1. Abstract
Objective
Bladder cancer is one of the most prominent malignancies affecting the urinary tract, characterized by a poor prognosis. Our previous research has underscored the pivotal role of m6A methylation in the progression of bladder cancer. Nevertheless, the precise relationship between N6-methyladenosine (m6A) regulation of long non-coding RNA (lncRNA) and bladder cancer remains elusive.
Methods
This study harnessed sequencing data and clinical records from 408 bladder cancer patients in the TCGA database. Employing R software, we conducted bioinformatics analysis to establish an m6A-lncRNA co-expression network. Analyzing the differences between high and low-risk groups, particularly at the immunological level, and subsequently investigating the primary regulatory factors of these lncRNA, validating the findings through experiments, and exploring their specific cellular functions.
Results
We identified 50 m6A-related lncRNA with prognostic significance through univariate Cox regression analysis. In parallel, we employed a LASSO-Cox regression model to pinpoint 11 lncRNA and calculate risk scores for bladder cancer patients. Based on the median risk score, patients were categorized into low-risk and high-risk groups. The high-risk cohort exhibited notably lower survival rates than their low-risk counterparts. Further analysis pointed to RBM15 and METTL3 as potential master regulators of these m6A-lncRNA. Experimental findings also shed light on the upregulated expression of METTlL3 and RBM15 in bladder cancer, where they contributed to the malignant progression of tumors. The experimental findings demonstrated a significant upregulation of METTL3 and RBM15 in bladder cancer specimens, implicating their contributory role in the oncogenic progression. Knockdown of METTL3 and RBM15 resulted in a marked attenuation of tumor cell proliferation, invasion, and migration, which was concomitant with a downregulation in the cellular m6A methylation status. Moreover, these results revealed that RBM15 and METTL3 function in a synergistic capacity, positing their involvement in cancer promotion via the upregulation of m6A modifications in long non-coding RNAs. Additionally, this study successfully developed an N-methyl-N-nitrosourea (MNU)-induced rat model of in situ bladder carcinoma, confirming the elevated expression of RBM15 and METTL3, which paralleled the overexpression of m6A-related- lncRNAs observed in bladder cancer cell lines. This congruence underscores the potential utility of these molecular markers in in vivo models that mirror human malignancies.
Conclusion
This study not only offers novel molecular targets,but also enriches the research on m6A modification in bladder cancer, thereby facilitating its clinical translation.
Keywords: RBM15 and METTL3, N6-methyladenosine (m6A), lncRNA, Bladder cancer (BC), Malignant progression
Graphical abstract
RBM15 and METTL3 promote m6A modification of lncRNA, facilitating malignant progression in bladder cancer.
1. Introduction
Bladder cancer (BC) is one of the three major malignant tumors within the urinary system, boasting some of the highest incidence and mortality rates among all cancers [1]. According to data from the American Cancer Association in 2021, the United States anticipated a staggering 1,898,160 new cancer cases and 608,570 cancer-related deaths, with BC accounting for 7% of the incidence and 4% of the mortality [2]. Among BC patients, muscle-invasive bladder cancer (MIBC) comprises roughly 25% of cases, marked by its clinical characteristics of rapid progression, swift metastasis, dismal prognosis, and frequent lymph node involvement [3].
Increasing researches have illuminated the pivotal role of m6A methylation in the initiation and progression of various tumors. Cheng et al. found that METTL3 was significantly upregulated in BC tissues, and that overexpression of METTL3 significantly promoted the growth and invasion of BC cells. In contrast, knockdown of METTL3 expression significantly reduced the proliferation, invasion, survival, and tumorigenicity of BC cells. The study also revealed the mechanism of the METTL3-mediated AFF4/NF-κβ/MYC signaling pathway in BC [4]. Chen downloaded the mRNA data of m6ARNA methylation regulators and the corresponding clinical and prognostic information from the Cancer Genome Atlas (TCGA) database, and analyzed the association between these data and clinicopathological variables in BC patients. They found that the m6A RNA methylation regulators METTL3, YTHDF1, and HNRNPC were highly expressed in BC and correlated with different clinicopathological variables in BC [5]. Han also showed that METTL3 was significantly expressed in BC tissues, and that METTL3 may play a carcinogenic role in BC by interacting with DGCR8 protein and regulating the formation of PRI-MIR221/222 in a manner dependent on m6A methylation modification. This is also the first comprehensive study to investigate the effect of METTL3 on tumor formation by regulating m6A methylation modification in non-coding RNA [6]. Long non-coding RNA (lncRNA), encoding more than 200 nucleotides, is widely recognized for its significant biological functions in diverse malignancies, including bladder cancer [7]. Yuxuan Song and colleagues have effectively developed a risk regression model based on immune-related lncRNA, identifying 8 specific lncRNA for calculating risk scores in patients with bladder cancer. This model not only provides prognostic markers but also identifies potential immunotherapy targets for MIBC [8]. Yao Zhan demonstrated the utility of urinary exosomal long non-coding RNA expression as an innovative non-invasive biomarker for diagnosing and predicting bladder cancer recurrence, highlighting the indispensable role of lncRNA in clinical prevention and treatment [9].
Current research indicates that lncRNAs can regulate the malignant progression of tumors through multiple pathways. lncRNAs are capable of recruiting chromatin-modifying complexes to the promoter regions of specific genes, thus modulating their expression. For instance, the lncRNA HOTAIR can bind to various chromatin-modifying enzymes, promoting protein-protein interactions, thereby affecting multiple cancer pathways such as epigenetic reprogramming, protein stability, and signal transduction [10]. lncRNAs can also affect the splicing, export, translation, and degradation of mRNA. Furthermore, lncRNAs can act as sponges for microRNAs (miRNAs), competitively binding to miRNAs to reduce their suppressive effects on target mRNAs, a mechanism known as the ceRNA hypothesis [11]. Additionally, lncRNAs can be transferred between cells through exosomes and other means, mediating intercellular communication within the tumor microenvironment and affecting the interactions between tumor cells and surrounding cells [12]. However, the relationship between m6A modification and lncRNA in bladder cancer, as well as its influence on tumor progression, remains elusive, with the precise regulatory mechanisms still in the early stages of exploration.
Based on existing research, we utilized a LASSO-Cox regression model to identify 11 lncRNA for calculating risk scores in bladder cancer (BC) patients. Furthermore, our investigation unveiled significant distinctions in the composition of infiltrating immune cells (TIICs) between the two patient groups. Notably, our findings highlighted the predominant regulatory influence of m6A 'writers,' specifically METTL3 and RBM15, on lncRNA. These proteins were found to be upregulated in bladder cancer, promoting the malignant progression of tumors. Collectively, our study reveals novel molecular targets and expands the realm of understanding concerning the intricate role of m6A modification in lncRNA within the context of bladder cancer, ultimately, facilitating its potential clinical application.
