Abstract
We have developed an in vitro procedure for packaging of recombinant adeno-associated virus (AAV). By using AAV replicative-form DNA as the substrate, it is possible to synthesize an infectious AAV particle in vitro that can be used to transfer a marker gene to mammalian cells. The packaging procedure requires the presence of both the AAV Rep and capsid proteins. Two kinds of in vitro products can be formed which facilitate DNA transfer. Both are resistant to heat and have a density in cesium chloride gradients that is indistinguishable from that of the in vivo-synthesized wild-type virus. This indicates that the particles formed have the appropriate protein-to-DNA ratio and a structure that shares the heat resistance of mature AAV particles. The two types of particles can be distinguished by their sensitivity to chloroform and DNase I treatment. The chloroform-resistant product is, by several criteria, an authentic AAV particle. In addition to having the correct density and being resistant to treatment with chloroform, DNase I, and heat, this particle is efficiently synthesized only if the AAV genome contains intact terminal repeats, which are known to be required for AAV packaging. It is also precipitated by a monoclonal antibody that recognizes mature virus particles but not bound by an antibody that recognizes monomeric or denatured capsid proteins. The chloroform-resistant species is not made when aphidicolin is present in the reaction mixture, suggesting that active DNA replication is required for in vitro packaging. In contrast, the chloroform-sensitive product has several features that suggest it is an incompletely assembled virus particle. It is sensitive to DNase I, does not require the presence of AAV terminal repeats, and is capable of transferring DNA that is theoretically too large to package. Sucrose gradient centrifugation of the in vitro-synthesized products reveals that the particles have sedimentation values between 60S and 110S, which is consistent with partially assembled and mature AAV particles. The in vitro packaging procedure should be useful for studying the mechanism by which a human icosahedral DNA virus particle is assembled, and it may be useful for producing recombinant AAV for gene therapy. The chloroform-sensitive particle may also be useful for transferring DNA that is too large to be packaged in mature recombinant AAV.
Adeno-associated virus (AAV) is a parvovirus and is composed of three structural proteins and a linear, single-stranded DNA (ssDNA) genome of approximately 4.7 kb (17). The particle has icosahedral symmetry and a diameter of 20 to 24 nm. The three capsid proteins of AAV, VP1, VP2, and VP3, have molecular masses of 87, 72, and 62 kDa, respectively, and a ratio of approximately 1:1:10 in the mature particle. All three capsid proteins are encoded by one of the two viral open reading frames, cap, and have overlapping amino acid sequences which differ only in the length of the N terminus. As expected, mutations that affect all three capsid proteins are defective for virus production (8, 28). These mutations are capable of synthesizing double-stranded replication intermediates but are not capable of accumulating ssDNA. Genetic studies also show that in the absence of VP1, VP2 and VP3 can encapsidate progeny ssDNA (8, 28). However, such virions appear to have lower infectivity, suggesting that VP1 is required for stability of the viral particle or for efficient infection. Expression of various combinations of the three capsid proteins in insect cells suggests that VP2 is required for some essential role in the assembly of viruslike empty particles (23). In addition, the major capsid protein, VP3, appears to require the presence of one of the minor capsid proteins for efficient nuclear localization (23). Virus assembly is believed to occur in the nucleus, and immunofluorescence studies indicate that the capsid proteins and the viral nonstructural Rep proteins colocalize in the nucleus (11, 30). Studies of the subcellular distribution of AAV capsid proteins suggest that the three capsid proteins form a variety of complexes with sedimentation coefficients of 10S to 180S, with major peaks at approximately 66S and 110S, the positions of empty and mature virus particles (31). Some of these complexes are associated with the Rep proteins (22, 30, 31).
The Rep proteins are a family of four overlapping proteins encoded by the other viral open reading frame, rep. They are required for AAV DNA replication and for the control of AAV gene expression (17). Genetic and biochemical experiments suggest that the two smaller Rep proteins, Rep52 and Rep40, are not required for viral double-stranded DNA (dsDNA) synthesis but are necessary for the accumulation of ssDNA and virus particles (1a), suggesting that Rep52 and Rep40 are involved in virus assembly.
The linear viral genome contains two inverted terminal repeats (TRs) of 145 bases (13). The TRs contain the viral origin of DNA replication and are required for viral packaging (15). It is not clear which sequences within the TR are required for packaging or what the immediate DNA precursor for packaging is.
In vivo pulse-labeling experiments (18) have suggested that newly synthesized AAV capsid proteins are rapidly assembled into empty capsids, which quickly associate with DNA. These intermediates are then converted to mature virions in a slow process that requires several hours. One of the intermediates identified was a 60S particle whose density was identical to that of mature particles but whose DNA component was sensitive to DNase. Inhibition of DNA synthesis with hydroxyurea inhibited packaging, suggesting that some step in DNA synthesis is necessary for packaging. However, addition of hydroxyurea after pulse-labeling suggested that maturation of AAV particles can occur in the absence of ongoing DNA synthesis.
Viral DNA packaging is an essential step in the virus life cycle, yet our knowledge of the mechanism is limited. To understand the mechanism of virus assembly, it is necessary to develop in vitro systems that are capable of assembling mature virions. Recently, significant progress has been made in the development of an in vitro system for the replication-coupled assembly of poliovirus (16). Here we report on an in vitro system for assembling AAV particles.
MATERIALS AND METHODS
Materials.
