Abstract
The transmembrane subunit (TM) of the avian leukosis and sarcoma virus (ALSV) envelope glycoprotein (Env) contains a stretch of conserved hydrophobic amino acids internal to its amino terminus (residues 21 to 42). By analogy with similar sequences in other viral envelope glycoproteins, this region has been proposed to be a fusion peptide. We investigated the role of this region by changing each of three hydrophobic residues (Ile-21, Val-30, and Ile-39) to glutamatic acid and lysine in the ALSV subgroup A Env. Like wild-type (wt) Env, all six mutant Env proteins were proteolytically processed, oligomerized, and expressed at the cell surface in a form that bound Tva, the ALSV subgroup A receptor. Like wt Env, Ile21Glu, Ile21Lys, Val30Glu, and Val30Lys changed conformation upon binding Tva, as assayed by sensitivity to thermolysin. Ile39Glu and Ile39Lys were cleaved by thermolysin in both the absence and presence of Tva. Although incorporated into virus particles at approximately equal levels, all mutant Envs were compromised in their ability to support infection. The mutants at residues 21 and 30 showed levels of infection 2 to 3 orders of magnitude lower than that of wt Env. The mutants at residue 39 were noninfectious. Furthermore, none of the mutants displayed activity in a cell-cell fusion assay. Our results support the contention that residues 21 to 42 of ALSV subgroup A Env constitute its fusion peptide.
Entry of enveloped viruses into host cells requires fusion of the viral envelope with a host cell membrane. This reaction is carried out by the spike envelope glycoproteins found on the viral surface and can occur at either low or neutral pH, depending on the virus (2, 16). Viruses that require low pH for fusion bind to receptors at the cell surface and are then internalized via the endocytic pathway. Biochemical and biophysical studies have shown that in the low-pH environment of the endosome, the corresponding viral fusion proteins undergo conformational changes that transform them into active fusion proteins (25). These changes include exposure of a previously buried hydrophobic region of the protein called the fusion peptide. Interaction of the fusion peptide with the endosomal membrane initiates the fusion event (16). Viruses that do not require low pH are thought to enter cells by fusing directly at the plasma membrane. Considerably less is known about the mechanisms of viral fusion proteins that function at neutral pH (18).
The avian leukosis and sarcoma viruses (ALSVs) are members of the retrovirus family and, like most retroviruses, fuse with target cells at neutral pH (11). ALSVs are divided into five major subgroups designated A through E based on host range, receptor binding, and interference patterns. The ALSV envelope (Env) glycoprotein is synthesized as a glycosylated precursor, Pr95, that is proteolytically cleaved during transport to the plasma membrane, yielding the mature surface (SU) and transmembrane (TM) subunits, with the SU subunit containing receptor binding and subgroup specificity determinants (29). The SU and TM subunits remain associated by a disulfide bond(s) and are oligomerized as trimers (5). For ALSV subgroup C (ALSV-C), it has been shown that posttranslational cleavage is required for infectivity (23).
The overall organization of ALSV Env is similar to those of the fusion proteins of other viruses, especially those of other retroviruses and members of the paramyxovirus and orthomyxovirus families. Like ALSV Env, the fusion proteins of retro-, paramyxo-, and orthomyxoviruses are proteolytically processed from a precursor protein into the mature receptor binding and TM subunits. The TM subunits of these fusion proteins generally contain a stretch of hydrophobic amino acids at their amino termini. In several cases, this region has been implicated in viral fusion activity and has thus been termed the fusion peptide. In general, fusion peptides (i) are 15 to 25 amino acids in length, (ii) are relatively hydrophobic, (iii) can be modeled as an alpha helix with the bulky hydrophobic residues lying on one face of the helix, and (iv) are rich in alanine and glycine (31). Where studied, the fusion peptide can be labeled by photoactivatable phospholipids in target membranes (4, 15), and mutations that introduce changes at hydrophobic residues impair or abolish fusion activity (8, 10, 32, 36). Fusion peptides are not always at the N terminus; internal fusion peptides have been identified in the Semliki Forest virus (SFV) and vesicular stomatitis virus (VSV) fusion proteins (20, 32, 36). Internal fusion peptides generally contain a helix-breaking residue such as proline near their centers (31). Synthetic fusion peptides adopt either α-helical or β-sheet structures in membranes (16).
