Abstract
The 441-nucleotide (nt) region (nt 5325 to 5766) around the splice acceptor (SA) site (nt 5491) was found to be necessary for high-level expression of gag-containing unspliced RNA of Moloney murine leukemia virus (M. Oshima, T. Odawara, T. Matano, H. Sakahira, K. Kuchino, A. Iwamoto, and H. Yoshikura, J. Virol. 70:2286–2295, 1996). Detailed genetic dissection of the 441-nt region revealed that the 5′-end 64 nt (nt 5325 to 5389) were necessary for high-level expression of the unspliced RNA when the spliced RNA was not produced, while the 3′-side 301 nt (nt 5466 to 5766) containing the SA site were necessary for producing spliced RNA. When the spliced RNA was produced, the unspliced RNA could be expressed at a high level even when the 5′-end 64 nt were absent. Probably the virus sequence ensuring the splicing could produce an RNA structure able to compensate for the function of the 5′-end 64-nt region responsible for the expression of the unspliced RNA.
Murine leukemia viruses produce unspliced and spliced RNAs. The unspliced RNA is packaged into virions as genomic RNA and is also used as mRNA encoding Gag precursor and Gag-Pol fusion proteins. The spliced RNA encodes Env protein. This scheme is maintained in all of the retroviruses, including lentiviruses and spumaviruses. Production of both unspliced and spliced RNAs in proper amounts is essential for the replication of retroviruses. Although cis elements affecting the efficiency of splicing have been reported previously (1, 2, 9, 14, 15, 19, 20), the detail of the regulation remains unknown.
We previously reported that G3.6, a mutant which was derived from Moloney murine leukemia virus (MLV) by deleting most of pol (nucleotides [nt] 2206 to 4600) and env (nt 4894 to 7674) and which consequently contained the Gag-coding region alone, produced only about 1/10 of the wild-type RNA level. The ligation of 441 nt containing a splice acceptor (SA) site to the 3′ end of the gag region restored the level of the unspliced message, simultaneously activating a cryptic splice donor site in the middle of the p30-coding region. Deletion of the 441 nt from the wild-type (wt) MLV resulted in the loss of the spliced RNA and concomitantly in a severe reduction of the unspliced RNA. The reduction of the RNA level was not due to the instability of RNA in the cytoplasm but was due to the reduced stability in the nucleus and/or inefficient nuclear-cytoplasmic transport of the RNA. The polyadenylation appeared unaffected by the mutation. We also found a region in gag which made expression of the unspliced RNA dependent on the region around the SA site (23). These experiments suggested that the 441-nt region around the SA site was crucial for producing enough gag region-containing unspliced RNA.
Since the 441-nt region contained the SA site, it remained unknown whether the splicing event itself was necessary for ensuring high-level expression of gag region-containing unspliced mRNA. The temperature shift experiments with a mutant whose splicing was temperature sensitive suggested that the splicing event itself was unnecessary (23) but that more crucial experiments were needed to arrive at this conclusion. In the present work, we tried to answer this question by examining the levels of unspliced RNA in various MLV mutants constructed by modifying the 441-nt region.
MATERIALS AND METHODS
Plasmids.
The infectious proviral MLV DNA used in this work originated from clone 48, which was a lambda clone containing an EcoRI DNA fragment from an MLV-infected mouse fibroblast line (3). This fragment was subcloned into EcoRI-cut pBR322 (pArMLV-48) (13, 21). The PstI sites in the cellular sequence, 1.7 kb upstream and 148 bases downstream from the 5′ and 3′ ends of the provirus in pArMLV-48, were used to subclone the proviral DNA into pSVKhmB-derived vector (23). Since the deletions and base substitutions were introduced into the provirus of this construct, all of the constructs used in this paper were flanked by about 1.7 kb and 148 bases of cellular DNA derived from the active site of MLV transcription. This is probably the reason why the transcript levels per proviral copy were similar for all of the transfected mouse cell clones (23). The simian virus 40 (SV40) promoter-driven hygromycin resistance gene was inserted in 0.2 kb downstream of the 3′ long terminal repeat (LTR) in the same orientation as the MLV provirus, and its expression served as an internal standard of the provirus expression (23). The mutations were generated by PCR amplification over the relevant regions. The oligonucleotide primers for PCR were synthesized by BEX Inc. (Tokyo, Japan). The structures of the mutant constructs are shown at the top of each figure. They were all checked by sequencing.
Cell transfection.
Subconfluent cultures of NIH 3T3 cells (2 × 105/6-cm-diameter dish) were transfected with 10 μg of plasmid DNA per dish by the standard calcium phosphate precipitation method (20). Selection with hygromycin (200 μg/ml) was started 48 h after transfection and continued for 3 weeks. A total of 50 to 100 colonies appeared in each dish. The culture medium employed was Eagle’s minimal essential medium supplemented with 7.5% fetal calf serum.
RNA analysis.