Ribonucleotides, deoxyribonucleotides, creatine phosphate, creatine phosphate kinase, and aphidicolin were purchased from Sigma or Pharmacia. Protein G-Sepharose was from Pharmacia. Ascites preparations of anti-Rep monoclonal antibodies (anti-78/68 and anti-52/40 [11]) and anti-capsid monoclonal antibodies (B1 and A20 [30]) were made by Rockland Inc. and purified on protein G-Sepharose prior to use. The ECL Western immunoblot detection kit was purchased from Amersham and used as suggested by the manufacturer. Guinea pig anti-AAV capsid protein polyclonal antibody was provided by R. J. Samulski (University of North Carolina). Cationic liposomes were prepared and used as previously described (5). Restriction endonucleases were purchased from New England BioLabs.
Viruses, plasmids, and cell culture.
293 cells were maintained in Dulbecco modified Eagle medium containing heat-inactivated calf serum and antibiotics. Only cells that had undergone fewer than 100 passages were used, and they were plated 1 day prior to transfection or infection. Plasmid dl63-87/45 was constructed by digesting pIM29+45, which has been renamed pIM45 (14), with ApaI and religating the resulting larger fragment. pTRBRLacZ (called pTRLacZ throughout this report) and pAB11 were recombinant AAV (rAAV) vectors which contained the Escherichia coli β-galactosidase (β-gal) gene (lacZ) under the control of the cytomegalovirus (CMV) immediate-early promoter (see Fig. 1). The two plasmids differ only in that pAB11 is missing an internal PstI site and contains a nuclear localization signal in the coding sequence of its lacZ gene. pAB11 was kindly supplied by R. J. Samulski, and its construction has already been described (6). pTRBRLacZ contains the 3.7-kb BamHI/HindIII fragment carrying the lacZ coding region from pCH110 (Pharmacia) ligated at the HindIII end to the 0.9-kb BamHI/HindIII CMV promoter fragment form pBS-CMV (Pharmacia) and cloned into the BglII site of pTRBR (24).
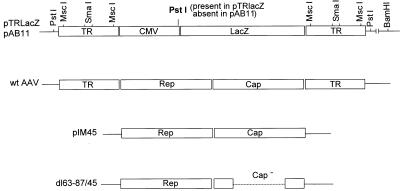
FIG. 1.
Diagrams of pTRLacZ, pAB11, wild-type (wt) AAV, pIM45, and dl63-87. pTRLacZ and pAB11 are recombinant plasmids which contain the lacZ coding sequence under the control of the CMV immediate-early promoter and the simian virus 40 early polyadenylation signal (not shown). pAB11 is different from pTRLacZ in that it is missing a PstI site near the junction of the CMV promoter and the left TR and contains a nuclear localization signal in the lacZ coding sequence. Both plasmids were used to generate substrates for in vitro packaging and contain only the AAV 145-bp TRs. Cut sites of certain restriction endonucleases in pTRLacZ and pAB11 are shown. These restriction fragments were used as substrates for the in vitro packaging experiments described in Table 2. pIM45 and dl63-87/45 were used to generate packaging extracts that contain AAV proteins. dl63-87/45 contains a deletion within the capsid coding region that is indicated by the dotted line. Both pIM45 and dl63-87/45 are missing the 145-bp wild-type AAV TRs and have no homologous overlap with pAB11 and pTRLacZ. The wild-type AAV genome is also shown for comparison. The solid lines indicate vector plasmid sequences.
rAAV containing the green fluorescence protein gene under the control of the CMV promoter was made from plasmid pTRBS-UF2 and purified by use of two successive cesium chloride gradients as previously described (32). Recombinant adenovirus AdΔE1GFP, containing the green fluorescence protein gene under the control of the CMV promoter, was made from plasmid pΔE1GFP as previously described (32). Adenovirus type 5 and wild-type AAV2 were prepared as previously described (15).
Preparation of cell extracts.
Ten 150-mm-diameter plates of 293 cells at approximately 60% confluency were transfected with 20 μg of plasmid DNA per plate by using cationic liposomes and infected with adenovirus type 5 at a multiplicity of infection (MOI) of 5. The cells were harvested at 48 h postinfection and washed with 20 ml of cold phosphate-buffered saline (PBS) and then 10 ml of cold hypotonic buffer (20 mM HEPES [pH 7.4]), 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT]). The cell suspension was centrifuged, and the cell pellet was resuspended in a final volume of 4.8 ml of hypotonic buffer and incubated on ice for 10 min. The cell suspension was Dounce homogenized (20 strokes with a type B pestle), and 0.2 ml of 5 M NaCl was added to raise the NaCl concentration to 0.2 M. The suspension was incubated on ice for 1 h, and the extract was cleared by centrifugation at 15,000 × g for 20 min. After dialysis against a buffer containing 20 mM Tris Cl (pH 7.4), 0.1 mM EDTA, 25 mM NaCl, 10% glycerol, and 1 mM DTT, the extract was stored at −80°C.
Depletion of Rep proteins from cell extracts.
Anti-78/68 monoclonal antibody was coupled to protein G-Sepharose as previously described (7). To immunoprecipitate Rep proteins, 3 volumes of cell extract was incubated twice with 1 volume of anti-78/68–protein G-Sepharose beads at 4°C for 1 h with rocking.
Immunoprecipitation of in vitro-synthesized recombinant UF2 virus particles.
Monoclonal antibody B1 or A20 (30) was added directly to the products of the in vitro packaging reaction, and the mixture was incubated for 30 min. The immune complexes were then precipitated with a 1:1 mixture of protein A-Sepharose and protein G-Sepharose, and the supernatant was tested for the presence of infectious virus by transduction assay for green fluorescence protein expression as described below.
Western analysis.
Ten microliters of each cell extract was electrophoresed on 8 to 14% polyacrylamide gradient gels. Proteins were transferred to nitrocellulose membranes, and the Rep and capsid proteins were detected with anti-52/40 monoclonal antibody or guinea pig anti-capsid polyclonal antibody.
