Abstract
Carbon materials are commonly used for soil carbon sequestration and fertilization, which can also affect crop growth by manipulating the rhizosphere bacterial community. However, the comparison of the differences between active carbon (e.g., organic fertilizers) and stable carbon (e.g., biochar) on rhizosphere microdomains is still unclear. Hence, a trial was implemented to explore the influence of control (CK, no fertilizer; NPK, chemical fertilizer), organic fertilizer (CF-O, organic fertilizer; CF-BO, biochar-based organic fertilizer) and biochar material (CF-B, perishable garbage biochar; CF-PMB, pig manure biochar) on the diversity, composition, and interaction of rice rhizosphere bacterial community through 16 S rRNA gene high-throughput sequencing. Our results demonstrate that organic fertilizer increases bacterial alpha-diversity compared to no-carbon supply treatment to the extend, whereas biochar has the opposite effect. The rhizosphere bacterial community composition showed pronounced variations among the various fertilization treatments. The relative abundance in Firmicutes decreased with organic fertilizer application, whereas that in Chloroflexi and Actinobacteria decreased with biochar application. Bacterial network analysis demonstrate that organic fertilizer enhances the complexity and key taxa of bacterial interactions, while biochar exhibits an opposing trend. The findings of our study indicate that organic fertilizer may contribute to a positive and advantageous impact on bacterial diversity and interaction in rice rhizosphere, whereas the influence of biochar is not as favorable and constructive. This study lays the foundation for elucidating the fate of the rhizosphere bacterial community following different carbon material inputs in the context of sustainable agricultural development.
Keywords: Rhizosphere bacterial community, Carbon materials, Bacterial network, Organic fertilizer, Biochar
1. Introduction
Plant health and productivity are fundamental for developing sustainable agriculture solutions [1]. The rhizosphere bacterial community is known as the second genome of plants [2]. The rhizosphere is a hotspot where plants and microorganisms intersect, playing a crucial role in plant-microbe interactions. Multiple functionalities are exhibited by rhizosphere bacterial communities, including nutrient solubilization, plant growth promotion and carbon sequestration [3], [4]. The composition of rhizosphere bacteria is shaped by selectively recruiting a subset of bacteria from the surrounding soil, which can have diverse impacts on the host plants, spanning from advantageous to detrimental[5]. A critical aspect of sustainable agriculture lies in regulating plant health through the lens of rhizosphere bacterial communities [6]. The regulation of rhizosphere bacterial communities composition and diversity is influenced by a myriad of factors [7], encompassing not only root secretions [8] but also exogenous material inputs.
While the extensive use of chemical fertilizers is integral to modern agricultural production [9], with a decline in rhizosphere microbial diversity [10]. Compared with biochar (a stable carbon material formed by pyrolysis), organic fertilizer is rich in a large amount of available nutrients and available organic matter, which can be directly decomposed and utilized by microorganisms. The combined use of organic and chemical fertilizers not only improves soil fertility but also enhances the diversity of rhizosphere bacterial communities and encourages the proliferation of copiotrophic populations [11]. The application of organic fertilizers is conducive to the formation of beneficial soil microbial banks to assist plants in recruiting these beneficial microorganisms in the rhizosphere (e.g. Bacillus, Paenibacillus, Haliangium, Streptomyces) [12]. Apart from the application of organic fertilizers, the strategic utilization of biochar has emerged as a prominent approach, poised to enhance the diversity and functionality of rhizosphere bacterial communities. For example, biochar can regulate rhizosphere bacterial diversity, effectively suppressing watermelon wilt [13]. Organic fertilizers and biochar are increasingly utilized in agriculture to address environmental and crop health issues arising from the prolonged or excessive use of chemical fertilizers [14], [15]. From the perspective of soil bacteria, organic fertilizers are considered as "active carbon", easily degradable to release accessible nutrients and energy. On the other hand, biochar is classified as "stable carbon", with its stable carbon structure posing challenges for direct breakdown and utilization by bacteria. Previous research has delved into the impact of organic fertilizers or biochar on rhizosphere bacterial communities [16], [17], [18]. These studies reveal intriguing changes in bacterial community composition, with organic fertilizers fostering beneficial genera like Acinetobacter and Pseudomonas [19], and biochar promoting the abundance of Acidobacteria [20]. A recent study comprehensively assessed the modulatory effects of biochar and compost on the bacterial community composition in maize soil [21]. However, there is a lack of comprehensive research on the combined impact of active (organic fertilizer) and stable (biochar) carbon (C) on the rice rhizosphere bacterial community, particularly in regard to bacterial interactions.
Network analysis has been employed to investigate microbial interactions in diverse environments [22], [23] and identify potential keystone species [24]. Research has revealed that the strategic utilization of organic fertilizers and biochar can effectively boost bacterial interactions, giving rise to intricate and stable bacterial communities within the plant rhizosphere [25], [26]. Organic fertilizers are more effective than no-compost, with higher available carbon content that mobilizes key Actinomycetes and Gemmatimonadetes taxa, resulting in a more complex microbial network [26]. The integrated comparison of bacterial network interactions and potential keystone species under the application of organic fertilizers and biochar is currently lacking.
To address the aforementioned issues, we conducted a pot experiment to investigate the changes in rice rhizosphere bacterial communities under different carbon strategies, including carbon-free supply (CK, no fertilizer; CF, chemical fertilizer), organic fertilizer (CF-O, chemical fertilizer and organic fertilizer; CF-BO, chemical fertilizer and biochar-based organic fertilizer) and biochar materials (CF-B, chemical fertilizer and perishable garbage biochar; CF-PMB, chemical fertilizer and pig manure biochar). Two months after rice cultivation, rhizosphere soil samples were collected to assess the bacterial diversity, composition, and interaction in rice rhizosphere through 16 S rRNA gene amplicon sequencing. We hypothesized that (1) the diversity and composition of the rhizosphere bacterial community respond differently to active and stable C materials, and (2) both organic fertilizer and biochar can also enhance the complexity, stability, and modularity of the bacterial network. The investigation of rhizosphere microorganisms in response to the application of organic fertilizers and biochar will shed light on the potential for achieving sustainable agricultural production.
2. Materials and methods
2.1. Soil collection and experimental design
The soil used for pot trial was collected from Tongxiang City, Zhejiang Province, China (120.618°E, 30.637°N). This region belongs to a typical rice growing area, characterized by a subtropical monsoon climate with an average annual temperature of 15–25 °C and annual rainfall ranging from 1000 to 1500 mm. The basic physicochemical properties of the soil are as follows: pH at 7.07, TC (total carbon) at 8.93 g kg−1, TN (total nitrogen) at 0.98 g kg−1, TP (total phosphorus) at 0.73 g kg−1, TK (total potassium) at 23.66 g kg−1.