Cells were cultured in 10-cm-diameter dishes at 37°C for 4 days, and the total RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform method (8). For Northern blot analysis, 5 μg of total RNA was electrophoresed in a formalin–1% agarose gel and blotted onto a Nitro-plus 2000 filter (Micron Separations, Inc., Westboro, Mass.). The ClaI-SacI fragment (nt 7674 to 8229 in the 3′ LTR region [Fig. 1]) of a cloned MLV, p8.2 (25), was used to detect all of the viral messages. The SmaI-HpaI fragment of pSVKhmB containing the hygromycin resistance gene (hygr) (23) was used for detecting the hygr gene message, which was used as an internal standard. For rough quantitation of RNA, the data were processed with the Bioimaging analyzer (Fuji Photo Co., Ltd., Tokyo, Japan). The radioactivity of a band corresponding to the unspliced RNA and that of the hygr transcript band were measured by a program in the analyzer, and the former value was divided by the latter. The ratio of the value thus obtained for a given sample to that obtained for wtΔSA was calculated. For obtaining the corresponding value for the wt, the radioactivity in the unspliced RNA band for the same amount of provirus copy was estimated for the wt and wtΔSA, and the ratio of the former to the latter was calculated. The ratios thus obtained for the wt were 2.3 for one experiment (Fig. 1) and 3.2 for another (Fig. 2). The quantitation by Northern blotting of the serially diluted sample showed that the relative amount of RNA expressed by the wt MLV in comparison with wtΔSA was about 10:1 (23). Therefore, the values 2.3 and 3.2, respectively, in the above experiments corresponded to 10-fold more RNA relative to the value 1.
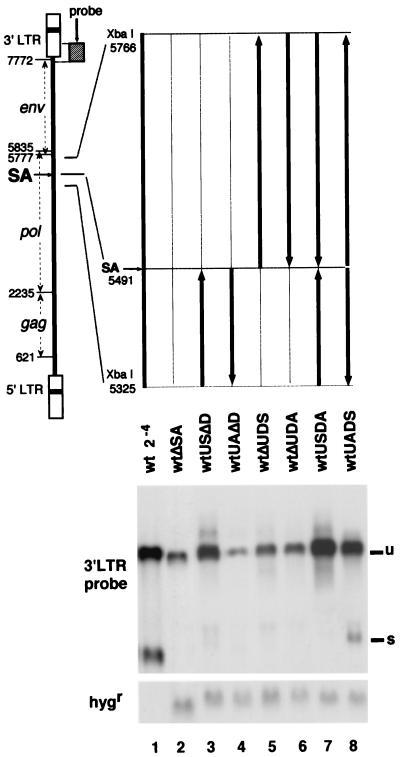
FIG. 1.
Genetic dissection of the 441-nt region (nt 5325 to 5766). Thick lines, viral genome; thin lines, deleted regions; arrows, 3′ direction of the viral genome. For example, wtUAΔD had the upstream XbaI-SA region (nt 5325 to 5491) in the inverted orientation and deletion of the downstream SA-XbaI region (nt 5491 to 5766). In Northern blot analysis, 5 μg of total cellular RNAs from the stable transfectants was loaded in each lane (lanes 2 to 8). Since the wt MLV-infected cells contained approximately 16-fold more proviral copies (23), the 16-fold-diluted RNA extract from the wt MLV-infected cells was applied to the gel for comparison (lane 1). The 3′ LTR probe (nt 7647 to 8229 [hatched box]) was used to detect the viral RNA, and the SmaI-HpaI fragment of pSVKhmB containing the hygr coding region was used to detect the reference hygr mRNA (23). The ratios of radioactivity in the unspliced RNA band to radioactivity in the corresponding band in wtΔSA (see Materials and Methods) were 2.2 for wtUSΔD (lane 3), 0.3 for wtUAΔD (lane 4), 1.1 for wtΔUDS (lane 5), 1.1 for wtΔUDA (lane 6), 6.9 for wtUSDA (lane 7), and 4.0 for wtUADS (lane 8). The value for 24-diluted wt was 3.2. Since the previous estimation indicated that the wt produced 10-fold more RNA per proviral copy than wtΔSA (23), the value 3.2 in this experiment roughly indicates 10-fold more RNA copies per provirus compared with the value 1. u, unspliced mRNA; s, spliced mRNA.
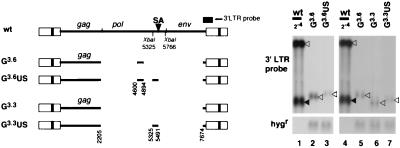
FIG. 2.
Effects of ligation of the fragment at nt 5325 to 5491 to G3.3 (G3.3US) or G3.6 (G3.6US) on transcription level. The probes used in Fig. 1 were used for Northern blot analysis. Open arrowhead, unspliced RNA; closed arrowhead, spliced RNA.
Immunoblotting.
Protein was extracted with radioimmunoprecipitation assay buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 0.5% sodium deoxycholate, 1% Triton X-100, 0.05% sodium dodecyl sulfate [SDS]) from the confluent cultures grown in 6-cm-diameter dishes. An aliquot of 8 μg of protein extract was electrophoresed through an SDS-10% polyacrylamide gel and blotted onto a Nitro-plus 2000 filter (Micron Separations, Inc.). Pr65gag was detected with anti-p30 monoclonal antibody R18-7 (7). Antibody bound to the filter was detected by using an enhanced chemiluminescence Western immunoblotting detection system (Amersham) and XAR-5 film (Eastman Kodak Co., Rochester, N.Y.). The polyacrylamide gels were stained with Coomassie brilliant blue to check the amounts of loaded proteins.