Preparation of pTRLacZ and pTR-UF2 replicative-form (RF) DNAs.
293 cells were cotransfected at 10 μg per 10-cm-diameter plate with pIM45 and pTRLacZ DNAs (3:1) or pTR-UF2 DNA by using cationic liposomes and infected with adenovirus at an MOI of 5. Rescued and replicated AAV(lacZ) or UF2 DNA was extracted 48 h later by using the method of Hirt as previously described (15). Briefly, Hirt supernatant DNA was treated with proteinase K and RNase to inactivate marker proteins and prevent the possibility of the marker protein message being translated in the in vitro packaging reaction. The Hirt supernatant, containing the AAV(lacZ) or UF2 DNA intermediate, was then extracted with phenol and chloroform and precipitated with ethanol to remove any remaining protein and prevent the possibility of pseudotransduction (i.e., marker protein transfer) as described by Alexander et al. (1). The concentration of AAV(lacZ) or UF2 DNA was determined by comparison with known amounts of AAV plasmid DNA following electrophoresis in an agarose gel and staining with ethidium bromide.
In vitro packaging of AAV DNA.
The complete reaction mixture contained (in 30 μl) 30 mM HEPES (pH 7.5); 7 mM MgCl2; 0.5 mM DTT; 0.1 mM each dATP, dGTP, dCTP, and dTTP; 4 mM ATP; 0.2 mM each CTP, GTP, and UTP; 40 mM creatine phosphate; 37.5-μg/ml creatine phosphokinase; 0.17-μg/ml pTRLacZ RF DNA; and 15 μl of cell extract. The reaction mixture was incubated at 37°C for 4 h. The products of the reaction were then incubated at 55°C for 30 min and extracted twice with an equal volume of chloroform, unless otherwise indicated.
Determination of the titer of in vitro-synthesized rAAV(lacZ) or recombinant UF2 virus.
The efficiency of the in vitro packaging reaction was assessed by infection of cells with aliquots of the packaged virus. The products of the packaging reaction were added to 293 cells in 96-well plates at 5 × 104 cells per well. The cells were coinfected with adenovirus type 5 to enhance the transient expression of the AAV transgenes (4, 31a). The cells were stained and counted for β-gal expression at 48 h postinfection by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as previously described (3) or counted for green fluorescence protein expression by fluorescence microscopy as previously described (32).
Determination of the titer of wild-type AAV by infectious-center assay.
The titer of wild-type AAV was determined by the infectious-center assay as previously described (15). Aliquots of virus stocks were used to infect 293 cells in a 96-well plate at a density of 5 × 104 cells per well. The cells were coinfected with adenovirus type 5 at an MOI of 5. After 30 h of incubation at 37°C, the cells were trypsinized and transferred onto nylon membranes with a filtration device. The membranes were wetted for 3 min on 3MM paper saturated with a solution containing 0.5 N NaOH and 1.5 M NaCl. This step was repeated once after the membranes had been blotted dry. The membranes were neutralized with 10 mM Tris Cl (pH 7.5) and 1.5 M NaCl and heated in a microwave oven for 5 min. The membranes then were hybridized with a wild-type AAV probe. Each spot on the membrane hybridizing to the probe represented one cell productively infected by AAV.
Cesium chloride gradient centrifugation.
The density of the AAV virions was determined by cesium chloride gradient centrifugation. In vitro- or in vivo-packaged AAV was treated at 55°C for 30 min and then added to 4 ml of a cesium chloride solution with a final refractive index of 1.3720. The solution was centrifuged in an SW50.1 rotor at 40,000 rpm for 20 h at 4°C. Two hundred-microliter fractions were taken from the top of the gradient, and the refractive index of each fraction was determined. Fractions were then dialyzed against PBS, and the titer of wild-type AAV or AAV(lacZ) was determined by the infectious-center assay or by staining for β-gal activity, respectively.
Sucrose gradient centrifugation.
A linear sucrose gradient of 15 to 30% (wt/wt) was prepared in 10 mM Tris Cl (pH 8.8). Peak fractions from the CsCl gradients were analyzed against PBS, adjusted to 100 ml, and loaded on top of the gradients. The gradients were centrifuged at 110,000 × g for 2.5 h at 20°C. Fractions were collected from the top of the gradients, dialyzed against PBS, and analyzed for wild-type or lacZ mutant virus as described for the CsCl gradients.
RESULTS
Capsid protein-mediated gene transfer.
Our approach to the development of an in vitro AAV packaging procedure was to isolate two types of extracts. One of these, the packaging extract, contained all of the AAV-encoded and host cell proteins. The other contained only the AAV RF DNA to be packaged.
The packaging extract was prepared from cells infected with adenovirus and transfected with AAV plasmid pIM45 (14). pIM45 contained all of the AAV coding sequences but was missing the AAV TRs, which are essential for viral DNA replication and encapsidation (Fig. 1). As a negative control, we made extracts from cells that had been infected with adenovirus and transfected with plasmid dl63-87/45. This plasmid contained an 1,103-base deletion within the capsid coding region which causes a frameshift mutation (Fig. 1). Our previous work had demonstrated that this mutation was completely defective for packaging but viable for AAV DNA replication (8). Finally, extracts were also made from cells that had been infected only with adenovirus.