Before the pot trial, the soil should be air-dried and sieved through a 5 mm mesh. Each pot (height 25 cm and internal diameter 15 cm) was filled with 1 kg of soil. This experiment comprised six treatment groups, designated as follows: the no fertilizer (CK), NPK fertilizer (CF), organic fertilizer plus NPK fertilizer (CF-O), biochar-based organic fertilizers plus NPK fertilizer (CF-BO), perishable waste biochar plus NPK fertilizer (CF-B), pig manure biochar plus NPK fertilizer (CF-PMB). The biochar-based organic fertilizer is prepared from 80% pig manure organic fertilizer and 20% perishable waste biochar. The composition and dosage of different treatments are listed in Table S1. The properties of various biochar and organic fertilizers are displayed in Table S2. The experimental design adopted a randomized design, with each treatment having 4 replicates. For each pot, fully mix the soil and fertilizer (including biochar) before adding them to the pot. Transplant three rice seedlings into each pot. Regular watering and control of disease and insect management were provided during the pot experiment. This study was conducted at the Greenhouse Base of Zhejiang University of Science and Technology, China. The greenhouse base maintains an average temperature of 25 °C with regular watering.
2.2. Sampling and measurement of soil and plant
Plant samples and rhizosphere soil were collected at the flowering stage (8-week growth after transplant). Rhizosphere soil samples were collected following the established method [25]. In brief, rhizosphere soil was obtained by gently washing the roots with water and then centrifuging (11 000g, 5 min), with the lower layer of soil defined as the rhizosphere soil. The collected rhizosphere soil samples stored at − 80 °C for subsequent DNA extraction within 24 h.
Plant samples were dried at 70 °C until a constant weight was reached to determine the biomass. Recognizing the prevalence of roots in the potted plants, we labeled the soil obtained after root removal as "proximal rhizosphere soil," which was then used for assessing soil physicochemical characteristics.
2.3. DNA extraction and high-throughput sequencing
Rhizosphere soil DNA was extracted using the FastDNA Spin Kit (MP Biomedicals). The quality of DNA extracts was assessed by a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The primer set 515 F (5′- GTGCCAGCMGCCGCGG-3′) and 907 R (5′- CCGTCAATTCMTTTRAGTTT-3′) was used to amplify the V4-V5 regions of the 16 S rRNA gene. Sequencing was performed by Genesky Biotechnologies Inc. (Shanghai, China) based on an Illumina NovaSeq 6000.
QIIME2 analysis platform (https://qiime2.org) was employed the to process the raw reads. Initially, the cutadapt plugin was used to trim adaptor and primer sequences. Quality control and identification of ASVs (p-max-ee = 2.0, p-trunc-q = 2, and p-chimera method = consensus) were performed using the DADA2 plugin [27]. Taxonomic assignments of ASV representative sequences were determined using a Naive Bayes Classifier (Ribosomal Database Project, RDP version 11.5). Subsequently, all samples were standardized to a depth of 54,839 sequences for downstream analysis, thus addressing potential sequencing depth variations among different samples. All raw data have been deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject ID PRJNA1000724.
2.4. Network analyses
The Molecular Ecological Network Analysis Pipeline (MENAP, http://ieg4.rccc.ou.edu/mena) was employed for the network analyses [24]. Considering the differences in carbon stability between organic fertilizer and biochar, as well as the sample size requirements for network construction, treatments are merged for network analysis. Three networks were constructed, including a network of carbon-free control group (CK and CF), an organic fertilizer group (CF-O and CF-BO), and a biochar group (CF-B and CF-PMB). A similarity matrix was computed using the Pearson correlation coefficient from the abundance data of OTUs present in at least 6 out of 8 samples. Subsequently, random matrix theory (RMT) was employed to automatically determine the similarity threshold for network construction. The network structure was visualized using Gephi software, showing distinct modules.
Closeness Centrality measures the average shortest path length between a node and others, reflecting faster information exchange for nodes with high closeness centrality. In bacterial networks, nodes with high closeness centrality hold broader influence, enhanced resource acquisition, and superior information transfer capabilities. Betweenness Centrality assesses how effectively a node serves as a bridge or essential intermediary for information transfer in the network, exerting significant influence on connectivity and data flow. In bacterial networks, nodes with high betweenness centrality play crucial functional and regulatory roles.
Potential keystone species were categorized based on their connectivity within modules (Zi) and among modules (Pi) as module hubs (Zi > 2.5, Pi ≤ 0.62), network hubs (Zi > 2.5, Pi > 0.62), connectors (Zi ≤ 2.5, Pi > 0.62), or peripherals (Zi ≤ 2.5, Pi ≤ 0.62) [28]. Negative:positive cohesion refers to the proportion of negative and positive interactions in a bacterial network. The absolute value of negative/positive cohesion was calculated to evaluate the network stability in different C material input treatment [29]. The network topological parameters of node and edge numbers, and betweenness and closeness centrality are used to assess bacterial network complexity, with higher these parameters representing greater network complexity.
2.5. Statistical analysis
The β-diversity of the rhizosphere bacterial communities in different treatment was analyzed using the Bray-Curtis distance and the weighted UniFrac distance [30], [31]. The dissimilarity in the distances was visualized by principal coordinate analysis plots (PCoA). Three non-parametric dissimilarity analyses (analysis of similarity (ANOSIM), multi-response permutation procedure (MRPP), and non-parametric multivariate ANOVA (Adonis)) were used to examine the variance among communities (using the "vegan" package of R). When the normality assumptions were met, one-way ANOVA and post LSD tests were conducted. LEfSe analysis was conducted using the online analyser at http://huttenhower.sph.harvard.edu/galaxy [32]. The samples were divided into 3 groups for LEfSe based on cluster analysis, including CK-CF, CFO-CFBO, and CFB-CFPMB (one treatment pair per group). The factorial Kruskal-Wallis test among different treatments was performed with an alpha value of 0.05. A logarithmic LDA score threshold of 3.5 was applied to identify discriminative features.
