Abstract
With the clinical success of immune checkpoint inhibitors (ICIs), cancer immunotherapy has become an important pillar of cancer treatment in various types of cancer. However, more than half of patients fail to respond to ICIs, even in combination, uncovering a limited window of clinical responses. Therefore, it is essential to develop more effective cancer immunotherapies and to define biomarkers for stratifying responders and nonresponders by exploring the immunological landscape in the tumor microenvironment (TME). It has become clear that differences in immune responses in the TME determine the clinical efficacy of cancer immunotherapies. Additionally, gene alterations in cancer cells contribute to the development of the immunological landscape, particularly immune suppression in the TME. Therefore, integrated analyses of immunological and genomic assays are key for understanding diverse immune suppressive mechanisms in the TME. Developing novel strategies to control immune suppression in the TME from the perspective of immunology and the cancer genome is crucial for effective cancer immunotherapy (immune-genome precision medicine).
Keywords: cancer immunotherapy, immune checkpoint inhibitors, tumor microenvironment, immune suppression, immune-genome precision medicine
1. Introduction
Currently, approximately half of Japanese people will develop malignant tumors (cancer) in their lifetime (https://ganjoho.jp/public/qa_links/report/statistics/2023_en.html, in Japanese). Furthermore, cancer accounts for approximately 20% of all deaths in Japan. Immunotherapy is attracting attention as the fourth pillar of cancer treatment following surgical resection, chemotherapy, and radiation therapy.1–4) Unlike conventional cancer therapies that directly attack cancer cells themselves, cancer immunotherapy does not target cancer cells but rather attacks and destroys cancer cells by activating the host immune system, namely, antitumor immune responses.2,3)
With the approval of sipuleucel-T, a dendritic cell therapy for prostate cancer, by the U.S. Food and Drug Administration (FDA), cancer immunotherapy entered a new era.5) In particular, the clinical effectiveness of blocking antibodies against immune checkpoint molecules, such as CTLA-4 and PD-1, which release the brake on T-cell responses, has been demonstrated in large-scale clinical trials.1–3) These antibodies are now widely used in the clinic to treat cancers, including malignant melanoma, non-small cell lung cancer, renal cell cancer, bladder cancer, head and neck cancer, liver cancer, breast cancer, and Hodgkin lymphoma.1) Clinical application of immune checkpoint inhibitors (ICIs) has revealed positive and negative aspects of cancer immunotherapy, i.e., durable clinical efficacy even in patients with advanced cancer compared with conventional anticancer agents6) but also challenges such as individual differences in therapeutic efficacy,7) emergence of resistance,8–12) and immune-related adverse events.13,14) Therefore, detailed elucidation of how the immune system is suppressed during cancer development and progression is key to achieving successful cancer immunotherapy.8,10–12,15)
We have recently shown that cell signaling in cancer cells delivered by gene alterations, such as epidermal growth factor receptor (EGFR) mutations10) and Ras homolog family member A (RHOA) mutations,11) contributes to the development of the immunosuppressive tumor microenvironment (TME) by recruiting and/or activating regulatory T (Treg) cells, indicating the importance of the united view of immunology and the cancer genome to understand diverse immune suppressive mechanisms in the TME.15–17) This review focuses on how diverse immune suppressive environments in the TME develop with interactions between cancer cells and immune cells for the successful development of cancer immunotherapy through combinations with cancer immunotherapy and molecular-targeted therapy against gene alterations that manipulate the immunological landscape in the TME (immune-genome precision medicine).
2. History of cancer immunology
The immune system discriminates between self and nonself components, through not reacting to self (autoimmune tolerance) and reacting to and eliminating nonself components.18–21) Because cancer cells form by accumulating genetic abnormalities, it has become clear that the immune system recognizes proteins derived from these genetic abnormalities as foreign substances (tumor antigens) and destroys them to inhibit cancer development and progression.22) However, it has long been debated whether the immune system can recognize and attack cancer cells that arise in the host as foreign substances.