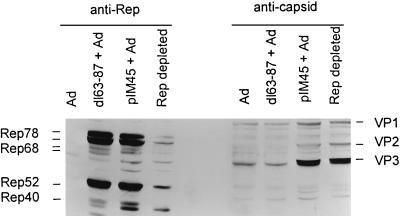
As expected, pIM45-derived extracts contained all of the AAV-encoded proteins: nonstructural proteins Rep78, Rep68, Rep52, and Rep40 and AAV capsid proteins VP1, VP2, and VP3 (Fig. 2). In contrast, extracts prepared from dl63-87/45-transfected cells did not have detectable levels of AAV capsid proteins but had normal levels of the Rep proteins. The absence of truncated capsid proteins in the dl63-87/45 extract was presumably due either to the instability of the proteins or to the absence of the epitopes required for antibody recognition. Extracts prepared from cells infected with adenovirus alone contained neither Rep nor capsid proteins.
FIG. 2.
Western analysis of in vitro packaging cell extracts. Extracts were prepared as described in Materials and Methods from 293 cells infected with adenovirus (Ad) alone or infected with adenovirus plus transfected with either dl63-87 or pIM45 plasmid DNA. Partial depletion of Rep proteins from the pIM45 + Ad extract (Rep-depleted lanes) was accomplished by incubating the extract with mouse anti-78/68 monoclonal antibody coupled to protein G-Sepharose beads. Ten microliters of each extract was electrophoresed on a polyacrylamide gel, transferred to a nitrocellulose membrane, and probed for Rep proteins (left side) by using the mouse anti-52/40 monoclonal antibody, which recognizes all four Rep proteins or for capsid proteins (right side) by using the guinea pig polyclonal anti-capsid antibody, which recognizes all three capsid proteins.
The substrate for the packaging reaction was AAV RF DNA generated from recombinant plasmid pTRLacZ or related plasmid pAB11 (Fig. 1). These substrates were prepared by Hirt precipitation from cells infected with adenovirus and cotransfected with the lacZ-containing plasmid and helper plasmid pIM45. pTRLacZ is an AAV vector which contains the E. coli β-gal marker gene (lacZ) under the control of the CMV immediate-early promoter. This genome was chosen for the packaging experiments for two reasons. First, it could be easily distinguished from any contaminating wild-type AAV. Second, it provided an easy method for measuring the efficiency of the in vitro packaging reaction. This was done by applying the products of the in vitro reaction to cells and then staining the infected cells for β-gal activity.
We did not know what cofactors might be required for AAV packaging. In addition, we were not certain whether some particular kind of RF species was the preferred substrate for AAV packaging or whether encapsidation required active DNA replication. For these reasons, the reaction conditions that we chose were essentially the same as those that we had shown previously to be optimum for in vitro AAV DNA replication (20). Furthermore, because authentic AAV particles are known to be chloroform resistant and heat stable, the products of the reaction were extracted with chloroform and heated at 55°C for 30 min prior to analysis.
When pTRLacZ DNA was incubated with the crude extract obtained from cells transfected with pIM45 and adenovirus, a significant number of the cells treated with the products of the reaction were capable of expressing the lacZ gene (Table 1). This was not true of reaction mixtures incubated with extracts derived from dl63-87/45 plus adenovirus or from adenovirus alone. Since the dl63-87/45 extract lacked only the capsid proteins (compared to the pIM45 extract), we concluded that the transfer and expression of the AAV vector DNA was being facilitated by some process that required AAV capsid proteins. The most likely possibility was that the pTRLacZ genome had been encapsidated into authentic AAV particles. Our previous experience with the pTRLacZ vector had been that, due to the intrinsic properties of the lacZ marker, the viral titer obtained by staining for β-gal activity was approximately 20-fold lower than the titer of infectious virus as determined by the infectious-center assay. Thus, the frequency of β-gal transduction obtained from the products of the in vitro packaging reaction suggested that as many as 105 infectious virus particles had been synthesized per ml of reaction mixture. Assuming a particle-to-infectivity ratio of 100:1, this would represent the production of 107 rAAV particles per ml of reaction mixture.
TABLE 1.
Capsid protein-dependent assembly of rAAVa
| Extractb | No. of blue cells
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| Ad | 0 | 0 | 0 |
| dl63-87 + Ad | 0 | 0 | 0 |
| pIM45 + Ad | 192 | 206 | 116 |
pTRLacZ DNA was packaged by using the extracts indicated. The products of the reaction were then treated with chloroform and heated at 55°C for 30 min. The yield of infectious units per 30-μl reaction mixture was determined by infection and expression of the transgene in 293 cells by using X-Gal staining as described in Materials and Methods.
Ad, adenovirus.
Substrate requirement for in vitro packaging.
One criterion for determining whether we had reconstituted a faithful in vitro packaging system is the substrate requirements for encapsidation. The cis-active AAV sequences that are required in vivo for both AAV DNA replication and packaging are contained within the 145-base TRs (2, 15, 26). Thus, if the in vitro packaging reaction faithfully mimicked the authentic in vivo reaction, it should be sensitive to modifications of the TR.
To determine whether the in vitro packaging reaction required an intact TR, we compared both RF ssDNA and dsDNA substrates with substrates that either contained deletions within the TRs or had additional sequences attached to each TR (Table 2). The modified substrates were generated by digesting pTRLacZ plasmid DNA or related plasmid pAB11 DNA with one of several restriction enzymes that cut within the TR or vector sequences of the plasmid but left the CMV and lacZ sequences intact (Fig. 1). The two plasmids differ only in that pAB11 is missing an internal PstI site and contains a nuclear localization signal in the coding sequence of its lacZ gene. In addition, we examined the effect of chloroform extraction on the products of the reaction.
TABLE 2.