3. Results
3.1. Alpha diversity of bacterial community in rice rhizosphere
The Chao1, Shannon diversity index, and phylogenetic diversity were used to estimate the α-diversity of the bacterial community in rice rhizosphere (Fig. 1). The three α-diversity indices demonstrated consistent trends among different fertilization treatments. The Chao1, Shannon diversity index, and phylogenetic diversity index showed their highest trends with the co-application of organic and chemical fertilizer (CF-O, CF-BO). Biochar application, particularly in the pig manure biochar (CF-PMB), exhibits a certain degree of decrease in the Chao1 and Shannon diversity index of rhizosphere bacteria when compared to organic fertilizer (CF-O). The non-fertilized treatment did not show statistically significant differences in the α-diversity indices compared to the other treatments. Taken together, the results indicate that organic fertilizer (CF-O, CF-BO) tended to promote α-diversity indices when compared to the non-carbon material treatment (CK, CF), while biochar (CF-B, CF-PMB) showed an opposite trend.
Fig. 1.
The α-diversity of the rhizosphere bacterial community in different fertilization treatment, including the Chao1 index (a) and Shannon diversity (b) and phylogenetic diversity (c). Different lowercase letters represent significant differences between the treatments (n = 4, LSD test, p < 0.05). CK, no fertilizer input; CF, chemical fertilizer; CF-O, organic fertilizer plus NPK fertilizer; CF-BO, biochar-based organic fertilizers plus NPK fertilizer; CF-B, perishable waste biochar plus NPK fertilizer; CF-PMB, pig manure biochar plus NPK fertilizer.
3.2. Beta diversity of bacteria community in rice rhizosphere
Principal coordinate analysis (PCoA) was employed to assess dissimilarities in bacterial ASV level community composition within the rhizosphere soil across different treatments using both Bray-Curtis and weighted UniFrac distances (Fig. 2). The results indicate significant variations in the composition of the bacterial community among the treatments (PERMANOVA, p < 0.01; ANOSIM, p < 0.01). The MRPP, ANOSIM, and ADONIS test the differences in community composition across all treatments (Table S3). Based on the Bray-Curtis and weighted UniFrac similarity indices, the ANOSIM test revealed R values ranging from 0.10 to 0.99 and − 0.02 to 0.93, respectively, for the 15 possible pairwise comparisons between groups. Regarding the CF-O and CF-BO treatments, no significant differences were observed, whereas all other pairwise comparisons among the treatments displayed significant variations. Given the similarity in bacterial communities between CF-O and CF-BO, along with the adoption of similar agronomic practices involving organic fertilizer input, we grouped these two treatments together for Lefse and network analysis (designated as the "Active C" group). Similarly, the CK and CF treatment exhibited the smallest R values (Bray-curtis, R=0.688, indicating the highest similarity), and both treatments involved no carbon input. Therefore, these two treatments were merged into a single group, designated as the "no-carbon material input" group. Likewise, the remaining two treatments, CF-B and CF-PMB, which involved the application of biochar, were combined into one group, designated as the "Stable C" group.
Fig. 2.
PCoA plots showing the variation in the community compositions of the bacteria in rhizosphere soil among treatments based on Bray-Curtis distance (a) and the weighted UniFrac metric (b). PERMANOVA and ANOSIM test of group differences according to the bacterial distance matrix. The relative abundances of rhizosphere bacteria at phyla level under different treatment (c). Different lowercase letters represent significant differences between the treatments (n = 4, LSD test, p < 0.05).
The composition of the rhizosphere bacterial community was assessed using the RDP Classifier to assign high-quality sequences to 32 phyla. Among these, nine phyla exhibited a relative abundance greater than 1%, contributing to 87.0–88.6% of the total relative abundance. Particularly, Proteobacteria accounted for 27.4–37.2%, Bacteroidetes accounted for 10.7–14.2%, and Firmicutes accounted for 6.6–13.3% across all treatments. Compared to the unfertilized CK treatment, the relative abundance of phylum Proteobacteria was consistently increased with the application of chemical fertilizer. Compared to the treatments without C input (CK, CF), the addition of organic fertilizer (CF-O) and biochar-based organic fertilizer (CF-BO) resulted in a decline in the relative abundance of phylum Firmicutes, while increasing the relative abundance of Actinobacteria. The addition of biochar (CF-B, CF-PMB) resulted in a decreasing trend in the relative abundance of Chloroflexi and Actinobacteria. At the bacterial genus level, Symbiobacterium, Ideonella, Geobacter, Flavisolibacter, Ornatilinea, and Bacillus dominate the majority of relative abundance (Fig. S2), with significant variations observed under different fertilization treatments. For example, organic fertilizer increased the relative abundance of Ornatilinea, Rhodobacter, Rhodococcus, and Mycobacterium, while decreasing the relative abundance of Symbiobacterium and Azospira. The abundance of Symbiobacterium, Deferrisoma, Azospira, and Ideonella increased with biochar treatment, while the relative abundance of Massilia, Parasegetibacter, Ornatilinea, and Oscillochloris decreased.
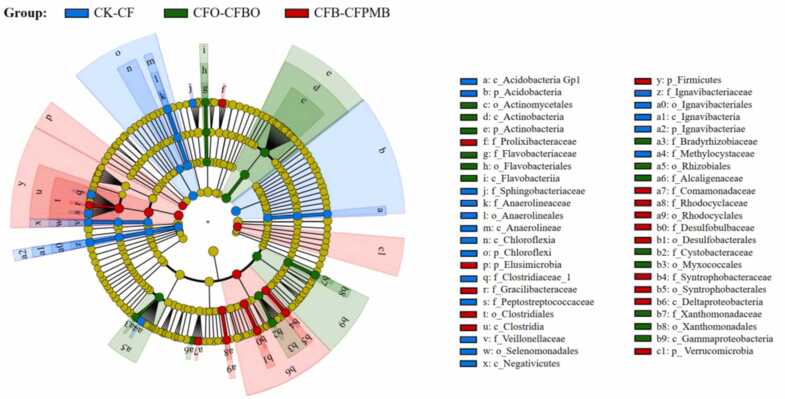
3.3. Biomarkers in different group
In the previous section, we divided the six treatments into three groups, and consequently, we utilized LEfSe analysis to discover biomarkers resulting from diverse C material applications (Fig. 3). The bacterial community biomarkers with a linear discriminant analysis (LDA) threshold of 3.5 for each group are shown in Fig. S3. The LEfSe analysis revealed distinctive biomarkers for each group. In the control group (CK-CF), Acidobacteria_Gp1 (belonging to Acidobacteria), Chloroflexia (including Chloroflexia and Anaerolineales), Selenomonadales (belonging to Firmicutes), and Ignavibacteriales (belonging to Ignavibacteriae) were identified as biomarkers. The organic fertilizer group (CFO-CFBO) exhibited biomarkers such as Actinobacteria (including Actinomycetales), Flavobacteriia (including Flavobacteriales), Rhizobiales, Myxococcales, and Gammaproteobacteria (including Xanthomonadales). In the biochar group (CFB-CFPMB), biomarkers included Clostridiales (belonging to Firmicutes), Elusimicrobia, Rhodocyclales, Deltaproteobacteria (including Desulfobacterales and Syntrophobacterales), and Verrucomicrobia.
