Abstract
The bicyclam AMD3100 is a potent and selective inhibitor of the replication of human immunodeficiency virus type 1 and type 2 (HIV-1 and HIV-2). It was recently demonstrated that the compound inhibited HIV entry through CXCR4 but not through CCR5. Selectivity of AMD3100 for CXCR4 was further indicated by its lack of effect on HIV-1 and HIV-2 infection mediated by the CCR5, CCR3, Bonzo, BOB, and US28, coreceptors. AMD3100 completely blocked HIV-1 infection mediated by a mutant CXCR4 bearing a deletion of most of the amino-terminal extracellular domain. In contrast, relative resistance to AMD3100 was conferred by different single amino acid substitutions in the second extracellular loop (ECL2) or in the adjacent membrane-spanning domain, TM4. Only substitutions of a neutral residue for aspartic acid and of a nonaromatic residue for phenylalanine (Phe) were associated with drug resistance. This suggests a direct interaction of AMD3100 with these amino acids rather than indirect effects of their mutation on the CXCR4 structure. The interaction of aspartic acids of ECL2 and TM4 with AMD3100 is consistent with the positive charge of bicyclams, which might block HIV-1 entry by preventing electrostatic interactions between CXCR4 and the HIV-1 envelope protein gp120. Other features of AMD3100 must account for its high antiviral activity, in particular the presence of an aromatic linker between the cyclam units. This aromatic group might engage in hydrophobic interactions with the Phe-X-Phe motifs of ECL2 or TM4. These results confirm the importance of ECL2 for the HIV coreceptor activity of CXCR4.
Bicyclams, a class of macrocyclic polyamines consisting of two cyclam units linked by an aliphatic bridge, were found to be potent and selective inhibitors of human immunodeficiency virus type 1 (HIV-1) and type 2 (HIV-2) replication (12). Bicyclams with an aromatic linker apparently had higher antiviral activity (5, 13). Among them, AMD3100 (also named JM3100 or SID791) had a 50% effective antiviral concentration (EC50) in the nanomolar range for different HIV-1 or HIV-2 strains and a very high (>100,000) selectivity index (ratio of EC50 to 50% cytotoxic concentration) (13). Furthermore, AMD3100 could inhibit HIV-1 production and CD4+ cell depletion in SCID mice engrafted with human tissues (11). It therefore represents an attractive candidate for the chemotherapy of HIV infection.
Based on a kinetic study of their antiviral activity, bicyclams were proposed to block HIV-1 entry into cells (12). The process of HIV-1 entry can be schematically divided into early steps, i.e., adsorption of virions to cells and interaction with cell surface components, in particular the CD4 receptor, and late steps, corresponding to fusion of the viral envelope with the cell membrane and transfer of the viral capsid into the cytoplasm (23, 30). Early and late steps are mediated by the viral envelope proteins (Env) gp120 and gp41, respectively. The apparent lack of inhibitory effect of bicyclams on cell-cell fusion mediated by HIV-1 Env led to the proposal that they blocked a postfusion step of HIV-1 entry, possibly uncoating (12). However, this mechanism did not seem consistent with the exclusive role of gp120 mutations in the phenotype of bicyclam-resistant mutants (15). Also, we found that AMD3100 could block the fusion of CD4+ cells induced by HIV-1 “from without,” i.e., before viral replication and de novo expression of Env, which suggested an effect on a step preceding virus-cell fusion (34, 35). The mechanism of action of bicyclams remained elusive until some new understanding was gained by the discovery of the role of the chemokine receptors CXCR4 and CCR5 in HIV entry.
Expression of CXCR4 or CCR5 in CD4+ cells allows their infection by T-cell-line-adapted (TCLA) HIV-1 or by primary-macrophage-tropic (PMT) HIV-1, respectively, and the formation of syncytia with cells expressing the corresponding type of Env. The interaction of HIV-1 gp120 with these chemokine receptors could be detected in the presence of CD4, suggesting that they behave as CD4-associated coreceptors for HIV-1 (reviewed in references 16 and 31). Several other chemokine receptors, and related orphan G protein-coupled receptors, are also capable of mediating HIV-1 entry into CD4+ cells (8, 14, 19, 28, 37, 38), but CCR5 and CXCR4 seem predominant in vivo. The former is used by most, possibly all, types of primary HIV-1 strains, while CXCR4 is used only by strains with a syncytium-inducing (SI) phenotype usually emerging at an advanced stage of infection (31). Both coreceptors are also used by HIV-2, with CXCR4 usage again being observed for TCLA and SI strains (22, 42). In spite of their close relationship with HIV-2, the simian immunodeficiency viruses (SIVs) from macaque and sooty mangabeys seem unable to use CXCR4 for CD4-dependent entry (7, 21). In contrast, cell-line-adapted feline immunodeficiency virus could use either human or cat CXCR4 as a receptor (45).