Substrate requirements for in vitro packaginga
| DNA substrate | No. of blue cells
|
|
|---|---|---|
| With chloroform | Without chloroform | |
| ds RF DNA | 120 | 300 |
| ss RF DNA | 178 | NDb |
| PstI pAB11 | 15 | 288 |
| SmaI pTRLacZ | 20 | 172 |
| MscI pTRLacZ | 8 | 116 |
| BamHI pTRLacZ | 14 | 192 |
Various DNA substrates, described in the text and Fig. 1, were packaged in vitro in the standard 30-μl reaction mixture as described in Materials and Methods. The products were heated at 55°C for 30 min and then tested for β-gal transduction either before or after treatment with chloroform. Shown is the number of blue cells produced by the product of each 30-μl reaction mixture.
ND, not done.
We discovered that when the products were chloroform extracted, only the in vivo-derived RF substrates which contained perfect TRs were efficiently packaged (Table 2, Hirt dsDNA and ssDNA). No preference was seen for AAV RF ssDNA, even though ssDNA rather than dsDNA genomes are packaged in AAV particles. Substrates that contained plasmid vector sequences attached to the ends of the TRs (BamHI substrate), even those that contained only 23 additional bases attached to each end of the lacZ genome (PstI pAB11 substrate), were not efficiently packaged (Table 2 and Fig. 1). These were approximately eightfold less efficient in transferring the β-gal gene to cultured cells. Similarly, substrates which had 46 bp (SmaI substrate) or 121 bp (MscI substrate) deleted from the ends of the 145-bp TRs were also poorly packaged.
In contrast, reaction products that were not treated with chloroform prior to testing for β-gal transfer activity behaved differently (Table 2). First, the products contained approximately twice the amount of transfer activity that was seen with chloroform extraction, suggesting that a significant amount of β-gal transfer activity was due to particles that were not stable in chloroform. Furthermore, the additional β-gal transfer activity was largely insensitive to the size of the DNA substrate. The plasmid substrate digested with BamHI was approximately twice the size of the wild-type AAV genome but was still efficiently transferred. Also, the additional β-gal transfer activity did not require an intact AAV TR sequence; substrates digested with either MscI or SmaI were efficiently transferred. Finally, the chloroform-sensitive β-gal transfer product was sensitive to digestion with DNase I (data not shown). Typically, DNase I treatment destroyed approximately 40% of the β-gal transfer activity in the in vitro-packaged products, while chloroform treatment destroyed approximately 50%. Both chloroform and DNase I treatments together did not remove more than 50% of the transfer activity. Thus, the two types of treatment appeared to remove the same type of product.
In vitro-synthesized particles had the same density as authentic wild-type AAV.
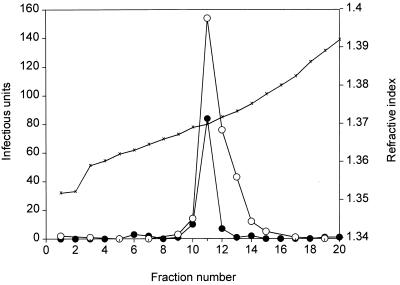
Another criterion for determining whether we had reconstituted a faithful in vitro packaging system was the density of the in vitro-synthesized pTRLacZ particle. The pTRLacZ genome was nearly the same size as the wild-type AAV genome (104% of the size of wild-type AAV). Since the density of the virus particle is a function of the protein and the DNA content of the particle, we predicted that the density of the in vitro-synthesized β-gal virus particles should be indistinguishable from that of wild-type AAV. To see if this was the case, we compared the density of in vitro-packaged β-gal transducing particles with that of authentic wild-type AAV that had been isolated from a cell culture by fractionating the two kinds of particles in parallel cesium chloride equilibrium density gradients. Both kinds of particles were treated with heat (55°C, 30 min) but not with chloroform. The distribution of wild-type AAV was determined by the infectious-center assay, and the β-gal particles were located by staining for β-gal activity following infection of 293 cell monolayers. As shown in Fig. 3, both types of particles produced a single peak of activity and the densities of the two types of particles were virtually the same. Both the wild-type and β-gal particles had a peak refractive index of 1.3698, equivalent to a density of 1.38 g/ml. Thus, although we could distinguish two kinds of particles among the products of the in vitro reaction, both appeared to have the same complement of protein and DNA, as judged by their density.
FIG. 3.
Density gradient centrifugation of in vitro-packaged pTRLacZ virus and wild-type AAV packaged in vivo. In vitro-packaged pTRLacZ virus and wild-type AAV produced in vivo were centrifuged in parallel cesium chloride density gradients as described in Materials and Methods. The pTRLacZ virus titer (solid circles) was determined with the β-gal staining method; the wild-type AAV titer (open circles) was determined by the infectious-center assay. The virus preparations were heated at 55°C for 30 min prior to centrifugation in CsCl but were not treated with chloroform. ×, refractive index.
Precipitation with structure-specific antibodies.
Two monoclonal antibodies are available which can recognize either soluble, unassembled capsid protein (B1) or an epitope that is present only in mature or partially assembled virus particles (A20) (30). When the chloroform-resistant products of the in vitro reaction were treated with the A20 antibody, all of the particles were precipitated (Table 3) or neutralized (data not shown). Treatment with the B1 antibody suggested that a small portion (approximately 30%) of the products were either not completely assembled or had denatured capsid protein associated with them. Control precipitation of purified authentic AAV virions demonstrated that mature particles were completely resistant to B1 antibody. Thus, the bulk of the chloroform-resistant fraction appeared to consist of completely mature virus particles.
TABLE 3.