Fig. 3.
LEfSe analysis of bacterial community in rhizosphere from different groups. LDA scores > 3.5. According to the similarity of the community matrix, the six treatments were divided into three groups, including no carbon application group (CK-CF), active carbon group (CFO-CFBO), and stable carbon group (CFB-CFPMB).
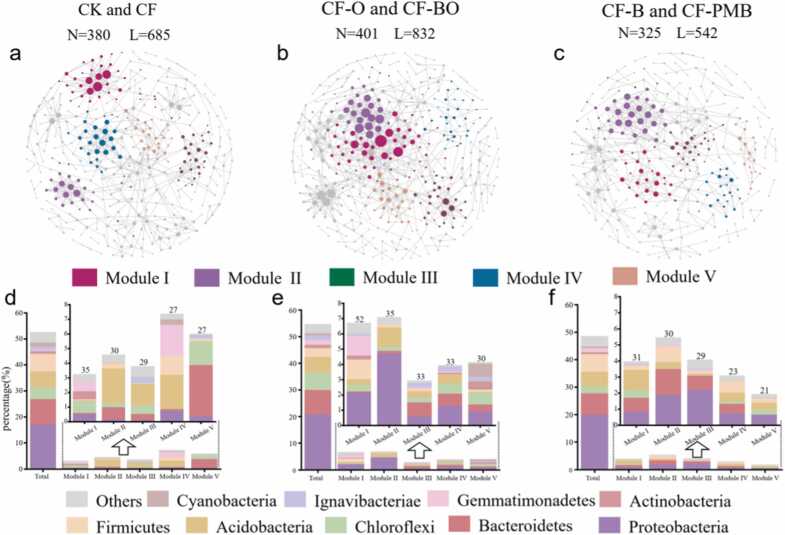
3.4. Co-occurrence network structure of bacterial community in rice rhizosphere
Considering the similarity of community composition and agronomic practices, as well as the increased replicates for constructing the network samples, we merged the four replicates of each of the six treatments into three groups with eight replicates each. These three groups were named as the carbon-free supply, organic fertilizer, and biochar group (Fig. 4). The organic fertilizer network exhibited a higher total number of nodes and links compared to the control network, and it also displayed higher average degree and clustering coefficient (Fig. 4, Table S4), indicating a greater complexity in the organic fertilizer network in comparison to the control group. Conversely, the network in biochar group appeared simpler than the control group (Fig. 4, Table S4). Regarding the number of links, the organic fertilizer network had 570 positive links, accounting for 68.5% of the total corresponding links, while the network in control and biochar treatments had 61.3% and 61.3% positive links, respectively. This finding implies that organic fertilizer could potentially promote positive bacterial cooperation.
Fig. 4.
Network analysis of rhizosphere bacterial communities based on the ASV level in three groups (a-c). The size of a node is proportional to the degree. N and L represent nodes and links, respectively. Composition of bacterial communities participating in the network and the relative abundance of bacterial ASVs in the first five modules (d-f). The numbers above the histogram represent the number of ASVs.
Furthermore, we performed in-depth analysis of the bacterial taxa participating in the entire network, including the five most prominent modules (Fig. 4). The number of bacterial taxa from different phyla participating in the entire network showed consistent trends to rhizosphere bacterial community composition among different treatments. The number of ASVs affiliated with the Chloroflexi within the network was enhanced by the addition of organic fertilizer. Module-specific analyses were conducted to detect shifts in the pattern of bacterial taxa involved in different modules. Notably, in the network without carbon materials (CK and CF), higher relative abundance of Acidobacteria was exhibited in modules II, III and IV, distinguishing them from other networks. The relative abundance of the Bacteroidetes and Chloroflexi was much higher in module V than in the other modules. Through module-specific analysis of the organic fertilizer network, it was observed that Proteobacteria had the highest relative abundance in all five modules. This suggests that Proteobacteria has stronger interactions with various bacterial taxa under the application of organic fertilizer. In contrast, in the biochar network modules, besides Proteobacteria, Actinobacteria also participated in the first four modules.
3.5. Cohesion and potential keystone species of network
The ratio of negative cohesion to positive cohesion was employed to assess the stability of bacterial community matrices in different groups (Fig. 5). The findings demonstrate that the absolute values of negative cohesion/positive cohesion are the least in the Control group. Both organic fertilizer and biochar increase the absolute values of negative cohesion/positive cohesion, but there is no significant difference in the values between the two groups. The ratio gradually approached 1, indicating a reasonable and dynamic shift in competition and synergistic relationships among bacterial communities within the organic fertilizer and biochar network. These results indicate that the input of C materials can strengthen the stability of the bacterial network.
Fig. 5.
The degree, closeness centrality and betweenness centrality values of the nodes in the different networks (a-c). The absolute value of positive/negative cohesion in the bacterial correlation network (d). Classification of nodes to identify potential keystone species within the correlation network (e).
The degree, closeness centrality (CC), and betweenness centrality (BC) value of each node can be employed to evaluate the total importance of bacteria in the network. The degree, CC of the majority of the nodes in the rhizosphere network associated with organic fertilizer (CF-O and CF-BO) were stronger than that of control group (CK and CF), while the trend of BC values is opposite. In comparison to the control group, biochar (CF-B and CF-PMB) has somewhat decreased the degree, CC, and BC values of nodes in the network. Moreover, potential keystone species are identified based on evaluating the connections of nodes within and outside the module, with Pi > 0.62 or Zi > 2.5 indicating potential keystone species. In the networks of the Control, organic fertilizer, and biochar groups, 13, 12, and 7 potential keystone species were recognized, respectively (Fig. 5, Table S5). The keystone networking roles are assumed by several nodes (e.g., Xanthomonadaceae, Comamonadaceae, Ignavibacteriaceae) in the Control group. Several pivotal nodes were detected in the network of the organic fertilizer group, such as Bacillaceae_1, Sphingomonadaceae, Desulfobulbaceae, Chitinophagaceae. In addition, the potential keystone species in the bacterial network of the biochar group include Ignavibacteriaceae, Saprospiraceae, Veillonellaceae, and so on.
