation signal. (A) Relative titers (normalized to that of RW 1) from three independent experiments, presented as the average with the standard deviation indicated. The average absolute titer of RW 1 from these three experiments was 3.5 × 102 CFU/5 × 104 FLK target cells. (B) Predicted stable RNA structure in region 2 that represents the secondary signal and general locations of the mutations made in each mutant (black bars with numbers) to disrupt the RNA stem structure. Detailed locations of mutations made are shown in Fig. 3. Numbers indicate coordinates relative to the proviral DNA or the genomic RNA (numbers in parentheses).
The titers of vector virus mutants allow the mutants to be grouped into two classes. The first class, consisting of four mutants (1047, 1052, 1078, and 1084), had a titer sixfold lower than that of the parental vector. A second class had 1 mutant (1070) with a titer that was two-thirds of that of the parental RW 1. The titers of these vectors (i.e., 1047, 1052, 1078, and 1084) indicate that disruption of the stem reduces the replication efficiency by a factor of 6 and supports the conclusion that mutation of the stem structure disrupted the secondary encapsidation signal.
The RNA encapsidation efficiency of a selected region 2 mutant (1052) was tested (Fig. 5B), and the amount of RNA in cells was found to be similar to that of RW 1 (Fig. 5B). In contrast, the amount of viral RNA detected in virus particles of 1052 was fourfold lower than that of RW 1 (Fig. 5B), indicating that the measured reduction in replication efficiency correlates well with the reduction in RNA encapsidation efficiency. Titers of vectors from helper cells were similar to that observed from FLK-BLV cells (data not shown). These results indicate that mutation of the stem in region 2 disrupts the secondary encapsidation signal.
Effects of compensatory mutations that restore the predicted RNA secondary structures on the replication efficiency of encapsidation mutants.
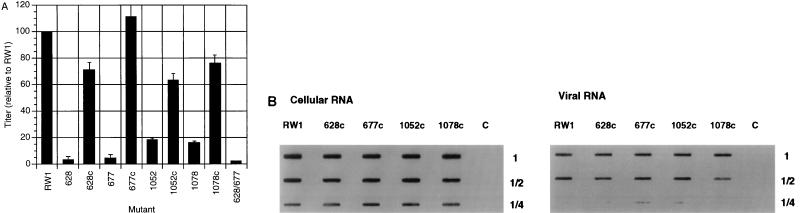
To confirm the correlation between disruption of predicted RNA secondary structures and the influence of the mutations on replication and RNA encapsidation efficiencies, compensatory mutations were made to restore the predicted RNA secondary structures (Fig. 7). Compensatory mutations were made in both the primary and secondary encapsidation signals, and these vectors were tested for replication efficiency relative to the parental vector, pRW 1. In region 1, compensatory mutants of 628 and 677 (628c and 677c) were made and tested; in region 2, compensatory mutants of 1052 and 1078 (1052c and 1078c) were made and tested (Fig. 7). The 628 mutant disrupts one of the predicted stem-loop structures in the primary encapsidation signal, while 677 disrupts the other predicted stem-loop structure. The compensatory mutant 628c replicated at a level about 70% of that of RW 1, indicating that the compensatory mutation increased replication efficiency (Fig. 7A). The replication of 628c was about 12-fold higher than that of 628. The mutant 677c replicated at a level comparable to that of RW 1 (an approximately 20-fold increase relative to that of 677), indicating that the compensatory mutation restored the replication efficiency of the vector. Replication of the compensatory mutant 1052c was restored to about three-fourths of that of RW 1, which was about a fourfold increase compared to the replication efficiency of 1052. Replication of the compensatory mutant 1078c was also restored to about three-fourths of that of RW 1, indicating an approximate fourfold increase compared to the replication efficiency of 1078 (Fig. 7A).
FIG. 7.
Replication of compensatory mutants. (A) Relative titers (normalized to that of RW 1) of vectors with compensatory mutations from three independent experiments, presented as the average with the standard deviation indicated. The average absolute titer of RW 1 from these three experiments was 3 × 102 CFU/5 × 104 FLK target cells. (B) Analysis of relative cytoplasmic and virion RNA levels. Cytoplasmic and virion RNA were purified and analyzed as described for Fig. 5. Lanes C, viral RNA from uninfected cells.