Antibody sensitivity of in vitro-synthesized productsa
| Antibody | No. of green cells
|
||
|---|---|---|---|
| In vitro UF2 | In vivo UF2 | In vivo AdΔE1GFP | |
| None | 468 | 198 | NDb |
| B1 | 320 | 214 | 126 |
| A20 | 2 | 0 | 118 |
pTR-UF2 DNA was packaged in the standard reaction mixture (60 μl), and the products were treated with chloroform and heated at 55°C for 30 min. Aliquots of the reaction mixture were then precipitated with the B1 or A20 monoclonal antibody, and the yield of virus resistant to antibody precipitation was measured by infection of 293 cells as described in Materials and Methods. In vivo-synthesized and cesium chloride-purified rAAV and adenovirus (Ad) were used as controls.
ND, not done.
Sucrose gradient centrifugation.
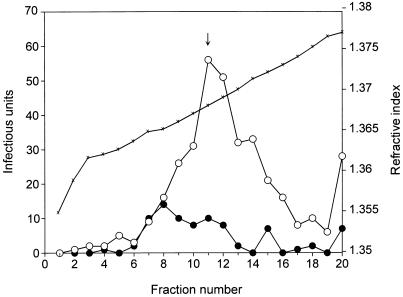
To compare the sedimentation profile of in vivo-synthesized wild-type particles with the products of the in vitro reaction, we applied the peak fractions of the wild-type and lacZ particles from the cesium chloride gradient shown in Fig. 3 to a 15 to 30% sucrose gradient (Fig. 4). In general, the sedimentation profiles of the two particle preparations were similar. Both contained species that sedimented at the position of mature 110S AAV particles. In addition, both preparations contained material that sedimented with either lower or higher sedimentation coefficients. The higher-molecular-weight species are likely to be aggregates of more than one AAV particle. The lower-molecular-weight species (60S to 110S) were consistent with packaging intermediates found in vivo in previous studies (19, 30).
FIG. 4.
Sucrose gradient centrifugation of in vitro-packaged pTRLacZ virus and wild-type AAV packaged in vivo. Peak fractions of the in vivo-packaged wild-type virus and the in vitro-packaged pTRLacZ virus obtained from the CsCl gradient shown in Fig. 3 were sedimented in linear 15 to 30% sucrose gradients. Virus titers were determined as described in the legend to Fig. 3. ↓, position of mature 110S AAV particles; ×, refractive index.
In vitro encapsidation requires Rep78 or Rep68 and active DNA synthesis.
We and others have shown previously that in vitro AAV DNA replication requires the presence of either Rep78 or Rep68 (10, 12, 20) and that DNA synthesis is inhibited by aphidicolin (15a). In addition, a mutant defective for the synthesis of Rep52 and Rep40 was found to be deficient in the accumulation of ssDNA and virus production, suggesting that the two smaller Rep proteins have a role in packaging (1a). To determine whether Rep78 or Rep 68 is required for in vitro packaging, the Rep proteins were depleted from the packaging extract by immunoprecipitation of the extract with anti-78/68 monoclonal antibody conjugated to protein G-Sepharose beads. This procedure was successful in reducing the Rep78 and Rep68 concentration by approximately 10-fold without significantly affecting the concentration of capsid protein in the extract (Fig. 2). Rep52 and Rep40, which were not recognized by the anti-78/68 antibody, were also partially depleted, presumably due to their interaction with the larger Rep proteins (21). When the depleted extract was tested for packaging activity, it was found to be significantly reduced in activity (approximately fourfold) compared to the complete extract (Table 4). Addition of aphidicolin (10 μg/ml) to reaction mixtures containing the complete extract reduced activity approximately ninefold. Finally, addition of aphidicolin to the depleted extract reduced the packaging activity even further, approximately 20-fold. We also measured the level of DNA synthesis under the conditions of reduced Rep concentration and aphidicolin treatment and found that the level of DNA synthesis was reduced to approximately the same extent as the level of in vitro packaging (data not shown). We concluded from these results that the presence of one or more of the Rep proteins and active DNA synthesis was required for in vitro AAV packaging.
TABLE 4.
Dependence of in vitro packaging on Rep and DNA synthesisa
| Extract | Aphidicolin | No. of blue cells |
|---|---|---|
| pIM45 | − | 120 |
| pIM45 | + | 13 |
| Rep depleted | − | 29 |
| Rep depleted | + | 6 |
pTRLacZ substrate was incubated with the indicated extract in the presence or absence of aphidicolin (10 μg/ml) in the standard packaging reaction mixture described in Materials and Methods. The products of the reaction were then heated at 55°C for 30 min, treated with chloroform, and assayed for β-gal transduction.
Cofactor requirements for in vitro packaging.
The divalent cation Mg2+ and ATP were essential for packaging activity (Table 5). Omission of the Mg ion completely eliminated packaging activity, while omission of ATP or the ATP-regenerating system, creatine phosphate and creatine phosphokinase, severely inhibited the packaging reaction (approximately 20-fold). The residual activity seen in the absence of ATP or the regenerating system presumably reflects the fact that the cell-free packaging extract contained substantial amounts of ATP. Since both Mg and ATP are necessary for AAV DNA replication, the requirement for these cofactors was not surprising. This also probably explains the modest decrease in activity seen when the four deoxynucleoside triphosphates were omitted from the reaction mixture (approximately 40%). Again, although the deoxynucleoside triphosphates are essential for DNA replication, the cell-free packaging extract is likely to have contained substantial amounts of these nucleotides. Surprisingly, the reaction was also partially dependent on the presence of the other three ribonucleoside triphosphates, UTP, CTP, and GTP. We previously showed that these are not required for in vitro AAV DNA replication (20). Thus, the reason for this dependence was not clear.
TABLE 5.