4. Discussion
Organic fertilizer amendments and biochar are widely utilized in agricultural applications to stimulate higher crop productivity[33], [34]. Both these materials contribute nutrients to the soil, but exhibit dissimilarities in terms of carbon stability. Organic fertilizers can serve as readily available carbon sources for soil bacteria (Table S6), while biochar, due to its production through high-temperature anaerobic pyrolysis, is characterized by its resistance to decomposition and stability. These two carbon materials are employed to respectively characterize active and stable carbon. Several previous studies have demonstrated that organic fertilizers and biochar exhibit the potential to enhance soil fertility and alleviate soil acidification [35], [36], which is in agreement with the findings of the our study (Fig. S1, Table S6). Nonetheless, what was unexpected is that our result revealed a reduction in rice biomass and nitrogen accumulation due to the application of biochar (results obtained from two pot experiments, Table S7). This observation implies a certain inhibitory impact of biochar on pre-flowering rice growth, which is in contrast with the findings of previous research [37], [38]. This phenomenon could potentially be attributed to the factors, such as the proportion of biochar to soil, soil characteristics, and potential biochar toxicity [39]. Within this context, our investigation has focused on evaluating the differential impacts of material inputs characterized by distinct attributes on the diversity, composition, and bacterial network of rhizosphere bacterial communities.
4.1. Distinct response of rhizosphere bacterial communities to organic fertilizer and biochar
As hypothesized, the active C (organic fertilizer) and stable C (biochar) have exerted distinct impacts on both the diversity and composition of bacterial communities in rice rhizosphere. In comparison with chemical fertilizers, organic fertilizers have demonstrated a propensity to elevate the diversity of rhizosphere bacteria to a certain extent, in accordance with previous study [40]. Organic fertilizers or biochar-based organic fertilizers have a large amount of carbon sources available to microorganisms to promote the propagation. In addition to carbon sources, various mineral elements in organic fertilizers can satisfy plant growth and indirectly provide nutrients for rhizosphere microorganisms. Our previous study also showed that rice associated with greater biomass had higher rhizosphere microbial diversity [25]. In addition, the increase in α-diversity index may be due to the introduction of exogenous bacteria by the organic fertilizer, because the composted organic fertilizer itself has abundant microorganisms. Given the difference in the characteristics of organic fertilizers and biochar, it is easy to understand that the risk of α-diversity decline under the application of biochar. First, compared with organic fertilizers, biochar is a stable carbon that cannot be directly utilized by plants or microorganisms. Second, it is noteworthy in this study, biochar, as compared to organic fertilizer, decreased the rice aboveground biomass (Fig. S1, Table S7). The reduction in aboveground biomass may lead to a decreased availability of carbon sources (e.g., rhizosphere exudates) required by microorganisms in rhizosphere. Another aspect to take into account is that biochar might contain certain contaminants (e.g., polycyclic aromatic hydrocarbons, heavy metal), which may reduce the number of bacteria that are not tolerant to contaminants [41], [42]. In addition, biochar can adsorb available C and nutrients, making them less available to rhizosphere bacteria, which may result in nutrient deficiencies for some bacteria [43]. Therefore, activated carbon had a better promoting effect than stable carbon in terms of rhizosphere bacterial α-diversity.
Distinct shifts in rhizosphere bacterial community composition were observed in response to varying C material inputs (Fig. 2), as expected from the first hypothesis. The PCoA plot showed that the two treatments without carbon materials input (CK and CF) had closer community composition, and similar scenarios were also seen for the organic fertilizer materials (CF-O and CF-BO) and biochar (CF-B and CF-PMB). Different carbon substrates possess the capability to modify soil macronutrient status, leading to a shift from oligotrophic to copiotrophic conditions. Proteobacteria decreased without any application of chemical fertilizers or C materials, because Proteobacteria can be assigned into copiotrophic taxa associated with substrates rich in resource availability [44]. The surge of copiotrophic groups (like Actinobacteria) occurred with the introduction of organic fertilizer. This coherence is consistent with the alterations in bacterial composition resulting from resource manipulation [45], as elucidated by the copiotrophic hypothesis. In the current study, biochar decreased the relative abundance of Chloroflexi and Actinobacteria (copiotrophic groups), contrary to previous study [46]. The possible reason is that the living environment of bacteria in this study is the rhizosphere, and biochar reduces the biomass of aboveground objects in this study, resulting in a decrease in rhizosphere sediments.
The bacterial markers Gammaproteobacteria and Rhizobiales in the organic fertilizer group can participate in the soil nitrogen cycle, where Rhizobiales can form a symbiotic relationship with plant roots, converting atmospheric nitrogen into ammonia or nitrogen compounds available to plants [47], [48]. This allows plants to have access to an adequate source of nitrogen, which helps to promote plant growth and development. In addition, Myxococcales bacteria have a unique lifestyle and social behaviour. They form complex multicellular aggregates in the soil. This social behaviour allows them to cooperate and help each other in a resource-limited environment. They also exhibit a degree of predatory behaviour, being able to break down and absorb the cells of other microorganisms such as bacteria and fungi [49]. The soil bacterial biomarkers in the biochar group are Clostridiales, Elusimicrobia, Rhodocyclales, Deltaproteobacteria and Verrucomicrobia, and these bacteria generally interact in a symbiotic manner with other bacteria to participate in the degradation of organic matter [50], [51], [52], [53]. Some genera of Clostridiales bacteria also have the ability to produce toxins, such as the botulinum toxin produced by Clostridium botulinum [54]. These toxins are a potential risk to human and animal health. This requires that in the future we should intensify our research into the scientific proportioning of biochar.