By analogy with other viral fusion proteins, a sequence in ALSV Env, residues 21 to 42 of the TM subunit, has been predicted to be its fusion peptide. Consistent with its suggested importance, this internal hydrophobic region is highly conserved among the different ALSV subgroups. Furthermore, Ebola virus, a member of the filovirus family whose envelope glycoprotein bears striking resemblance to the TM subunit of ALSV Env, contains a candidate fusion peptide at an analogous location (9). The candidate fusion peptide of ALSV Env, however, has not yet been subjected to experimental analysis.
We tested the role of the candidate fusion peptide of the Env of ALSV-A (Env A) by mutating three of its hydrophobic residues to charged residues. The effects of these mutations on the processing, cell surface expression, oligomerization, and receptor binding of Env as well as the ability of the mutant Envs to mediate infection and cell-cell fusion were examined. Our findings are fully consistent with the prediction that this hydrophobic region serves as the fusion peptide of ALSV Env.
MATERIALS AND METHODS
Recombinant DNA and plasmids.
Plasmids pCB6-Env A, pCB6-Env C, pCB6-Env Acl, and pCB6-Tva have been described previously (12, 13). Mutations within the putative fusion region were created by oligonucleotide-directed site-specific mutagenesis (17) on single-stranded preparations of Env A cDNA which had been cloned into the BamHI site of Bluescript. The oligonucleotides used were Glu 1 (5′CTTTGGGGTCCTACAGCTCGAGAATTTGCATCTATCTTAGCCCCGGGG3′), Glu 2 (5′GCATCTATCTTAGCCCCGGGGGAAGCTGCAGCGCAAGCCTTAAGA3′), Glu 3 (5′CAAGCCTTAAGAGAAGAAGAGAGGCTAGCCTGTTGGTCCG3′), Lys 1 (5′CTTTGGGGTCCTACAGCAAGAAAATTTGCATCGATCTTAGCCCCGGGG3′), Lys 2 (5′GCATCTATCTTAGCCCCGGGGAAAGCTGCAGCGCAAGCCTTAAGA3′), and Lys 3 (5′CAAGCCTTAAGAGAAAAAGAGCGCCTAGCCTGTTGGTCCG3′). The oligonucleotides were designed to encode the altered amino acid as well as a new restriction site for rapid detection of mutants. Mutant plasmids were sequenced to confirm the fidelity of mutagenesis and then subcloned into the BamHI site of pCB6. The murine leukemia virus (MLV) Gag-Pol expression plasmid pHIT60 and the lacZ expression plasmid pHIT111 were gifts from Alan Kingsman. The pOS8 plasmid, containing lacZ under the T7 promoter, was a gift from Bernard Moss.
Antibodies.
The rabbit polyclonal antibodies against the carboxy-terminal cytoplasmic tails of Env A and Env C have been described previously (12). The rabbit polyclonal antiserum anti-Ngp37 was raised against a peptide corresponding to the first 17 residues of the amino terminus of the TM subunit. Antibodies were affinity purified against the corresponding peptide coupled to a SulfoLink column (Pierce Chemical Company, Rockford, Ill.) according to the manufacturer’s instructions. The rat anti-MLV Gag monoclonal antibody was obtained from culture supernatants of a hybridoma cell line purchased from the American Type Culture Collection. Donkey anti-rabbit (Amersham, Arlington Heights, Ill.) and goat anti-rat (Jackson ImmunoResearch Laboratories, West Grove, Pa.) secondary antibodies coupled to horseradish peroxidase (HRP) were used for Western blot analyses.
Cells, transfections, production of pseudotyped viruses, and virus titration.
