Abstract
Group A Streptococcus isolates of the recently described M1UK clade have emerged to cause human infections in several European countries and elsewhere. Full-genome sequence analysis of M1 isolates discovered a close genomic relationship between some isolates from Scotland and the majority of isolates from Iceland causing serious infections in 2022 and 2023. Phylogenetic analysis strongly suggests that an isolate from or related to Scotland was the precursor to an M1UK variant responsible for almost all recent M1 infections in Iceland.
Keywords: Molecular Epidemiology, Population Genomics, Infectious Disease, Phylogenetics, Streptococcus
A new emergent genetic variant of type emm1 group A Streptococcus (GAS) known as M1UK was recently discovered during a period of increased scarlet fever activity and invasive infection notifications in England [1]. The M1UK variant over-expresses streptococcal pyrogenic exotoxin A (SpeA, also known as scarlet fever toxin), a GAS virulence factor [1-3]. The M1UK variant has been identified in several European and North American countries, commonly in conjunction with surges in GAS infections occurring in 2022 and 2023 [4-14]. While investigating recent surges in severe invasive GAS (iGAS) infections in Scotland and Iceland using whole genome sequencing (WGS), we discovered a very close genetic relationship between several M1UK from Scotland and all M1UK isolates from Iceland. Given the geographical separation, this finding was unexpected. The GAS population genomic data are consistent with transmission of a single M1UK strain from Scotland to Iceland, where it spread extensively, causing nearly all M1 invasive infections in 2022 and 2023.
iGAS infection surge
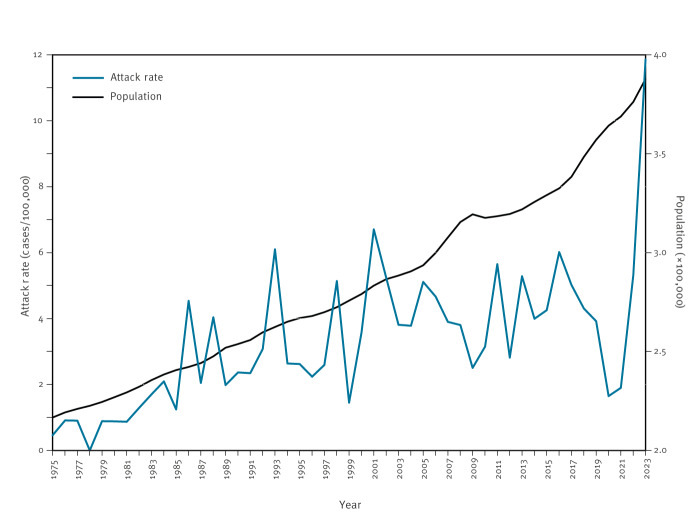
In Iceland where current iGAS surveillance encompasses the entire populace, iGAS infections increased strongly in the latter half of 2022, extending into the first half of 2023 (Figure 1). The population-adjusted iGAS attack rate for the first half of 2023 (11.9/100,000) was 2.7-fold higher than the mean (4.4/100,000) for the 20 years preceding the COVID-19 pandemic (2000–2019). The increase in iGAS infections was greatest for children younger than 6 years, in whom it increased 8.2-fold from a mean of 1.7 cases per year for 2000–2019 to 14 cases in the first half of 2023. Although the surge involved multiple emm types, M1 was most prevalent, causing 48 of 75 iGAS infections in 2023 [10]. Although less comprehensive surveillance information is available for Scotland, a similar substantial increase in iGAS infections in 2022 and 2023 occurred in central Scotland, with M1 isolates being most prevalent and young children disproportionately affected [6,15].
Figure 1.