The observation by W.B. Coley that malignant tumors regress in some patients who experience erysipelas infection marked the beginning of attempts of applying inflammation and immunity to cancer therapy (Coley toxin).23–25) In the early 20th century, P. Ehrlich postulated that cancers occur spontaneously in vivo and that the immune system is able to recognize and protect against them.26) If the immune system does not eliminate the abnormal cells (cancer cells) that continually appear in the body, cancer can occur with alarming frequency. This idea was taken over by F.M. Burnet and L. Thomas, and in the 1960s, they proposed the concept of “cancer immunosurveillance”, which states that “while genetic alterations frequently occur in cells and malignant cells appear, these dangerous malignant cells are recognized as non-self components and eliminated by the immune system”.27–29)
On the other hand, clarification of the immunological tolerance mechanism by P. B. Medawar et al. caused skepticism regarding the existence of cancer immunosurveillance.30,31) In addition, O. Stutman et al. reported that there was no difference in chemical carcinogenesis between wild-type mice and thymus-deficient nude mice, in which T cells are theoretically not present.32) Moreover, clinical results with Coley’s toxin were insufficient. Consequently, the presence of antitumor immune responses was questioned, and cancer immunology research temporarily regressed.
With advances in experimental techniques such as gene modification technology, effects of the lack of various immune-related genes on cancer development have been examined in animal models, and the existence of cancer immunosurveillance was demonstrated. Cancer development and progression accelerated in mice deficient in recombination-activating genes (RAGs), in which T and B lymphocytes are absent,33) and in mice lacking molecules involved in antitumor immune responses, such as interferon (IFN)-γ and/or perforin.34–36) Furthermore, in 1991, the first human cancer antigen, MZ2-E (so-called MAGE-1), was identified by T. Boon et al.37) However, the key question remains as to why abnormal cells (cancer cells) are not eliminated by the immune system in an immunologically healthy host. To answer this question, immune responses during cancer development were summarized by R.D. Schreiber, M.J. Smyth, and L.J. Old as “Cancer immunoediting”.38)
3. Immune responses during cancer development and progression
3.1. Cancer immunoediting.
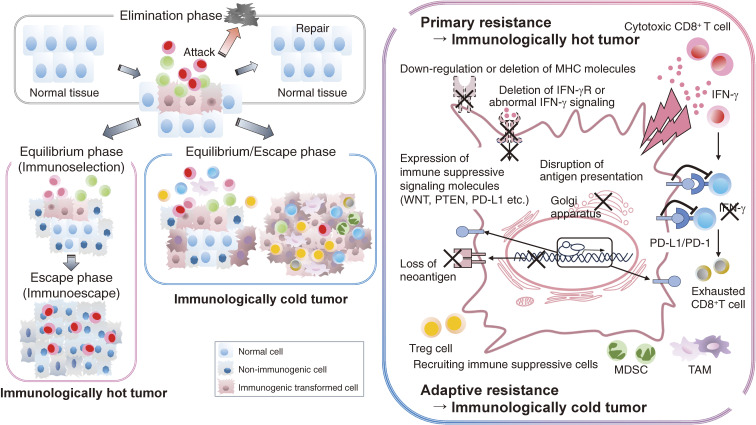
Involvement of the immune system during cancer development is divided into three phases in cancer immunoediting: elimination phase, equilibrium phase, and escape phase.38,39) Understanding this concept is critical for developing effective cancer immunotherapies.
When abnormal cells (cancer cells) with genetic alterations appear due to various stimuli, such as exposure to ultraviolet light, radiation, or chemical substances, they are repaired by the self-healing function of the cells. The chronic inflammatory response induced by these stimuli contributes to the emergence of abnormal cells (cancer cells), and neutrophils are considered to play a key role.40) If the self-healing function does not operate, the immune system, including natural killer (NK) cells, NKT cells, and CD8+ T cells, recognizes aberrant components, such as abnormal proteins derived from gene alterations as foreign and attacks them (elimination phase, formerly called cancer immunosurveillance). In studies on tumor development using genetically engineered mice, it has been shown that in mice deficient in NK cells and NKT cells, tumor development is promoted. There are studies in which involvement of T cells in tumor development cannot be sufficiently proven.41,42) Therefore, in cancer immunity, the innate immune system, especially NK cells, may play an important role in the early stage of cancer development, when abnormal cells (cancer cells), as foreign substances, have just appeared in the body as in infectious immunity.
Cancer cells with low immunogenicity that are suitable for survival in environments where the immune system is normally present are selected in a Darwinian natural selection manner (immunoselection), which allows them to evade attack by the immune system and survive (equilibrium phase).38,39) Then, cancer cells establish an immunosuppressive environment that inhibits antitumor immune responses by incorporating immunosuppressive cells and immunosuppressive molecules, which originally suppress inappropriate or excessive immune responses to maintain immune homeostasis (immunoescape).38,39) Consequently, cancer cells escape attack by the immune system and vigorously proliferate (escape phase), becoming a clinical cancer (Fig. 1).