4.2. Organic fertilizers and biochar exert distinct influences on rhizosphere bacterial networks
Within the rhizosphere environment, microorganisms forge elaborate networks of associations, as opposed to isolated existence. The study of bacterial networks unveils interactions among bacteria in rhizosphere under different C material inputs. The results from this study emphasize the effect of organic fertilizer and biochar on reshaping the network structure of rhizosphere microbiota. The incorporation of organic fertilizer resulted in a complexification of the rhizosphere bacterial network, as observed by the higher nodes and links within the organic fertilizer network compared to the control group (Fig. 4, Table S4). In contrast, the biochar group exhibited an opposing behavior compared with organic fertilizer group. It's worth noting that both biochar and organic fertilizers increased stability indicators of rhizosphere bacterial networks when compared to no carbon materials, as evidenced by the absolute value of negative/positive cohesion. These findings do not entirely align with our second hypothesis. The results of previous studies align with our findings regarding the organic fertilizer network, demonstrating the capacity of organic fertilizers to enhance the complexity and stability of microbial networks [55], [56], [57]. Organic fertilizers contribute to the enhancement of the available ecological niche in the rhizosphere by both introducing available carbon sources and resources, as well as indirectly promoting rice root exudates. This leads to the promotion of positive bacterial interactions (Table S4). The rhizosphere harbored a combination of organic and inorganic nutrients, supporting microbial growth across different trophic levels and mitigating competition. Additionally, organic fertilizers promote the formation of modularity, amplifying the significance of copiotrophic bacteria within the symbiotic systems (Fig. 4e). Research shows intricate networks bolster environmental resilience[58]. Enhanced complexity, tied to organic fertilizer, suggests that bacteria in rhizosphere could better endure stress via diverse taxonomic complement. The use of organic fertilizer has the potential to mold a more interconnected community and boost microbial functional capacity, potentially indicating increased ecosystem multifunctionality. However, it is surprising that biochar reduces the complexity of the microbial network, which is contrary to previous study [59]. Microbial complexity may be related to its own microbial diversity [57], and the reduction in rhizosphere α-diversity under biochar application in this study may be an explanation for the reduction in network complexity. Reduced network complexity may result from enhanced resource limitation (e.g., reduced availability of water, soil carbon and nutrients) that impaired microbial diversity and network complexity. Another possible explanation is that biochar compete with the ecological niche of rhizosphere microorganisms, because biochar has a “house” that is conducive to the growth of microorganisms [60]. Fortunately, biochar contributes to the stability of the bacterial network, which highlights the ability of biochar to resist changes in high-stress environments in promoting bacterial interactions. Thus, carbon materials amendments have the potential to enhance the resilience of bacterial communities against external stresses, thereby contributing to mitigating soil quality decline by fostering a more interconnected community structure.
The present top five module of network results show that Proteobacteria (copiotrophic) dominated interactions in the organic fertilizer and biochar networks compared to the no cabon materials input. This implies that Proteobacteria might play a fundamental role in facilitating information transfer and energy exchange between different species. To further identify potential keystone species, we identified a small number of module hubs (i.e. nodes highly connected within a module) and connectors (i.e. nodes linking different modules together) within rhizosphere modules under different C materials input. Potential keystone bacteria can exert a profoundly influence on interactions within the bacterial community and contribute to network stability [61], and they might promote resource cycling in the rhizosphere. Twelve potential keystone genera were detected in the bacterial network associated with organic fertilizer, in which Bacillaceae_1, Sphingomonadaceae had the function of promoting plant growth [62]. In addition, some potential keystone genera capable of participating in C or N cycling have been detected in the biochar network, e.g., Ignavibacteriaceae, Saprospiraceae, Veillonellaceae [63].
5. Conclusions
The changes of rice rhizosphere bacterial community under different carbon material input were revealed from the perspective of community composition and species interaction. Compared with no carbon material application, organic fertilizer promoted the increase of rice rhizosphere α-diversity, while biochar showed the opposite trend. Both organic manure and biochar significantly reshaped the rhizosphere bacterial community composition. For the rhizobacterial interaction network, organic fertilizer increased its complexity and stability, while biochar decreased its complexity and increased its stability. Overall, organic fertilizers can contribute to sustainable agricultural production by affecting rhizosphere bacterial networks and plant growth. While biochar is an excellent material for fertilizing soil and fixing carbon, its potential dangers to rhizosphere microorganisms are worth examining.
Funding statement
This work was financially supported by the Key Research and Development Program of Zhejiang Province (2021C03025), the National Key Research and Development Program of China (2022YFE0196000), the National Natural Science Foundation of China (42207350), the Zhejiang Province Natural Science Foundation (LQ23D030003) and Fundamental Research Funds for the Zhejiang University of Science and Technology (2023QN051, 2022yjsjg05).
CRediT authorship contribution statement
Zongkun Yang, Xin Cui, Xiaoge Fan, Investigation, original draft, Writing – review & editing, Software, Visualization. Yefeng Ruan, Zhennan Xiang, Investigation. Lingfei Ji, Han Gao, Writing – review & editing. Min Zhang, Shengdao Shan, Supervision. Wenbo Liu, Conceptualization, Methodology, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.03.012.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Porter J.R., Semenov M.A. Crop responses to climatic variation. Philos Trans R Soc B Biol Sci. 2005;360:2021–2035. doi: 10.1098/rstb.2005.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berendsen R.L., Pieterse C.M.J., Bakker P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Rui J., Mao Y., Yannarell A., Mackie R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem. 2014;68:392–401. doi: 10.1016/j.soilbio.2013.10.017. [DOI] [Google Scholar]
