Abstract
Comparisons among sequences predicted to encode the major late promoter (MLP) of adenoviruses from a wide variety of host species show that an inverted CAAT box is among the most highly conserved transcription elements found in the putative MLPs. The high degree of conservation suggests that the CAAT box plays an important role in the function of the MLP in vivo, an idea supported by a previous mutational analysis of the core CCAAT sequence. To address the importance of the CAAT box, in terms both of quantitative levels of transcription and of specificity, a further set of mutations was created and examined in the context of the viral genome. One mutation, CAAT5, contains individual changes at five positions, four of which correspond to invariant residues in a CAAT box consensus derived either by computer analysis or empirically. The CAAT5 mutation had no discernible phenotype by itself but when coupled with the previously described USF0 mutation, which disrupts binding of the upstream stimulating factor (USF) but is otherwise phenotypically silent, gave rise to virus with a severe replication deficiency. Nuclear run-on assays showed that transcription initiation at the mutant MLP was significantly reduced compared with that of the wild type or the virus containing CAAT5 alone. Replication of the double mutant was lower than that of the previously described USF0::CCCAT virus, suggesting that the additional mutations in the CAAT box had further lowered the binding of transcription factor CP1 (also called CBF, NF-Y). Replacement of the CAAT box by an ATF binding site or an OCT1 binding site had no phenotypic effect in an otherwise wild-type background, but replacement in a USF0::CCCAT background led to only partial restoration of the wild-type phenotype. The failure to restore the functional redundancy normally exhibited by the CAAT box and the proximal upstream activating element is consistent with the idea that in the adenovirus MLP the CAAT box is preferred over others as the distal transcriptional element.
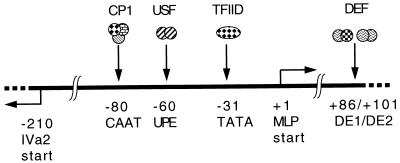
The major late promoter (MLP) of human adenoviruses belonging to subgroup C is one of the most intensively studied examples of a eukaryotic polymerase II promoter. Early studies, using in vitro transcription and plasmid-borne transfection assays, defined the requirements for specific cis-acting sequences for quantitative and accurate transcription initiation (reviewed in reference 2). These studies identified several transcriptional elements and allowed the isolation of two cellular transcriptional activating factors (7, 8, 31, 39). More recently, genetic systems were developed to examine the function of the MLP in the context of the viral genome, either at ectopic sites (25) or at the normal genomic location (4, 36). Together, the results strongly support the promoter structure shown in Fig. 1. The MLP contains two basal elements, namely, a TATA box and an initiator element (INR) (43), and two upstream activating elements, namely, the proximal upstream activating element (UPE) that binds the helix-loop-helix transcription factor USF (16, 39) and an inverted CAAT box that binds the heterotrimeric transcription factor CP1 (also called CBF or NF-Y) (7, 29). Three further activating elements are located downstream of the transcription start site and within the first intron of the major late transcription unit, elements DE1, DE2b, and DE2a, which bind DEF-A and DEF-B (20, 32). Recently, DEF-A and DEF-B have been shown to consist of a heterodimer and a homodimer, respectively, of the virus-specified protein IVa2 (48).
FIG. 1.
Genetic organization of the MLP of human adenovirus. The elements are shown with their cognate binding factors. The region is located at ∼16.8 map units from the left-hand end of the viral genome with the MLP start site at bp 6030. The divergent promoter IVa2 is also shown. The region lies within the coding sequences of the essential DNA polymerase gene, which is encoded on the opposite strand.
Previous genetic analysis of the UPE showed that it plays an important role in the function of the MLP in the course of a normal viral infection, although this role was evident only when mutations were created in both the UPE and at least one other transcriptional element (26, 37). The analysis also suggested that the CAAT box contributes to MLP function, but the phenotypic effects of mutation in the element were less marked than those observed with UPE mutations. Nevertheless, recent sequence analyses of potential MLPs from adenoviruses isolated from diverse mammalian species demonstrated that the CAAT box was conserved in all viruses examined, implying its functional significance (44). This implication was one of the reasons that led us to reconsider the importance of this more distal upstream element in viral infection, and in particular the possibilities that the CAAT box plays a specific role in transcription from the MLP and that the element cannot be substituted by other common activating sequences.
The inverted CCAAT box is located about 80 nucleotides (nt) upstream of the transcription start site and binds the cellular factor CP1 (7, 29). CP1 (CBF) consists of three subunits, CBF-A, CBF-B, and CBF-C, all of which are needed for DNA binding (40). There is a high degree of sequence identity with segments of the Saccharomyces cerevisiae HAP3, HAP2, and HAP5 polypeptides, respectively, (30, 41, 52), and the conserved regions of CBF-A and CBF-C have sequence similarities to the histone fold motifs of histones H2B and H2A, respectively (40). The transcriptional activation properties of CBF have been demonstrated in vitro by using several promoters, including the MLP (29), but the mechanism by which activation is achieved is unknown, although it is presumed that CP1 must interact with one or more proteins in the preinitiation complex.
To investigate the functional significance of the CAAT box in the correct genomic context, several mutations were created in the CAAT box alone or in combination with mutations in other transcriptional elements and analyzed for their effects during the viral life cycle. The mutations were designed with two questions in mind. First, a multiple mutation was created in the CAAT box, on the assumption that this might achieve a greater reduction in the binding of CP1 compared to that of the single point mutation analyzed previously, so that the importance of the element to quantitative transcription from the MLP could be more readily assessed. Second, the CAAT box was replaced by other transcription factor binding sites to determine if the element has a specific role in MLP function. The results presented in this study show that the CAAT box has an important role in viral replication and suggest that there is some degree of specificity to the CAAT box function. They also confirm the previous observation that there is functional redundancy between the two upstream activating elements (36) and support the suggestion (37) that there is a functional interaction between the CAAT box and the TATA box.
MATERIALS AND METHODS
Creation of mutations in the MLP of human adenovirus. (i) M13 mutagenesis.