Group A streptococcus invasive infections per 100,000 population in Iceland, January 1975–June 2023 (n = 495)
Whole genome sequencing
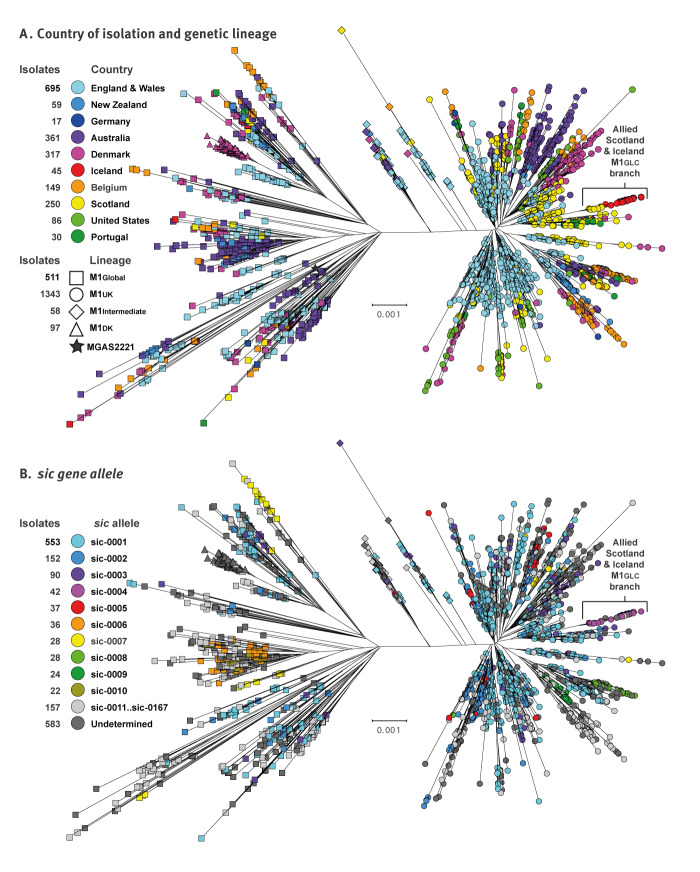
The GAS M1 isolates from Scotland (n = 250; collected 2014–2023) and Iceland (n = 45, collected between November 2022 and November 2023) were cultured from normally sterile sources such as blood or deep tissue, and identified by standard microbiological laboratory procedures at Landspitali (the National University Hospital of Iceland) in Reykjavik or at the Scottish Microbiology Reference Laboratory in Glasgow, where isolates are referred as part of the National invasive GAS surveillance programme. Most Icelandic patients were from the Reykjavik region. Two isolates were cultured from patients hospitalised in Akureyri, the largest community in northern Iceland. The genomes of all isolates from Iceland and Scotland were characterised by Illumina short-read sequencing [16]. In addition, all 45 Icelandic and 10 of the Scottish isolates were characterised using Oxford Nanopore Technology long-read sequencing [16]. To place the strains from Scotland and Iceland within a larger phylogenetic context, we compared them with publicly available WGS data for 1,714 M1 isolates collected in nine countries on three continents between 2005 and 2023 (19 years) (Table). Individual isolate characteristics are listed in Supplementary Table S1. Most of these isolates (n = 1,958, 97%) were collected after the 2010 emergence of the earliest known M1UK isolate. The phylogenetic tree based on the WGS data (Figure 2A) shows two major branches with an M1Global (n = 511) and an M1UK clade (n = 1,343) separated by a subset of genetically intermediate isolates (n = 58 M1Int). Of the isolates from Iceland, 41 belonged to the M1UK clade, and all 41 were closely related to one another and nine phylogenetically allied isolates from Scotland. These 50 allied isolates were genetically distinct and located on a separate branch from all other M1UK isolates. The isolates from Scotland were proximal to the isolates from Iceland, showing the Scottish isolates to be the evolutionary precursors.