RT-PCR and sequencing.
Reverse transcriptase-PCR (RT-PCR) was performed with the whole-RNA extracts (24). The primers used for the RT-PCR around the splice junction were ENcom (nt 6037 to 6018 [5′-AGATGGTCCATGGTGGGCTA-3′]) and Kpnp (nt 28 to 50 [5′-CCCGGGTACCCGTGTATCCAATA-3′]). The amplified fragments were subcloned into the SmaI site of pBluescriptII and were sequenced by using a Taq dye terminator cycle sequencing kit (Perkin-Elmer) and an ABI 373 A sequencer.
RESULTS
Different roles played by the 5′ and 3′ halves of the 441-nt region.
The wt MLV-infected cells produced 24-fold more RNA per cell than the cells transfected with the replication-incompetent MLV proviruses, such as Δwt, fswt, or GE6.4 (23). Since the former contained 24-fold more proviral copies per cell than the latter, levels of RNA expression per proviral copy were considered approximately the same for both (23). Therefore, to compare RNAs or proteins expressed by the mutant provirus with those expressed by the wt virus, the samples from the wt MLV-infected cells were diluted 24-fold and applied to the gels. For comparison among the mutants, the expression of SV40 promoter-driven hygr, which was inserted downstream of the provirus (23), was used as an internal standard.
The deletion of the 441 nt around SA (nt 5325 to 5766) from the wt resulted in a severe reduction of the unspliced RNA (23) (Fig. 1, lane 2). The reduction was estimated to be about 10-fold, as shown in the previous report (23). In order to narrow down the responsible region, the 441-nt region was first divided into two smaller regions, upstream and downstream of SA, and each region was deleted or inverted. The RNA expression levels from these transfectants were compared (Fig. 1). Deletion of the 5′ half of the 441-bp region abolished the production of the spliced RNA and reduced the unspliced RNA to the level of the mutant with the whole 441-nt deletion, wtΔSA (compare lanes 5 and 6 with lane 2). Deletion or inversion of the 3′ half of the region also abolished splicing but did not reduce the level of unspliced RNA when the 5′ half of the region was present in the correct orientation (lanes 3 and 7). Inversion of the 5′ half in the absence of the 3′ half greatly reduced the unspliced RNA level (lanes 4). These observations suggested (i) that the splicing required both the 5′ and 3′ halves and (ii) that high-level expression of the unspliced RNA required the intact sequence of the 5′ half of the 441 nt when the splicing was absent.
When the 3′-half region was retained intact and the 5′ half was inverted, the spliced RNA was produced and, at the same time, the unspliced RNA was produced at a high level (Fig. 1, lane 8). In our previous experiments, expression of unspliced RNA could be restored by the 441-nt fragment ligated in the antisense orientation immediately downstream of the Gag-coding region which simultaneously activated the silent SA signal (23). In both cases, the region which was found responsible for producing the unspliced RNA at a high level was in the inverted orientation, but the spliced RNA was produced. Therefore, it was speculated that a secondary structure ensuring the production of spliced RNA could compensate the 5′ half’s function in restoring high-level expression of the unspliced RNA.
G3.6 was obtained by deleting nt 4894 to 7674 from GE6.4 to remove the env region (23), and G3.3 was obtained by further deletion of the remaining nt 4600 to 4894 in the pol region (Fig. 2, left panel). Both G3.6 and G3.3 had only the Gag-coding region and produced Gag-coding RNA at a low level (Fig. 2, right panel, lanes 2, 5, and 6). Ligation of the 441 nt to the 3′ end (nt 4894) of gag (Fig. 2) in G3.6 restored the level of the unspliced RNA, concomitantly resulting in production of the spliced RNA (23). In order to know if the 5′ half of the 441 nt alone had a signal sufficient for the production of unspliced RNA at a high level, it was inserted immediately downstream of gag both in G3.6 and G3.3 to obtain G3.6US and G3.3US, respectively. The levels of RNA produced by G3.6US and by G3.3US were not increased in comparison with those for G3.6 and G3.3, indicating that the 5′-half region of the 441 nt alone was incapable of restoring the transcript level.
Detailed dissection of the 5′-half region.