2. Materials and methods
2.1. Data collection and correlation analysis
The RNA sequencing data of 408 patients diagnosed with MIBC and 19 healthy subjects, all obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The original count of RNA sequencing data was transformed into one-millionth transcript (TPM) value, and further log2 transformation (log2(TPM+1)) for subsequent analysis, Clinical information, including gender, age, pathological stage, and TNM stage, was concurrently collected. Subsequently, the RNA data were categorized into protein coding genes and lncRNA genes, guided by human genome annotations. The evaluation of associations between 24 m6A methylation regulatory genes and lncRNA employed Pearson correlation coefficient analysis. LncRNA demonstrating an absolute correlation coefficient surpassing 0.4 and a P-value below 0.001 were identified as m6A-associated lncRNA. A total of 743 m6A-related lncRNA were ultimately chosen for downstream bioinformatics investigations.
2.2. Establishment of the prognostic gene signature
In this study, we used univariate Cox regression analysis to analyze m6A-related lncRNA, and 50 prognostic m6A-related lncRNA were screened by P < 0.01.
2.3. Establishment and validation of the risk signature
In this investigation, we employed the entire TCGA dataset, which encompassed a total of 408 samples. These samples were randomly partitioned into two subsets: a training set consisting of 204 samples and a test set also comprising 204 samples. The primary purpose of the training set was to formulate a predictive model for m6A-related lncRNA, while the test set served to validate the efficacy of the developed model. Utilizing 50 previously identified m6A-related lncRNA with prognostic significance, we conducted lasso Cox regression analysis with the R software package "glmnet," employing a penalty parameter determined through 10-fold cross-validation. In the analysis, the horizontal axis represents the log-transformed lambda (log λ) values, while the vertical axis corresponds to the performance metrics for each λ value. The mean and standard error of multiple model evaluation metrics are displayed using error bars. In Fig. S2a, two vertical dashed lines are observed; the left dashed line corresponds to the optimal lambda value (lambda.min), where a higher concordance index (c-index) is preferable and a lower deviance is desirable. The right dashed line indicates the lambda value (lambda.1se) of the model within one standard error of the optimal metric, representing a balance between model complexity and performance. The corresponding penalty value at the lowest point of the vertical dashed line in Fig. S2a (i.e., the lambda value at the nadir of the curve) can be extracted. Subsequently, the equivalent penalty value in Fig. S2b is located, and the number of intersecting points at this penalty value indicates the number of variables included in the final model. The vertical coordinate at the intersection points represents the regression coefficients for these variables. Our analysis revealed that 11 m6A-related lncRNA exhibited statistically significant correlations with overall survival (OS) in MIBC patients sourced from the TCGA dataset. Subsequently, we conducted time-dependent receiver operating characteristic (ROC) analysis, comparing the area under the curve (AUC), and decision curve analysis (DCA) between high-risk and low-risk groups. This culminated in the establishment of an 11-m6A-related lncRNA risk model. The risk score was computed using the following formula: Risk score = coef (lncRNA1) x expr (lncRNA1) + coef (lncRNA2) x expr (lncRNA2) + …. + coef (lncRNA11) x expr (lncRNA11), where "coef" represents the coefficients associated with lncRNAs related to survival, and "expr" represents the expression levels of the respective lncRNAs. Subsequently, we categorized individuals into low- and high-risk subgroups based on the median risk score, as outlined by Ref. [13].
2.4. Acquisition of immunogenomic signatures
Initially, we procured the Infiltration Estimation data for the TCGA dataset from the TIMER2.0 database (http://timer.comp-genomics.org/). Concurrently, we integrated this data with the "immune.gmt" annotation file containing immune gene sets using the R packages "GSEABASE" and "GSVA." This integration allowed us to conduct an in-depth analysis aimed at discerning potential disparities in immune-related functions between the high-risk and low-risk groups.
2.5. Cell culture
Human uroepithelial cells (SV-HUC-1) and human embryonic kidney cells (293T) were procured from the American Type Culture Collection (Manassas, VA, USA). The human bladder cancer cell lines, namely EJ1, J82, UMUC3, and 5637, were sourced from the Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China).
2.6. siRNA transfection
In this study, all siRNAs were synthesized by Sangon Biotech (Shanghai, China). To prepare a 10 μM solution, the synthesized siRNAs were dissolved in RNase-free water. The target cells were promptly subjected to transfection with small interfering RNAs (siRNAs) according to the Lipofectamine® RNAiMAX Transfection Reagent protocol, as stipulated by Invitrogen (CA, USA). Detailed sequences of the specific siRNA oligonucleotides utilized in this procedure are furnished in Table S1.
2.7. Cell proliferation, invasion, migration experiments, immunohistochemistry (IHC), western blotting, dot blot
See supplementary information.
2.8. Methylated RNA immunoprecipitation (MeRIP)
The MeRIP assay was executed based on a protocol delineated in a previous study. Initially, anti-m6A primary antibodies, sourced from Synaptic Systems, were allowed to incubate with Pierce™ Protein A/G Magnetic Beads, a product of Thermo Scientific, for a duration of 3 h at 4 °C. Following this, mRNA underwent fragmentation, facilitated by the RNA fragmentation kit from Ambion. The fragmented mRNA was then combined with the pre-prepared antibody-bead complex, allowing for overnight incubation at 4 °C. Subsequent to this, the captured RNA was subjected to a rigorous washing process, repeated five times, and was eluted employing an m6A nucleotide solution. The final purification of the RNA was accomplished using the Oligo Clean & Concentrator kit provided by Zymo.
2.9. RNA isolation and qRT-PCR
Total RNA extraction was carried out in strict accordance with the manufacturer's instructions for TRIzol (Invitrogen). Subsequently, cDNA synthesis was executed utilizing the PrimeScript™ RT Reagent Kit, featuring gDNA Eraser (TaKaRa). For quantitative real-time PCR (qPCR), the Fast SYBR Green PCR Master Mix by Applied Biosystems was employed, and the qPCR procedure was conducted on a Step-One Fast Real-time PCR System, also from Applied Biosystems. Data analysis was conducted utilizing the 2-ΔΔCT method, and primer sequences are available in Table S4.
2.10. Animal experiments
All protocols pertaining to animal experiments adhered rigorously to the guidelines outlined in Directive 2010/63/EU of the European Parliament, which governs the protection of animals employed for scientific purposes. The study obtained formal approval from the Institutional Animal Care and Use Committee at Sun Yat-sen University Affiliated First Hospital in China. Informed consent was also obtained ([2017]257). Forty Sprague-Dawley (SD) rats, aged 6–8 weeks, were procured from the Guangdong Provincial Experimental Animal Center and randomly divided into two groups, each consisting of 20 rats. These rats were provided with ample water and food in a Specific Pathogen-Free (SPF) environment for one week before undergoing transurethral bladder injections under isopentane anesthesia. The modeling group received N-methyl-N-nitrosourea (MNU) injections (2mg/each), while the control group received PBS treatment. The injections were administered at weekly intervals, totaling four injections. After 10 weeks, the rats were euthanized, and their bladder tissues were excised for paraffin embedding, sectioning, and hematoxylin and eosin (HE) staining.