While bicyclams efficiently neutralized TCLA HIV-1 and HIV-2, they had a weaker antiviral activity on PMT HIV-1 and were unable to block infection by SIVs from macaques or sooty mangabeys (13). These properties suggested a selective antiviral activity of bicyclams on HIV-1 and HIV-2 strains by using the CXCR4 coreceptor. In recent studies, AMD3100 was indeed found to block HIV-1 infection via CXCR4 but not via CCR5 (17, 40, 41). The compound also dose-dependently inhibited the binding of the anti-CXCR4 monoclonal antibody 12G5 and the intracellular calcium flux induced by the natural ligand of CXCR4, the stromal cell-derived factor (SDF-1) chemokine (17, 40, 41). Bicyclams were therefore proposed to selectively bind CXCR4, thereby preventing its functional interaction with gp120 and its HIV coreceptor function.
We have further addressed the selectivity of AMD3100 by testing its possible effect on HIV-1 or HIV-2 infection mediated by coreceptors other than CXCR4 and CCR5. Also, we sought to identify the domain(s) of CXCR4 interacting with AMD3100 by analyzing a series of CXCR4 mutants for their ability to mediate HIV-1 infection in the presence of AMD3100. Our analysis was concentrated on the second extracellular loop (ECL2), which seems important for the HIV-1 coreceptor activity of CXCR4 (4).
MATERIALS AND METHODS
Bicyclam.
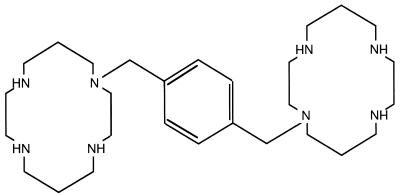
AMD3100, the octahydrochlorodihydrate salt of phenylbis(methylene)-bis-(1,4,8,11-tetraazacyclotetradecane) (Fig. 1), was obtained from G. Henson (AnorMED, Langley, British Columbia, Canada) as dry powder and stored as a 1-mg/ml water solution.
FIG. 1.
Structure of the bicyclam AMD3100.
Cell lines and virus strains.
The human cell lines HeLa P5 (37) and U373MG-CD4 (24) are stably transfected with Escherichia coli lacZ under transcriptional control of the HIV-1 long terminal repeat (LTR). HeLa P5 cells express CCR5 and CXCR4, while U373MG-CD4 cells do not seem to express functional HIV-1 or HIV-2 coreceptors. U373MG-CD4 cells were stably transfected with chemokine receptor expression vectors (see below) and a vector allowing selection with puromycin. Individual clones were screened for their ability to fuse with cells expressing HIV-1 Env, as described previously (37). All cell lines were propagated in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with antibiotics (60 μg of penicillin per ml, 100 μg of streptomycin per ml) and 10% fetal calf serum. HIV-1 stocks were produced by transient-transfection HeLa cells with cloned HIV-1LAI (33), HIV-1NDK (43), HIV-189.6 (10), and HIV-1ADA (44) proviruses. The latter was actually a recombinant HIV-1LAI provirus with env from HIV-1ADA (37). HIV-2ROD was produced by infected cells (6). The infectivity of viral stocks was determined as described below in HeLa P5 (HIV-1ADA) or in the parental CCR5-negative HeLa P4 cell line (9).
Plasmids.
The cDNAs of Bonzo and BOB (14) were obtained from D. Littman (Skirball Institute, New York, N.Y.), and the rat CXCR4 cDNA was from R. Duman (Yale University, New Haven, Conn.). The cDNAs of other chemokine receptors used in this study have been described previously (36, 37, 42). All chemokine receptors were expressed from the cytomegalovirus immediate-early promoter. The RRHR and HHRH CXCR4 chimeras, previously named M and N, and the Δ4-36 human CXCR4 have been described previously (4). Other human CXCR4 mutants were obtained by oligonucleotide-directed mutagenesis on a single-stranded template. Mutants were screened for the creation of restriction enzyme sites and checked by sequencing (3).
Infections.
HeLa P5 or U373MG-CD4 cells stably expressing coreceptors were infected in subconfluent 12-well trays. Infections of U373MG-CD4 cells transiently expressing chemokine receptors were performed in 6-well trays 24 h after transfection by a standard calcium phosphate precipitation technique. All infections were performed by adding fresh culture medium containing approximately 1,000 infectious units of HIV-1 or HIV-2 per well and AMD3100 at the indicated concentration. Cells were fixed with 0.5% glutaraldehyde 24 (HeLa P5) or 48 (U373MG-CD4) h after infection and stained with the β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described previously (20). Blue foci were scored under ×20 magnification.
Flow cytometry.
COS cells were cotransfected with a CXCR4 expression vector and a green fluorescent protein (GFP) expression vector (EGFPN-1; Clontech, Palo Alto, Calif.) in a 6:1 ratio. Cells were detached with phosphate-buffered saline (PBS) containing 1 mM EDTA 36 to 48 h after transfection. About 106 cells were stained with the 12G5 anti-CXCR4 monoclonal antibody (22) (obtained from the NIH AIDS Reagent Program) and then with a secondary phycoerythrin (PE)-coupled anti-mouse antibody (Dako, Glostrub, Denmark). Incubations (1 h at 4°C) with 12G5 (6 μg/ml) and secondary antibody (16 μg/ml) were performed in PBS containing 1% fetal calf serum. Cells were fixed in PBS with 2% formaldehyde and analyzed for red and green fluorescence on an Epics Elite cytofluorometer (Becton Dickinson, San Jose, Calif.). Cells were considered positive for GFP or for PE when their green or red fluorescence was higher than 3 (arbitrary units). The fluorescence of more than 99% of mock-transfected cells was below this threshold. For cells cotransfected with GFP and CXCR4 vectors, the mean intensities of fluorescence ranged from 30 to 50 (green) and from 50 to 200 (red).