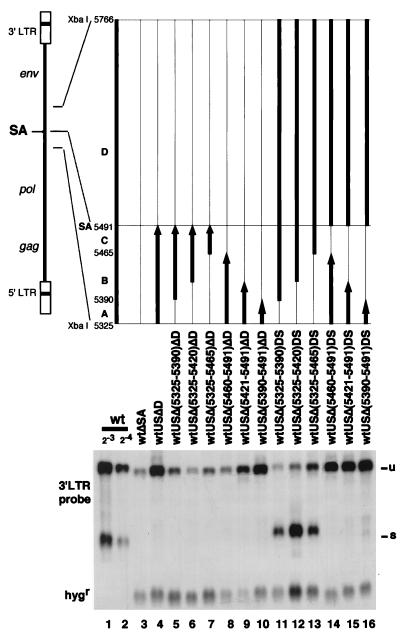
Since the above experiments suggested that the 5′ half of the 441-nt region (nt 5325 to 5490) was responsible for high-level production of the unspliced RNA, we tried to narrow down the responsible region further. The 441-nt region was divided into four regions, i.e., region A (nt 5325 to 5389), region B (nt 5390 to 5464), region C (nt 5465 to 5490), and region D (nt 5491 to 5765). The regions A, B, and C are upstream of SA, and region D is downstream. The constructs without region D produced no spliced RNA (Fig. 3, lanes 4 to 10). Deletion of region A in the absence of region D resulted in low-level expression of the unspliced RNA (lanes 5 to 7), indicating that region A was essential for high-level expression of the unspliced RNA in the absence of splicing. In the constructs which had region A but not region D (lanes 8 to 10), the presence of region B instead suppressed the increase in the level of the unspliced RNA attributable to region A (lanes 8 and 9).
FIG. 3.
Detailed dissection of the region upstream of SA in the 441-nt region. Coding of the mutants is such that wtUSΔ(5325-5390)ΔD stands for a mutant with deletion of nt 5325 to 5390 in the upstream region of SA and a whole deletion (nt 5491 to 5766) of the downstream region of SA. The ratios of radioactivity in the unspliced RNA band to radioactivity in the corresponding band in wtΔSA (see Materials and Methods) were 2.6 for wtUSΔD (lane 4), 0.9 for wtUSΔ(5325-5390)ΔD (lane 5), 0.7 for wtUSΔ(5325-5420)ΔD (lane 6), 0.8 for wtUSΔ(5325-5465)ΔD (lane 7), 1.3 for wtUSΔ(5460-5491)ΔD (lane 8), 3.8 for wtUSΔ(5421-5491)ΔD (lane 9), 2.9 for wtUSΔ(5390-5491) (lane 10), 0.8 for wtUSΔ(5325-5390)DS (lane 11), 0.5 for wtUSΔ(5325-5420)DS (lane 12), 1.2 for wtUSΔ(5325-5465)DS (lane 13), 3.0 for wtUSΔ(5460-5491) (lane 14), 3.0 for wtUSΔ(5421-5491)DS (lane 15), and 3.4 for wtUSΔ(5390-5491)DS (lane 16). The value for 24-diluted wt was 2.3. Therefore, the value 2.3 indicates roughly 10-fold more RNA copies per provirus compared with the value 1 in this assay. See the legend to Fig. 1 for other abbreviations.
In the constructs with region D (Fig. 3, lanes 11 to 16), deletion of region C immediately upstream of the SA site or larger upstream deletion resulted in loss of the spliced RNA (lanes 14 to 16). The constructs with region C in addition to region D all produced unspliced RNA and spliced RNA (lanes 11 to 13). Among them, wtUSΔ(5325-5465)DS expressed a high level of the unspliced RNA (lane 13). The presence of region B instead suppressed expression of the unspliced RNA (compare lanes 11 to 12 with lane 13). These observations suggested that (i) region C together with region D downstream of the SA site was indispensable for the production of the spliced RNA, (ii) generation of the spliced RNA could compensate for the function of region A (nt 5325 to 5389) at least partly in producing the unspliced RNA (lane 13), and (iii) the presence of region B instead suppressed the increase in the level of unspliced RNA attributable to the generation of spliced RNA (lanes 11 and 12). Comparison of lanes 8 to 10 with lanes 14 to 16 indicated that the retention of region D elevated the basal level of the production of the unspliced RNA in the absence of splicing.
Base substitution mutants in the 441-nt region.