2.11. Statistical analysis
All experiments were meticulously performed in triplicate, except where explicitly specified otherwise. The resultant data are expressed as the mean ± standard error of the mean (SEM). Assessment of normality and homogeneity of variance was executed through the utilization of the Shapiro-Wilk and Brown-Forsythe tests, respectively. In cases involving the comparison of two groups, an unpaired two-tailed t-test was employed. When comparisons involved more than two groups, an analysis of variance (ANOVA) was initially conducted, followed by Tukey's test for post hoc comparisons. The significance level was set at p < 0.05.
3. Result
3.1. Identification of 50 prognostic m6A-related lncRNA
To identify m6A-related lncRNA implicated in the progression MIBC, we obtained RNA sequencing data from 408 MIBC patients and 19 healthy subjects, along with their corresponding clinical information (Table S2), from the TCGA database. By leveraging data from 24 m6A methylation enzymes, we meticulously executed a rigorous screening process, ultimately leading to the discovery of 743 m6A-related lncRNA. (Fig. S1a). These lncRNA exhibited a correlation coefficient exceeding 0.4 and a P < 0.001. Subsequently, we further screened for genes with prognostic value from these 743 genes, using a P-value of less than 0.01 as the criterion, ultimately identifying 50 m6A-related lncRNAs with prognostic significance.(Fig. S1c). Furthermore, we performed an analysis of the expression levels of these 50 lncRNA in both tumor tissues and healthy tissues. Employing a significance threshold of P < 0.05, we observed statistically significant differential expression among these 50 genes (see Fig. S1b).
3.2. Construction and evaluation of a risk model based on m6A-related lncRNA in MIBC patients
LASSO-penalty Cox analysis is a widely employed method for multiple regression analysis, particularly useful when dealing with high-dimensional data featuring poor correlation and prominent predictive values. This approach helps mitigate overfitting, effectively identifying the most influential predictors and generating prognostic indicators for predicting clinical outcomes. Within the graph, the vertical dotted line has been employed to denote the initial-rank value of the logarithm (Log), which corresponds to the minimum segmented likelihood deviation. Our study's methodology encompassed a series of well-defined steps. In the preliminary phase, we performed univariate Cox regression analysis to sift through a pool of 743 m6A-related long non-coding RNAs (lncRNA) and identify 50 lncRNA with prognostic significance. We selected 11 m6A-related lncRNA for further multivariate analysis. To evaluate the prognosis risk of MIBC patients, we divided the 408 MIBC dataset into a training set and a test set, each containing 204 samples. Using the 11 m6A-related lncRNA independently associated with OS, we constructed a risk model in the training set, following the identification of these 50 m6A-related long non-coding RNAs (lncRNA), we proceeded to validate their prognostic significance in the test set. Based on the median prognostic risk value, we stratified the cohort into two distinct groups: the low-risk and high-risk groups. Fig. 1b vividly depicts the distribution of risk levels within these groups, while Fig. 1c visually represents the survival status, survival time of patients in both groups, and the relative expression levels of the 11 m6A-related lncRNA for each patient. Our Kaplan-Meier survival analysis unequivocally revealed that the high-risk group experienced significantly shorter overall survival (OS) in comparison to the low-risk group (P < 0.001, Fig. 1d). To gauge the predictive performance of our risk score model, we conducted time-dependent receiver operating characteristic (ROC) analysis, which demonstrated an area under the curve (AUC) of 0.706 in the training cohort and 0.660 in the test cohort (Fig. 1a). Furthermore, for a more comprehensive assessment of the model's prognostic utility, we calculated the risk score for each patient within the entire test set using a consistent formula. Principal component analysis (PCA) underscored marked distinctions between the high-risk and low-risk groups (Fig. S2c). Fig. S3a displayed the AUC of the risk score predicting OS at various time intervals: 0.685 at 1 year, 0.675 at 2 years, 0.672 at 3 years, and 0.664 at later stages based on ROC analysis. Importantly, this risk model exhibited superior predictive ability compared to clinical features (Fig. S3b and c). Significantly, our findings revealed a distinct pattern: the low-risk group predominantly consisted of samples characterized by lower grades and less advanced stages, whereas the high-risk group displayed a contrasting trend. Remarkably, this observation corresponds with the less favorable prognosis observed among patients belonging to the high-risk group (Fig. S3d). In Fig. S3e, KCNQ1OT1, SNHG16, and AC097359.2 were identified as high-risk genes, whereas AC097641.2, EHMT2-AS1,AC006160.1/MED28-DT, AC116914.2/MYOSLID, AC104564.3, BDNF-AS, AC007686.3/TMEM105, and AC012615.6 were categorized as low-risk genes. In addition, the study successfully established a model that integrates clinical variables and genes associated with m6A-related lncRNAs (Fig. S4).
Fig. 1.
Prognostic evaluation of the risk model based on the 11 m6A-related lncRNA in the TCGA dataset. (a) Receiver Operating Characteristic (ROC) curves illustrating the performance of the risk score. (b) Distribution of risk scores derived from the m6A-related lncRNA model; Visualization of survival status and survival duration disparities between the high-risk and low-risk groups. (c) Heatmap displaying clustering analysis results, indicating the expression levels of the 11 m6A-related lncRNA for each patient in the training set. (d) Kaplan-Meier survival analysis focusing on MIBC outcomes within the high-risk and low-risk groups in the training set.
3.3. Evaluation of MIBC immune microenvironment distribution and immunotherapy response using the risk model of m6A-Related lncRNA
In pursuit of a more comprehensive understanding of the variations in immune characteristics between the high-risk and low-risk groups, our initial step involved acquiring immune cell infiltration estimations for Muscle-Invasive Bladder Cance (MIBC) patients from the TIMER2.0 database. This extensive evaluation encompassed data derived from seven distinct software tools, namely TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC, which we subsequently analyzed. The results revealed noteworthy distinctions between the two groups. Macrophages (M0, M1, M2), regulatory T cells (Tregs), cancer-associated fibroblasts, immune scores, stroma scores, and microenvironment scores were significantly elevated in the high-risk group. Conversely, in the low-risk group, natural killer (NK) cells, T cells (CD4+), T follicular helper cells, monocytes, and activated myeloid dendritic cells were more prominent (Fig. S5a). Simultaneously, an assessment of immune function distribution between the two groups indicated that immune activity was markedly higher in the high-risk group compared to the low-risk group (Fig. S5b). These findings collectively underscored the significant association between survival prognosis disparities and immune function between the two groups. Moreover, the study revealed that patients in the high-risk group exhibited reduced sensitivity to PD-1 monoclonal antibody therapy (Fig. S6).