Figure 1.
Modification of the cancer immunoediting hypothesis. The relationship between cancer cells and the immune system is divided into three phases: elimination phase, equilibrium phase, and escape phase. Immune selection (primary resistance) and immune escape (adaptive resistance), which are involved in the equilibrium and escape phases of cancer immune editing, become activated, leading to carcinogenesis. In humans, these mechanisms are thought to operate in overlapping rather than sequential order.
3.2. Immune response during cancer development and progression.
Proteins derived from genetic alterations in cancer cells are recognized as newly emerged foreign components (antigens) for the body [so called tumor-specific antigens (TSAs) = neoantigen]. Because these neoantigens are suitable targets of the immune system, especially CD8+ T cells,43) it is thought that cancer cells with many genetic alterations (= many neoantigens) recruit and activate many CD8+ T cells in the TME.22) On the other hand, one can envision that cancer cells with many genetic alterations (highly antigenic cancer cells) are easily recognized as foreign substances and should have been eliminated during the cancer development process. Thus, during the equilibrium phase, cancer cells that shed these tumor antigens are selected to form nonantigenic cancer cells. In addition, highly antigenic cancer cells sometimes have additional genetic abnormalities in the antigen-presentation mechanism or receptors and the related pathways for effector molecules, such as IFN-γ and the downstream molecules including JAK1/2 and IRF1.44) More generally, highly antigenic cancer cells simultaneously invoke additional immunosuppressive mechanism(s) that engage immunosuppressive cells45) and immunosuppression-related molecules.46,47)
Therefore, the following two immunological mechanisms are essential during cancer development.
(1) Cancer cells escape the immune system and proliferate by losing highly antigenic tumor antigens and/or harboring genetic abnormalities that are directly involved in immune suppression (primary resistance).
(2) Cancer cells actively utilize immunosuppressive mechanisms such as immune-suppressive cells, including Treg cells, bone marrow-derived immunosuppressor cells (MDSCs), and tumor-associated macrophages (TAMs), as well as immunosuppressive molecules, including immune checkpoint molecules, to inhibit antitumor immune responses (adaptive resistance).
3.3. Immunologically hot and cold cancers.
The two mechanisms, 1) primary resistance and 2) adaptive resistance, are thought to concurrently occur during cancer development in humans rather than progressing in a stepwise manner, as shown in mouse models. Hence, the mechanisms used for escape from the immune system differ in each patient. In other words, each “clinical cancer” might harbor a unique immune suppressive mechanism in the TME (Fig. 1).
Cancer in which adaptive resistance is dominant generally becomes an immunologically hot cancer, in which many neoantigens are present; consequently, CD8+ T cells that recognize these antigens accumulate abundantly, although they are suppressed by immunosuppressive molecules, including PD-1, and/or immunosuppressive cells, including Treg cells, MDSCs, and TAMs.48,49) Therefore, it is expected that removing the immunosuppressive components by inhibitors, including antibodies, can restore effective antitumor immune responses. Among the immunosuppressive mechanisms against effector T cells such as CD8+ T cells, the PD-1–PD-1 ligand (PD-L1) interaction is the most well studied.46) When CD8+ T cells are elicited against neoantigens and attack cancer cells, PD-L1 expression is induced upon IFN-γ exposure produced by CD8+ T cells in cancer cells to escape from attack.48) Accordingly, PD-L1 expression by cancer cells is a sign of previous attack by effector cells, such as tumor antigen-specific CD8+ T cells; therefore, PD-1/PD-L1 inhibitors are likely to resurge CD8+ T cells in cancers harboring a high frequency of PD-L1 expression, resulting in favorable clinical responses.