RESULTS
Blocking of HIV-1 entry via CXCR4.
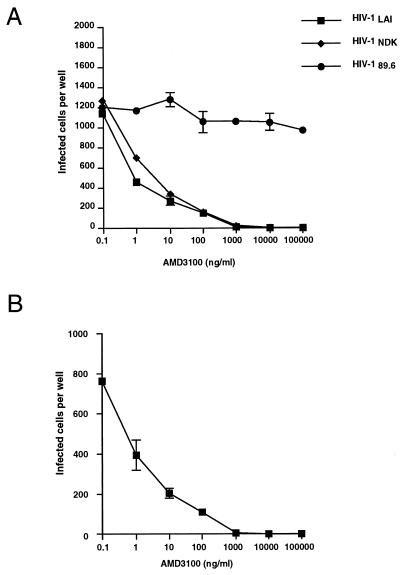
The antiviral effects of AMD3100 were first determined in the CD4+ cell line HeLa P5, expressing both the CXCR4 and CCR5 coreceptors. These cells are stably transfected with an E. coli β-galactosidase gene (lacZ) inducible by the HIV-1 or HIV-2 protein Tat. Infectivity can be determined by staining cells with a β-galactosidase substrate (X-Gal) after a single cycle of HIV reverse transcription, which is particularly useful for studying antiviral agents blocking the initial steps of the virus life cycle (27, 35). Infections of HeLa P5 cells with HIV-1LAI (TCLA), HIV-1NDK (TCLA), or HIV-189.6 (primary SI) were performed in the presence of AMD3100 at concentrations ranging from 0.1 ng/ml to 100 μg/ml. For HIV-1LAI and HIV-1NDK, infectivity was markedly reduced at AMD3100 concentrations of 1, 10, and 100 ng/ml and completely blocked at higher concentrations (Fig. 2A). For these strains, the EC50 of AMD3100 was in the range of 1 ng/ml. In contrast, there was no apparent inhibition of HIV-189.6 infection by AMD3100 at the lower concentrations and only a minor effect at the higher concentrations, indicating the absence of cytotoxicity in this assay. A similar antiviral effect of AMD3100 towards HIV-1LAI was observed in CXCR4+ U373MG-CD4 cells (Fig. 2B). These results were consistent with the ability of dualtropic strains, such as HIV-189.6, to infect cells either via CXCR4 or via CCR5, while the TCLA HIV-1 cannot use CCR5, and with the selective blocking of the CXCR4 pathway by AMD3100.
FIG. 2.
Effects of AMD3100 on HIV-1 infectivity in CD4+ LTRlacZ cell lines. (A) HeLa P5 cells (CXCR4+, CCR5+) were infected with HIV-1LAI (TCLA), HIV-1NDK (TCLA), or HIV-189.6 (primary SI, dualtropic). (B) CXCR4+ U373MG-CD4 cells were infected with HIV-1LAI. Infections were performed in 12-well trays by adding fresh medium with approximately 1,000 infectious units of HIV-1 and AMD3100 at the indicated concentration. Infection was scored 24 h later (48 h for U373MG-CD4 cells) by staining with X-Gal and counting blue-stained cells. Numbers >200 were extrapolated from randomly selected fields. Results represent the mean number of infected cells in duplicate wells.
Effects on other coreceptors.