Cofactor requirements for in vitro packaginga
| Reaction conditionsb | No. of blue cells |
|---|---|
| Complete | 199 |
| No Mg2+ | 0 |
| No GTP, CTP, or UTP | 123 |
| No dNTPs | 120 |
| No ATP | 16 |
| No CP or CPK | 14 |
| No ATP, CP, or CPK | 10 |
pTRLacZ DNA was packaged under standard reaction conditions in which one or more components of the reaction were omitted. The products of the reaction were then heated at 55°C for 30 min and treated with chloroform. The amount of pTRLacZ virus produced was determined by infection and expression of the transgene in 293 cells.
CP, creatine phosphate; CPK, creatine phophokinase. dNTPs, deoxynucleoside triphosphates.
DISCUSSION
We have demonstrated that it is possible to package in vitro an rAAV genome carrying a marker gene. The particles made in this fashion can be used to transduce mammalian cells in vivo. Taken together, our results suggest that the in vitro reaction produces a mixture of virus particles that consist of both mature virions and partially assembled intermediates. At least two kinds of in vitro products can be formed which facilitate DNA transfer of a marker gene. These can be distinguished by their sensitivity to chloroform and DNase I treatment. The chloroform-resistant product is, by several criteria, an authentic AAV particle. First, the in vitro-synthesized particle is resistant to treatment with chloroform, DNase I and heat. Second, the particle is synthesized only in the presence of AAV capsid proteins. Third, only rAAV that contains an intact terminal repeat is efficiently packaged. This is consistent with in vivo genetic studies that have demonstrated the TR to be a cis-active element required for both DNA replication and viral packaging (2, 15, 26). Fourth, the in vitro packaging reaction requires the presence of one or more of the Rep proteins. This is also consistent with the in vivo genetic data and reflects either a need for Rep78 and Rep68 in DNA replication (20) or a possible direct role of Rep52 and Rep40 in packaging (1a). Fifth, the particles have a density, and therefore a protein-to-DNA ratio, that is identical to that of wild-type AAV. Sixth, at least a portion of the in vitro-synthesized product has a molecular weight that is indistinguishable from that of in vivo-synthesized wild-type virus. Finally, most of the chloroform-resistant particles are bound by an antibody that recognizes mature virus and not bound by an antibody that recognizes monomeric or denatured capsid proteins.
The chloroform-sensitive product can also transfer a marker gene to mammalian cells, but it has several features that suggest that it is either an incompletely assembled virus particle or a DNA molecule that is nonspecifically associated with AAV capsid proteins. As already mentioned, it is sensitive to chloroform and DNase I. In addition, although its formation requires the presence of AAV capsid proteins, it facilitates the transfer of DNA molecules that are much larger than those that can be packaged in authentic AAV particles in vitro. Like many other icosahedral particles, AAV particles can package DNA molecules up to 105% of full-length AAV in size with no significant reduction in viral yield or stability. Yet, the chloroform-sensitive in vitro product facilitated transfer of a molecule nearly two times the size of the normal genome. Finally, the chloroform-sensitive product did not require an intact AAV TR and, thus, appears to be capable of transferring any DNA molecule to mammalian cells.
Although the structure of the chloroform- and DNase I-sensitive β-gal transfer products was not clear, the most likely possibility was that these were due to incomplete particle formation or a nonspecific association of capsid proteins with DNA, irrespective of its size or the composition of the TRs. In vivo studies of AAV packaging have identified a number of possible packaging intermediates that are consistent with the size, density, antibody sensitivity, and DNase sensitivity of the in vitro products described here. Wistuba et al. (31) identified a number of different capsid complexes in addition to completely assembled empty particles. Some of these (notably in the 60S-to-110S range) were recognized by monoclonal antibody A20 but not by B1. Myers and Carter (18) found evidence for a partially assembled intermediate of approximately 60S (as opposed to 110S for mature particles) which had the same density as mature particles but contained DNA which was DNase sensitive. It is worth noting that the properties of this intermediate are similar to those of the chloroform- and DNase I-sensitive particles that are made in the in vitro assay described here. We note, however, that although it appears that the chloroform-sensitive particles synthesized in the in vitro reaction have the same density as mature particles, we have not ruled out completely the possibility that such particles are unstable in CsCl and were lost during CsCl centrifugation.
Mechanism of AAV packaging.
Relatively little is known about the mechanism of AAV packaging, and it is hoped that the packaging system described here will be useful for studying the mechanism of this process. Only one other in vitro packaging system is available for a eukaryotic virus; Wimmer and his colleagues (16) have demonstrated that poliovirus can be packaged in a coupled in vitro translation-and-replication system.
In general, we can imagine at least two mechanisms for virus assembly. It is possible that preformed empty capsids are made first and then serve as the precursor for packaging. Alternatively, packaging may be a process intimately associated with ongoing DNA replication. As a new ssDNA molecule is displaced, a complex of capsid proteins may bind to a TR and the complete capsid particle may assemble around the DNA molecule. It is also possible that a preformed empty capsid may associate with an ssDNA molecule as it is being displaced. In vivo studies of capsid assembly intermediates have not clearly distinguished between these two possibilities. Myers and Carter (18) suggested, on the basis of pulse-labeling experiments, that empty capsids were rapidly assembled and served as precursors for mature particles. Association of the newly synthesized progeny DNA strands with empty capsids appeared to be rapid and was followed by a slow maturation process that occurred over several hours. However, Myers and Carter (18) also reported that hydroxyurea, a DNA synthesis inhibitor, reduced but did not eliminate virus packaging when the inhibitor was added after pulse-labeling and severely inhibited packaging when the label was added after the inhibitor. This suggested that some key step in packaging requires DNA synthesis but that other steps can be uncoupled from ongoing DNA replication. The fact that our in vitro reaction appeared to be sensitive to inhibition by aphidicolin was consistent with these observations.