When primary resistance is mainly involved, cancers often develop an immunologically cold TME, which is not suitable for current cancer immunotherapies, particularly ICIs, and complementary strategies need to be considered to induce CD8+ T-cell responses against a limited number of cancer antigens. Moreover, it is becoming clear that cancer cells themselves directly suppress antitumor immune responses through signals derived from driver gene alterations. In the equilibrium phase, in addition to selection of cancer cells that have shed highly antigenic molecules, cancer cells with driver gene alterations that directly suppress antitumor immune responses are selected. We have recently shown that EGFR mutations in non-small cell lung cancer10) and RHOA mutations in gastric cancer are associated with inhibition of CD8+ T-cell infiltration11) through recruitment and activation of Treg cells in the TME. WNT/β-catenin activation has been shown to induce ATF3 transcription repressor, which suppresses transcription of the CCL4 gene, a crucial chemokine for the recruitment of antigen-presenting cells, thereby inhibiting the infiltration of antigen-presenting cells such as dendritic cells into the TME.12,50) Consequently, infiltration and activation of effector T cells such as CD8+ T cells are prevented. Notably, in non-small cell lung cancer, WNT/β-catenin activation is frequently observed in cancers with a high tumor mutation burden.12) Therefore, in the presence of WNT/β-catenin abnormalities, CD8+ T cells that recognize tumor antigens derived from genetic alterations cannot infiltrate into the TME, even if they are induced, which may allow cancer cells that would normally be eliminated due to high levels of gene alterations to become “clinical cancer”. In hematological malignancies, gene amplification occurs at chromosome 9p23-24 (locus of PD-L1/L2) in Hodgkin lymphomas,51) and increased expression of PD-L1 due to structural abnormalities in the 3′ untranslated region of the PD-L1 gene is observed in adult T-cell lymphoma/leukemia.52) In such cases, molecular-targeted therapy, such as EGFR tyrosine kinase inhibitors (EGFR-TKIs), can change the TME from immunologically cold to hot by reducing Treg cells in the TME.10,53) Hence, we may need to consider the additional functions of some molecular-targeted therapies as immunomodulators (Fig. 2).
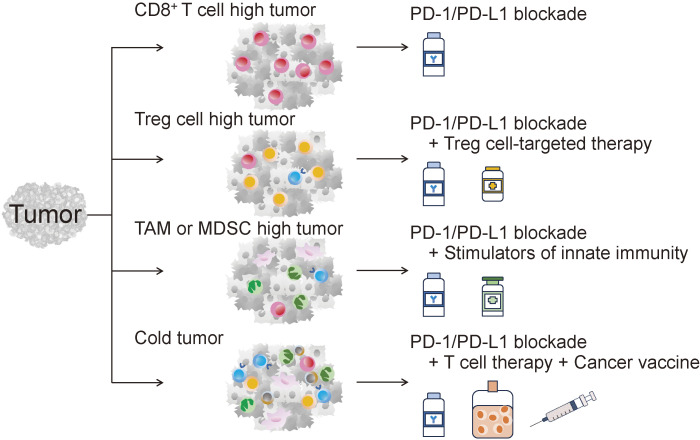
Figure 2.
Effects of genetic alterations in cancer cells on the immune system. A summary of the effects of genetic abnormalities in cancer cells on the immune system. Some genetic alterations in cancer cells are recognized by the immune system, especially CD8+ T cells, as neoantigens. On the other hand, they are also directly involved in the infiltration and activation of immunosuppressive cells in the TME through various mechanisms.
4. Antitumor immune responses in cancer immunotherapy (Fig. 3)
Figure 3.
The concept of immune-genome precision medicine. The TME differs for each patient. Therefore, it is important to clarify the key mechanisms suppressing tumor antigen-specific CD8+ T cells in each cancer patient. If Treg cells are a key mechanism for immunosuppression, Treg cell-targeted therapy needs to be combined with PD-1 blockade. In TAM-rich cancer, use of innate immunity-targeted therapy is reasonable. For immunologically cold tumors, comprehensive cancer immunotherapy may be needed.