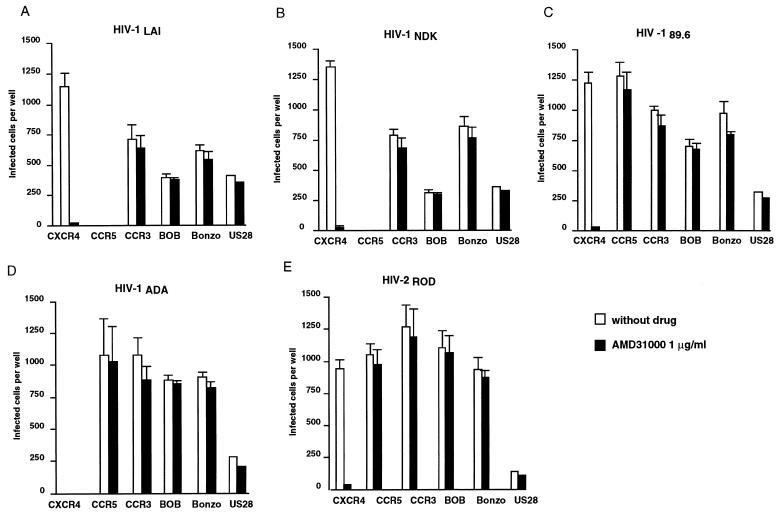
We next compared the effects of AMD3100 on HIV-1 and HIV-2 infection mediated by the chemokine receptors CXCR4, CCR5, CCR3, and US28 and by the orphan receptors Bonzo (also designated STRL33) and BOB (also designated gpr15). These coreceptors were stably expressed (except US28) in U373MG-CD4 cells, which are naturally resistant to HIV-1 or HIV-2 infection (24, 37). All viral strains tested could infect U373MG-CD4 cells expressing coreceptors, with variable efficiency, but not mock-transfected parental cells (less than 10 infected cells per well) (data not shown). As expected, the TCLA strains HIV-1LAI and HIV-1NDK infected CXCR4+ cells but not CCR5+ cells, while the opposite was seen with the PMT strain HIV-1ADA (Fig. 3). We already observed that the TCLA strain HIV-2ROD could use CXCR4 or CCR5 with similar efficiency, like primary SI and dualtropic HIV-1 (42). In this study, cells expressing CCR3, Bonzo, BOB, or US28 could be infected by all viral strains tested. The promiscuous coreceptor activity of Bonzo, BOB, and US28 was in agreement with that described in previous reports (14, 28, 37). The ability of TCLA HIV-1 to use CCR3 is not unprecedented (1). When infections were performed in the presence of 1 μg of AMD3100 per ml, HIV-1 or HIV-2 infectivity was abolished in CXCR4+ cells, while virus titers were identical or only slightly lower in cells expressing other coreceptors (Fig. 3). Increasing AMD3100 concentrations did not result in a detectable antiviral activity (data not shown). This experiment confirmed the apparent selectivity of AMD3100 for CXCR4.
FIG. 3.
Selective inhibition of CXCR4-mediated HIV entry by AMD3100. U373MG-CD4 cells stably expressing the HIV coreceptor CXCR4, CCR5, CCR3, Bonzo, or BOB or transiently expressing US28 were infected with HIV-1LAI, HIV-1NDK, HIV-189.6, HIV-1ADA (PMT), or HIV-2ROD (TCLA), in the absence or presence of 1 μg of AMD3100 per ml. Infections were performed and scored as described in the legend to Fig. 2.
Effects of mutations in CXCR4.
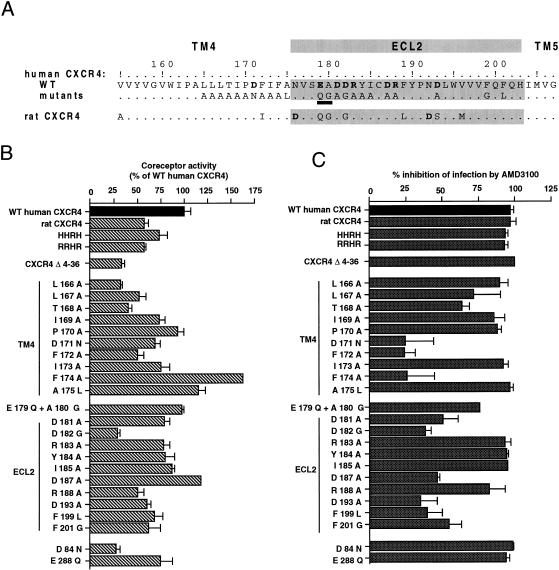
In the next series of experiments, different forms of CXCR4 (Fig. 4A) were expressed by transient transfection in U373MG-CD4 cells. The infectivity of HIV-1LAI was determined in the absence of drug (Fig. 4B) or in the presence of 1 μg of AMD3100 per ml, a concentration completely blocking infection mediated by wild-type (WT) human CXCR4 (Fig. 4C). A complete antiviral effect was observed when infection was mediated by rat CXCR4 and by the HHRH and RRHR chimeras, corresponding to exchanges of ECL2 domains between human (H) and rat (R) CXCR4. Therefore, the amino acid differences between rat and human CXCR4 sequences, principally found in the extracellular domains, did not seem to prevent the interaction of rat CXCR4 with AMD3100. Infection mediated by the Δ4-36 human CXCR4 mutant, in which most of the amino-terminal extracellular domain is deleted, was relatively less efficient than infection mediated by WT CXCR4 but was completely blocked by AMD3100. It confirmed that the amino-terminal domain was not absolutely required for the coreceptor activity of CXCR4, at least with HIV-1LAI. This domain also seems dispensable for interaction with AMD3100.
FIG. 4.
Effect of mutations in CXCR4 on HIV-1 infectivity and sensitivity to AMD3100. (A) Partial amino acid (aa) sequence of WT human and rat CXCR4. All mutations in CXCR4 correspond to single aa substitutions, except EA to QG in ECL2 (underlined). Charged aa are shown in boldface type. (B) Infection of U373MG-CD4 cells transiently expressing the indicated forms of CXCR4 with HIV-1LAI. Results are shown as a percentage, relative to WT human CXCR4, which yielded about 850 infected cells per well. HHRH and RRHR are human CXCR4-rat CXCR4 chimeras. Δ4-36 is a deletion between the corresponding aa of human CXCR4. (C) Percentage of inhibition of infectivity in parallel infections performed with 1 μg of AMD3100 per ml. Cells were infected 24 h after transfection in six-well trays, as described in the legend to Fig. 2. Results are the mean of duplicate infected wells.