Stable NIH 3T3 cell lines were established by transfection of pCB6 constructs, using the calcium phosphate precipitation method (33), and by selection of Geneticin-resistant colonies as described previously (12, 13). Single-cell clones of the fusion peptide mutant cell lines were obtained by limiting dilution. All stable NIH 3T3 cell lines and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% supplemented calf serum (SCS; HyClone, Ogden, Utah) and 500 mg of Geneticin per liter. To induce higher levels of protein expression, cells were treated with sodium butyrate overnight: 5 mM for Env Acl; 10 mM for Tva, Glu 1, Glu 2, Lys 1, and Lys 2; 15 mM for Glu 3; 20 mM for Lys 3; and 25 mM for Env A.
The three-plasmid transfection method was used to produce replication-incompetent (MLV) ALSV pseudotyped virus (26). 293T cells in 10-cm-diameter plates were transfected by the calcium phosphate precipitation method when ∼50 to 70% confluent with a total of 30 μg of DNA consisting of 10 μg each of pHIT60 (Gag-Pol), pHIT111 (lacZ), and pCB6-Env. All of these plasmids contain both a cytomegalovirus promoter and a simian virus 40 ori. Cells were treated with 10 mM sodium butyrate 24 h after transfection. Viral supernatants were harvested 48 h posttransfection, and supernatants were centrifuged at 1,500 × g for 10 min at room temperature to remove cell debris.
To assay for ALSV Env-mediated infection, serial dilutions of the pseudotype viral supernatants were added to Tva-expressing 3T3 cells plated on six-well dishes at a density of 2 × 105 cells per well the day before the infection; 48 h postinfection, cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Boehringer Mannheim, Indianapolis, Ind.) as previously described (24) to identify infected cells.
Envelope incorporation into virus.
To check incorporation of wild-type (wt) and mutant Envs into (MLV) ALSV pseudotyped viruses, 293T cell viral supernatants were centrifuged at 1,500 × g for 10 min at room temperature to remove cell debris. Viral supernatants (8 ml) were centrifuged at 4°C through 20% sucrose for 2 h at 100,000 × g in a Beckman SW41 rotor. Pellets were resuspended in Laemmli sample buffer, boiled, analyzed on a Western blot probed with either the anti-A tail or anti-MLV Gag antibody, and visualized by enhanced chemiluminescence (ECL; Amersham). All blots were exposed in the linear range. To confirm Env incorporation, wt, Glu 1, Glu 2, and Glu 3 viral pellets were resuspended in 200 μl of TE buffer (5 mM Tris-hydrochloride, 1 mM EDTA [pH 8.6]) with brief sonication. The samples were then layered onto a 15 to 60% linear sucrose gradient in TE buffer and centrifuged at 4°C for 3 h at 250,000 × g in a Beckman SW41 rotor. Fractions of 500 μl were collected, precipitated with chloroform-methanol (16a), and analyzed on a Western blot probed with the anti-A tail antibody and a donkey anti-rabbit-HRP secondary antibody. Blots were then incubated for 30 min at 70°C in stripping buffer (2% sodium dodecyl sulfate [SDS], 100 mM 2-mercaptoethanol, a 62.5 mM Tris [pH 6.8]) to remove primary and secondary antibodies from the membranes. Membranes were reprobed with secondary antibody, incubated with ECL detection reagents, and exposed to film to ensure removal of any potential cross-reacting antibodies. Blots were then reprobed with the anti-MLV Gag antibody and a goat anti-rat-HRP secondary antibody and visualized by ECL.
Cell surface labeling, immunoprecipitation, coimmunoprecipitation, and thermolysin digestion assay.
After induction with sodium butyrate for 16 to 18 h, cells expressing either Env or Tva were labeled with the membrane-impermeant biotinylation reagent NHS-LC-biotin (Pierce). Cells were lysed, and proteins were immunoprecipitated or coimmunoprecipitated with the anti-A tail antibody as described previously (13). Immunoprecipitates were washed and processed for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) or were used in a thermolysin digestion assay as described previously (13). The soluble Tva (sTva) used in the thermolysin assay was a gift from Paul Bates. Following electrophoresis, proteins were transferred to nitrocellulose and probed with HRP coupled to streptavidin (Pierce) to detect biotinylated proteins. The HRP signal was detected by ECL.