Table. Group A streptococcus epidemiological surveillance cohorts, 2005–2023 (n = 2,009).
| Country | M1 isolates | Collection period | Bioproject(s) |
|---|---|---|---|
| England & Wales [1] | 695 | 2009–2016 | PRJEB12015 and PRJEB17673 |
| Australia [2] | 319 | 2005–2020 | PRJNA872282 |
| Belgium [13] | 149 | Jan 2020–May 2023 | PRJNA10033449 |
| Denmark [10] | 317 | Jan 2018–Feb 2023 | PRJEB62579 and PRJEB62635 |
| Germany [14] | 17 | Oct 2022–Apr 2023 | PRJEB64404 |
| New Zealand [11] | 59 | 2018–2019 | PRJNA985396 |
| Portugal [9] | 30 | Sep 2022–May 2023 | PRJEB65018 |
| United States [12] | 86 | 2019–2021 | PRJNA395240 |
| Australia [5] | 42 | 2007–2021 | PRJNA996294 |
| Scotland (this study) | 250 | 2014–2023 | PRJNA1076228 |
| Iceland (this study) | 45 | 2022–2023 | PRJNA1076228 |
Figure 2.
Phylogenetic relationships among contemporary group A streptococcus M1 isolates, 2005–2023 (n = 2,009)
Phylogeny for 2,009 GAS M1 isolates by inferred neighbour-joining based on 8,599 core chromosomal SNPs identified by Illumina short-read whole genome sequencing read mapping relative to reference strain MGAS2221 (NCBI accession number CP043530). Panel A: isolates are coloured by country of isolation and shaped by M1 genetic lineage. Panel B: isolates are coloured by the 10 most prevalent sic gene alleles in descending order.
With current short read sequencing technology it is problematic to assemble reads containing multiple different repeat domains, such as the sic gene, and accurately map them to a reference strain containing such repeat structures. The variants labeled ‘undetermined’ are isolates in which a consensus assembly of the sic gene was not obtained using the Illumina short-read-length sequencing data. This is likely because the sic gene in these isolates had more repeats or a more complex set of repeats that confounded the SPAdes assembler.
SIC genetic diversity
The sic gene encodes the extracellular streptococcal inhibitor of complement protein that contributes to enhanced evasion of immune functions. It is the most highly polymorphic gene in M1 GAS, a feature that has been exploited in epidemiological and population genetical studies [17-22]. To seek additional evidence supporting the hypothesis that the closely allied M1 isolates from Iceland and Scotland shared a very recent common ancestor, we analysed allelic variation in the sic gene using both Illumina assemblies for all 2,009 isolates and hybrid sequenced/assembled closed genomes for the allied isolates. If the phylogenetically allied isolates share a very recent common ancestor, we reasoned that they would have the same unique sic allele. In the 2,009 M1 strains, BLASTN search identified sic assembled on a single contig for 1,430 strains (71.2%). There were 167 sic alleles encoding 167 SIC variants (alleles and variants listed individually in Supplementary Table S2, and given for individual isolates in Supplementary Table S1), consistent with the rapidly evolving hypervariable nature of this gene and protein. Importantly, although 167 alleles were identified, the majority (38 of 50; 76%) of the closely related isolates from Scotland and Iceland had the same sic allele (designated sic0004), and these were the only isolates to have this allele (Figure 2B). Thus, our sic data provided additional molecular evidence that the phylogenetically allied isolates from Scotland and Iceland were related by recent descent.
Discussion
Although the actual transmission sources and routes are unknown, based on the WGS data currently available, the most parsimonious explanation for the close genetic relationship between the allied M1UK isolates posits direct introduction of a single isolate from Scotland to Iceland, probably by an individual with pharyngitis or an asymptomatic carrier. Iceland is a major tourist destination, with ca 1.9 million foreigners visiting for example in 2022, including 13% from the United Kingdom [23]. Identification of the unexpected genetic link between the isolates from these two countries was made possible by WGS analysis of comprehensive longitudinal samples available from Iceland and Scotland, stressing the importance of reference and public health laboratories in providing new actionable information about isolate dissemination and transmission routes.