3.4. RBM15 and METTL3 are key regulatory genes for these risk-associated lncRNA
To further investigate the regulatory factors of these risk-associated lncRNA, the study analyzed m6A regulatory factors that exhibited correlations with the expression of these 11 lncRNA. The findings revealed that RBM15 can modulate six of these risk-lncRNA, specifically KCNQ1OT1, SNHG16, AC097359.2, AC097641.2,AC006160.1/MED28-DT, and AC007686.3/TMEM105. In parallel, METTL3 exerts regulatory control over four lncRNA, namely EHMT2-AS1, AC116914.2/MYOSLID, AC104564.3, and AC012615.6. VIRMA regulates SNHG16, YTHDF3 regulatesAC006160.1/MED28-DT, YTHDC1 regulates AC104564.3, BDNF-AS, and AC012615.6, while YTHDC2 regulates AC012615.6, and these interactions are all characterized by a positive regulatory relationship (Table S3). RBM15 regulates the highest number of downstream target genes along with METTL3, and it is believed to be a key regulatory factor in the modulation of lncRNA m6A modification, according to the study (Fig. 2a and b). Pairwise analysis of the TCGA gene set revealed significant overexpression of RBM15 and METTL3 in bladder cancer (Fig. 2c). Moreover, these two genes exhibited significant overexpression across a wide spectrum of malignant tumors, encompassing lung cancer, kidney cancer, gastric cancer, and various other tissues (Fig. 2d and e).
Fig. 2.
METTL3 and RBM15 stand out as pivotal genes involved in m6A modification within the context of bladder cancer. (a) Examination of the correlation between the 11 m6A-related lncRNA and their respective m6A regulatory factors. (b) Correlation coefficients between 11 m6A-related lncRNAs and their respective m6A regulatory factors. (c) Demonstrated upregulation of METTL3 and RBM15 in the TCGA dataset. (d) Elevated expression of METTL3 (e) and RBM15 (f) observed across various malignant tumors.
3.5. RBM15 and METTL3 exhibited high expression levels in the primary bladder cancer tissues of rats and bladder cancer cells
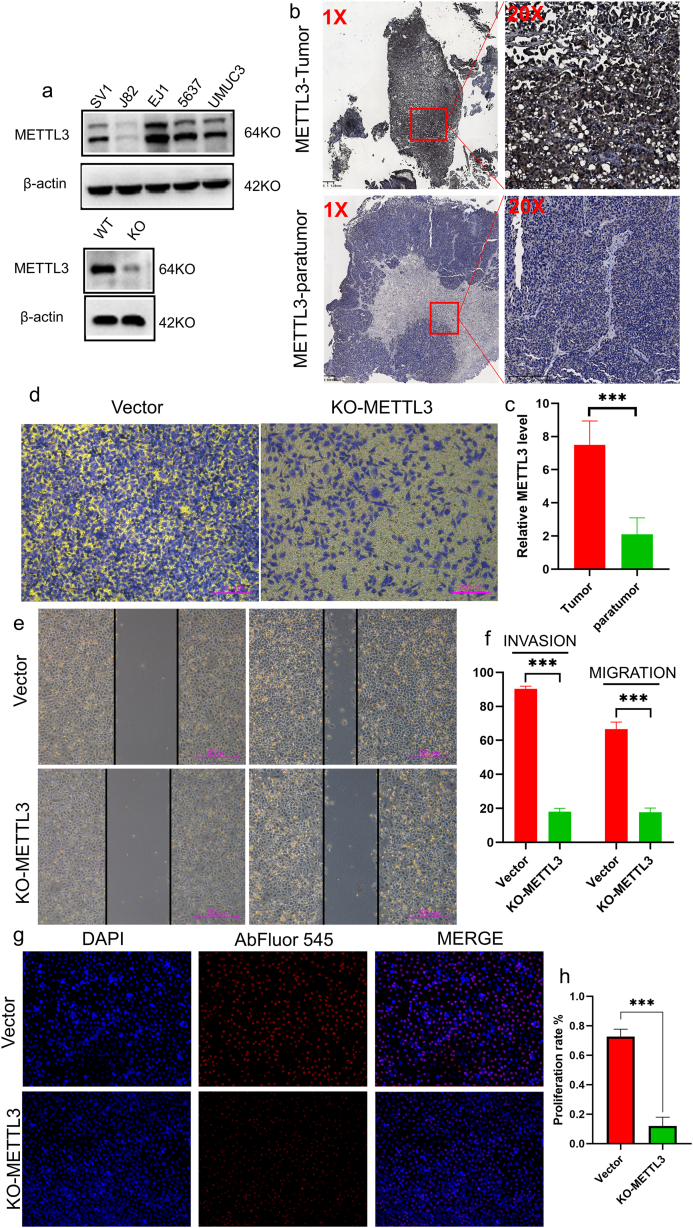
To further validate the expression levels of RBM15 and METTL3 in bladder cancer, this study initially established an in-situ bladder cancer model in rats for verification. Following a 10-week induction period using MNU to promote bladder carcinogenesis in rats, the animals were euthanized, and bladder tissues were collected for paraffin embedding and sectioning (Fig. 3a). Histopathological examination with HE staining revealed evident epithelial hyperplasia and disordered subcutaneous tissue in the bladders of the MNU treatment group rats. Microscopically, nuclear lobulation and atypical hyperplasia of tumor cells were observed (Fig. 3b). Immunohistochemical analysis further demonstrated significant overexpression of METTL3 and RBM15 in these tumor cells (Fig. 3c and d). Our experiments also revealed that RBM15 is notably overexpressed in bladder cancer cell lines, particularly in 5637 cells, whereas METTL3 exhibits predominant overexpression in EJ1 cells (Fig. 4a and Fig. 5a). Notably, the immunohistochemical analysis of tissue sections from bladder cancer patients corroborated the results observed in cell experiments (Fig. 4b and c and Fig. 5b and c).
Fig. 3.
RBM15 and METTL3 exhibited high expression levels in bladder cancer. (a) Schematic Diagram of Rat Urinary Tract Injection of MNU for In Situ Bladder Cancer Model Establishment. (b) In the HE staining display of rat bladder, nuclear lobulation and atypical hyperplasia of tumor cells were observed. (c) METTL3 was notably overexpressed in bladder cancer. (d) RBM15 demonstrated significant overexpression in bladder cancer.
Fig. 4.
RBM15 is highly expressed in bladder cancer and promotes tumor proliferation, invasion and migration. (a). RBM15 overexpression in bladder cancer cell lines, especially 5637 cells. (b). RBM15 overexpression in paraffin sections of human bladder cancer and (d) two-way ANOVA followed by Tukey's test, ***P < 0.001. After knocking down RBM15, the invasion (c) and migration (e) ability of 5637 cells decreased, two-way ANOVA followed by Tukey's test, ***P < 0.001 (f). (g). The proliferation of 5637 cells decreased after knocking down RBM15, two-way ANOVA followed by Tukey's test, ***P < 0.001 (h).
Fig. 5.