The methods of M13-based mutagenesis of the MLP by the techniques of Kunkel (21) have been described in detail previously (36). Individual M13 virus isolates arising from the mutagenesis were screened for those containing the mutation by DNA sequencing using the dideoxy method (38). The oligonucleotides used to produce nucleotide substitutions within the MLP region were as follow: 9403-5C2 (to create five point mutations in the CAAT box in the wild-type background; MLP antisense), 5′-GGC CTA CAC CTA tAA gCC cAT aAC aTT CCT TGA TGC CG-3′; 9502-5C2U (to create the same five point mutations in the CAAT box in the USF0 background; MLP antisense), 5′-CAC CTA tAA gCC cAT aAC aTT CCT TGA TGC CG-3′; 9207-AT2 (to replace the CAAT box with an ATF binding site in the wild-type background; MLP antisense), 5′-GGC CTA CAC CTA CAA cga cgT CAC CTT CCT TGA TGC CG-3′; 9209-AT2U (to replace the CAAT box with an ATF binding site in the USF0 background; MLP antisense), 5′-AGC ATA CAC CTA CAA cga cgT CAC CTT CCT TGA TGC CG-3′; and 9409-OC2 (to replace the CAAT box with an OCT1 binding site in both the wild-type and USF0 backgrounds; MLP antisense), 5′-CCT ACA AAC CcA Ttt gCa TCC TTG ATG CC-3′. The mutated nucleotides are in lowercase.
PCR mutagenesis.
Some mutations were created by PCR using Taq DNA polymerase and a PCR Core kit (Boehringer Mannheim) as described by the manufacturer. Briefly, two complementary oligonucleotides were designed for each mutation. First, two separate PCRs were performed with an MLP-containing plasmid that contains SP6 and T7 RNA polymerase promoters flanking the MLP. The T7 primer and one of the two complementary oligonucleotides were used as PCR primers in one reaction, and the SP6 primer and the other oligonucleotide were used in the other reaction. Then the PCR products were used as templates in the second PCR using the T7 and SP6 primers. The cycling condition was as follows: 1 min at 94°C; 30 cycles of 1 min at 94°C, 2 min at 55°C, and 3 min at 72°C; and 7 min at 72°C. The PCR products were cloned into a plasmid vector, and the mutations were confirmed by sequencing performed with dye terminators on an ABI 373A machine in the Columbia Cancer Center DNA facility. The oligonucleotides used to produce nucleotide substitutions within the MLP region were as follow: 9608-27T1 (to mutate the TATA box; MLP sense), 5′-GGG GGG CTA TAg AAG GGG GTG GGG GCG CG-3′; and 9608-27T2 (to mutate the TATA box; MLP antisense), 5′-CGC GCC CCC ACC CCC TTc TAT AGC CCC CC-3′. The mutated nucleotides are shown in lowercase.
Oligonucleotides were purchased from Oligos Etc., Genosys, or the oligonucleotide-synthesizing facility, Comprehensive Cancer Center, Columbia University.
Testing of MLP mutations by overlap recombination.
After both types of mutagenesis, the XhoI-to-HindIII MLP fragment was cloned into pMR2, which contains adenovirus type 5 (Ad5) DNA sequences extending from the left end to bp 9523. Overlap recombination with DNA-protein complex (DNA-PC) from virus LLX1 was done according to published procedures (36, 50). Briefly, the DNA-PC was cleaved with PaeR7I and ClaI, and the unpurified mixed fragments were cotransfected with the pMR2-derived plasmids into human A549 cells in a direct plaque assay. Overlap between the large right terminal fragment of LLXI extending from bp 8254 and the plasmid sequence, which extends through bp 9523 will generate a mutant virus genome. The presence of the mutations in the resulting viruses was confirmed by cloning the XhoI-to-HindIII MLP fragment into a plasmid containing SP6 and T7 RNA polymerase promoters, followed by sequencing with dye terminators. Mutant viruses were plaque purified once before further analysis. It should be emphasized that the use of mixed fragments of LLX1 DNA-PC in the overlap reaction allows recovery of virus genomes that have experienced more than one recombination event, a likely source of the rare CAAT5 viruses arising in transfections with the CAAT5::TATA27 mutation-containing plasmids that failed to give rise to the expected virus genotype (see Results).
Measurement of viral replication cycles.
Growth curves were performed on human A549 cells, derived from a small cell carcinoma of the lung (13). Cells were grown to confluency in 35-mm-diameter dishes in Dulbecco modified Eagle medium plus 10% supplemental calf serum (HyClone). Infections were performed by removing the medium and adding 0.2 ml of virus at a multiplicity of infection (MOI) of 10 PFU per cell and incubating the dishes at 37°C for 1 h, with periodic shaking. The plates were then overlaid with infecting fluid (22). The infected cells were harvested at intervals by freezing individual dishes, and virus was liberated by repeated freezing and thawing. Titration was performed on A549 cells by fluorescent focus assay (35).
Measurement of DNA replication.
Viral DNA was extracted, by a modification (50) of the Hirt technique (18), from A549 cells infected identically to those used for the growth curves. The DNA was digested with XhoI, run on an 1% agarose gel, and transferred to nitrocellulose (Schleicher & Schuell). The filter was blocked for 2 h in a solution containing 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate, 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 100 μg of carrier DNA (salmon sperm DNA) per ml and then probed with the MLP fragment, which had been labeled by random priming synthesis (11) by using [α-32P]dATP (3,000 Ci/mmol; NEN). The filter was washed first with 2× SSC–0.5% sodium dodecyl sulfate for 5 min at room temperature, with 2× SSC–0.1% sodium dodecyl sulfate for 15 min at room temperature, and then with 0.1× SSC-0.5% sodium dodecyl sulfate for 2 h at 65°C, air dried, and exposed to a Fuji RX film.
Nuclear run-on transcription assay.