The M1UK variant terminology was initially used to describe a lineage that differed from many contemporary circulating M1 strains by 27 shared single nucleotide polymorphisms (SNPs; 26 core and one phage) [1]. This number of SNPs is similar to the magnitude of core genome SNPs differentiating two clades of M1 strains causing human infections in the last decade of the 20th century and first decade of the 21st century [24]. Similarly, an emergent lineage of isolates has been identified in Denmark (n = 97), which differed from all other M1Global and M1UK isolates in Denmark by 15 SNPs in the core genome and was designated M1DK [10]. In the present analysis, we found that all 41 Iceland M1 isolates with the 27 SNPs characteristic of the M1UK lineage differed from all other M1UK (n = 1,293, omitting the nine closely related Scottish isolates) and M1Global (n = 511) isolates by 15 and 43 SNPs in the core genome, respectively. The details of these individual SNPs are appended in Supplementary Table S3. We designated the M1UK sublineage causing infections in Iceland as M1GLC to reflect the historical relationship between the Gaelic populations in Scotland and Iceland.
The genomic data suggest that virtually all M1GLC infections in Iceland were caused by descendants of a single bacterial cell very recently introduced into Iceland from elsewhere, probably Scotland. However, because GAS can be carried asymptomatically and causes large numbers of pharyngitis cases, and because Iceland has a very robust tourism industry, it is not possible to prove the progenitor’s geographical source. The epidemiological data show that the introduction of the M1GLC sublineage into Iceland resulted in rapid dissemination countrywide (data not shown) and an increase in iGAS infections, particularly among young children. We speculate that social distancing practices implemented during the COVID-19 pandemic reduced exposure to GAS, resulting in a reduction in herd immunity that contributed to the 2022 and 2023 surge in iGAS infections. Consistent with this idea, very few cases of iGAS disease occurred in Iceland during the COVID-19 pandemic, from 2020 to early 2022, suggesting that exposure to GAS was less prevalent in Iceland before the surge in iGAS infections between late 2022 and early 2023. Also consistent is the disproportionate number of infections during the surge occurring in young children as they constitute the population with the least prior exposure and developed herd immunity, making them more susceptible to infection on exposure. Given that it is possible to rapidly sequence the genomes of thousands of bacterial isolates for an affordable cost and that Iceland has a small population, strain emergence and dissemination patterns can, with appropriate sampling strategies, be analysed in near real-time, analogous to work done on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Conclusion
Our study reveals a public health challenge posed by the emergence and international transmission of Streptococcus pyogenes M1 infections between Iceland and Scotland that affected young children disproportionately. Nearly all isolates were of the M1UK lineage, which has a high morbidity and mortality, and the derivative M1GLC sub-lineage, underscoring the need for effective surveillance and response strategies. Although our work and others’ demonstrates the public health value of WGS in identifying transmission events, the effectiveness of such efforts is limited, as iGAS infections are not notifiable in all European countries. In addition to the absence of co-ordinated European surveillance programmes capable of real-time data collection and analysis, this limits the capacity for comprehensive assessment and timely public health interventions. Addressing these gaps is crucial for managing iGAS emergence and spread more effectively.
Ethical statement
Epidemiological and genomic data presented in this study were obtained as part of public health protection surveillance programmes in Scotland and Iceland, and therefore approval by an ethical approval board was not needed for this study.
Funding statement
Research in the laboratory of JMM is supported in part by the Fondren Foundation. Logistical support for retrieval and shipping of Scottish isolates was funded in part by a grant from the Glasgow Children’s Hospital Charity.
Data availability
Whole genome sequencing data determined for this investigation was submitted to the National Center for Biotechnology Information Sequence Read Archive and closed genomes were submitted to Genbank, both under Bioproject accession PRJNA1076228.