METTL3 is highly expressed in bladder cancer and promotes tumor proliferation, invasion and migration. (a). METTL3 overexpression in bladder cancer cell lines, especially 5637 cells. (b). METTL3 overexpression in paraffin sections of human bladder cancer and (d) two-way ANOVA followed by Tukey's test, ***P < 0.001. After knocking down METTL3, the invasion (c) and migration (e) ability of 5637 cells decreased, two-way ANOVA followed by Tukey's test, ***P < 0.001 (f). (g). The proliferation of 5637 cells decreased after knocking down METTL3, two-way ANOVA followed by Tukey's test, ***P < 0.001 (h).
3.6. RBM15 and METTL3 synergistically drive the progression of bladder cancer cells
To further elucidate the biological functions of RBM15 and METTL3, experiments were conducted in bladder cancer cell lines as part of the study. Subsequently, upon silencing RBM15 expression in 5637 cells, we observed a significant reduction in cell proliferation, invasion, and migration abilities (Fig. 4d–h). Similarly, after knocking out METTL3 in EJ1 cells, the malignancy of the cells significantly decreased (Fig. 5d–h). Furthermore, when both cell lines underwent joint knockout of METTl3 and RBM15, the malignant characteristics of the cells were further diminished, led to decreased capabilities in terms of proliferation, invasion, and migration. (Fig. 6a–g and Fig. 7a–c). These findings suggest that RBM15 and METTL3 may collaboratively promote the malignant progression of bladder cancer. Our study further investigated changes in m6A modification levels in EJ1 and 5637 cells after knocking out METTL3 and RBM15 using anti-m6A antibody dot blot hybridization. The results demonstrated that following the knockout of METTL3 and RBM15, the m6A modification levels of RNA (lncRNA) significantly decreased. This indicates that METTL3 and RBM15 may facilitate the onset and development of bladder cancer by jointly regulating RNA modification levels (Fig. 7d). Additionally, the study measured the expression levels of these six long non-coding RNAs (lncRNAs) in bladder cancer cell lines, and the results were found to be consistent with the sequencing data from TCGA database (Fig. S7).
Fig. 6.
Simultaneously knocking down METTL3 and RBM15 cells showed a more significant decrease in malignant phenotype. Knocking out METTL3 and RBM15 simultaneously in (a) Ej1 and (b) 5637 cell lines. The simultaneous knockout of METTL3 and RBM15 cells showed a more significant decrease in (d) migration and (e) invasion ability in the 5637 cells. The simultaneous knockout of METTL3 and RBM15 cells showed a more significant decrease in (f) migration and (g) invasion ability in the EJ1 cells, two-way ANOVA followed by Tukey's test, ***P < 0.001(c).
Fig. 7.
Knockout of METTL3 and RBM15 leads to a reduction in m6A modification levels in bladder cancer. Simultaneous knockout of METTL3 and RBM15 resulted in a more pronounced decrease in proliferation ability in both (a) 5637 and (b) EJ1 cells, as determined by two-way ANOVA followed by Tukey's test, with a highly significant ***P < 0.001 (c). (d) Furthermore, the m6A modification level in tumor cells decreased following the knockdown of METTL3 and RBM15. Particularly, when METTTL3 and RBM15 were simultaneously knocked out, the reduction in m6A modification levels was even more significant.
3.7. RBM15 and METTL3 independently positively regulate m6A modification levels in their respective risk-associated lncRNA
Prior research has revealed that RBM15 and METTL3 can independently regulate the m6A modification levels of their respective risk-associated lncRNA, and both act as positive regulators. Among the six downstream lncRNA regulated by RBM15, gene sequences were identified for four genes - KCNQ1OT1, SNHG16, AC007686.3/TMEM105, and AC006160.1/MED28-DT, with the exception of AC097359.2 and AC097641.2. On the other hand, among the four genes downstream regulated by METTL3, gene sequences were identified for EHMT2-AS1 and AC116914.2/MYOSLID, while AC104564.3 and AC012615.6 lacked gene sequences. In light of this, experimental validation analysis was conducted. The results revealed a significant increase in m6A modification levels for KCNQ1OT1, SNHG16, AC007686.3/TMEM105, and AC006160.1/MED28-DT in the 5637 cell line, with a corresponding significant decrease after the knockdown of RBM15 (Fig. 8a). A similar trend was observed in EJ1, EHMT2-AS1 and AC116914.2/MYOSLID exhibited reduced modification abundance following the downregulation of METTL3 expression levels (Fig. 8b). Additionally, predictions of m6A modification sites for these six lncRNA were carried out using the m6Avar (http://m6avar.renlab.org) and SRAMP (http://www.cuilab.cn/sramp) websites (Fig. 8c and Table S5). This indicates that these lncRNAs indeed possess the potential to be regulated by m6A methylation, corroborating our findings from another perspective, namely that METTL3 and RBM15 can indeed modulate the m6A methylation levels of these genes (Fig. 7d). This also provides targets for further in-depth exploration of the regulatory mechanisms in subsequent studies.
Fig. 8.
RBM15 and METTL3 are capable of regulating the abundance of m6A modifications in lncRNA. (a) In the 5637bladder cancer cell line, KCNQ1OT1, SNHG16, AC007686.3/TMEM105, AC006160.1/MED28-DT exhibited elevated levels of m6A modifications, and these levels decreased following RBM15 knockdown. (b) Similarly, in the EJ1 bladder cancer cell line, EHMT2-AS1 and AC116914.2/MYOSLID demonstrated increased m6A modification levels, which subsequently decreased upon METTL3 knockdown. (c) Predictions were generated regarding the potential m6A modification sites for these six lncRNA.
4. Discussion
MIBC is a type of urinary malignancy known for its heightened sensitivity to the immune system [14]. In recent years, extensive research efforts have been dedicated to unraveling the origins, progression, and therapeutic strategies of MIBC. These studies have shed light on the distinct clinical characteristics and outcomes observed in various subtypes of MIBC, particularly in the context of immune aspects within molecular subcategories. Within these subtypes, an increasing number of investigations have focused on elucidating the distinct attributes of long non-coding RNAs (lncRNA) for predicting survival rates and assessing responses to immune-based therapies in MIBC patients [15,16]. This research direction holds significant importance in shaping the direction of future investigations and therapeutic approaches for MIBC.