Differences with human CXCR4 in ECL2 account for the inability of rat CXCR4 to mediate HIV-1NDK or HIV-2ROD infection (4). Also, ECL2 contains the epitope (or part of it) of the 12G5 monoclonal antibody (4) which can inhibit HIV-1 and HIV-2 infection (22). Both elements suggest that ECL2 is directly involved in the HIV coreceptor activity of CXCR4. We have tested the effect of AMD3100 on HIV-1LAI infection mediated by CXCR4 mutants bearing amino acid substitutions in ECL2 (Fig. 4A). All of the ECL2 mutants tested allowed infection of U373MG-CD4 cells, indicating that they were folded and transported to the cell surface. The lower efficiency of infection mediated by certain mutants, in particular D182G, might be due to a lower cell surface expression or to a less efficient interaction with gp120. The effects of ECL2 mutations on the coreceptor activity of CXCR4 will be studied in detail elsewhere (3). AMD3100 completely blocked infection mediated by the R183A, Y184A, and I185A mutants and markedly reduced infection mediated by the R188A and the E179Q + A180G mutants. Infection mediated by the other ECL2 mutants, D181A, D182G, D187A, D193A, F199L, and F201G, was also inhibited but to a lower extent. For WT CXCR4, a similar reduction of infectivity was observed with 100- to 1,000-fold-lower AMD3100 concentrations (Fig. 2). Substitutions of neutral and nonaromatic amino acids for aspartic acid and phenylalanine, respectively, can therefore be considered to confer an important degree of resistance to AMD3100.
We have also tested the effect of mutations in the membrane-spanning domain TM4, which immediately precedes ECL2. All TM4 mutants could mediate HIV-1 infection, although with a variable efficiency. A markedly reduced antiviral effect of AMD3100 was observed when infection was mediated by the D171N, F172A, and F174A mutants. Infection mediated by other TM4 mutants was completely neutralized. There was, therefore, a striking similarity between ECL2 and TM4 for the type of mutations associated with resistance to AMD3100.
Negatively charged amino acids are also found in the TM2 and TM7 domains of CXCR4. In TM2, their replacement by a neutral amino acid resulted in a nonfunctional CXCR4 in the case of the D93N mutant (data not shown) or markedly reduced the HIV-1 coreceptor activity in the case of the D84N mutant (Fig. 4B). Infection mediated by this CXCR4 mutant, or by the E288Q mutant in TM7, was completely blocked by AMD3100 at 1 and 10 μg/ml (Fig. 4C).
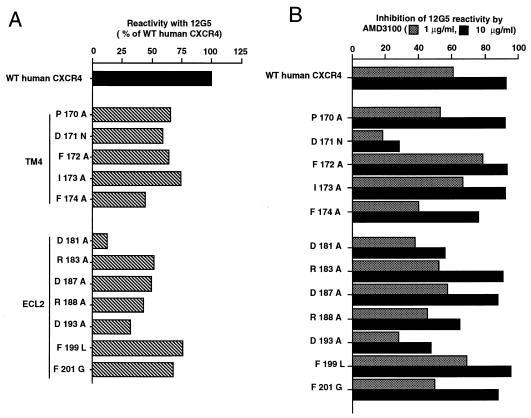
Reactivity with 12G5.
Flow cytometry was used to test the reactivity with the 12G5 antibody of WT CXCR4 and CXCR4 mutants transiently expressed in COS cells (Fig. 5A). Parallel experiments were performed in the presence of two AMD3100 concentrations (Fig. 5B). The E179Q + A180G and D182G mutations apparently abolished the reactivity of CXCR4 with 12G5 (data not shown). Since these CXCR4 mutants have detectable coreceptor activity, and are therefore expressed at the cell surface, the corresponding mutations must disrupt the 12G5 epitope. The other ECL2 and TM4 mutations tested allowed reactivity with 12G5, but the resulting fraction of 12G5+ cells was always lower than that with WT CXCR4. These mutations might modify the accessibility of the 12G5 epitope or exert an indirect effect on the expression of CXCR4 at the cell surface. Experiments with other antibodies and epitope-tagged forms of these CXCR4 mutants will be necessary to sort between these possibilities.
FIG. 5.
Reactivity of WT CXCR4 and CXCR4 mutants with the 12G5 monoclonal antibody and effect of AMD3100. COS cells were cotransfected with expression vectors for the indicated form of CXCR4 and for GFP and grown either in the absence (A) or in the presence (B) of AMD3100 at 1 or 10 μg/ml. Cells were stained 36 h after transfection with 12G5 and with a PE-coupled secondary antibody. The measurement for 12G5 reactivity was the percentage of PE-positive cells in the GFP-positive fraction (see Materials and Methods). (A) 12G5 reactivity of mutant CXCR4, shown relative to WT human CXCR4. (B) Percentage of inhibition of 12G5 reactivity by AMD3100. One representative experiment is shown.