From the viewpoint of cancer immunotherapy, it is important to clarify the key mechanisms suppressing tumor antigen-specific CD8+ T cells in each cancer patient.54) Because the PD-1/PD-L1 pathway is thought to be central to antitumor immunosuppression, PD-1/PD-L1 inhibitors are an important therapeutic option.46) Other immune checkpoint molecules and immune suppressive cells, such as Treg cells, MDSCs, and TAMs, may also play a role.45) For example, in liver metastases, the level of lactic acid is increased due to hypermetabolism of cancer cells in the liver, which in turn induces activation of Treg cells because these cells, but not effector T cells, can utilize lactic acid for their activation.8) In this case, one can envision the importance of combining Treg cell-targeted therapy in addition to PD-1/PD-L1 inhibitors.55–57) Therefore, it is crucial to extend TME analysis from PD-L1 expression, which is currently measured in the clinic, to multifaceted immunosuppressive mechanisms. Accordingly, the key mechanism suppressing antitumor immunity in each patient can be uncovered, leading to determination of the optimal cancer treatment.7)
It is difficult to treat immunologically cold tumors with PD-1/PD-L1 inhibitors alone because activation of CD8+ T cells is not sufficient. In this case, it is necessary to intentionally generate a flow to stimulate innate immunity to acquired immunity, similar to the immune responses induced during infectious immunity.58) Many studies have been conducted to activate the innate immune response against immunologically cold tumors, yet sufficient clinical efficacy has not been achieved.59) One of the reasons is that induction/activation of the innate immune response often not only induces/activates NK cells and dendritic cells, which are important for the antitumor immune response, but also activates neutrophils and other myeloid suppressor cells that inhibit effective antitumor immunity.60) Therefore, it is important to appropriately stimulate the flow from innate immunity to acquired immunity, which can efficiently activate CD8+ T cells. Alternatively, use of adoptive immunotherapy should be considered, as is currently the case with chimeric antigen receptor (CAR)-T-cell therapy, which has been successfully used in hematological malignancies.61)
In another type of immunologically cold tumor in which cancer cells directly suppress antitumor immune responses through signals derived from gene alterations, combination therapy of immunotherapy and molecular-targeted therapy may be effective.10,11) Currently, genetic abnormalities in cancer cells are comprehensively measured in clinical practice with panel tests.62) Thus, it is expected that more effective treatment choices may be applicable based on this information, namely, choosing the optimal combination treatment with ICIs and molecular-targeted therapy. Furthermore, it is also possible that some drugs that have not shown sufficient therapeutic efficacy as molecular-targeted agents may be revived as immunomodulators by re-examining them from the viewpoint of modulating immune responses.
5. Conclusions
Optimal cancer immunotherapy for both immunologically cold and hot tumors varies between individual patients. Therefore, it is important to know the key suppressive mechanism in each patient to select the optimal treatment strategy. The ideal cancer immunotherapy requires appropriate analysis of the TME focusing on the immunosuppressive mechanism(s). While the concept of “cancer precision medicine” has been utilized in molecular-targeted therapy, it is important to expand the concept to cancer immunotherapy and develop “immune-genome precision medicine”.
On the other hand, it has become clear that treatment resistance sometimes occurs during the clinical course of cancer immunotherapy. This is an obstacle to long-term clinical efficacy (tail plateau on Kaplan–Meier curve), one of the good points of cancer immunotherapy based on immunological memory.63) Deletion of tumor antigens and abnormalities in antigen presentation and IFN-γ signaling have been reported to be acquired during cancer immunotherapy (acquired resistance).64,65) Because these abnormalities are similar to those of primary resistance that cancer cells acquire during cancer development to escape attack, it needs to be examined whether cancer immunotherapy induces the appearance of new cancer cells or selects already existing cancer cells. Therefore, as mentioned above, it is essential to fully examine the TME to choose the optimal treatment strategy during the clinical course for each patient.
Acknowledgments
I would like to thank Drs. S. Kumagai, H. Nishinakamura, and Y. Maeda for helpful discussions. This study was supported by Grants-in-Aid for Scientific Research (A grant no. 22H00455, S grant no. 17H06162) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; by the Moonshot Research and Development Program (grant no. 22zf0127009h0001); by Projects for Cancer Research by Therapeutic Evolution (P-PROMOTE, no. 22ama221301h0001); by the Development of Technology for Patient Stratification Biomarker Discovery grant (no. 19ae0101074s0401); by Practical Research for Innovative Cancer Control (no. 22ck0106724h0001) from the Japan Agency for Medical Research and Development (AMED); by the National Cancer Center Research and Development Fund (no. 28-A-7 and 31-A-7); by the Yasuda Foundation; and by the Kobayashi Foundation for Cancer Research.
Conflicts of interests
H.N. received research funding and honoraria from Ono Pharmaceutical, MSD, Bristol Myers Squibb, and Chugai Pharmaceutical, honoraria from Amgen, and research funding from Taiho Pharmaceutical, Daiichi-Sankyo, Kyowa Kirin, Zenyaku Kogyo, Oncolys BioPharma, Debiopharma, Asahi-Kasei, Sysmex, Fujifilm, SRL, Astellas Pharmaceutical, Sumitomo Dainippon Pharma, and BD Japan outside of this study. He serves as a board member as a founder of Sustainable Cell Therapeutics and Cellian-Biclo outside of this study.