The techniques for preparation of nuclei, nuclear transcription, and analysis of labeled nascent RNA by hybridization to M13 single-stranded probes on nylon membranes have been described in detail previously (36) and were used with some modifications based on a published protocol (15). Briefly, A549 cells grown in 175-cm2 flasks were prepared 1 day before infection. The confluent monolayers were infected at an MOI of 10 focus-forming units (FFU) per cell with the various viruses and incubated for 36 h at 37°C. The infected monolayers (approximately 5 × 107 cells) were washed three times with ice-cold phosphate-buffered saline and resuspended in 4 ml of Nonidet P-40 (NP-40) lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40). The cells were kept on ice for 5 min and centrifuged for 5 min at 500 rpm in a model CR6000 IEC refrigerated centrifuge, and the nuclear pellet was resuspended in 4 ml of NP-40 buffer. They were centrifuged again, and the pellet was resuspended in 200 μl of glycerol storage buffer (50 mM Tris-HCl [pH 8.3], 5 mM MgCl2, 0.1 mM EDTA, 50% glycerol). Approximately half of each of the nuclear samples was used immediately for the transcription reaction, which was essentially as described before (36) except that incubation was at 37°C for 30 min. Following incorporation of [32P]UTP (100 μCi, 800 Ci/mmol; NEN), RNA was isolated and samples containing 1.5 × 107 cpm were hybridized by standard procedures to nylon membranes (Nytran; Schleicher & Schuell). The membrane was prepared on a slot blot apparatus to which had been added individual samples of 5 μg of single-stranded DNAs from two M13 clones. The M13 glyceraldehyde 3-phosphate dehydrogenase (GAPDH) clone used as an internal control is complementary to the sense orientation of the 1.3-kb PstI fragment of the rat GAPDH cDNA (12); the M13 L1 clone (36) is complementary to the Ad5 L1 region between the HindIII sites at bp 11565 and extending through bp 13651. Hybridization was at 65°C for 48 h, followed by extensive washing of the membrane and exposure on Fuji RX film. Radioactivity bound to the filter was quantitated with a PhosphorImager (Molecular Dynamics).
RESULTS
A five-point mutation in the MLP CAAT box.
The sequence analysis of MLPs from a set of adenoviruses, isolated from a diverse group of mammalian species, showed an absolute conservation of a CAAT box at an appropriate distance (60 to 80 nt) upstream of the predicted start site of transcription (44). This observation strongly suggests an important functional role for this element. In addition, previous genetic analysis of the CAAT box showed that a single point mutation in the 5′ A residue had significant phenotypic effects when coupled with otherwise transcriptionally silent mutations in the UPE or TATA box (36, 37). However, the effects were not as profound as those with a double UPE::TATA mutation, which was lethal (37). This finding suggests either that the CAAT box is not as important to the functioning of the MLP as the UPE or that the single point mutation is not as deleterious to CP1 binding as the four-point mutation in the UPE is to the binding of USF.
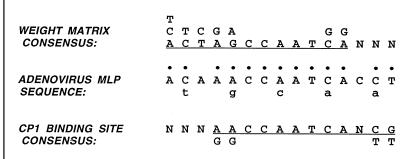
Others have shown experimentally that sequences outside the canonical CAAT box affect the binding of CP1 in vitro (3, 7), and a consensus outside the core CAAT sequence has been derived from a compilation of eukaryotic promoters (5). To test the importance of the MLP CAAT box further, we made mutations in 5 nt in and around the CAAT motif (Table 1) while maintaining the amino acid sequence of the essential viral DNA polymerase, encoded on the opposite strand. Figure 2 compares the newly created alterations to the previous experimental consensus and database compilation. Three of the five mutations are transversions to nucleotides not present in either consensus. Most importantly, a recent selection experiment, using purified CP1 and random oligonucleotides, showed that besides the invariant central CCAAT, the adjacent downstream residue was predominantly a C, although A was obtained ∼17% of the time. This finding suggests that the C may be important for high-affinity binding. The other two mutations are transitions in nucleotides that can vary in either consensus. The latter changes may have no effect on binding, although the 5′ A-to-G transition is in a nucleotide shown to display methylation interference (7).
TABLE 1.
Mutations in the elements of the adenovirus MLP
| Virus name | Sequencea
|
||
|---|---|---|---|
| CAAT boxb | UPEc | TATA boxd | |
| Wild type | AAGGTGATTGGTTTGT | GGCCACGTGACC | TATAAAA |
| TATA27 | TATAgAA | ||
| USF0 | aGCtACaTGgCC | ||
| CCCAT | AAGGTGATgGGTTTGT | ||
| vBS4 | AAtGTtATgGGcTTaT | ||
| vBS5 | AAtGTtATgGGcTTaT | aGCtACaTGgCC | |
| vBS1 | AAGGTGAcgtcgTTGT | ||
| vBS2 | AAGGTGAcgtcgTTGT | aGCtACaTGgCC | |
| vBS6 | AtGcaaATgGGTTTGT | ||
| vBS8 | AtGcaaATgGGTTTGT | aGCtACaTGgCC | |
| [vBS10]e | AAtGTtATgGGcTTaT | TATAgAA | |
Mutations are indicated in lowercase. The substitutions in the CAAT box in vBS1 and vBS2 create an ATF consensus (24) (underlined) and cause three changes in the amino acid sequence of the DNA polymerase coded on the strand opposite the MLP (Lys to Asn, Pro to Asp, and Ile to Val). The substitutions in the CAAT box in vBS6 and vBS8 create an OCT1 consensus (46) and cause two amino acids changes (Thr to Cys and Phe to Ile). Other mutations maintain the amino acid sequence of the DNA polymerase. Mutations in viruses TATA27, USF0, and CCCAT have been described previously (36, 37).
The CAAT box is a canonical sequence, CCAAT (5), and is inverted with respect to the start site of transcription.
The UPE is a typical E box and contains the site of highest affinity, RYCACGTGRY (1).
The TATA box is a canonical sequence, TATA(T/A)AA (5).
This mutation could not be recovered in virus.
FIG. 2.