In the case of WT CXCR4 and also for most mutants tested, the reactivity with 12G5 was markedly reduced in the presence of AMD3100 (Fig. 5B). The fraction of 12G5+ cells was also reduced, but to a lower extent, when cells expressed the D181A and D193A mutants (ECL2) and, moreover, the D171N mutant (TM4). These mutants might interact less efficiently with AMD3100, thereby explaining their relative resistance to its antiviral effect. However, the 12G5 reactivity of certain drug-resistant mutants, for example, D187A and F172A, was markedly reduced by AMD3100 (Fig. 5B). There was not, therefore, an absolute correlation between the ability of AMD3100 to compete with 12G5 binding and its antiviral activity.
DISCUSSION
These experiments confirmed that the bicyclam AMD3100 blocked HIV-1 and HIV-2 infection mediated by the CXCR4 coreceptor but not by the CCR5 coreceptor. The inability of PMT HIV-1 and SIV to use CXCR4 is the likely explanation of their resistance to bicyclams. In some experiments, the infection of activated peripheral blood mononuclear cells (PBMC) by PMT HIV-1 could be blocked by AMD3100 and other bicyclams, although higher concentrations of the bicyclams were generally required in these infections than in infections with TCLA strains (13). This observation is difficult to reconcile with the apparent lack of effect of AMD3100 on CCR5-mediated HIV-1 entry and with the essential role of the CCR5 pathway, which is shown by the resistance of PBMC from CCR5-negative individuals to infection by PMT HIV-1 (29, 39). In this study, bicyclams might have exerted indirect effects on the replication of PMT HIV-1, possibly through their interaction with CXCR4. Antiviral effects of AMD3100 on PMT HIV-1 were not observed in other recent studies (17, 40, 41).
Besides CCR5 and CXCR4, a growing list of chemokine receptors and related orphan receptors were found to behave as HIV-1 or HIV-2 coreceptors, at least under certain experimental conditions (8, 14, 19, 28, 37, 38). Here we found that AMD3100 did not inhibit HIV-1 or HIV-2 infection mediated by the CCR3, US28, BOB, or Bonzo coreceptors. The latter (Bonzo) is expressed, at least at the RNA level, in cell lines and activated PBMC (28). Since infection by TCLA HIV-1 and HIV-2 is completely blocked by AMD3100 and other bicyclams (12, 13, 41), it can be inferred that Bonzo or other bicyclam-resistant coreceptors are not functional in these cells. Unless bicyclams can block other HIV coreceptors, it seems that CXCR4 has a predominant role in the infection of PBMC and T-cell lines by TCLA HIV-1. Most primary HIV-1 able to use CXCR4 are in fact dualtropic and can also use CCR5 (31). Their ability to use alternative coreceptors in vivo might be addressed by testing the effect of bicyclams in CCR5-negative PBMC.
A direct interaction with CXCR4 was strongly suggested by the ability of AMD3100 to compete with the SDF-1 chemokine and with the 12G5 monoclonal antibody (17, 41). Also in support of this interaction was our finding that several mutations in CXCR4 markedly reduced the antiviral effect of AMD3100, although no single mutation could totally abolish this effect. Our study was initially focused on the second extracellular loop (ECL2) of CXCR4, which bears at least part of the 12G5 epitope and seems to play an important role in the HIV coreceptor activity (4). The substitution of a neutral amino acid for any of the four aspartic acids of ECL2 resulted in a marked resistance to AMD3100, while the substitution of glutamine for a glutamic acid (E179Q) had a lesser effect. The other ECL2 mutations associated with resistance to AMD3100 were replacements of two phenylalanines that formed a Phe-X-Phe motif near the TM5 junction by nonaromatic amino acids. The other aromatic acid of ECL2, a tyrosine at position 184, was apparently dispensable to the antiviral effect of AMD3100. The TM4 domain contains a negatively charged amino acid (aspartic acid) and two phenylalanines, also forming a Phe-X-Phe motif. The mutation of these amino acids also conferred relative resistance to AMD3100. Other mutations in TM4 had no such effect.
Some amino acids of ECL2 or TM4 that are apparently critical for a complete antiviral effect of AMD3100 for example, the aspartic acids D182 and D193 and the phenylalanine F172, are not conserved in rat CXCR4 (Fig. 4A). The reason for their apparent lack of importance in the rat CXCR4 context, or in the HHRH chimera, is unclear. The presence of two aspartic acids in rat CXCR4, instead of neutral amino acids at positions 176 and 192 in human CXCR4, might be compensatory. Other domains of rat CXCR4 might also play a role in its interaction with AMD3100.