AAV packaging requires information contained within the TR. The simplest possibility is that a specific sequence within the TR is recognized by one or more capsid proteins to initiate packaging. This would explain why sequence modifications that we introduced in the TRs of our in vitro substrates eliminated the formation of chloroform-resistant particles. An alternative possibility is that the Rep protein, which is covalently attached to the TR during replication, is itself the signal for packaging. The mutant substrates used in these studies would not distinguish between these possibilities.
Potential uses of the in vitro packaging system.
We showed previously that AAV can be used to stably transfer foreign DNA to mammalian cells, and we suggested that AAV may be useful for human gene therapy (9). One of the difficulties in applying AAV to gene therapy is the cumbersome method currently used for isolating recombinant AAV stocks (15, 25). In addition to producing rAAV titers that are significantly lower than those of wild-type AAV stocks, the method also produces stocks which are contaminated with adenovirus. Furthermore, all AAV vectors produced in cell culture are potentially prone to contamination with adventitious viruses, as well as wild-type rAAV. The in vitro method described here produces rAAV in a cell-free reaction that contains only one kind of DNA substrate, and therefore, there is no possibility of contamination with adventitious agents, adenovirus, or wild-type rAAV. Although the method produces AAV vectors at a level that is approximately 1/10 of the level currently seen with cell culture techniques, we anticipate that the method can be improved significantly as the individual components involved in the reaction are purified and identified.
Another difficulty with applying AAV vectors to gene therapy is the fact that the recombinant genome is limited to approximately 5 kb. In this respect, the chloroform-sensitive products generated in the in vitro packaging reaction provide a means by which molecules with a size significantly larger than that of the AAV genome can be transferred to mammalian cells. Since these molecules can be engineered to have the AAV TRs, it should be possible to transfer foreign DNA to human cells by use of the chloroform-sensitive particle in a way that leads to integration and, therefore, results in stable transduction. Furthermore, it may be possible to achieve site-specific AAV vector integration only in the presence of the Rep protein (27, 29). Since we and others have shown that the products of in vitro DNA replication contain a covalently attached Rep molecule (12, 22, 30, 31), it is possible that foreign DNA transferred in this way may integrate site specifically into chromosome 19, as has been seen with wild-type AAV molecules. Alternatively, the absence of a size constraint would allow the incorporation of a functional rep gene, as well as the transgene, into an AAV vector, thus ensuring site-specific integration.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of General Medical Sciences (RO1 GM3572302).
We thank Ti-hua Ni and Sergei Zolotukhin for assistance with various technical problems, as well as many helpful discussions. We also thank S. Zolotukhin for constructing pTRLacZ and J. A. Kleinschmidt for providing A20 and B1 monoclonal antibodies.
REFERENCES
- 1.Alexander I E, Russell D W, Miller A D. Transfer of contaminants in adeno-associated virus vector stocks can mimic transduction and lead to artificial results. Hum Gene Ther. 1997;8:1911–1920. doi: 10.1089/hum.1997.8.16-1911. [DOI] [PubMed] [Google Scholar]
- 1a.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 2.de la Maza L M, Carter B J. Molecular structure of adeno-associated virus variant DNA. J Biol Chem. 1980;255:3194–3203. [PubMed] [Google Scholar]
- 3.Dhawan J, Pan L C, Pavlath G K, Travis M A, Lanctot A M, Blau H M. Systemic delivery of human growth hormone by injection of genetically engineered myoblasts [see comments] Science. 1991;254:1509–1512. doi: 10.1126/science.1962213. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao X, Huang L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem Biophys Res Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 6.Goodman S, Xiao X, Donahue R E, Moulton A, Miller J, Walsh C, Young N S, Samulski R J, Nienhuis A W. Recombinant adeno-associated virus-mediated gene transfer into hematopoietic progenitor cells. Blood. 1994;84:1492–1500. [PubMed] [Google Scholar]
- 7.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 8.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermonat P L, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong G, Ward P, Berns K I. Intermediates of adeno-associated virus DNA replication in vitro. J Virol. 1994;68:2011–2015. doi: 10.1128/jvi.68.3.2011-2015.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter L A, Samulski R J. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J Virol. 1992;66:317–324. doi: 10.1128/jvi.66.1.317-324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 13.Lusby E, Fife K H, Berns K I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol. 1980;34:402–409. doi: 10.1128/jvi.34.2.402-409.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.McLaughlin, S. K., and M. Muzyczka. Unpublished data.
- 16.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 17.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 18.Myers M W, Carter B J. Assembly of adeno-associated virus. Virology. 1980;102:71–82. doi: 10.1016/0042-6822(80)90071-9. [DOI] [PubMed] [Google Scholar]
- 19.Myers M W, Laughlin C A, Jay F T, Carter B J. Adenovirus helper function for growth of adeno-associated virus: effect of temperature-sensitive mutations in adenovirus early gene region 2. J Virol. 1980;35:65–75. doi: 10.1128/jvi.35.1.65-75.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira D J, Muzyczka N. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate Rep protein activation of the adeno-associated virus p19 promoter. J Virol. 1997;71:1747–1756. doi: 10.1128/jvi.71.3.1747-1756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad K M, Trempe J P. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology. 1995;214:360–370. doi: 10.1006/viro.1995.0045. [DOI] [PubMed] [Google Scholar]
- 23.Ruffing M, Zentgraf H, Kleinschmidt J A. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol. 1992;66:6922–6930. doi: 10.1128/jvi.66.12.6922-6930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan J, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1995;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samulski R J, Chang L S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 27.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 28.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Zhou, X., and N. Muzyczka. Unpublished data.
- 32.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]