Sequence of the CAAT box of the MLP compared with the consensus derived either from a weight matrix analysis (5) of eukaryotic promoters or a consensus (7) from promoters shown experimentally to bind CP1. The single bases shown above the weight matrix consensus correspond to those found in at least 20% of the promoters analyzed. The single bases shown below the CP1 binding consensus are alternative residues found in more than one promoter of high affinity. The dots indicate those residues in the adenovirus MLP that correspond to one or both consensus sequences. Bases shown in lowercase below the MLP sequence are the mutations in CAAT5.
The mutagenesis was performed in both wild-type and UPE mutation-containing plasmids, because it has been shown previously that mutations in single transcriptional elements usually have no phenotypic effects (26, 36, 37). The mutant MLP sequences were transferred to virus LLX1, using methods described in detail before (36). Cotransfections with both plasmids yielded virus, but the plaques of the CAAT5::USF0 mutant virus (vBS5) were much smaller than those of virus with the single-mutant CAAT5 (vBS4), and the individual infected cells displayed a distinctive cytopathic effect typical of MLP mutants with deficient replicative cycles.
Replicative cycles of viruses with mutations in the CAAT box.
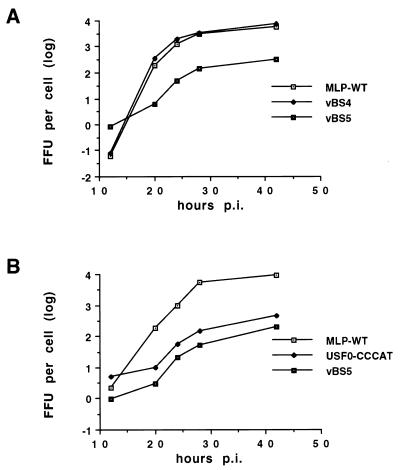
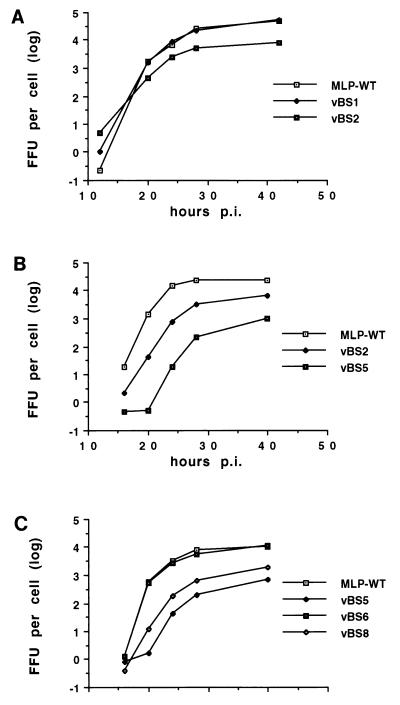
Human A549 cells were infected with the individual viruses at an MOI of 10, and samples were taken at intervals to measure viral replication in one-step growth curves. As shown in Fig. 3A, mutant virus vBS4 replicated with efficiency equal to that of the wild type, with almost identical eclipse periods and final viral yields. In contrast, the replication of mutant virus vBS5, with mutations in both the CAAT box and the UPE, was significantly less than that of the single-mutant virus, the final yield being approximately 20-fold lower at the final time point. The replication phenotype of the USF0 virus has been shown previously to be identical to that of the wild type (26, 36). These results suggest that the mutations in the CAAT box affect the binding of CP1 to the CAAT box and confirm previous evidence that the MLP has redundant upstream activating elements (36). The replicative abilities of vBS5 were compared to those of the previously described double mutant USF0::CCCAT (Fig. 3B). The reduction in viral replication of vBS5 was significantly greater than that of USF0::CCCAT. This replicative deficiency was correlated with a correspondingly smaller plaque size during plaque isolation, a slower cytopathic effect during infection, and a lower yield of the viral stock of the vBS5 compared to USF0::CCCAT (data not shown). Taken together, these results strongly suggest that the five-point mutation in the CAAT box has a greater effect on CP1 binding than the single point mutation previously described.
FIG. 3.
Replication of viruses with the wild-type MLP (MLP-WT) or mutations in the CAAT box of the MLP. A549 cells in monolayer culture were infected at an MOI of 10 FFU per cell, and samples were taken at intervals postinfection (p.i.). Virus was titrated by fluorescent focus assay. Yields per infected cell were calculated and are plotted on a log10 scale.
Rate of transcription initiation during infection with viruses containing the CAAT5 mutation.
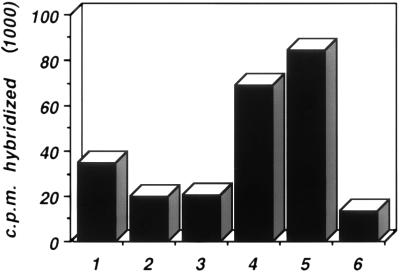
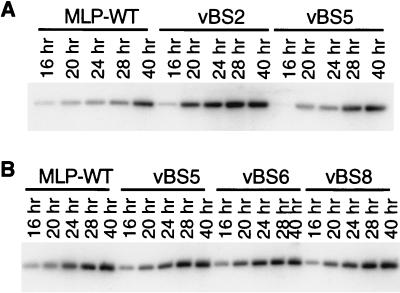
The growth defect of vBS5 is expected to arise from defective transcription from the mutant MLP. To demonstrate this unequivocally, however, it is necessary to measure transcription initiation. Nuclei were isolated from cells infected with the wild type, vBS4, and vBS5 at late times postinfection and incubated in vitro to allow previously initiated RNA polymerases to elongate. RNA was isolated and hybridized to single-strand sequences specific for either the LI region of the major late transcription unit or the GAPDH gene as a housekeeping cellular gene control. The counts bound to each probe were quantitated with a PhosphorImager (Fig. 4). The levels of hybridization, and thus incorporation in nuclei, of the RNA from cells infected with the double mutant vBS5 (lane 6) were significantly lower than those in the wild-type- or single-mutant (vBS4)-infected cell nuclei (lanes 4 and 5). The reduction in transcription for the double mutant compared to that of the single mutant was sixfold. The transcription of the single mutant (lane 5) was slightly better than that of the wild-type virus (lane 4), but this is probably within experimental variation. Although the reduction in transcription initiation in the nuclei from the double mutant (6-fold) is less than the reduction in final viral yield (20-fold), the results show that the replication deficiency of the double mutant is correlated with a reduction in transcription initiation.