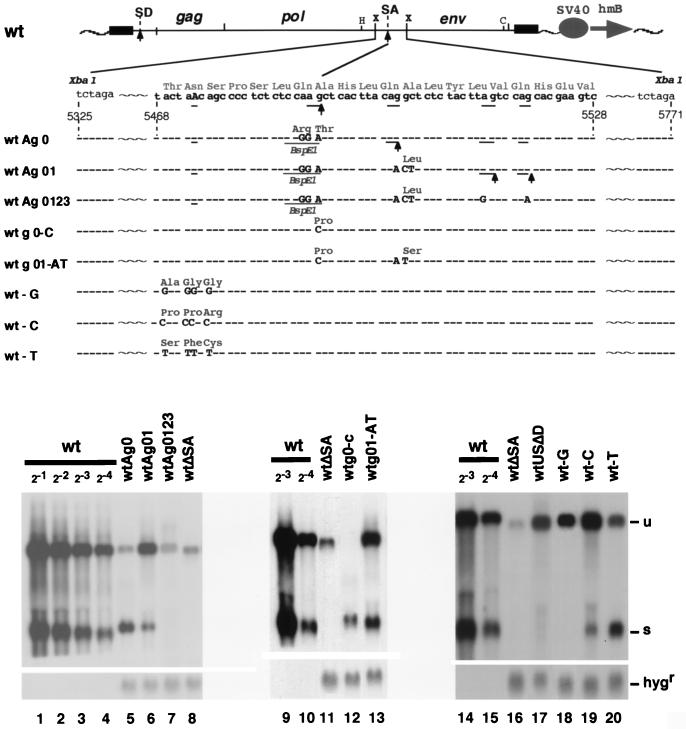
Since the above experiments dealt with deletion mutants which affected gene structure substantially, we asked whether base substitutions could result in high-level expression of unspliced mRNA without producing spliced RNA. The data are summarized in Fig. 4. A base change from CAA GCT (GlnAla) to CGG ACT (ArgThr) at the splice boundary (wtAg0 in Fig. 4) shifted the SA site to the next downstream AG. A further base change from CAG GCT (GlnAla) to CAA CTT (GlnLeu) at the second SA site (wtAg01) resulted in splicing by using the two further downstream AG’s (Fig. 4, lane 6; the two SA sites which were determined by RT-PCR amplification of the transcripts and their sequencing are indicated by upward arrowheads in the upper panel). The point mutations of all four potential SA sites (wtAg0123) resulted in the loss of the spliced RNA but also in the reduction of unspliced RNA (Fig. 4, lane 7). The base change from CAA GCT (GlnAla) to CAA CCT (GlnPro) at the first SA (wtg0-C) or the further base change from CAG GCT (GlnAla) to CAA TCT (GlnSer) at the second SA (wtg01-AT) retained the splicing of the transcripts (Fig. 4, lanes 12 and 13). Thus, deprivation of the downstream SA sites by a base change could not produce mutants expressing unspliced RNA at a high level without producing spliced RNA. It was noted that the mutation in the first AG, which relocated the SA site to the next downstream AG, resulted in relatively more spliced RNA (Fig. 4, lanes 5 and 12).
FIG. 4.
Point mutants around the SA site in the 441-nt region. Lowercase letters, nucleotides in the wt; capital letters, mutated nucleotides. Amino acid changes are also indicated above the nucleotide sequences. Upward arrowheads, SA sites determined by RT-PCR amplifications of transcripts with Kpnp (nt 28 to 50) and ENcom (nt 6027 to 6018) primers and their sequencing. Absence of the spliced RNA in wtAg0123 and wt-G transcripts was checked by the failure of RT-PCR amplification of the corresponding DNA fragment. SD, splice donor; H, HindIII; C, ClaI; X, XbaI; SV40, SV40 promoter. See the legend to Fig. 1 as well as for other abbreviations.
We constructed base change mutants by substituting four potential branching point A residues in the region upstream of the SA site. The base substitution with T’s (wt-T) did not change the splice pattern (Fig. 4, lane 20), and substitution with C’s (wt-C) shifted RNA expression to the unspliced message (Fig. 4, lane 19), while retaining a small amount of spliced message. The base substitution with G’s (wt-G) (Fig. 4, lane 18) abolished the spliced RNA (confirmed by failure of RT-PCR amplification of the fragment expected for the spliced RNA) while ensuring high-level expression of unspliced RNA.
Gag protein produced by some of the mutants.
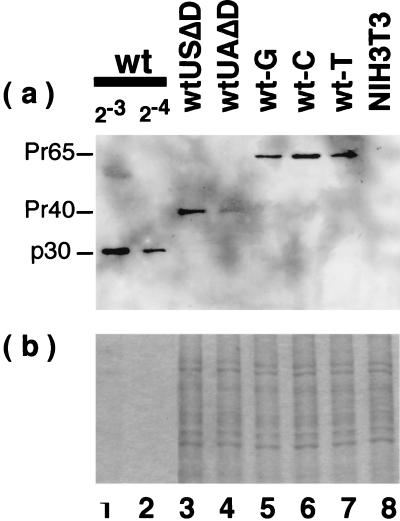
In order to confirm that the mRNA levels of the mutants were reflected in the protein levels, we compared the levels of Gag products expressed by some of the constructs (Fig. 5). In the wt MLV-infected cells which were actively producing virions, Gag precursor protein was processed into p30 (in Fig. 5, only p30 is visible due to dilution of the sample; less diluted cell extracts gave the Pr65gag band clearly [23]). wtUSΔD- and wtUAΔD-transfected cells produced partially processed Gag p41, which was observed as a major product in Δwt with a 306-nt deletion (nt 3229 to 3535) or ΔwtdSA with a 4-nt insertion (nt 3705) in the pol region (23). wt-G, wt-C, and wt-T produced unprocessed Pr65gag, but the cleavage into p30 was undetectable. Since the gag region and most of the pol region were intact in these mutant constructs and, consequently, the protease region was also intact, how the anomalous processing of the Gag products was produced in these mutants is obscure. Probably, interaction between Gag and Gag-Pol fusion proteins was affected by deletion of or base changes in the proteins associated with the nucleic acid mutations, and, as a result, interaction of the protease with Gag was affected. This point is interesting but is not explored further in the present report. As expected, wtUAΔD with a low level of the unspliced RNA produced a low level of Gag protein, while other constructs (wtUSΔD, wt-G, wt-C, and wt-T) with a high level of unspliced mRNA produced a high level of Gag (Fig. 5).