Prior research has highlighted the critical role of m6A methylation modifications in bladder cancer [17]. Key factors implicated in the regulation of m6A modifications, including METTL3, METTL14, WTAP, ALKBH5, and FTO, have been extensively documented in the existing literature [[18], [19], [20]]. Furthermore, our prior investigations have revealed the regulatory pathway of METTL3/YTHDF1-3/ITGA6, which actively promotes the migration and invasion of bladder cancer [21]. Serving as the most prevalent post-transcriptional modification in eukaryotic mRNA and lncRNA, m6A modification exerts wide-ranging regulatory effects on mRNA transcription, splicing, translation, as well as the structure and function of lncRNA [22]. Building upon these insights, we proceeded to conduct an analysis of m6A-related lncRNA with the aim of forecasting prognosis and immune therapy responses in bladder cancer patients [23]. Regrettably, this study did not delve deeper into the specifics of molecular regulatory mechanisms or the identification of novel regulatory elements. In our current research, we have delved further into the analysis of regulatory factors associated with m6A-related lncRNA and have identified RBM15, in addition to METTL3, as playing a pivotal role. Existing literature suggests that RBM15, counted among the key regulatory factors in m6A methylation modification, exerts control over various biological processes, including hematopoietic cell homeostasis, selective mRNA splicing, and X chromosome inactivation mediated by Xist RNA [24]. Contemporary studies have further illuminated RBM15's significant role in promote cancer progression. For instance, Wang's study detected a substantial increase in the overall mRNA m6A methylation levels in LSCC (Laryngeal Squamous Cell Carcinoma) patients. As a "writer" of methyltransferase, RBM15 was notably upregulated in LSCC and was associated with an unfavorable prognosis. A series of in vitro and in vivo experiments demonstrated that the downregulation of RBM15 led to reduced proliferation, invasion, migration, and other malignant characteristics of LSCC, while overexpression of RBM15 had the opposite effect [25]. Zeng's study revealed that RBM15 can boost the proliferation, clonal formation, migration, invasion, and epithelial-mesenchymal transition of ccRCC (Clear Cell Renal Cell Carcinoma) cells. Furthermore, it was observed that RBM15's enhancement was triggered by EP300/CBP, resulting in abundant histone 3 acetylation modifications of the RBM15 promoter, which, in turn, bolstered the stability of CXCL11 mRNA in an m6A-dependent manner. Both in vitro and in vivo experiments found that RBM15 promoted macrophage infiltration and M2 polarization by fostering the secretion of CXCL11 in ccRCC cells [26]. Nevertheless, the precise mechanism of RBM15's action in bladder cancer remains elusive. Additionally, prior studies have indicated that specific lncRNA, modified by m6A regulatory factors, have been utilized to varying degrees as potential prognostic biomarkers in conditions such as low-grade glioma, colon cancer, clear cell carcinoma of the kidney, and lung adenocarcinoma [[27], [28], [29], [30]]. These studies have either directly classified subtypes or predicted prognosis based on this information.
Literature reports have explored the mechanisms involving m6A-lncRNAs in the onset and progression of MIBC. Xie's research, for instance, has revealed that the upregulation of m6A-mediated lncRNA BLACAT3, facilitated by YBX3 nuclear translocation and the subsequent enhancement of NCF2 transcription, contributes to angiogenesis and hematogenous metastasis in bladder cancer [31]. Similarly, Lu's study has suggested that N6-methyladenosine-related non-coding RNAs could potentially serve as prognostic indicators and indicators of immune therapy responsiveness in bladder cancer, a concept reinforced by our previous research [32]. These studies collectively underscore a notable correlation between m6A-lncRNAs and the advancement of bladder cancer. However, it is important to note that these investigations have primarily relied on bioinformatics data for predicting treatment outcomes in bladder cancer and have not provided an in-depth exploration of the underlying molecular mechanisms. In this investigation, our primary objective entailed the establishment of an independent model grounded in m6A-related lncRNA to meticulously dissect their prognostic attributes and immune implications. By harnessing comprehensive data encompassing 11 m6A-related lncRNA, we adeptly engineered a predictive model tailored to gauge the overall survival (OS) of MIBC patients, employing the analytical prowess of Lasso Cox regression analysis. Notably, within this ensemble of lncRNA, the triumvirate comprising CNQ1OT1, SNHG16, and AC097359.2 surfaced as high-risk determinants, whereas the remaining eight were designated as low-risk counterparts. Subsequently, following the stratification of MIBC patients into high-risk and low-risk subsets predicated upon intermediate risk scores, our observations unveiled distinctly contrasting clinical outcomes, with the high-risk group manifesting a notably unfavorable prognosis. Moreover, the high-risk group demonstrated notably heightened immune activity, including enhanced checkpoint activity, robust cytolytic responses, pronounced proinflammatory reactions, as well as an array of T cell-related activities, including inhibition, stimulation, and type II interferon responses. Existing studies have indicated that certain lncRNAs indeed modulate tumor immunity, thereby influencing patient outcomes. For example, Hui successfully established a ceRNA network of the long non-coding RNA (SBF2-AS1)-miRNA (has-miR-582-5p)-mRNA (HNRNPA2B1). Targeting HNRNPA2B1 protein, the top eight small molecules with the highest affinity were identified. The study successfully developed an HNRNPA2B1 protein-based prognostic risk scoring model for immune-therapy-associated long non-coding RNAs and determined its significant correlation with bladder cancer immune cell infiltration and immune therapy response [33]. Zheng's research showed that the long non-coding RNA BCCE4 in bladder cancer enhances smoking-related PD-L1/PD-1 interaction through the modulation of the miR-328-3p-USP18 signaling pathway. This aligns with our research findings [34]. Subsequent analysis uncovered that these high-risk lncRNA were under the regulation of RBM15 and METTL3. METTL3 and RBM15 may modulate bladder cancer immune responses by regulating these lncRNAs. Previous research has demonstrated that METTL3 can promote immune surveillance by inhibiting YTHDF2-mediated degradation of NLRC5 mRNA in endometrial carcinoma [35]. Moreover, METTL3-mediated m6A methylation can regulate the progression of ovarian cancer by recruiting myeloid-derived suppressor cells [36]. Meanwhile, Research indicates that RBM15 can act as a prognostic target for various tumors and may be associated with the distribution of the immune microenvironment [37,38]. Although some studies suggest that RBM15 can promote the malignant progression of tumors, it is regrettable that the regulatory mechanisms at the immune level have not been thoroughly investigated experimentally [39]. Our experimental findings indicated elevated expression levels of RBM15 and METTL3 in tumors, and upon their downregulation, we observed a reduction in tumor proliferation, invasion, and migration capabilities. Dot blot experiments complemented this by demonstrating a decrease in the m6A modification levels of lncRNA following RBM15 and METTL3 knockdown. Furthermore, our MERIP experiments elucidated that RBM15 downstream regulated the m6A modification levels of KCNQ1OT1, SNHG16, AC007686.3/TMEM105, and AC006160.1/MED28-DT, while METTL3 was primarily responsible for modulating EHMT2-AS1 and AC116914.2/MYOSLID m6A abundances. Among these lncRNA, KCNQ1OT1 and SNHG16 have been documented in the literature as oncogenic factors in the progression of bladder cancer, whereas the roles of the other four genes have not been previously reported. Simultaneously, this study employed database predictions to identify potential m6A modification sites within these lncRNA, providing valuable insights for future research endeavors. One limitation of our study is the absence of in-depth exploration into the m6A modification sites of lncRNA regulated by RBM15 and METTL3. Nevertheless, our research not only sheds light on the functions of m6A-regulated lncRNA in bladder cancer but also underscores the notion that m6A modification may not be governed by single factors alone but potentially by a collaborative network of multiple factors. This concept could be pivotal in unraveling the intricate role of m6A modification in bladder cancer and serves as a reference point for the investigation of m6A modification in lncRNA.