The RNA encapsidation efficiency of each compensatory mutant (i.e., 628c, 677c, 1052c, and 1078c) was tested (Fig. 7B). The expression of viral RNA in cells for the compensatory mutants 628c, 677c, 1052c, and 1078c was measured, and the amount detected for each mutant was similar to that for RW 1 (Fig. 7B). The amount of viral RNA detected in virus particles of each compensatory mutant was found to be similar to that of RW 1 (Fig. 7B). These data indicate that the measured increase in replication efficiency of each of these compensatory mutants correlates to the increase in RNA encapsidation efficiencies and is comparable to that of RW 1. These data indicate that compensatory mutations which restore the predicted RNA secondary structures in the primary or secondary encapsidation signal results in vectors with replication and RNA encapsidation efficiencies similar to that of the parental vector containing the wild-type primary or secondary encapsidation signal. This observation indicates that the encapsidation signal is largely recognized by secondary or tertiary structures.
Replacement of the BLV encapsidation signal region with the encapsidation signal region of either HTLV-1 or HTLV-2.
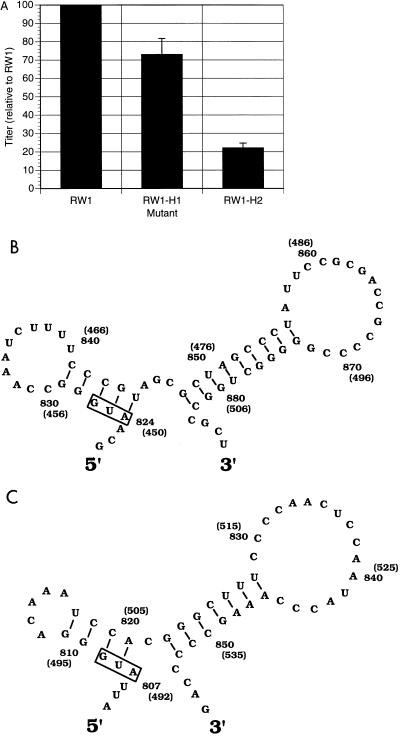
To test for conservation of function among the encapsidation signals in the BLV/HTLV genus, we constructed derivatives of pRW 1 where the BLV encapsidation signal region (containing both regions 1 and 2) was replaced with the encapsidation signal region of either HTLV-1 or HTLV-2. These derivatives were then replicated in parallel with RW 1 to determine their relative replication efficiencies. Figure 8 indicates that the replication efficiency of the RW 1 derivative containing the HTLV-1 encapsidation signal in place of the BLV signal (RW1-H1) was approximately three-fourths of that of RW 1. The replication and RNA encapsidation efficiency of the RW 1 derivative with the HTLV-2 encapsidation signal (RW1-H2) was one-fifth of that of the parental vector (Fig. 8A). Since RW1-H1 and RW1-H2 replicated, we analyzed the predicted stable RNA secondary structures in the HTLV-1 and HTLV-2 sequences included in these vectors, specifically just downstream of the HTLV-1 and HTLV-2 gag start codons. Two stable stem-loop structures were predicted for each (Fig. 8B). The HTLV-2 structures more closely resemble the two BLV stem-loops. That these structures resemble the primary encapsidation signal of BLV provides further evidence that the BLV encapsidation signal is largely recognized by secondary or tertiary structures. Taken together, these data indicate that the encapsidation signal regions of either HTLV-1 or HTLV-2 can lead to BLV replication and suggest some level of conservation of encapsidation signal function.
FIG. 8.
Replication of BLV vectors with either the HTLV-1 or HTLV-2 genome region predicted to contain the encapsidation signal in place of the BLV encapsidation signal region. (A) Relative titers (normalized to that of RW 1) from three independent experiments, presented as the average with the standard deviation indicated. The average absolute titer of RW1 from these three experiments was 3 × 102 CFU/5 × 104 FLK target cells. RW1-H1 is RW 1 with the predicted HTLV-1 encapsidation signal region in place of the BLV region containing the primary and secondary encapsidation signals; RW1-H2 is pRW 1 with the predicted HTLV-2 encapsidation signal region in place of the BLV encapsidation region. (B and C) RNA secondary structures for the primary encapsidation signals of HTLV-1 (B) and HTLV-2 (C). Structures were predicted by computer-assisted analysis of suboptimal and minimal free-energy foldings. The regions shown are directly downstream of the HTLV-1 or HTLV-2 gag start codon. The gag AUG is boxed. Nucleotide numbering is based on the complete HTLV-1 (47) or HTLV-2 (48) proviral DNA sequence.