Acknowledgements
We thank all clinical diagnostic laboratories in Iceland and Scotland; Alma Amaya, Wendy Barragan, Merissa Dias, Ryan Gadd, Regan Mangham, Eleanor Nichols, Matthew Ojeda Saavedra, Jordan Pachuca, Sindy Pena, and Kristina Reppond contributed to bacterial culture and genome sequencing; and Dilzi Mody provided logistical support. Sasha Pejerrey and Heather McConnell provided editorial support. In the Scottish Microbiology Reference Laboratory, Glasgow, Kevin Scott, Diane Lindsay, and Roisin Ure contributed to bacterial identification confirmation, emm typing, and logistics.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Conceptualisation: JMM, KGK, AS, RJO, SWL, SBB. Strain culturing, banking, cataloguing, and shipping: HE, RL, TW, AS, KGK. Sequencing, data collections, sequence data analysis: RJO, SWL, SBB. Analysis: all authors. Original draft preparation: JMM. Writing, reviewing, and editing: all authors. All authors read, provided input and approved the final manuscript.
References
- 1. Lynskey NN, Jauneikaite E, Li HK, Zhi X, Turner CE, Mosavie M, et al. Emergence of dominant toxigenic M1T1 Streptococcus pyogenes clone during increased scarlet fever activity in England: a population-based molecular epidemiological study. Lancet Infect Dis. 2019;19(11):1209-18. 10.1016/S1473-3099(19)30446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davies MR, Keller N, Brouwer S, Jespersen MG, Cork AJ, Hayes AJ, et al. Detection of Streptococcus pyogenes M1UK in Australia and characterization of the mutation driving enhanced expression of superantigen SpeA. Nat Commun. 2023;14(1):1051. 10.1038/s41467-023-36717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li HK, Zhi X, Vieira A, Whitwell HJ, Schricker A, Jauneikaite E, et al. Characterization of emergent toxigenic M1UK Streptococcus pyogenes and associated sublineages. Microb Genom. 2023;9(4):mgen000994. 10.1099/mgen.0.000994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aboulhosn A, Sanson MA, Vega LA, Segura MG, Summer LM, Joseph M, et al. Increases in group A streptococcal infections in the pediatric population in Houston, TX, 2022. Clin Infect Dis. 2023;77(3):351-4. 10.1093/cid/ciad197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butler TAJ, Story C, Green E, Williamson KM, Newton P, Jenkins F, et al. Insights gained from sequencing Australian non-invasive and invasive Streptococcus pyogenes isolates. Microb Genom. 2024;10(1):001152. 10.1099/mgen.0.001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davies PJB, Russell CD, Morgan AR, Taori SK, Lindsay D, Ure R, et al. Increase of severe pulmonary infections in adults caused by M1UK Streptococcus pyogenes, central Scotland, UK. Emerg Infect Dis. 2023;29(8):1638-42. 10.3201/eid2908.230569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Gier B, Marchal N, de Beer-Schuurman I, Te Wierik M, Hooiveld M, de Melker HE, et al. Increase in invasive group A streptococcal (Streptococcus pyogenes) infections (iGAS) in young children in the Netherlands, 2022. Euro Surveill. 2023;28(1):2200941. 10.2807/1560-7917.ES.2023.28.1.2200941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Demczuk W, Martin I, Domingo FR, MacDonald D, Mulvey MR. Identification of Streptococcus pyogenes M1UK clone in Canada. Lancet Infect Dis. 2019;19(12):1284-5. 10.1016/S1473-3099(19)30622-X [DOI] [PubMed] [Google Scholar]