FIG. 5.
Viral Gag proteins expressed by the mutants. See the legends to Fig. 1 and 4 for the structures of the mutant proviruses. (Upper panel) Western blot with anti-p30 antibody; (bottom panel) Coomassie blue stain of the gel applied with the same amount of protein as that for the upper panel.
DISCUSSION
Generation of both spliced and unspliced RNAs at a proper ratio is essential for the replication of all of the retroviruses. In mouse and avian leukemia viruses, the unspliced RNA encodes Gag and Gag-Pol (it is also used as genomic RNA), and the spliced RNA encodes Env. In human T-cell leukemia virus type 1 or human immunodeficiency virus type 1 (HIV-1), there are two classes of spliced RNAs in addition to the unspliced RNA: a singly spliced RNA encoding Env (Vif also in the case of HIV-1) and multispliced RNAs encoding accessory regulatory proteins, such as Tax or Tat and Rex or Rev, etc. Rev or Rex translated from the multispliced RNA is known to interact with the viral sequence RRE to ensure expression of the unspliced and singly spliced RNAs (18). The Rev-RRE interaction requires splice sites (6) and may involve small nuclear RNAs which are involved in splicing (16, 17). Thus, expression of the unspliced and singly spliced RNAs appeared to be regulated by the virus-coded accessory protein and cellular factor(s) involved in the splicing. This regulation is reported to operate in nuclear-cytoplasmic transport or stabilization of these RNAs (6, 12, 22). If there is a regulation of the unspliced and singly spliced RNAs versus multiply spliced RNAs, it is reasonable to suppose a presence of similar regulation of the unspliced RNA encoding Gag and Gag-Pol versus singly spliced RNA encoding Env, and such a regulation must exist in all retroviruses. Actually, cis elements affecting the efficiency of splicing and the stability of unspliced RNA have been reported for avian and some mammalian retroviruses (1, 2, 5, 9, 10, 11, 14, 15, 19, 20, 23, 26).
We previously reported that the 441-nt sequence around SA (nt 5326 to 57666) was necessary for high-level expression of unspliced gag-containing RNA of MLV (23). The previous experiments, however, did not clarify whether the phenomenon of splicing and the high-level expression of the unspliced RNA could be dissociated. In this report, we dissected the 441-nt region in more detail.
Analyses of mutants in this report showed that splicing and high-level expression of the unspliced RNA could actually be dissociated. Deletion or inversion of the sequence spanning from the SA site (nt 5491) down to the 3′ end of the 441-nt sequence (nt 5766) (region D in Fig. 6) completely eliminated the spliced RNA while retaining high-level expression of the unspliced RNA. Meanwhile, deletion or inversion of the sequence in the region upstream of SA in the 441-nt region, i.e., nt 5325 to 5491, drastically reduced the level of the unspliced RNA (an exception was wtUADS which produced both spliced and unspliced RNAs; see below for discussion) (Fig. 1), i.e., the upstream region of SA (nt 5325 to 5491) in the 441-nt region was required for high-level expression of the unspliced RNA in the absence of splicing. However, when this region alone was ligated to the 3′ end of gag of the Gag-encoding proviruses (G3.6US and G3.3US) (Fig. 2), unlike the whole 441-nt region (23), the level of the unspliced RNA was not elevated, indicating that the 5′-half region alone was not functional.
FIG. 6.
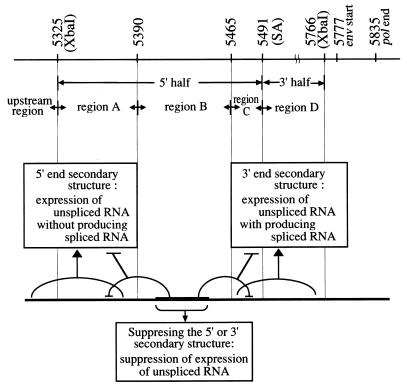
Model explaining the different patterns of RNA expression by the constructs. The region between the two XbaI sites (nt 5325 to 5766) is the 441-nt region. The hypothetical secondary structures are symbolized by the loops connecting the two regions involved.
The region upstream of SA in the 441-nt region was divided into three regions, i.e., A, B, and C (Fig. 6) and analyzed in more detail. We found that the 5′-end 64-nt (nt 5325 to 5390) region A was indispensable for high-level expression of the unspliced RNA (Fig. 2, compare lanes 5 and 10). However, some constructs which were lacking in region A but produced the spliced RNA, such as wtUSΔ(5325-5465)DS (Fig. 3, lane 13) and wtUADS (Fig. 1, lane 8), produced the unspliced RNA at a high level. This suggested that the splicing could compensate, at least partly, the function of region A, which ensured high-level expression of the unspliced RNA in the absence of splicing. This supposition was also supported by high-level expression of the unspliced RNA by GinvSA (23), which had the whole 441-nt region in the inverted orientation but produced spliced RNA.