DISCUSSION
Based on a previous deletion analysis study where we had mapped the RNA regions that overlap with the primary and secondary encapsidation signals of BLV, a global minimum-energy optimal folding for the entire BLV RNA was analyzed. Two stable stem-loop structures in the gag gene (just downstream of the gag start codon) were predicted to overlap with the region mapped to contain the primary encapsidation signal, while one stable stem-loop structure was predicted in gag (in the capsid domain) to overlap with the region mapped to contain the secondary encapsidation signal. We introduced a series of mutations into these predicted structures to study the sequence and structural information contained within these regions. Mutants were screened for replication and RNA encapsidation efficiencies.
Mutations that disrupted either or both of the predicted stem-loop structures in the primary encapsidation signal reduced the replication efficiency by a factor of 7 or 40, respectively. RNA encapsidation efficiencies for each mutant were determined by comparing the level of viral RNA in the cytoplasm and that in virus particles to RNA levels of the parental vector, RW 1. Levels of viral RNA in the cytoplasm for each mutant tested were similar to that for the parental vector, but a 7- to 40-fold reduction in virion RNA was observed for mutants with mutations in either or both of the predicted stem-loop structures relative to RW 1, respectively. The phenotype of the mutant with both stem-loop structures disrupted was similar (Fig. 5) to that of a previously characterized mutant, Δ 551-698, that contained a deletion of the entire primary encapsidation signal region (34). Several mutants were tested that had disruptions in the predicted stem-loop structure in the secondary signal. These mutants were found to have a reduction, by a factor of 6, in replication efficiency, and one mutant (i.e., 1052) was found to have a fourfold reduction in RNA encapsidation efficiency, indicating that the efficiency of RNA encapsidation was largely responsible for the reduction in virus replication. These results indicate a structure-function relationship between (i) the predicted RNA secondary structures in the primary and secondary encapsidation regions and (ii) the efficiency of replication and RNA encapsidation.
The reduction in virus titers that we observed correlates well, in general, with the measured reductions in RNA encapsidation efficiency. However, this does not preclude the possibility that other cis-acting elements that overlap with the encapsidation signal are influenced by the introduced mutations or that the cis elements we have mapped affect more than one step in BLV replication. For example, a cis-acting element has been shown for HIV-1 to influence both RNA packaging and synthesis of proviral DNA (39).
To confirm these structure-function relationships, we created in several of the mutants a series of compensatory mutations that would be expected to restore the predicted RNA secondary structures disrupted by the mutations initially introduced. These compensatory mutants led to levels of replication and RNA encapsidation comparable to those of virus containing the wild-type encapsidation signal (Fig. 7). This finding indicates that these mutants confirm the biological significance of the tested RNA secondary structures in BLV replication and RNA encapsidation. Therefore, they represent the primary and a secondary encapsidation signal of BLV.
A derivative of RW 1 that contains mutations that change bases on the opposite side of the stem mutated by 677 (i.e., 659) had a similar effect on virus titer (Fig. 4), indicating that the observed increase in virus titer and RNA encapsidation of 677c is not due to the 659 mutations alone but rather a result of restoration of the secondary structure in this region. (It should be noted that 677c was created by introducing the 659 mutations into 677.) This is also evident for the BLV secondary signal stem-loop structure by analyzing the replication of 1052, 1078, 1052c, and 1078c.
To determine if the encapsidation signal region of BLV could be replaced by an encapsidation signal from other members of the BLV/HTLV genus, we constructed vectors in which the BLV encapsidation region was replaced with sequences near the 5′ end of either HTLV-1 or HTLV-2 that we predicted would contain the encapsidation regions of these viruses. The HTLV-1 sequences were derived from pHTLV-1-CMVneo (13), a vector constructed from a provirus cloned from the lymphocytic cell line CS-1; HTLV-2 sequences were from pH6neo (11, 48). The sequences from either HTLV-1 or HTLV-2 used to replace the BLV encapsidation signal region spanned from just downstream of the primer binding site to the central region of the gag gene. Replacement of the BLV RNA region containing the primary and secondary encapsidation signals with a similar region from HTLV-1 led to replication at three-fourths of that of the parental virus, while replacement with a similar region from HTLV-2 led to replication at a level one-fifth of that of the parental virus. The primary sequences of BLV, HTLV-1, and HTLV-2 are well conserved, and computer-assisted analysis of the predicted stable RNA secondary structures in HTLV-1 and HTLV-2 reveals two stem-loop structures located at the beginning of the gag open reading frame that resemble the primary BLV signal (Fig. 8B). Further studies will reveal if these predicted structures play a role in BLV and HTLV RNA encapsidation.