FIG. 4.
Transcription in nuclei isolated from cells infected with various viruses. Experimental details are given in Materials and Methods. A549 cells in monolayer culture were infected with viruses at 10 FFU per cell for 36 h, and nuclei were isolated. Individual filters with slots containing single-stranded DNA from M13 clones corresponding to GAPDH (lanes 1 to 3) or Ad5 L1 region (lanes 4 to 6) were hybridized to RNA isolated from nuclei of infected cells. Results are for nuclei of cells infected with wild-type MLP (lanes 1 and 4), with vBS4 (the single CAAT5 mutant) (lanes 2 and 5), and with vBS5 (the CAAT5::USF0 double mutant) (lanes 3 and 6). Labeled RNA present on the filters was quantitated by PhosphorImager analysis and is presented in arbitrary units.
Genomes containing the CAAT5 and TATA27 mutations cannot be recovered as virus.
Early biochemical evidence had suggested that there is an interaction between the factors that bind to the UPE and the TATA box in the adenovirus MLP (39), and previous genetic evidence corroborated this suggestion, because it was impossible to create viruses with combined mutations in the UPE and the TATA box (37). Previous genetic evidence also suggested that there was a functional interaction between the CAAT box and TATA box because the double-mutant virus TATA27::CCCAT is severely deficient in replication (37). We were interested to see if the TATA27::CAAT5 double mutation could be constructed, because the evidence cited above suggested that the binding of CP1 was more impaired in CAAT5 than in CCCAT, and the corresponding phenotype would be expected to be more severe. Three attempts were made to build the TATA27::CAAT5 double mutant virus, and a few plaques were isolated. Analysis of the DNAs from three independent viral clones with the enzyme XhoI, which discriminates between plasmid and LLX1 DNA (36), showed that recombination between the mutation-containing plasmid and the right-end viral DNA fragment had taken place. However, the three virus isolates had a wild-type replication phenotype, and sequencing of the MLP region demonstrated that the TATA box was the wild-type sequence instead of the expected TATAGAA. The viral genomes contained the CAAT5 mutation, which has no overt phenotype. These results suggest that a mutant genome containing multiple mutations in the CAAT box and a single mutation in the TATA box is lethal and confirm that there is an important functional interaction between the CAAT and TATA boxes. In addition, the apparent lethality of the TATA27::CAAT5 mutation, as opposed to the deficient but nonlethal phenotype of the TATA27::CCCAT mutation, further supports the idea that CAAT5 binds CP1 less well than does CCCAT.
The apparent reversion of the TATA box in the three plaque isolates whose DNA sequence was analyzed may have been caused by one of two mechanisms. A true reversion at the TATA box could have taken place during the expansion of the original plaque on the assay plate, although the number of genomes and the expected mutation rate for a specific transition perhaps make this unlikely. We have observed such reversions before in the severely defective mutant TATA27::CCCAT, but only after serial passages (36a). A more likely alternative is that a secondary recombination took place between the recombinant genome produced in the initial overlap reaction and a fragment of wild-type viral DNA present in the transfected cell (see Materials and Methods for the likely origin of this fragment).
Replacement of the MLP CAAT box by other transcription elements.
The conservation of the CAAT box in all MLPs sequenced to date and the empirical evidence shown above and earlier strongly suggest that it is functionally important to the MLP. However, neither the experimental data nor the sequence conservation indicates if the CAAT box plays a specific role in MLP function. One way to answer this question is to attempt to replace the CAAT box with other activating elements to see if they can function in its place with equal efficiency.
The CAAT box was replaced with an ATF binding site or an OCT1 binding site, both of which are widely used in cellular and viral genes (23, 24, 46). As well as creating these mutations in an otherwise wild-type background, we also combined them with the previously described USF0 mutation (Table 1). In both replacement mutations, the changes necessary to create the desired transcription element were made so that alterations to the DNA polymerase encoded on the opposite strand were minimized. Viral replication was measured in one-step growth curves (Fig. 5). As shown in Fig. 5A, vBS1, which contains the ATF binding site in place of the CAAT box in the wild-type UPE background, replicated with efficiency equal to that of the wild-type virus, with identical eclipse periods. In contrast, the replication of the mutant virus vBS2, containing the ATF binding site replacement in the USF0 background, was considerably less than that of wild-type virus (Fig. 5A and B) but was significantly greater than that of vBS5 (Fig. 5B). This result suggests that an ATF binding site is able to replace CAAT box function, at least partially. Note also that although vBS1 contains unavoidable alterations to the DNA polymerase, the wild-type phenotype suggests that these changes do not affect DNA replication. Replacement of the CAAT box with the OCT1-binding site showed similar results (Fig. 5C). vBS6, which contains the OCT1 binding site in place of the CAAT box in the wild-type background, replicated like the wild-type virus. In contrast, replication of the mutant virus vBS8, containing the OCT1 binding site replacement in the mutant UPE background, was significantly less than that of the vBS6 and wild-type viruses, but again replication was greater than that of vBS5. These results show that the OCT1 binding site is also capable of partial substitution of the CAAT box. The implications of the partial functional replacement of the CAAT box by ATF and OCT1 binding sites will be considered further in Discussion.
FIG. 5.
Replication of viruses with an ATF binding site or OCT1 binding site replacing the CAAT box. A549 cells in monolayer culture were infected with the various viruses at an MOI of 10 FFU per cell and processed as described in the legend to Fig. 3. (A and B) Two experiments involving ATF replacement viruses; (C) one experiment with the OCT1 replacement viruses.
Viral DNA replication in mutant and wild-type infection.