- 4.Marschner P., Crowley D., Yang C.H. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil. 2004;261:199–208. doi: 10.1023/B:PLSO.0000035569.80747.c5. [DOI] [Google Scholar]
- 5.Singh B.K., Millard P., Whiteley A.S., Murrell J.C. Unravelling rhizosphere–microbial interactions: opportunities and limitations. Trends Microbiol. 2004;12:386–393. doi: 10.1016/j.tim.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Chepsergon J., Moleleki L.N. Rhizosphere bacterial interactions and impact on plant health. Curr Opin Microbiol. 2023;73 doi: 10.1016/j.mib.2023.102297. [DOI] [PubMed] [Google Scholar]
- 7.Tiquia S.M., Lloyd J., Herms D.A., Hoitink H.A.J., Michel F.C. Effects of mulching and fertilization on soil nutrients, microbial activity and rhizosphere bacterial community structure determined by analysis of TRFLPs of PCR-amplified 16S rRNA genes. Appl Soil Ecol. 2002;21:31–48. doi: 10.1016/S0929-1393(02)00040-9. [DOI] [Google Scholar]
- 8.Sasse J., Martinoia E., Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018;23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Tilman D., Cassman K.G., Matson P.A., Naylor R., Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. doi: 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- 10.Kavamura V.N., Hayat R., Clark I.M., Rossmann M., Mendes R., Hirsch P.R., et al. Inorganic nitrogen application affects both taxonomical and predicted functional structure of wheat rhizosphere bacterial communities. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang R., Hou R., Li J., Lyu Y., Hang S., Gong H., et al. Effects of different fertilizers on rhizosphere bacterial communities of winter wheat in the North China Plain. Agronomy. 2020;10:93. doi: 10.3390/agronomy10010093. [DOI] [Google Scholar]
- 12.Qiu M., Zhang R., Xue C., Zhang S., Li S., Zhang N., et al. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol Fertil Soils. 2012;48:807–816. doi: 10.1007/s00374-012-0675-4. [DOI] [Google Scholar]
- 13.Ling N., Deng K., Song Y., Wu Y., Zhao J., Raza W., et al. Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol Res. 2014;169:570–578. doi: 10.1016/j.micres.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Chivenge P., Vanlauwe B., Six J. Does the combined application of organic and mineral nutrient sources influence maize productivity? A meta-analysis. Plant Soil. 2011;342:1–30. doi: 10.1007/s11104-010-0626-5. [DOI] [Google Scholar]
- 15.European Commission, Joint Research Centre, Institute for Environment and Sustainability . Publications Office; LU: 2010. Biochar application to soils: a critical scientific review of effects on soil properties, processes and functions. [Google Scholar]
- 16.Jin X., Zhang J., Shi Y., Wu F., Zhou X. Green manures of Indian mustard and wild rocket enhance cucumber resistance to Fusarium wilt through modulating rhizosphere bacterial community composition. Plant Soil. 2019;441:283–300. doi: 10.1007/s11104-019-04118-6. [DOI] [Google Scholar]
- 17.Pietikäinen J., Kiikkilä O., Fritze H. Charcoal as a habitat for microbes and its effect on the microbial community of the underlying humus. Oikos. 2000;89:231–242. doi: 10.1034/j.1600-0706.2000.890203.x. [DOI] [Google Scholar]
- 18.Saito M. Charcoal as a micro-habitat for VA mycorrhizal fungi, and its practical implication. Agric Ecosyst Environ. 1990;29:341–344. doi: 10.1016/0167-8809(90)90298-R. [DOI] [Google Scholar]
- 19.Zhang X., Zhang R., Gao J., Wang X., Fan F., Ma X., et al. Thirty-one years of rice-rice-green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol Biochem. 2017;104:208–217. doi: 10.1016/j.soilbio.2016.10.023. [DOI] [Google Scholar]
- 20.Khodadad C.L.M., Zimmerman A.R., Green S.J., Uthandi S., Foster J.S. Taxa-specific changes in soil microbial community composition induced by pyrogenic carbon amendments. Soil Biol Biochem. 2011;43:385–392. doi: 10.1016/j.soilbio.2010.11.005. [DOI] [Google Scholar]
- 21.Bello A., Liu W., Chang N., Erinle K.O., Deng L., Egbeagu U.U., et al. Deciphering biochar compost co-application impact on microbial communities mediating carbon and nitrogen transformation across different stages of corn development. Environ Res. 2023;219 doi: 10.1016/j.envres.2022.115123. [DOI] [PubMed] [Google Scholar]
- 22.Li D., Ni H., Jiao S., Lu Y., Zhou J., Sun B., et al. Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome. 2021;9:20. doi: 10.1186/s40168-020-00978-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan M.M., Guo X., Wu L., Zhang Y., Xiao N., Ning D., et al. Climate warming enhances microbial network complexity and stability. Nat Clim Chang. 2021;11:343–348. doi: 10.1038/s41558-021-00989-9. [DOI] [Google Scholar]
- 24.Deng Y., Jiang Y.-H., Yang Y., He Z., Luo F., Zhou J. Molecular ecological network analyses. BMC Bioinform. 2012;13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W., Ling N., Guo J., Ruan Y., Zhu C., Shen Q., et al. Legacy effects of 8-year nitrogen inputs on bacterial assemblage in wheat rhizosphere. Biol Fertil Soils. 2020;56:583–596. doi: 10.1007/s00374-020-01435-2. [DOI] [Google Scholar]
- 26.Yan T., Xue J., Zhou Z., Wu Y. Effects of biochar-based fertilizer on soil bacterial network structure in a karst mountainous area. CATENA. 2021;206 doi: 10.1016/j.catena.2021.105535. [DOI] [PubMed] [Google Scholar]
- 27.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olesen J.M., Bascompte J., Dupont Y.L., Jordano P. The modularity of pollination networks. Proc Natl Acad Sci USA. 2007;104:19891–19896. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez D.J., David A.S., Menges E.S., Searcy C.A., Afkhami M.E. Environmental stress destabilizes microbial networks. ISME J. 2021;15:1722–1734. doi: 10.1038/s41396-020-00882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray J.R., Curtis J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol Monogr. 1957;27:326–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 31.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M., Zhang S., Liu L., Wu L., Ding X. Combined organic amendments and mineral fertilizer application increase rice yield by improving soil structure, P availability and root growth in saline-alkaline soil. Soil Tillage Res. 2021;212 doi: 10.1016/j.still.2021.105060. [DOI] [Google Scholar]
- 34.Kumar A., Joseph S., Tsechansky L., Schreiter I.J., Schüth C., Taherysoosavi S., et al. Mechanistic evaluation of biochar potential for plant growth promotion and alleviation of chromium-induced phytotoxicity in Ficus elastica. Chemosphere. 2020;243 doi: 10.1016/j.chemosphere.2019.125332. [DOI] [PubMed] [Google Scholar]
- 35.Biederman L.A., Harpole W.S. Biochar and its effects on plant productivity and nutrient cycling: a meta-analysis. GCB Bioenergy. 2013;5:202–214. doi: 10.1111/gcbb.12037. [DOI] [Google Scholar]
- 36.Diacono M., Montemurro F. In: Sustainable agriculture volume 2. Lichtfouse E., Hamelin M., Navarrete M., Debaeke P., editors. Springer Netherlands; Dordrecht: 2011. Long-term effects of organic amendments on soil fertility; pp. 761–786. [DOI] [Google Scholar]