Sequences involved in dimerization of retroviral RNA (the dimer linkage sequence [DLS]) have been identified in cell-free systems. The DLS for several retroviruses, with the exception of RSV, has been previously mapped in vivo to a region overlapping the encapsidation region (8, 14, 24, 36, 42, 50). The BLV DLS appears to be near the 5′ end of the viral RNA (24, 25). This region has been found to contain sequences or structures important for cell-free binding of the RNA to viral matrix-associated proteins, nucleocapsid protein, or the nucleocapsid domain of the Gag polyprotein precursor. Our results indicate that a structure important for cell-free dimer formation (nucleotides 445 to 574 in the proviral DNA) does not overlap with the primary encapsidation signal, but that a structure important for cell-free matrix-associated protein binding (nucleotides 628 to 682 in the proviral DNA) (24, 25) does overlap with the primary signal.
Analysis of the predicted folding indicates that the structures predicted for the primary and secondary encapsidation signals were also present when the entire BLV genome is simultaneously folded. The fact that the encapsidation signal structures were preserved in the secondary structure of the entire BLV genome strengthened their potential biological relevance. Our data indicate that these structures function as the encapsidation signal. In addition, these structures are located in the gag open reading frame, which suggests that there is selective pressure for simultaneously maintaining not only an efficient viral RNA for encapsidation but also an efficient mRNA template for translation of the gag gene.
It is remarkable that there are no very long range interactions as described for other RNA virus complete genome foldings (40), which can adopt a “fish tail” configuration. Such a configuration facilitates a close proximity between the 5′ and 3′ ends. It is interesting that this is not observed with BLV, as this close proximity of 5′ and 3′ ends could be envisioned as being important for the strand transfer process during reverse transcription. Since two copies of the retroviral genomic RNA are encapsidated into virus particles, it would also be interesting to see what potential stable RNA structures form between two RNA molecules. Currently, methods to evaluate this are not readily available.
We have identified the RNA secondary structures for the primary encapsidation signal of BLV in addition to a secondary encapsidation signal in the gag gene. The approximate twofold reduction in replication efficiency of mutant 559 indicates that other secondary encapsidation signals are located upstream of the primary signal. This conclusion is supported by the work of Kurg et al. (29), who showed that mutation or deletion in the same region resulted in a three- or fivefold reduction in packaging efficiency of BLV RNA, respectively.
The RNA secondary structures that we have shown to be the encapsidation signal of BLV resemble structures found in HTLV-1 and HTLV-2, suggesting some degree of conservation in this signal among members of the HTLV/BLV genus of the Retroviridae family. Further characterization of these RNA structures will aid in identifying the retroviral proteins that interact with retroviral RNA to initiate the encapsidation process.
ACKNOWLEDGMENTS
We thank Xiao-Juan Bi, Brad Seufzer, and Nicolas Strehl for superior technical assistance and Jean-Yves Sgro for help in analyzing the global secondary structure of the full-length BLV RNA. We also thank Dan Loeb and Nito Panganiban for helpful suggestions; Ann Palmenberg and Jean-Yves Sgro for stimulating conversations; and Ann Palmenberg, Jean-Yves Sgro, and Shiaolan Yang for critical reviews of the manuscript.
This work was supported by grants CA22443 and CA07175 from the Public Health Service. L.M.M. was supported by NRSA viral oncology training grant CA09075-17, by NIH postdoctoral fellowship F32 AI08763-01, and by State of Nebraska Cancer and Smoking Related Disease program grants 288151 and 288177.