5. Conclusion
In summary, our study provides valuable insights for predicting the prognosis of MIBC patients and may pave the way for further exploration into the relationship between m6A-related lncRNA and immune infiltration. We have delved deeply into the oncogenic roles of m6A regulatory factors METTL3 and RBM15 in bladder cancer, as well as their regulation of downstream lncRNA, offering a reference for investigating the multifaceted roles of various m6A regulatory factors in bladder cancer. Additionally, this research identifies potential novel molecular targets for clinical assessment and treatment.
Funding
This research was supported by the National Natural Science Foundation of P. R. China (NSFC 82073047, 82272995), China Postdoctoral Science Foundation (2019M653194). Guangdong Basic and Applied Basic Research Fund Joint Fund (Guangdong Dongguan) Project (2021A1515111029)
Availability of data and materials
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
All protocols pertaining to animal experiments adhered rigorously to the guidelines outlined in Directive 2010/63/EU of the European Parliament, which governs the protection of animals employed for scientific purposes. The study obtained formal approval from the Institutional Animal Care and Use Committee at Sun Yat-sen University Affiliated First Hospital in China ([2017]257).
Data availability statement
The bladder cancer sequencing dataset used in the article, along with its clinical information, was downloaded from the TCGA public dataset (https://portal.gdc.cancer.gov/).
CRediT authorship contribution statement
Yapeng Huang: Writing – review & editing, Writing – original draft, Methodology, Funding acquisition, Formal analysis, Data curation. Yifan Lv: Writing – original draft, Data curation, Conceptualization. Baotong Yang: Validation, Funding acquisition, Formal analysis. Shike Zhang: Writing – review & editing, Validation. Bixia liu: Formal analysis, Data curation, Conceptualization. Chengcheng Zhang: Project administration, Methodology, Data curation, Conceptualization. Wenyu Hu: Resources, Project administration, Conceptualization. Lujing Jiang: Supervision, Resources, Project administration, Methodology. Cong Chen: Methodology, Formal analysis. Ding Ji: Formal analysis, Data curation. Chang Xiong: Formal analysis. Yaoming Liang: Methodology, Investigation, Data curation. Mingrui Liu: Formal analysis. Xiaoling Ying: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Funding acquisition, Data curation. Weidong Ji: Writing – review & editing, Supervision, Software, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28165.
Contributor Information
Xiaoling Ying, Email: 2023991001@gzhmu.edu.cn.
Weidong Ji, Email: jiweidong@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hu X., Zhang Y.S., Liu Y.C., Wang N., Zeng X.T., Zhang L.L. Emerging photodynamic/sonodynamic therapies for urological cancers: progress and challenges. J. Nanobiotechnol. 2022 Oct 4;20(1):437. doi: 10.1186/s12951-022-01637-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA - Cancer J. Clin. 2021 Jan;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Kulkarni G.S., Black P.C., Sridhar S.S., Kapoor A., Zlotta A.R., Shayegan B., Rendon R.A., Chung P., van der Kwast T., Alimohamed N., Fradet Y., Kassouf W. Canadian Urological Association guideline: muscle-invasive bladder cancer. Can Urol Assoc J. 2019 Jan 31;13(8):230–238. doi: 10.5489/cuaj.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng M., Sheng L., Gao Q., Xiong Q., Zhang H., Wu M., Liang Y., Zhu F., Zhang Y., Zhang X., Yuan Q., Li Y. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019 May;38(19):3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 5.Chen M., Nie Z.Y., Wen X.H., Gao Y.H., Cao H., Zhang S.F. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci. Rep. 2019 Dec 20;39(12) doi: 10.1042/BSR20192892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J., Wang J.Z., Yang X., Yu H., Zhou R., Lu H.C., Yuan W.B., Lu J.C., Zhou Z.J., Lu Q., Wei J.F., Yang H. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019 Jun 22;18(1):110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quan J., Pan X., Zhao L., Li Z., Dai K., Yan F., Liu S., Ma H., Lai Y. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. OncoTargets Ther. 2018 Oct 4;11:6415–6424. doi: 10.2147/OTT.S167853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y., Jin D., Chen J., Luo Z., Chen G., Yang Y., Liu X. Identification of an immune-related long non-coding RNA signature and nomogram as prognostic target for muscle-invasive bladder cancer. Aging (Albany NY) 2020 Jun 24;12(12):12051–12073. doi: 10.18632/aging.103369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhan Y., Du L., Wang L., Jiang X., Zhang S., Li J., Yan K., Duan W., Zhao Y., Wang L., Wang Y., Wang C. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol. Cancer. 2018 Sep 29;17(1):142. doi: 10.1186/s12943-018-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajagopal T., Talluri S., Akshaya R.L., Dunna N.R. HOTAIR LncRNA: a novel oncogenic propellant in human cancer. Clin. Chim. Acta. 2020 Apr;503:1–18. doi: 10.1016/j.cca.2019.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Herman A.B., Tsitsipatis D., Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell. 2022 Jun 16;82(12):2252–2266. doi: 10.1016/j.molcel.2022.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu M., Zhou C., Weng J., Chen Z., Zhou Q., Gao J., Shi G., Ke A., Ren N., Sun H., Shen Y. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J. Exp. Clin. Cancer Res. 2022 Aug 19;41(1):253. doi: 10.1186/s13046-022-02458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong W., Liang L., Gu Y., Qi Z., Qiu H., Yang X., Zeng W., Ma L., Xie J. Immune-related lncRNA to construct novel signature and predict the immune landscape of human hepatocellular carcinoma. Mol. Ther. Nucleic Acids. 2020;22:937–947. doi: 10.1016/j.omtn.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzano R.G., Catalan-Latorre A., Brugarolas A. RB1 and TP53 co-mutations correlate strongly with genomic biomarkers of response to immunity checkpoint inhibitors in urothelial bladder cancer. BMC Cancer. 2021 Apr 20;21(1):432. doi: 10.1186/s12885-021-08078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu G., Chen Z., Danilova I.G., Bolkov M.A., Tuzankina I.A., Liu G. Identification of miR-200c and miR141-mediated lncRNA-mRNA crosstalks in muscle-invasive bladder cancer subtypes. Front. Genet. 2018 Sep 28;9:422. doi: 10.3389/fgene.2018.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinaldetti S., Worst T.S., Rempel E., Kriegmair M.C., Hartmann A., Porubsky S., Bolenz C., Erben P. Subtype specific expression and survival prediction of pivotal lncRNA in muscle invasive bladder cancer. Sci. Rep. 2020 Nov 24;10(1) doi: 10.1038/s41598-020-77252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F., Zheng Z., Chen W., Li D., Zhang H., Zhu Y., Mo Q., Zhao X., Fan Q., Deng F., Han C., Tan W. Regulation of cisplatin resistance in bladder cancer by epigenetic mechanisms. Drug Resist. Updates. 2023 May;68 doi: 10.1016/j.drup.2023.100938. [DOI] [PubMed] [Google Scholar]