Non-standard abbreviation list
- ICIs
immune checkpoint inhibitors
- TME
tumor microenvironment
- Treg cell
regulatory T cell
Profile
Hiroyoshi Nishikawa was born in Mie Prefecture in 1970. He graduated from Mie University School of Medicine in 1995. After residency studies in internal medicine for 3 years, he entered graduate school to learn new cancer treatments with a focus on the immune system and received his PhD degree from the Mie University Graduate School of Medicine in 2002. He studied the role of immune suppressive mechanisms, particularly regulatory T cells in tumor immunity working in Dr. Lloyd J. Old’s group at Memorial Sloan Kettering Cancer Center (New York, N.Y.) and then in Dr. Shimon Sakaguchi’s group at Osaka University. He is now Chief of the Division of Cancer Immunology, Research Institute/Exploratory Oncology Research and Clinical Trial Center (EPOC), at the National Cancer Center Japan and cross-appointed as Professor and Chairman of the Department of Immunology, Nagoya University Graduate School of Medicine. He established a unique research style in which phenomena revealed by integrated analyses based on human genome data and comprehensive immune response data are universalized in mouse models. His research clarifies the control mechanism(s) of immune tolerance and immune surveillance in tumor immunity. He is also working to develop novel cancer immunotherapies targeting regulatory T cells.
References
- 1).Twomey J.D., Zhang B. (2021) Cancer immunotherapy update: FDA-approved checkpoint inhibitors and companion diagnostics. AAPS J. 23, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Drake C.G., Lipson E.J., Brahmer J.R. (2014) Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat. Rev. Clin. Oncol. 11, 24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Pardoll D.M. (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Kochenderfer J.N., Rosenberg S.A. (2013) Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol. 10, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Kantoff P.W., Higano C.S., Shore N.D., Berger E.R., Small E.J., Penson D.F., et al. (2010) Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med. 363, 411–422. [DOI] [PubMed] [Google Scholar]
- 6).Johnson D.B., Nebhan C.A., Moslehi J.J., Balko J.M. (2022) Immune-checkpoint inhibitors: long-term implications of toxicity. Nat. Rev. Clin. Oncol. 19, 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Kumagai S., Togashi Y., Kamada T., Sugiyama E., Nishinakamura H., Takeuchi Y., et al. (2020) The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat. Immunol. 21, 1346–1358. [DOI] [PubMed] [Google Scholar]
- 8).Kumagai S., Koyama S., Itahashi K., Tanegashima T., Lin Y.T., Togashi Y., et al. (2022) Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell 40, 201–218.e9. [DOI] [PubMed] [Google Scholar]
- 9).Kamada T., Togashi Y., Tay C., Ha D., Sasaki A., Nakamura Y., et al. (2019) PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 9999–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Sugiyama E., Togashi Y., Takeuchi Y., Shinya S., Tada Y., Kataoka K., et al. (2020) Blockade of EGFR improves responsiveness to PD-1 blockade in EGFR-mutated non-small cell lung cancer. Sci. Immunol. 5, eaav3937. [DOI] [PubMed] [Google Scholar]
- 11).Kumagai S., Togashi Y., Sakai C., Kawazoe A., Kawazu M., Ueno T., et al. (2020) An oncogenic alteration creates a microenvironment that promotes tumor progression by conferring a metabolic advantage to regulatory T cells. Immunity 53, 187–203.e188. [DOI] [PubMed] [Google Scholar]
- 12).Takeuchi Y., Tanegashima T., Sato E., Irie T., Sai A., Itahashi K., et al. (2021) Highly immunogenic cancer cells require activation of the WNT pathway for immunological escape. Sci. Immunol. 6, eabc6424. [DOI] [PubMed] [Google Scholar]
- 13).Yasuda Y., Iwama S., Sugiyama D., Okuji T., Kobayashi T., Ito M., et al. (2021) CD4+ T cells are essential for the development of destructive thyroiditis induced by anti-PD-1 antibody in thyroglobulin-immunized mice. Sci. Transl. Med. 13, eabb7495. [DOI] [PubMed] [Google Scholar]
- 14).Kobayashi T., Iwama S., Sugiyama D., Yasuda Y., Okuji T., Ito M., et al. (2021) Anti-pituitary antibodies and susceptible human leukocyte antigen alleles as predictive biomarkers for pituitary dysfunction induced by immune checkpoint inhibitors. J. Immunother. Cancer 9, e002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Kumagai S., Koyama S., Nishikawa H. (2021) Antitumour immunity regulated by aberrant ERBB family signalling. Nat. Rev. Cancer 21, 181–197. [DOI] [PubMed] [Google Scholar]