When the region downstream of the SA site (region D) was left intact, deletion from the 5′ end of the 441-nt region to nt 5465 (regions A and B) did not abolish splicing (Fig. 3, lanes 11 to 13), but the further deletions of 27 nt of the SA site completely abolished splicing (Fig. 1, lane 5). The 32-nt deletion immediately upstream of the SA site (nt 5460 to 5491) alone could abolish splicing even though the downstream potential SA sites (Fig. 2, lane 14; see also Fig. 4, upper panel) remained intact. Therefore, region C immediately upstream of the SA site together with its downstream region D was found to be essential for splicing.
The presence of region B, either in the absence (Fig. 3, lanes 8 and 9) or the presence of region D (Fig. 3, lanes 11 and 12), lowered the expression of unspliced RNA in comparison with its absence (lanes 10 and 13, respectively). It appeared that region B suppressed expression of the unspliced RNA both in the region A-dependent, splicing-independent expression and the region A-independent, splicing-dependent expression of the unspliced RNA. Retention of the region D downstream of the SA site appeared to increase the level of the unspliced RNA in the constructs which did not produce spliced RNA (Fig. 3, compare lanes 8 to 10 and lanes 14 to 16).
The potential branching point A residue at nt 5473 (Fig. 4) was present in the middle of region C immediately upstream of SA. The base substitution of the A residue and three other A residues nearby with G residues (wt-G in Fig. 4) resulted in the abolishment of splicing while retaining the production of unspliced mRNA at a high level. The base substitution from A to C resulted in a strong shift to the unspliced mRNA while retaining a small amount of spliced mRNA (wt-C in Fig. 4). The base substitution from A to T did not affect the splice pattern (wt-T in Fig. 4). Since an increase in G or C residues results in more G-C pairs, which are more stable than A-T pairs, substitution of A with G or C will result in a higher secondary structure disrupting the local RNA conformation, but substitution of A with T will not. Therefore, after substitution of A’s with G’s or C’s, the secondary structure around the branching point will be greatly changed.
Based on the above observations, we propose, among others, the following hypothesis. The 5′-half region could be divided into three regions, i.e., A, B, and C (Fig. 6). We postulate secondary structures formed by region A with its upstream (5′-end secondary structure), those formed by region C with its downstream region D (3′-end secondary structure), those formed by the middle part region B with the 5′ end region A, and those formed by region B with the 3′ end region C. Region B is thus able to competitively inhibit formation of the 5′-end and the 3′-end secondary structures (Fig. 6, bottom). The 5′-end secondary structure is postulated to be responsible for producing unspliced RNA in the absence of splicing, and the 3′-end secondary structure is postulated to be responsible for splicing and concomitant production of unspliced RNA. If either region A or C is deleted but region B is retained, region B will form secondary structures with the remaining region C or A, which will prevent formation of the 3′ end or the 5′-end secondary structures. This well explains why the transcript levels of wtUSΔ(5460-5491)ΔD and wtUSΔ(5325-5390)DS were lower than the transcript levels of their derivatives obtained by further deletion of region B (Fig. 2, lanes 8 and 11, respectively).
The above hypothesis (as summarized in Fig. 6) can also explain the base substitution mutants. As discussed above, the base substitution of A residues with G or C residues in the potential branching points in region C can change the local folding structure of RNA. The change in the folding structure of region C may result in formation of a stable secondary structure between regions C and B. This will prevent formation of the 3′-end secondary structure necessary for splicing. Since the 5′-end region A is free to interact with the upstream region to form the 5′-end secondary structure, high-level expression of the unspliced RNA in the absence of splicing is ensured. wtAg0123, whose four successive potential SA sites were inactivated, produced no spliced RNA. It produced unspliced RNA only at a low level even though region A-C, which is hypothesized to ensure high-level production of unspliced RNA, was retained intact (Fig. 4, lane 7). It appeared that the mutations in the four candidate SA sites not only abolished splicing but also suppressed expression of the unspliced RNA. Here, three G residues were changed to A residues and one A residue was changed to a G residue. These base substitutions may result in a rather drastic change in the RNA folding structure as discussed above. As a consequence, the mutated region downstream of SA and region C just upstream of it may form a stable secondary structure which is unfavorable for splicing. Region B is then free to interact with region A and prevents formation of the 5′-end secondary structure which is necessary for production of the unspliced RNA. This may explain why the transcript level of wtAg0123 was low.
What is the mechanism which ensures production of unspliced RNA through the cis elements present in the 441-nt region? The level of RNA will be determined by the efficiency of transcription initiation, stability in the nucleus, efficiency of nuclear-cytoplasmic transport, and stability in the cytoplasm. The low level of transcript in the MLV with a deletion in the 441-nt region was found to be due not to instability in the cytoplasm (23) or to inefficient transcription initiation (since the same LTR was used as a promoter for all of the constructs). Therefore, the low level of expression must be caused by intranuclear instability and/or inefficient nuclear-cytoplasmic transport. The genes with introns are transcribed and spliced in the nucleus and are transported to the cytoplasm through the nuclear pores. Transcription and splicing appear to be closely linked; in Drosophila embryos, splicing was shown to occur cotranscriptionally (4). Transcripts containing introns have to complete splicing before they are exported into the cytoplasm; otherwise, the useless transcripts will be exported and will accumulate in the cytoplasm. Therefore, transcription, splicing, and nuclear export are considered to be linked to each other in an orderly fashion. If a transcript cannot interact with splicing and export machineries, it will be degraded.