Although mutations in the MLP usually are designed so that there are no changes to the amino acid sequence of the DNA polymerase (26, 36, 37), the ATF replacement (vBS1 and vBS2) and OCT1 replacement (vBS6 and vBS8) caused changes of three and two amino acids, respectively, in the viral DNA polymerase encoded on the strand opposite to the MLP strand. Although the single-mutant viruses showed no evidence of replication deficiency (Fig. 5), and therefore no suggestion that the changes to the DNA polymerase had any phenotypic consequences, it was important to demonstrate that the levels of viral DNA synthesis were unaffected in the double mutants. Viral DNA was extracted from infected cells by a modified Hirt procedure, and the amounts of accumulated DNA were measured by Southern hybridization. As shown in Fig. 6, there are no substantial differences in timing or total accumulation of viral DNA. Thus, there is no evidence that the replacement mutations affect DNA replication either directly by altering the DNA polymerase or by feedback mechanisms to regulate the rate of DNA replication in response to lowered MLP transcription.
FIG. 6.
Viral DNA accumulation in cells infected with mutant viruses. A549 cells in monolayer culture were infected at an MOI of 10 FFU per cell and harvested at the times indicated. DNA was extracted by a modification of the Hirt technique and examined by Southern blotting after XhoI digestion, using labeled adenovirus DNA probe. Results shown in panels A and B are from independent experiments.
DISCUSSION
This study originated from sequence analyses of potential MLPs from a diverse set of mammalian adenoviruses, which showed an absolute conservation of two sequences predicted to be functional transcription elements, namely, the basal TATA box and the distal upstream activating element, the CAAT box. Recently, the complete genomic sequence for the avian adenovirus CELO (6) and two further mammalian viruses have been published (references 33 and 51 and references therein). In all cases, an element corresponding to a CAAT box can be found in the region expected to contain the MLP. In contrast, sequences corresponding to the proximal element, the UPE, are not absolutely conserved (44), as they can be replaced by a functional SP1 site in mouse adenovirus type 1 (45), and the predicted INR element and the three downstream activating elements are also not well conserved.
The absolute conservation of the CAAT box could arise for two reasons. The element might play an important and specific role in the natural host environment of all mammalian viruses examined, so that there is selection to maintain the identity of the element. Alternatively, the ancestral progenitor to all currently known mammalian adenoviruses might have acquired a particular activating sequence by chance, and the activating function per se has been subject to conservative selection. In the latter case, replacement by any functional activating element would be permitted, but because this requires several concerted nucleotide changes to create a new transcriptional element, it has not happened in the time available since the divergence from the ancestral progenitor. The experiments reported in this paper were designed to (i) reexamine the importance of the CAAT box to the function of the MLP and (ii) investigate the specificity of its function.
Previous results with a single point mutation in the CAAT box showed that its function was redundant with that of the UPE, but the transcriptional defects in combination with mutations in basal elements were less severe than those observed with combinations between the UPE disruption and basal element mutations (26, 37). This could arise either because the CAAT box is intrinsically less important than the UPE to MLP function or because the single A-to-C transition does not interfere completely with the binding of CP1 to the DNA. To address the latter possibility, multiple point mutations were introduced into the CAAT box in a variety of viral genetic backgrounds. The results are summarized in Table 2. In every case, the phenotype of double mutations that contain the CAAT5 mutation is more severe than that of the corresponding CCCAT double mutation. This is most clearly seen in the inability to create viable virus in which the CAAT5 mutation was coupled to the TATA27 mutation, whereas the TATA27::CCCAT mutant could be recovered, even though it has severe transcriptional deficiencies (37). Taken together, these results confirm that the CAAT box is important in viral infection, that there is a functional redundancy between the two upstream activating elements, the CAAT box and the UPE element, and that there is some functional interaction between the CAAT box and the TATA element. Furthermore, the more defective transcriptional phenotype of the CAAT5 double mutants than of the CCCAT double mutants strongly suggests that the binding of CP1 has been more severely disrupted in the former. Attempts to examine this in vitro by electrophoretic mobility shift assays using CAAT-box-containing oligonucleotides and crude cellular extracts were unsuccessful, even with the wild-type sequence (37a). It may be necessary to test the idea with the purified CP1 heterotrimer, as has been done by Bi et al. (3), particularly if the difference in binding affinities to CCCAT and CAAT5 is of small magnitude.
TABLE 2.
Comparison of phenotypes of CAAT5 and CCCAT double mutations
| Virus | Mutant sequence present at element
|
Viral growth | Figure and/or reference | |||
|---|---|---|---|---|---|---|
| CAAT | UPE | TATA | INR | |||
| CCCAT | CCCAT | + | + | + | Wild type | 37 |
| vBS4 | CAAT5 | + | + | + | Wild type | Fig. 3A |
| USF0::CCCAT | CCCAT | USF0 | + | + | 5–10× lower | 36 and Fig. 3B |
| vBS5 | CAAT5 | USF0 | + | + | 20–30× lower | Fig. 3B and 5 |
| TATA27::CCCAT | CCCAT | + | TATA27 | + | 100× lower | 37 |
| [vBS10] | CAAT5 | + | TATA27 | + | Lethal | This report |
| CCCAT::INR-5 | CCCAT | + | + | INR-5 | Wild type | 26 |
| vBS11 | CAAT5 | + | + | Start-1 | Small plaque | Preliminary data |