- 16).van Weverwijk A., de Visser K.E. (2023) Mechanisms driving the immunoregulatory function of cancer cells. Nat. Rev. Cancer 23, 193–215. [DOI] [PubMed] [Google Scholar]
- 17).Spranger S., Gajewski T.F. (2018) Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 18, 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Goodnow C.C., Sprent J., Fazekas de St Groth B., Vinuesa C.G. (2005) Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435, 590–597. [DOI] [PubMed] [Google Scholar]
- 19).Cheng M., Anderson M.S. (2018) Thymic tolerance as a key brake on autoimmunity. Nat. Immunol. 19, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Pace L., Tempez A., Arnold-Schrauf C., Lemaitre F., Bousso P., Fetler L., et al. (2012) Regulatory T cells increase the avidity of primary CD8+ T cell responses and promote memory. Science 338, 532–536. [DOI] [PubMed] [Google Scholar]
- 21).Maeda Y., Nishikawa H., Sugiyama D., Ha D., Hamaguchi M., Saito T., et al. (2014) Detection of self-reactive CD8+ T cells with an anergic phenotype in healthy individuals. Science 346, 1536–1540. [DOI] [PubMed] [Google Scholar]
- 22).Schumacher T.N., Schreiber R.D. (2015) Neoantigens in cancer immunotherapy. Science 348, 69–74. [DOI] [PubMed] [Google Scholar]
- 23).Coley W.B. (1991) The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin. Orthop. Relat. Res. 3–11. [PubMed] [Google Scholar]
- 24).Coley W.B. (1891) II. Contribution to the Knowledge of Sarcoma. Ann. Surg. 14, 199–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Starnes C.O. (1992) Coley’s toxins in perspective. Nature 357, 11–12. [DOI] [PubMed] [Google Scholar]
- 26).Strebhardt K., Ullrich A. (2008) Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer 8, 473–480. [DOI] [PubMed] [Google Scholar]
- 27).Thomas L. (1982) On immunosurveillance in human cancer. Yale J. Biol. Med. 55, 329–333. [PMC free article] [PubMed] [Google Scholar]
- 28).Burnet F.M. (1971) Immunological surveillance in neoplasia. Transplant. Rev. 7, 3–25. [DOI] [PubMed] [Google Scholar]
- 29).Burnet F.M. (1967) Immunological aspects of malignant disease. Lancet 1, 1171–1174. [DOI] [PubMed] [Google Scholar]
- 30).Billingham R.E., Brent L., Medawar P.B. (1956) The antigenic stimulus in transplantation immunity. Nature 178, 514–519. [DOI] [PubMed] [Google Scholar]
- 31).Billingham R.E., Brent L., Medawar P.B. (1953) Actively acquired tolerance of foreign cells. Nature 172, 603–606. [DOI] [PubMed] [Google Scholar]
- 32).Stutman O. (1974) Tumor development after 3-methylcholanthrene in immunologically deficient athymic-nude mice. Science 183, 534–536. [DOI] [PubMed] [Google Scholar]
- 33).Koebel C.M., Vermi W., Swann J.B., Zerafa N., Rodig S.J., Old L.J., et al. (2007) Adaptive immunity maintains occult cancer in an equilibrium state. Nature 450, 903–907. [DOI] [PubMed] [Google Scholar]
- 34).Nakajima C., Uekusa Y., Iwasaki M., Yamaguchi N., Mukai T., Gao P., et al. (2001) A role of interferon-γ (IFN-γ) in tumor immunity: T cells with the capacity to reject tumor cells are generated but fail to migrate to tumor sites in IFN-γ-deficient mice. Cancer Res. 61, 3399–3405. [PubMed] [Google Scholar]
- 35).van den Broek M.E., Kägi D., Ossendorp F., Toes R., Vamvakas S., Lutz W.K., et al. (1996) Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 184, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Kaplan D.H., Shankaran V., Dighe A.S., Stockert E., Aguet M., Old L.J., et al. (1998) Demonstration of an interferon γ-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 95, 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).van der Bruggen P., Traversari C., Chomez P., Lurquin C., De Plaen E., Van den Eynde B., et al. (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 38).Schreiber R.D., Old L.J., Smyth M.J. (2011) Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570. [DOI] [PubMed] [Google Scholar]
- 39).Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. (2002) Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 3, 991–998. [DOI] [PubMed] [Google Scholar]
- 40).Greten F.R., Grivennikov S.I. (2019) Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 51, 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Smyth M.J., Thia K.Y., Street S.E., Cretney E., Trapani J.A., Taniguchi M., et al. (2000) Differential tumor surveillance by natural killer (NK) and NKT cells. J. Exp. Med. 191, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Smyth M.J., Crowe N.Y., Godfrey D.I. (2001) NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int. Immunol. 13, 459–463. [DOI] [PubMed] [Google Scholar]