- 18.He J., Liu F., Zhang Z. Functions of N6-methyladenosine in cancer metabolism: from mechanism to targeted therapy. Biomark. Res. 2023 Apr 13;11(1):40. doi: 10.1186/s40364-023-00483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao T., Sun D., Long K., Lemos B., Zhang Q., Man J., Zhao M., Zhang Z. N6-methyladenosine upregulates ribosome biogenesis in environmental carcinogenesis. Sci. Total Environ. 2023 Jul 10;881 doi: 10.1016/j.scitotenv.2023.163428. [DOI] [PubMed] [Google Scholar]
- 20.Tang J., Zhang J., Lu Y., He J., Wang H., Liu B., Tu C., Li Z. Novel insights into the multifaceted roles of m6A-modified LncRNAs in cancers: biological functions and therapeutic applications. Biomark. Res. 2023 Apr 17;11(1):42. doi: 10.1186/s40364-023-00484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin H., Ying X., Que B., Wang X., Chao Y., Zhang H., Yuan Z., Qi D., Lin S., Min W., Yang M., Ji W. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019 Sep;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T., Feng Y.L., Wang R.Y., Yang S., Ge Y.L., Zhang T.Y., Li J., Li C.Y., Ruan Y., Luo B., Liang G.Y. Long-term MNNG exposure promotes gastric carcinogenesis by activating METTL3/m6A/miR1184 axis-mediated epithelial-mesenchymal transition. Sci. Total Environ. 2024 Feb 25;913 doi: 10.1016/j.scitotenv.2023.169752. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Zhu B., He M., Cai Y., Ying X., Jiang C., Ji W., Zeng J. N6-Methylandenosine-Related lncRNAs predict prognosis and immunotherapy response in bladder cancer. Front. Oncol. 2021 Aug 11;11 doi: 10.3389/fonc.2021.710767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016 Sep 15;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Tian L., Li Y., Wang J., Yan B., Yang L., Li Q., Zhao R., Liu M., Wang P., Sun Y. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J. Exp. Clin. Cancer Res. 2021 Feb 26;40(1):80. doi: 10.1186/s13046-021-01871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X., Chen K., Li L., Tian J., Ruan W., Hu Z., Peng D., Chen Z. Epigenetic activation of RBM15 promotes clear cell renal cell carcinoma growth, metastasis and macrophage infiltration by regulating the m6A modification of CXCL11. Free Radic. Biol. Med. 2022 May 1;184:135–147. doi: 10.1016/j.freeradbiomed.2022.03.031. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N., Zhang H., Wu W., Zhou R., Li S., Wang Z., Dai Z., Zhang L., Liu F., Liu Z., Zhang J., Luo P., Liu Z., Cheng Q. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics. 2022 Aug 8;12(13):5931–5948. doi: 10.7150/thno.74281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S., Shen X. Long noncoding RNAs: functions and mechanisms in colon cancer. Mol. Cancer. 2020 Nov 28;19(1):167. doi: 10.1186/s12943-020-01287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Feng Y.C., Gan Y., Teng L., Wang L., La T., Wang P., Gu Y., Yan L., Li N., Zhang L., Wang L., Thorne R.F., Zhang X.D., Cao H., Shao F.M. LncRNA MILIP links YBX1 to translational activation of Snai1 and promotes metastasis in clear cell renal cell carcinoma. J. Exp. Clin. Cancer Res. 2022 Aug 26;41(1):260. doi: 10.1186/s13046-022-02452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan J., Fang S., Tian H., Zhou C., Zhao X., Tian H., He J., Shen W., Meng X., Jin X., Gong Z. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol. Cancer. 2020 Jan 15;19(1):9. doi: 10.1186/s12943-020-1133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J., Zhang H., Wang K., Ni J., Ma X., Khoury C.J., Prifti V., Hoard B., Cerenzia E.G., Yin L., Zhang H., Wang R., Zhuo D., Mao W., Peng B. M6A-mediated-upregulation of lncRNA BLACAT3 promotes bladder cancer angiogenesis and hematogenous metastasis through YBX3 nuclear shuttling and enhancing NCF2 transcription. Oncogene. 2023 Aug 23;42(40):2956–2970. doi: 10.1038/s41388-023-02814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M., Zhan H., Liu B., Li D., Li W., Chen X., Zhou X. N6-methyladenosine-related non-coding RNAs are potential prognostic and immunotherapeutic responsiveness biomarkers for bladder cancer. EPMA J. 2021 Oct 21;12(4):589–604. doi: 10.1007/s13167-021-00259-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui P., Ni F., Zheng L., Jia L., Wang Z. Identification of immunotherapy-related lncRNA signature for predicting prognosis, immunotherapy responses and drug candidates in bladder cancer. BMC Cancer. 2023 Apr 18;23(1):355. doi: 10.1186/s12885-023-10828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng R., Gao F., Mao Z., Xiao Y., Yuan L., Huang Z., Lv Q., Qin C., Du M., Zhang Z., Wang M. LncRNA BCCE4 genetically enhances the PD-L1/PD-1 interaction in smoking-related bladder cancer by modulating miR-328-3p-USP18 signaling. Adv. Sci. 2023 Oct;10(30) doi: 10.1002/advs.202303473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan L., Zhang J., Zhang J.H., Liu X.J., Guo B., Chen J.H., Tang Z.H., Wang W.Y., Wang Q.Y., Wei B., Cao Y.X. METTL3 facilitates immunosurveillance by inhibiting YTHDF2-mediated NLRC5 mRNA degradation in endometrial cancer. Biomark. Res. 2023 Apr 21;11(1):43. doi: 10.1186/s40364-023-00479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J., Ling D., Shi L., Li H., Peng M., Wen H., Liu T., Liang R., Lin Y., Wei L., Zhang G., Chen S. METTL3-mediated m6A methylation regulates ovarian cancer progression by recruiting myeloid-derived suppressor cells. Cell Biosci. 2023 Nov 6;13(1):202. doi: 10.1186/s13578-023-01149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia M., Wang S., Ye Y., Tu Y., Huang T., Gao L. Effect of the m6ARNA gene on the prognosis of thyroid cancer, immune infiltration, and promising immunotherapy. Front. Immunol. 2022 Nov 1;13 doi: 10.3389/fimmu.2022.995645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z., Ju Q., Ji J., Li Y., Zhao Y. N6-Methyladenosine methylation regulator RBM15 is a potential prognostic biomarker and promotes cell proliferation in pancreatic adenocarcinoma. Front. Mol. Biosci. 2022 Feb 9;9 doi: 10.3389/fmolb.2022.842833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H., Zhang H., Mao X., Liu S., Xu W., Zhang Y. RBM15 promates the proliferation, migration and invasion of pancreatic cancer cell lines. Cancers. 2023 Feb 8;15(4):1084. doi: 10.3390/cancers15041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The bladder cancer sequencing dataset used in the article, along with its clinical information, was downloaded from the TCGA public dataset (https://portal.gdc.cancer.gov/).