The second question to be addressed was the specificity of the CAAT box to the function of the MLP. In general, specificity of a particular element can be examined by replacing it with other upstream activating elements, a method previously used in transient transfection assays to gain an understanding of the transactivational abilities of adenovirus E1A (47) and simian virus 40 T antigen (14). Accordingly, a consensus ATF binding site and OCT1 binding site were used to replace the MLP CAAT box in both a wild-type and a USF0 background to see if other transcription factor binding sites, widely used in cellular and viral genes, can substitute for the CAAT box function. It should be pointed out that one or more ATF binding sites are found in the promoters of several adenovirus genes including E2 (early), E3, and E4 (23). The use of OCT1 sites as transcriptional activating sites has not been demonstrated, although an OCT1 site has been shown to be important for the initiation of DNA replication (reference 49 and references therein). Because of the functional redundancy of the two upstream activating elements, the critical test is the test of function in a USF0 genetic background. The results presented in Fig. 5 and summarized in Table 3 show that the ATF and OCT1 binding sites are capable of partial replacement of CAAT box function in this background, whereas the five-point mutation in the CAAT box leads to a severe deficiency in viral transcription and replication. The intermediate level of functional replacement could arise for several reasons. First, the choice of replacement elements might not be optimal because of lower specific activities of the cognate transcription factors compared with that of CP1. This seems unlikely because all three factors are universally and abundantly expressed (17, 42, 46). Second, the position of the replacement elements might not be ideal in relation to the TATA box, and thus the stereochemistry of the protein-protein interactions between the upstream factors and the basal machinery might not be fully functional. This possibility is more difficult to assess because the mechanisms by which the three proteins transactivate are not fully understood, although it has been shown that ATF interacts with TFIID (19). Third, the CAAT box might indeed be the optimal transcriptional element at this position because of a specific contribution to the establishment of the preinitiation complex, for example, by a stronger recruitment of the basal machinery than that exerted by ATF or OCT1. At present, we cannot make a distinction between these and other possibilities, but we note in passing that the problem of determining the structure of the optimal promoter, or indeed of identifying promoters in mammalian genomes in general, is much more difficult than in prokaryotes, where there is a robust algorithm for determining promoter strength (34). Finally, it should be pointed out that earlier studies suggested that the CAAT box might be important not so much as an activating element but rather as a sequence necessary for “insulating” the MLP from transcription initiated further to the left on the adenovirus genome (9, 10). Transcription proceeding from early genes located to the left of the MLP might interfere with its transcription, and it is possible that CP1 is a better insulator than either ATF or OCT1. Resolution of this question awaits evidence as to the mechanism of CP1 action in vitro.
TABLE 3.
Genotypes and phenotypes of mutant viruses
| Virus name | CAAT box | UPE | TATA box | Viral growtha | RNA levelb |
|---|---|---|---|---|---|
| Wild type | + | + | + | Wild type | Wild type |
| TATA27 | + | + | Mutant | Wild type | Wild typec |
| USF0 | + | Mutant | + | Wild type | Wild typec |
| vBS4 | Mutant | + | + | Wild type | Wild type |
| vBS5 | Mutant | Mutant | + | 20× lower | 6× lower |
| vBS1 | ATF | + | + | Wild type | NDd |
| vBS2 | ATF | Mutant | + | 3× lower | ND |
| vBS6 | OCT1 | + | + | Wild type | ND |
| vBS8 | OCT1 | Mutant | + | 10× lower | ND |
| [vBS10] | Mutant | + | Mutant | Lethal | ND |
Genetic analysis has also shown that replacement of the UPE by either an ATF binding site or an SP1 site is fully functional in a CCCAT background (30a), suggesting that the UPE is less stringent in its sequence requirements than is the CAAT box. As mentioned earlier, the mouse adenovirus MAV-1 also has a functional SP1 site in the MLP. However, the replacements in the human virus are not completely equivalent to the original UPE because they show a reduction in viral replication of some 18-fold when coupled with the TATA27 mutation (30a), whereas TATA27 by itself has no phenotype (37). Perhaps USF, the factor that binds to the UPE, is better able than either SP1 or ATF to recruit the basal machinery to the mutant TATA box present in TATA27.
Regardless of the precise role of the CAAT box to MLP function and the mechanism by which CP1 acts, the data presented in this report demonstrate the usefulness of the combination of comparative sequence analysis and a mutational approach. The sequence comparisons, which point to an important role for the conserved CAAT box, are completely congruent with the mutational analysis which demonstrates its important role in the correct viral context. Adenovirus is particularly amenable to this kind of dual approach because of the wide range of eukaryotic species that harbor viruses that belong to this family. This allows wide-ranging phylogenetic inferences to be drawn. Equally important is the ease of genetic manipulation, which allows specific experimental questions of function to be addressed. Recent precedents for this dual approach are the comparisons of VA RNAs from many adenovirus types (28) and the structure-function analyses, which are partly based on the derived consensus for important structural domains (27). It is to be expected that more such comparisons will be forthcoming as the complete sequences of many adenovirus species and serotypes become available.
ACKNOWLEDGMENTS
We thank Patricia Munz for assistance with cell culture and the virology group at Columbia University for advice and suggestions.
This work was supported by grant R01 GM49070 from the NIGMS and by core grant CA13696 from the NCI to the Columbia Comprehensive Cancer Center.