- 43).Wölfel T., Hauer M., Schneider J., Serrano M., Wölfel C., Klehmann-Hieb E., et al. (1995) A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269, 1281–1284. [DOI] [PubMed] [Google Scholar]
- 44).Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q., et al. (2016) Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Togashi Y., Shitara K., Nishikawa H. (2019) Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat. Rev. Clin. Oncol. 16, 356–371. [DOI] [PubMed] [Google Scholar]
- 46).Iwai Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., Minato N. (2002) Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U.S.A. 99, 12293–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Leach D.R., Krummel M.F., Allison J.P. (1996) Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736. [DOI] [PubMed] [Google Scholar]
- 48).Spranger S., Spaapen R.M., Zha Y., Williams J., Meng Y., Ha T.T., et al. (2013) Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci. Transl. Med. 5, 200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Saito T., Nishikawa H., Wada H., Nagano Y., Sugiyama D., Atarashi K., et al. (2016) Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684. [DOI] [PubMed] [Google Scholar]
- 50).Spranger S., Bao R., Gajewski T.F. (2015) Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235. [DOI] [PubMed] [Google Scholar]
- 51).Roemer M.G., Advani R.H., Ligon A.H., Natkunam Y., Redd R.A., Homer H., et al. (2016) PD-L1 and PD-L2 genetic alterations define classical hodgkin lymphoma and predict outcome. J. Clin. Oncol. 34, 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Kataoka K., Shiraishi Y., Takeda Y., Sakata S., Matsumoto M., Nagano S., et al. (2016) Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 534, 402–406. [DOI] [PubMed] [Google Scholar]
- 53).Tanaka A., Nishikawa H., Noguchi S., Sugiyama D., Morikawa H., Takeuchi Y., et al. (2020) Tyrosine kinase inhibitor imatinib augments tumor immunity by depleting effector regulatory T cells. J. Exp. Med. 217, e20191009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Ahmadzadeh M., Johnson L.A., Heemskerk B., Wunderlich J.R., Dudley M.E., White D.E., et al. (2009) Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 114, 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Maeda Y., Wada H., Sugiyama D., Saito T., Irie T., Itahashi K., et al. (2021) Depletion of central memory CD8+ T cells might impede the antitumor therapeutic effect of Mogamulizumab. Nat. Commun. 12, 7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Sugiyama D., Nishikawa H., Maeda Y., Nishioka M., Tanemura A., Katayama I., et al. (2013) Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl. Acad. Sci. U.S.A. 110, 17945–17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Itahashi K., Irie T., Yuda J., Kumagai S., Tanegashima T., Lin Y.T., et al. (2022) BATF epigenetically and transcriptionally controls the activation program of regulatory T cells in human tumors. Sci. Immunol. 7, eabk0957. [DOI] [PubMed] [Google Scholar]
- 58).Demaria O., Cornen S., Daëron M., Morel Y., Medzhitov R., Vivier E. (2019) Harnessing innate immunity in cancer therapy. Nature 574, 45–56. [DOI] [PubMed] [Google Scholar]
- 59).Myers J.A., Miller J.S. (2021) Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 18, 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).van den Boorn J.G., Hartmann G. (2013) Turning tumors into vaccines: co-opting the innate immune system. Immunity 39, 27–37. [DOI] [PubMed] [Google Scholar]
- 61).Watanabe K., Nishikawa H. (2021) Engineering strategies for broad application of TCR-T- and CAR-T-cell therapies. Int. Immunol. 33, 551–562. [DOI] [PubMed] [Google Scholar]
- 62).Kohno T., Kato M., Kohsaka S., Sudo T., Tamai I., Shiraishi Y., et al. (2022) C-CAT: The national datacenter for cancer genomic medicine in Japan. Cancer Discov. 12, 2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Tokunaga A., Sugiyama D., Maeda Y., Warner A.B., Panageas K.S., Ito S., et al. (2019) Selective inhibition of low-affinity memory CD8+ T cells by corticosteroids. J. Exp. Med. 216, 2701–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Zaretsky J.M., Garcia-Diaz A., Shin D.S., Escuin-Ordinas H., Hugo W., Hu-Lieskovan S., et al. (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Rosenthal R., Cadieux E.L., Salgado R., Bakir M.A., Moore D.A., Hiley C.T., et al. (2019) Neoantigen-directed immune escape in lung cancer evolution. Nature 567, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]