REFERENCES
- 1.Abrahams J P, van den Berg M, van Batenburg E, Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990;18:3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam M A, Miller A D. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. J Virol. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armentano D, Yu S-F, Kantoff P W, von Ruden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronoff R, Linial M. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag gene. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieth E, Gabus C, Darlix J-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruccoleri R E, Heinrich G. An improved algorithm for nucleic acid secondary structure display. Comput Appl Biosci. 1988;4:167–173. doi: 10.1093/bioinformatics/4.1.167. [DOI] [PubMed] [Google Scholar]
- 10.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen I S Y, McLaughlin J, Gasson J C, Clark S C, Golde D W. Human T-cell leukemia virus type II transforms normal human lymphocytes. Proc Natl Acad Sci USA. 1983;80:7006–7009. doi: 10.1073/pnas.80.22.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clavel F, Orenstein J M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J Virol. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copeland K F T, Haaksma A G M, Derse D, Heeney J L. Detection of human T-cell leukemia virus 1 permissive cells using cell lines producing selectable recombinant virions. J Virol Methods. 1994;50:219–226. doi: 10.1016/0166-0934(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 14.Darlix J-L, Gabus C, Nugeyre M-T, Clavel F, Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 15.Derse D, Martarano L. Construction of a recombinant bovine leukemia virus vector for analysis of virus infectivity. J Virol. 1990;64:401–405. doi: 10.1128/jvi.64.1.401-405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Embretson J E, Temin H M. Lack of competition results in efficient packaging of heterologous murine retroviral RNAs and reticuloendotheliosis virus encapsidation-minus RNAs by the reticuloendotheliosis virus helper cell line. J Virol. 1987;61:2675–2683. doi: 10.1128/jvi.61.9.2675-2683.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freier S M, Kierzek R, Jaeger J A, Sugimito N, Caruthers M H, Neilson T, Turner D H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallis B, Linial M, Eisenman R. An avian oncovirus mutant deficient in genomic RNA: characterization of the packaged RNA as cellular messenger RNA. Virology. 1979;94:146–161. doi: 10.1016/0042-6822(79)90445-8. [DOI] [PubMed] [Google Scholar]
- 20.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi T, Shisuo T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 22.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh I, Kyushiki H, Sakamoto Y, Ikawa Y, Yoshinaka Y. Bovine leukemia virus matrix-associated protein MA(p15): further processing and formation of a specific complex with the dimer of the 5′ terminal genomic RNA fragment. J Virol. 1991;65:6845–6855. doi: 10.1128/jvi.65.12.6845-6855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh I, Yasunaga T, Yoshinaka Y. Bovine leukemia virus RNA sequences involved in dimerization and specific gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J Virol. 1993;67:1830–1839. doi: 10.1128/jvi.67.4.1830-1839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koyama T, Harada F, Kawai S. Characterization of Rous sarcoma virus mutant defective in packaging its own genomic RNA: biochemical properties of mutant TK15 and mutant-induced transformants. J Virol. 1984;51:154–162. doi: 10.1128/jvi.51.1.154-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurg A, Sommer G, Metspalu A. An RNA stem-loop structure involved in the packaging of bovine leukemia virus genomic RNA in vivo. Virology. 1995;211:434–442. doi: 10.1006/viro.1995.1425. [DOI] [PubMed] [Google Scholar]
- 30.Lever A, Gottlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luban J, Goff S P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann R, Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985;54:401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 34.Mansky L M, Krueger A E, Temin H M. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J Virol. 1995;69:3282–3289. doi: 10.1128/jvi.69.6.3282-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansky L M, Temin H M. Lower mutation rate of bovine leukemia virus relative to that of spleen necrosis virus. J Virol. 1994;68:494–499. doi: 10.1128/jvi.68.1.494-499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marquet R, Baudin F, Gabus C, Darlix J-L, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osborne W R A, Miller A D. Design of vectors for efficient expression of human purine nucleoside phosphorylase in skin fibroblasts from enzyme-deficient humans. Proc Natl Acad Sci USA. 1988;85:6851–6855. doi: 10.1073/pnas.85.18.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paillart J C, Berthoux L, Ottmann M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmenberg, A. C., and J.-Y. Sgro. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol., in press.
- 41.Parolin C, Dorfman T, Palu G, Gottlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prats A-C, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pugatsch T, Stacey D W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983;128:505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- 44.Rein A. Retroviral RNA packaging: a review. Arch Virol Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 45.Rizvi T A, Panganiban A T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagata N, Yasunaga T, Tsuzuku-Kawamura J, Ohishi K, Ogawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci USA. 1985;82:677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Sgro, J. Personal communication.
- 48.Shimotohno K, Takahashi Y, Shimizu N, Gojobori T, Chen I S Y, Golde D W, Miwa M, Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: a new open reading frame for the protease gene. Proc Natl Acad Sci USA. 1985;82:3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorge J, Ricci W, Hughes S H. cis-acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983;48:667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundquist W, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Temin H M. Studies on carcinogenesis by avian sarcoma viruses. VIII. Glycolysis and cell multiplication. Int J Cancer. 1968;3:273–282. doi: 10.1002/ijc.2910030213. [DOI] [PubMed] [Google Scholar]
- 52.Van der Maaten M, Miller J. Replication of bovine leukemia virus in monolayer cell cultures. Bibl Haematol. 1976;43:360–362. doi: 10.1159/000399166. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe S, Temin H M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5′ long terminal repeat and the start of the gag gene. Proc Natl Acad Sci USA. 1982;79:5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1985. [Google Scholar]
- 55.Yang S, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 57.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]