REFERENCES
- 1.Bendall A J, Molloy P L. Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berk A J. Adenovirus promoters and E1a transactivation. Annu Rev Genetics. 1986;20:45–79. doi: 10.1146/annurev.ge.20.120186.000401. [DOI] [PubMed] [Google Scholar]
- 3.Bi W M, Wu L, Coustry F, de Crombrugghe B, Maity S N. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 4.Brunet L J, Babiss L E, Young C S H, Mills D R. Mutations in the adenovirus major late promoter: effects upon viability and transcription during infection. Mol Cell Biol. 1987;7:1091–1100. doi: 10.1128/mcb.7.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 6.Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chodosh L A, Baldwin A S, Carthew R W, Sharp P A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988;53:11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- 8.Chodosh L A, Carthew R W, Sharp P A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986;6:4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly S, Manley J L. A CCAAT box sequence in the adenovirus major late promoter functions as part of an RNA polymerase II termination signal. Cell. 1989;57:561–571. doi: 10.1016/0092-8674(89)90126-8. [DOI] [PubMed] [Google Scholar]
- 10.Connelly S, Manley J L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989;9:5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 12.Fort P, Marty L, Piechaczyk M, El Sabrouty S, Dani C, Jennteur P, Blanchard J M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigene family. Nucleic Acids Res. 1985;13:1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giard D J, Aaronson S A, Todaro G J, Arnstein P, Kersey J H, Dosik H, Parks W P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 14.Gilinger G, Alwine J C. Transcriptional activation by simian virus 40 large T antigen: requirements for simple promoter structures containing either TATA or initiator elements with variable upstream factor binding sites. J Virol. 1993;67:6682–6688. doi: 10.1128/jvi.67.11.6682-6688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg M E, Bender T P. Identification of newly transcribed RNA, unit 4.10. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene, Wiley Interscience; 1987. [DOI] [PubMed] [Google Scholar]
- 16.Gregor P D, Sawadogo M, Roeder R G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 17.Hai T, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Horikoshi M, Hai T, Lin Y-S, Green M R, Roeder R G. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 20.Jansen-Durr P, Mondésert G, Kedinger C. Replication-dependent activation of the adenovirus major late promoter is mediated by the increased binding of a transcription factor to sequences in the first intron. J Virol. 1989;63:5124–5132. doi: 10.1128/jvi.63.12.5124-5132.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence W C, Ginsberg H S. Intracellular uncoating of type 5 adenovirus deoxyribonucleic acid. J Virol. 1967;1:851–867. doi: 10.1128/jvi.1.5.851-867.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K A W, SivaRaman L, Thimmappaya B, Hurst H C, Jones N C, Green M R. A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci USA. 1987;84:8355–8359. doi: 10.1073/pnas.84.23.8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y-S, Green M R. Interaction of a common cellular transcription factor, ATF, with regulatory elements in both E1A- and cyclic AMP-inducible promoters. Proc Natl Acad Sci USA. 1988;85:3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logan J, Shenk T. In vivo identification of sequence elements required for normal function of the adenovirus major late transcriptional control region. Nucleic Acids Res. 1986;14:6327–6335. [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Reach M D, Minaya E, Young C S H. The initiator element of the adenovirus major late promoter has an important role in transcription initiation in vivo. J Virol. 1997;71:102–109. doi: 10.1128/jvi.71.1.102-109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Mathews M B. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J Virol. 1993;67:6605–6617. doi: 10.1128/jvi.67.11.6605-6617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y L, Mathews M B. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J Virol. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maity S N, Golumbek P T, Karsenty G, de Crombrugghe B. Selective activation of transcription by a novel CCAAT binding factor. Science. 1988;241:582–585. doi: 10.1126/science.3399893. [DOI] [PubMed] [Google Scholar]
- 30.Maity S N, Vuorio T, de Crombrugghe B. The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc Natl Acad Sci USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Minaya, E., and C. S. H. Young. Unpublished data.
- 31.Miyamoto N G, Moncollin V, Egly J M, Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985;4:3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mondesert G, Tribouley C, Kedinger C. Identification of a novel downstream binding protein implicated in late-phase-specific activation of the adenovirus major late promoter. Nucleic Acids Res. 1992;20:3881–3889. doi: 10.1093/nar/20.15.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison M D, Onions D E, Nicolson L. Complete DNA sequence of canine adenovirus type 1. J Gen Virol. 1997;78:873–878. doi: 10.1099/0022-1317-78-4-873. [DOI] [PubMed] [Google Scholar]
- 34.Mulligan M E, Hawley D K, Entriken R, McClure W R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philipson L. Adenovirus assay by fluorescent cell-counting procedure. Virology. 1961;15:263–268. doi: 10.1016/0042-6822(61)90357-9. [DOI] [PubMed] [Google Scholar]
- 36.Reach M, Babiss L E, Young C S H. The upstream factor binding site is not essential for activation of transcription from the adenovirus major late promoter. J Virol. 1990;64:5851–5860. doi: 10.1128/jvi.64.12.5851-5860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Reach, M., E. Minaya, and C. S. H. Young. Unpublished data.
- 37.Reach M, Xu L-X, Young C S H. Transcription from the adenovirus major late promoter uses redundant activating elements. EMBO J. 1991;10:3439–3446. doi: 10.1002/j.1460-2075.1991.tb04908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Reach, M., and C. S. H. Young. Unpublished data.
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawadogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 40.Sinha S, Kim I S, Sohn K Y, de Crombrugghe B, Maity S N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol. 1996;16:328–337. doi: 10.1128/mcb.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha S, Maity S N, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NF-Y, allows formation of a protein-DNA complex with CBF-A and CBF-B, and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinha S, Maity S N, Seldin M F, de Crombrugghe B. Chromosomal assignment and tissue expression of CBF-C/NFY-C, the third subunit of the mammalian CCAAT-binding factor. Genomics. 1996;37:260–263. doi: 10.1006/geno.1996.0555. [DOI] [PubMed] [Google Scholar]
- 43.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 44.Song B, Hu S-L, Darai G, Spindler K R, Young C S H. Conservation of DNA sequence in the predicted major late promoter regions of selected mastadenoviruses. Virology. 1996;220:390–401. doi: 10.1006/viro.1996.0327. [DOI] [PubMed] [Google Scholar]
- 45.Song B W, Young C S H. Functional characterization of the major late promoter of mouse adenovirus type 1. Virology. 1997;235:109–117. doi: 10.1006/viro.1997.8677. [DOI] [PubMed] [Google Scholar]
- 46.Sturm R A, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 47.Taylor I C A, Kingston R E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990;10:165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tribouley C, Lutz P, Staub A, Kedinger C. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J Virol. 1994;68:4450–4457. doi: 10.1128/jvi.68.7.4450-4457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Leeuwen H C, Rensen M, Van der Vliet P C. The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J Biol Chem. 1997;272:3398–3405. doi: 10.1074/jbc.272.6.3398. [DOI] [PubMed] [Google Scholar]
- 50.Volkert F C, Young C S H. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983;125:175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]
- 51.Vrati S, Brookes D E, Boyle D B, Both G W. Nucleotide sequence of ovine adenovirus tripartite leader sequence and homologues of the IVa2, DNA polymerase and terminal proteins. Gene. 1996;177:35–41. doi: 10.1016/0378-1119(96)00266-1. [DOI] [PubMed] [Google Scholar]
- 52.Vuorio T, Maity S N, de Crombrugghe B. Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast HAP3 transcription factor. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]