Two types of mechanisms could explain the relative resistance to AMD3100 conferred by mutations in the ECL2 and TM4 domains of CXCR4. The first hypothesis is that these mutations affect amino acids directly involved in the interaction of CXCR4 with AMD3100. This will be discussed in further detail later. Alternatively, the mutations might modify the overall structure of the extracellular domains of CXCR4, indirectly affecting its interaction with AMD3100. The latter mechanism could be consistent with the scattering of amino acids apparently critical for the antiviral effect of AMD3100 throughout ECL2 and with their presence in a membrane-spanning domain, which is a priori less accessible to the drug. However, the loop structure of ECL2 might bring these residues closer together. Also, spatial models of G protein-coupled receptors predict the assembly of membrane-spanning domains into a cylindrical structure with a pocket accessible to small ligands (2). Also consistent with indirect effects on the CXCR4 structure, most mutations in ECL2 or TM4 reduced the reactivity with 12G5. However, the 12G5 epitope is probably conformational (22) and might therefore be affected by slight modifications of the spatial structure of CXCR4 which have little effect on the interaction with gp120 and coreceptor activity. Indeed, all the CXCR4 mutants tested could mediate HIV-1 infection. It will be of interest to test their reactivity with other ligands, such as SDF-1 and other antibodies. Until the interaction of CXCR4 with its different ligands is known in its molecular details, indirect effects of mutations on the spatial structure of CXCR4 cannot be formally ruled out. However, the finding that resistance to AMD3100 was associated with the same type of mutations in ECL2 and TM4, substitutions for aspartic acid and phenylalanine, represents a strong argument in favor of a direct interaction of these amino acids with the AMD3100 bicyclam. Also, a role for these amino acids seems consistent with available structure-function data on bicyclams and with our current view of the mechanism of the HIV coreceptor activity of CXCR4.
Bicyclams are strongly basic at physiological pH due to the presence of four primary amines in each cyclam unit and might therefore engage in electrostatic interactions with negatively charged domains of CXCR4, in particular ECL2 (5 acidic residues; net charge, −3). The homologous domain of CCR5 has a net positive charge (+5). The accumulation of basic amino acids in the variable domains of gp120, in particular V3, is associated with a switch of HIV-1 to the SI phenotype (26) and therefore to usage of the CXCR4 coreceptor. In the context of HIV-1NDK, replacing V3 with the homologous domain from HIV-1LAI restored the ability to infect CD4+ cells expressing rat CXCR4 (36). We also observed that the RRHR chimera (rat CXCR4 with ECL2 from human CXCR4) mediated HIV-1NDK infection (4). These results are compatible with a direct interaction between V3 and ECL2. The binding of AMD3100 to ECL2 might therefore prevent its interaction with V3. A similar mechanism might also account for the selective blocking of CXCR4-mediated HIV-1 entry by a polyarginine peptide, ALX40-4C (18), and by an 18-mer peptide with a +8 net charge (32).
Bicyclams with an aromatic linker, a phenylene in AMD3100, have higher antiviral activity (5, 13). This aromatic group might interact with the aromatic groups of the phenylalanines in TM4 and ECL2, thus explaining their critical role. The tyrosine of ECL2 (Y184) was apparently not required for interaction with AMD3100. The predominant role of phenylalanines might be due to their position and/or to their organization in a Phe-X-Phe motif. This could be addressed by testing the antiviral activity of bicyclams with a nonaromatic linker against HIV-1 infection mediated by CXCR4 mutants. Finally, other features of CXCR4 are also likely to play a role in its sensitivity to bicyclams. Indeed, CCR3 has negatively charged ECL2 (net charge, −5) and a Phe-X-Phe motif in TM4, yet it mediates AMD3100-resistant HIV entry.
We recently observed that a peptide corresponding to the amino terminus of SDF-1, and several analogs of this peptide, could selectively interact with CXCR4 and block HIV-1 entry (25). Basic amino acids and two phenylalanines seemed important to the antiviral activity of these peptides. It will be of interest to test their effect on HIV-1 infection that is mediated by AMD3100-resistant CXCR4 mutants and a possible synergy with bicyclams.
AMD3100 is the first small-molecular-weight, nonpeptidic molecule that blocks HIV-1 infection by preventing its access to a cellular coreceptor. In vivo activity of AMD3100 was observed in SCID mice engrafted with human tissues (11). This bicyclam is therefore a candidate molecule for antiviral strategies aimed at blocking HIV-1 entry. Given the selectivity of AMD3100 for CXCR4, a direct beneficial effect of this bicyclam, for example, in terms of virus load, would be expected at later stages of HIV-1 infection, when SI strains are prevalent. However, AMD3100 and other compounds blocking CXCR4 might be useful at earlier stages of HIV-1 infection, in preventing the emergence of viral strains using this abundant and ubiquitous coreceptor. Antiviral strategies should therefore associate inhibitors of the CCR5 and CXCR4 coreceptors.
ACKNOWLEDGMENTS
We thank G. Henson, R. Duman, and D. Littman for gifts of reagents and I. Bouchaert, E. Gomas, and C. Tréboute for technical assistance.
This work was supported by the Agence Nationale de Recherche sur le SIDA.