- 37.Liu Y., Lu H., Yang S., Wang Y. Impacts of biochar addition on rice yield and soil properties in a cold waterlogged paddy for two crop seasons. Field Crops Res. 2016;191:161–167. doi: 10.1016/j.fcr.2016.03.003. [DOI] [Google Scholar]
- 38.Oladele S.O., Adeyemo A.J., Awodun M.A. Influence of rice husk biochar and inorganic fertilizer on soil nutrients availability and rain-fed rice yield in two contrasting soils. Geoderma. 2019;336:1–11. doi: 10.1016/j.geoderma.2018.08.025. [DOI] [Google Scholar]
- 39.Lyu H., He Y., Tang J., Hecker M., Liu Q., Jones P.D., et al. Effect of pyrolysis temperature on potential toxicity of biochar if applied to the environment. Environ Pollut. 2016;218:1–7. doi: 10.1016/j.envpol.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Ma K., Wang Y., Jin X., Zhao Y., Yan H., Zhang H., et al. Application of organic fertilizer changes the rhizosphere microbial communities of a gramineous grass on Qinghai–Tibet plateau. Microorganisms. 2022;10:1148. doi: 10.3390/microorganisms10061148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta T., Kwon E., Bhattacharya S.S., Jeon B.H., Deep A., Uchimiya M., et al. Polycyclic aromatic hydrocarbons and volatile organic compounds in biochar and biochar-amended soil: a review. GCB Bioenergy. 2017;9:990–1004. doi: 10.1111/gcbb.12363. [DOI] [Google Scholar]
- 42.Li X., Qu C., Bian Y., Gu C., Jiang X., Song Y. New insights into the responses of soil microorganisms to polycyclic aromatic hydrocarbon stress by combining enzyme activity and sequencing analysis with metabolomics. Environ Pollut. 2019;255 doi: 10.1016/j.envpol.2019.113312. [DOI] [PubMed] [Google Scholar]
- 43.Gong H., Tan Z., Zhang L., Huang Q. Preparation of biochar with high absorbability and its nutrient adsorption–desorption behaviour. Sci Total Environ. 2019;694 doi: 10.1016/j.scitotenv.2019.133728. [DOI] [PubMed] [Google Scholar]
- 44.Fierer N., Bradford M.A., Jackson R.B. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 45.Ling N., Chen D., Guo H., Wei J., Bai Y., Shen Q., et al. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma. 2017;292:25–33. doi: 10.1016/j.geoderma.2017.01.013. [DOI] [Google Scholar]
- 46.Sheng Y., Zhu L. Biochar alters microbial community and carbon sequestration potential across different soil pH. Sci Total Environ. 2018;622–623:1391–1399. doi: 10.1016/j.scitotenv.2017.11.337. [DOI] [PubMed] [Google Scholar]
- 47.Lefèvre C.T., Viloria N., Schmidt M.L., Pósfai M., Frankel R.B., Bazylinski D.A. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. 2012;6:440–450. doi: 10.1038/ismej.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black M., Moolhuijzen P., Chapman B., Barrero R., Howieson J., Hungria M., et al. The genetics of symbiotic nitrogen fixation: comparative genomics of 14 rhizobia strains by resolution of protein clusters. Genes. 2012;3:138–166. doi: 10.3390/genes3010138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W., Luo X., Ye X., Chen Y., Wang H., Wang L., et al. Predatory Myxococcales are widely distributed in and closely correlated with the bacterial community structure of agricultural land. Appl Soil Ecol. 2020;146 doi: 10.1016/j.apsoil.2019.103365. [DOI] [Google Scholar]
- 50.Nakasaki K., Hirai H., Mimoto H., Quyen T.N.M., Koyama M., Takeda K. Succession of microbial community during vigorous organic matter degradation in the primary fermentation stage of food waste composting. Sci Total Environ. 2019;671:1237–1244. doi: 10.1016/j.scitotenv.2019.03.341. [DOI] [Google Scholar]
- 51.Chen J., Long S., Liu X., Pavlostathis S.G. Long-term evaluation of the effect of peracetic acid on a mixed anoxic culture: organic matter degradation, denitrification, and microbial community structure. Chem Eng J. 2021;411 doi: 10.1016/j.cej.2021.128447. [DOI] [PubMed] [Google Scholar]
- 52.McKew B.A., Dumbrell A.J., Taylor J.D., McGenity T.J., Underwood G.J.C. Differences between aerobic and anaerobic degradation of microphytobenthic biofilm-derived organic matter within intertidal sediments. FEMS Microbiol Ecol. 2013;84:495–509. doi: 10.1111/1574-6941.12077. [DOI] [PubMed] [Google Scholar]
- 53.Ding J., Zhang Y., Wang M., Sun X., Cong J., Deng Y., et al. Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol Ecol. 2015;24:5175–5185. doi: 10.1111/mec.13384. [DOI] [PubMed] [Google Scholar]
- 54.Popoff M.R., Bouvet P. Genetic characteristics of toxigenic Clostridia and toxin gene evolution. Toxicon. 2013;75:63–89. doi: 10.1016/j.toxicon.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z., Guo Q., Feng Z., Liu Z., Li H., Sun Y., et al. Long-term organic fertilization improves the productivity of kiwifruit (Actinidia chinensis Planch.) through increasing rhizosphere microbial diversity and network complexity. Appl Soil Ecol. 2020;147 doi: 10.1016/j.apsoil.2019.103426. [DOI] [Google Scholar]
- 56.Tang Q., Xia Y., Ti C., Shan J., Zhou W., Li C., et al. Partial organic fertilizer substitution promotes soil multifunctionality by increasing microbial community diversity and complexity. Pedosphere. 2023;33:407–420. doi: 10.1016/j.pedsph.2022.06.044. [DOI] [Google Scholar]
- 57.Yang L., Sun R., Li J., Zhai L., Cui H., Fan B., et al. Combined organic-inorganic fertilization builds higher stability of soil and root microbial networks than exclusive mineral or organic fertilization. Soil Ecol Lett. 2022;5 doi: 10.1007/s42832-022-0142-6. [DOI] [Google Scholar]
- 58.Banerjee S., Walder F., Büchi L., Meyer M., Held A.Y., Gattinger A., et al. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019;13:1722–1736. doi: 10.1038/s41396-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J., Deem L.M., Crow S.E., Deenik J.L., Penton C.R. Biochar application influences microbial assemblage complexity and composition due to soil and bioenergy crop type interactions. Soil Biol Biochem. 2018;117:97–107. doi: 10.1016/j.soilbio.2017.11.017. [DOI] [Google Scholar]
- 60.Lehmann J., Rillig M.C., Thies J., Masiello C.A., Hockaday W.C., Crowley D. Biochar effects on soil biota – a review. Soil Biol Biochem. 2011;43:1812–1836. doi: 10.1016/j.soilbio.2011.04.022. [DOI] [Google Scholar]
- 61.Greenblum S., Turnbaugh P.J., Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boss B.L., Wanees A.E., Zaslow S.J., Normile T.G., Izquierdo J.A. Comparative genomics of the plant-growth promoting bacterium Sphingobium sp. strain AEW4 isolated from the rhizosphere of the beachgrass Ammophila breviligulata. BMC Genom. 2022;23:508. doi: 10.1186/s12864-022-08738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su C., Zhang M., Lin L., Yu G., Zhong H., Chong Y. Reduction of iron oxides and microbial community composition in iron-rich soils with different organic carbon as electron donors. Int Biodeterior Biodegrad. 2020;148 doi: 10.1016/j.ibiod.2019.104881. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material