- 9. Gouveia C, Bajanca-Lavado MP, Mamede R, Araújo Carvalho A, Rodrigues F, Melo-Cristino J, et al. Sustained increase of paediatric invasive Streptococcus pyogenes infections dominated by M1UK and diverse emm12 isolates, Portugal, September 2022 to May 2023. Euro Surveill. 2023;28(36):2300427. 10.2807/1560-7917.ES.2023.28.36.2300427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johannesen TB, Munkstrup C, Edslev SM, Baig S, Nielsen S, Funk T, et al. Increase in invasive group A streptococcal infections and emergence of novel, rapidly expanding sub-lineage of the virulent Streptococcus pyogenes M1 clone, Denmark, 2023. Euro Surveill. 2023;28(26):2300291. 10.2807/1560-7917.ES.2023.28.26.2300291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lacey JA, Bennett J, James TB, Hines BS, Chen T, Lee D, et al. A worldwide population of Streptococcus pyogenes strains circulating among school-aged children in Auckland, New Zealand: a genomic epidemiology analysis. Lancet Reg Health West Pac. 2024;42:100964. 10.1016/j.lanwpc.2023.100964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y, Nanduri SA, Van Beneden CA, Beall BW. M1UK lineage in invasive group A streptococcus isolates from the USA. Lancet Infect Dis. 2020;20(5):538-9. 10.1016/S1473-3099(20)30279-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rodriguez-Ruiz JP, Lin Q, Lammens C, Smeesters PR, van Kleef-van Koeveringe S, Matheeussen V, et al. Increase in bloodstream infections caused by emm1 group A Streptococcus correlates with emergence of toxigenic M1UK, Belgium, May 2022 to August 2023. Euro Surveill. 2023;28(36):2300422. 10.2807/1560-7917.ES.2023.28.36.2300422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolters M, Berinson B, Degel-Brossmann N, Hoffmann A, Bluszis R, Aepfelbacher M, et al. Population of invasive group A streptococci isolates from a German tertiary care center is dominated by the hypertoxigenic virulent M1UK genotype. Infection. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holdstock V, Twynam-Perkins J, Bradnock T, Dickson EM, Harvey-Wood K, Kalima P, et al. National case series of group A streptococcus pleural empyema in children: clinical and microbiological features. Lancet Infect Dis. 2023;23(2):154-6. 10.1016/S1473-3099(23)00008-7 [DOI] [PubMed] [Google Scholar]
- 16. Beres SB, Olsen RJ, Long SW, Eraso JM, Boukthir S, Faili A, et al. Analysis of the genomics and mouse virulence of an emergent clone of Streptococcus dysgalactiae Subspecies equisimilis. Microbiol Spectr. 2023;11(2):e0455022. 10.1128/spectrum.04550-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoe N, Nakashima K, Grigsby D, Pan X, Dou SJ, Naidich S, et al. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999;5(2):254-63. 10.3201/eid0502.990210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoe NP, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, et al. Rapid selection of complement-inhibiting protein variants in group A Streptococcus epidemic waves. Nat Med. 1999;5(8):924-9. 10.1038/11369 [DOI] [PubMed] [Google Scholar]
- 19. Hoe NP, Kordari P, Cole R, Liu M, Palzkill T, Huang W, et al. Human immune response to streptococcal inhibitor of complement, a serotype M1 group A Streptococcus extracellular protein involved in epidemics. J Infect Dis. 2000;182(5):1425-36. 10.1086/315882 [DOI] [PubMed] [Google Scholar]
- 20. Hoe NP, Vuopio-Varkila J, Vaara M, Grigsby D, De Lorenzo D, Fu YX, et al. Distribution of streptococcal inhibitor of complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 group A Streptococcus infection. J Infect Dis. 2001;183(4):633-9. 10.1086/318543 [DOI] [PubMed] [Google Scholar]
- 21. Matsumoto M, Hoe NP, Liu M, Beres SB, Sylva GL, Brandt CM, et al. Intrahost sequence variation in the streptococcal inhibitor of complement gene in patients with human pharyngitis. J Infect Dis. 2003;187(4):604-12. 10.1086/367993 [DOI] [PubMed] [Google Scholar]
- 22. Stockbauer KE, Grigsby D, Pan X, Fu YX, Mejia LM, Cravioto A, et al. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci USA. 1998;95(6):3128-33. 10.1073/pnas.95.6.3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Icelandic Tourist Board. Tourism in Iceland in Figures, January 2023: Summary for the year 2022. 2023. Reykjavík: Ferðamálastofa; 2023. Available from: https://www.ferdamalastofa.is/en/moya/news/tourism-in-figures-january-2023
- 24. Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA. 2014;111(17):E1768-76. 10.1073/pnas.1403138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.