REFERENCES
- 1.Alkhatib G, Berger E A, Murphy P M, Pease J E. Determinants of HIV-1 coreceptor function on CC chemokine receptor. Importance of both extracellular and transmembrane/cytoplasmic regions. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin J M. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994;6:180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 3.Brelot, A., N. Heveker, B. Labrosse, and M. Alizon. 1998. Unpublished data. [DOI] [PMC free article] [PubMed]
- 4.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridger G J, Skerlj R T, Thornton D, Padmanabhan S, Martellucci S A, Henson G W, Abrams M J, Yamamoto N, De Vreese K, Pauwels R, De Clercq E. Synthesis and structure-activity relationships of phenylenebis(methylene)-linked tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J Med Chem. 1995;38:368–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 6.Brun-Vézinet F, Rey M A, Katlama C, Girard P M, Roulot D, Yéni P, Lenoble L, Clavel F, Alizon M, Gadelle S, Madjar J J, Harzic M. Lymphadenopathy-associated virus type 2 in AIDS and AIDS-related complex. Lancet. 1987;i:128–132. doi: 10.1016/s0140-6736(87)91967-2. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collman R, Balliet J, Gregory S, Friedman H, Kolson D, Nathanson N, Srinivasan A. An infectious clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datema R, Rabin L, Hingenbergs M, Moreno M B, Warren S, Linquist V, Rosenwirth B, Seiffert J, McCune J M. Antiviral efficacy of the anti-human immunodeficiency virus bicyclam SDZ SID 791 (JM 3100), an inhibitor of infectious cell entry. Antimicrob Agents Chemother. 1996;40:750–754. doi: 10.1128/aac.40.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Clercq E, Yamamoto N, Pauwels R, Baba M, Schols D, Nakashima H, Balzarini J, Debyser Z, Murrer B A, Schwartz D, Thornton D, Bridger G, Fricker S, Henson G, Abrams M, Picker D. Potent and selective inhibition of human immunodeficiency virus (HIV)-1 and HIV-2 replication by a class of bicyclams interacting with a viral uncoating event. Proc Natl Acad Sci USA. 1992;89:5286–5290. doi: 10.1073/pnas.89.12.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq E, Yamamoto N, Pauwels R, Balzarini J, Witvrouw M, De Vreese K, Debyser Z, Rosenwirth B, Peichl P, Datema R, Thornton D, Skerlj R, Gaul F, Padmanbhan S, Bridger G, Henson G, Abrams M. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob Agents Chemother. 1994;38:668–674. doi: 10.1128/aac.38.4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 15.De Vreese K, Kofler-Mongold V, Leutgeb C, Weber V, Vermeire K, Schacht S, Anné J, De Clercq E, Datema R, Werner G. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J Virol. 1996;70:689–696. doi: 10.1128/jvi.70.2.689-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 17.Donzella G A, Schols D, Lin S W, Esté J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 18.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S-H, Goetz M B, Daar E S, Doms R W, O’Brien W A. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2B as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Hazan U, Alizon M. Detection of cell fusion mediated by the envelopes of human retroviruses by transactivation of a reporter gene. Methods Mol Genet. 1995;7:218–236. [Google Scholar]
- 21.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endres M J, Clapham P R, Marsh M, Ahuja M, Davis Turner J, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 23.Freed E O, Martin M A. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 24.Harrington R D, Geballe A P. Cofactor requirements for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heveker N, Montes M, Germeroth L, Amara A, Trautmann A, Alizon M, Schneider-Mergener J. Dissociation of the signalling and antiviral properties of SDF-1-derived small peptides. Curr Biol. 1998;8:369–376. doi: 10.1016/s0960-9822(98)70155-1. [DOI] [PubMed] [Google Scholar]
- 26.Kuiken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrosse B, Pleskoff O, Sol N, Jones C, Hénin Y, Alizon M. Resistance to a drug blocking human immunodeficiency virus type 1 entry (RPR103611) is conferred by mutations in gp41. J Virol. 1997;71:8230–8236. doi: 10.1128/jvi.71.11.8230-8236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao S, Alkhatib G, Liao F, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlman H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 30.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. The HIV-cell fusion reaction. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press Inc.; 1993. pp. 233–289. [Google Scholar]
- 31.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 32.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 34.Pleskoff, O., and M. Alizon. Unpublished results.
- 35.Pleskoff O, Seman M, Alizon M. Amphotericin B derivative blocks human immunodeficiency virus type 1 entry after CD4 binding: effect on virus-cell fusion but not on cell-cell fusion. J Virol. 1995;69:570–574. doi: 10.1128/jvi.69.1.570-574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleskoff O, Sol N, Labrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pleskoff O, Tréboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 38.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Doranz B J, Collman R G, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapouméroulie C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 40.Schols D, Esté J A, Henson G, De Clercq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor Fusin/CXCR4. Antiviral Res. 1997;35:147–156. doi: 10.1016/s0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- 41.Schols D, Struyf S, Van Damme J, Esté J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sol N, Ferchal F, Braun J, Pleskoff O, Tréboute C, Ansard I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spire B, Sire J, Zachar V, Rey F, Barré-Sinoussi F, Galibert F, Hampe A, Chermann J-C. Nucleotide sequence of HIV1-NDK, a highly cytopathic strain of the human immunodeficiency virus, HIV1. Gene. 1989;81:275–284. doi: 10.1016/0378-1119(89)90188-1. [DOI] [PubMed] [Google Scholar]
- 44.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus type 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]