Our work showed that in MLV, the sequence ensuring the production of unspliced RNA and the sequence ensuring splicing were present in different viral genomic elements. This suggested that each element was interacting with different machineries or enzymes. However, the constructs lacking in the former region produced unspliced RNA at a high level if they also produced the spliced RNA [such as wtUAUS and wtUSΔ(5325-5465)DS]. This indicated a possible interaction between the processes of the splicing and extranuclear transport and/or stabilization of the unspliced RNA. To explain these phenomena, we speculate as follows. In wt MLV, interaction of the 3′-end secondary structure with the splicing enzymes exposed the 5′-end secondary structure to the enzymes necessary for nuclear export of the unspliced RNA. In mutants lacking in the 5′-end region but which produced unspliced RNA at a high level while producing spliced RNA, an interaction of the 3′-end region with the splicing enzymes changed the conformation of the unspliced RNA so that it was more accessible to the enzymes necessary for nuclear export.
In contrast to other intron-containing genes, the MLV gene must produce both spliced and unspliced RNAs. We have already found a sequence in the gag gene which makes production of unspliced RNA dependent on the production of spliced RNA (23). How the gag gene region affects the 441-nt region responsible for extranuclear transport and/or intranuclear stabilization of RNA remains to be elucidated. Although the model postulated here is highly speculative, it may be useful for designing future experiments.
ACKNOWLEDGMENTS
This work was supported in part by a grant-in-aid from the Japanese Ministry of Health and Welfare to H.Y.
We thank M. Oyane for editorial assistance.
REFERENCES
- 1.Armentano D, Yu S F, Kantoff P W, von Ruden T, Anderson W F, Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987;61:1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacheler L, Fan H. Isolation of recombinant DNA clones carrying complete integrated proviruses of Moloney murine leukemia virus. J Virol. 1981;37:181–190. doi: 10.1128/jvi.37.1.181-190.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 5.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 7.Chesebro B, Britt W, Evans L, Wehrly K, Nishio J, Cloyd M. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of friend MCF and Friend ecotropic murine leukemia virus. Virology. 1983;127:134–148. doi: 10.1016/0042-6822(83)90378-1. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid-guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cochrane A W, Jones K S, Beidas S, Dillon P J, Skalka A M, Rosen C A. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J Virol. 1991;65:5305–5313. doi: 10.1128/jvi.65.10.5305-5313.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grone M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 13.Jones D S, Nemoto F, Kuchino Y, Masuda M, Yoshikura H, Nishimura S. The effect of specific mutations at and around the gag-pol gene junction of Moloney murine leukemia virus. Nucleic Acids Res. 1989;17:5933–5945. doi: 10.1093/nar/17.15.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz R A, Kotler M, Skalka A M. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988;62:2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz R A, Skalka A M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kjems J, Sharp P. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6 · U5 small nuclear ribonucleoprotein in spliceosome assembly. J Virol. 1993;67:4769–4776. doi: 10.1128/jvi.67.8.4769-4776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X B, Heimer J, Rekosh D, Hammarskjold M L. U1 small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remains unspliced. Proc Natl Acad Sci USA. 1990;87:7598–7602. doi: 10.1073/pnas.87.19.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luciw P A. Human immunodeficiency viruses and their replication. In: Fields B N, editor. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1995. pp. 1881–1952. [Google Scholar]
- 19.McNally M T, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNally M T, Gontarek R R, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 21.Odawara T, Yoshikura H, Oshima M, Tanaka T, Jones D S, Nemoto F, Kuchino Y, Iwamoto A. Analysis of Moloney murine leukemia virus revertants mutated at the gag-pol junction. J Virol. 1991;65:6376–6379. doi: 10.1128/jvi.65.11.6376-6379.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen H S, Cochrane A W, Rosen C. Intraction of cellular factors with intragenic cis-acting repressive sequences within the HIV genome. Virology. 1992;191:709–715. doi: 10.1016/0042-6822(92)90246-l. [DOI] [PubMed] [Google Scholar]
- 23.Oshima M, Odawara T, Matano T, Sakahira H, Kuchino K, Iwamoto A, Yoshikura H. Possible role of splice acceptor site in expression of unspliced gag-containing message of Moloney murine leukemia virus. J Virol. 1996;70:2286–2295. doi: 10.1128/jvi.70.4.2286-2295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiki R K, Bugawan T L, Horn G T, Mullis K B, Erlich H A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986;324:163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 25.Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra S W, Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci USA. 1980;77:3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahir A R, Lew K A, Murphy E C, Jr, Schmidt R D. Role of Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE) in the propagation of MPMV vectors by genetic complementation using homologous/heterologous env genes. Virology. 1996;224:517–532. doi: 10.1006/viro.1996.0558. [DOI] [PubMed] [Google Scholar]