Analysis of oligomer formation.
To determine the oligomeric state of the Env proteins, cell lysates, prepared as described in reference 14, containing biotinylated Env proteins were layered onto a 10 to 30% linear sucrose gradient in HEPES-buffered saline containing 40 mM octylglucoside and centrifuged in a Beckman SW41 rotor for 17 h at 275,000 × g at 4°C. The gradient was fractionated and processed for the detection of Env proteins as described previously (14).
Fusion assay.
The fusion assay used is a modified version of that reported by Nussbaum et al. (22). 293T cells were cotransfected as described above with pOS8 and a pCB6-Env vector or pCB6-Tva; 16 to 18 h before the fusion assay, six-well dishes of Tva- or Env-expressing 3T3 cells were infected with the modified vaccinia virus Ankara (MVA) (34) at a multiplicity of infection of approximately 10 for 30 min at 37°C in DMEM supplemented with 2% SCS. Cells were then washed twice with DMEM–10% SCS and incubated for 16 to 18 h at 31°C in DMEM–10% SCS supplemented with 100 μg of cytosine β-d-arabinofuranoside (Ara-C; Sigma, St. Louis, Mo.) per ml. Uninfected Env- or Tva-expressing 293T cells were lifted off the dish with calcium- and magnesium-free phosphate-buffered saline (PBS), centrifuged, resuspended in DMEM–10% SCS containing 100 μg of Ara-C per ml, and then added to vaccinia virus-infected Tva- or Env-expressing cells. The cell mixtures were incubated at 37°C for 12 h. Cells were analyzed for β-galactosidase activity either by staining with X-Gal or by measuring β-galactosidase activity as described below. MVA was a gift from Bernard Moss.
Analysis of β-galactosidase activity.
Following the fusion assay, cells were lifted off the dish, pelleted, and lysed in 50 μl of lysis buffer (1% Nonidet P-40 in 130 mM NaCl–20 mM HEPES [pH 7.4]). A 5- to 10-μl aliquot of the cell lysate was diluted in 90 μl of lysis buffer containing 0.1 mg of 4-methylumbelliferyl galactoside (MUG; Molecular Probes, Eugene, Oreg.) per ml and incubated at 37°C for 4 min. The reactions were quenched with 2 ml of 0.2 M glycine (pH 10.0), and the fluorescence in each sample was determined in an LS-5B fluorimeter (Perkin-Elmer, San Jose, Calif.) with the excitation and emission wavelengths set at 365 and 450 nm, respectively.
Flow cytometry.
Approximately 2 × 105 transiently transfected 293T cells were incubated on ice with the anti-Ngp37 antibody in a total volume of 100 μl of PBS–2% fetal calf serum (FCS) for 30 min. Cells were washed twice in PBS and then incubated with fluoresceinated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories) in 100 μl of PBS–2% FCS on ice. After 30 min, cells were washed twice with PBS, resuspended in PBS containing 2% paraformaldehyde, and analyzed at the University of Virginia FACS (fluorescence-activated cell sorting) Core Facility, using a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
RESULTS AND DISCUSSION
Design of mutant Env proteins.
Sequences are available for the Env proteins of four of the five major ALSV subgroups. Analysis of these sequences revealed a highly conserved hydrophobic region near the amino terminus of the TM subunit (amino acids 21 to 42; Fig. 1A and B). This region is also strikingly similar in relative location and overall hydrophobicity to the predicted fusion peptide of the Ebola virus envelope glycoprotein (9). By analogy with other viral fusion proteins, this region is predicted to be the fusion peptide of ALSV Env. Similar to many fusion peptides, this region displays a hydrophobic face when modeled as an alpha helix (Fig. 1C). To examine the importance of this region, we engineered six mutations that replaced the hydrophobic residues Ile-21, Val-30, and Ile-39 (marked as positions 1, 2, and 3) on the hydrophobic face of the modeled helix with the charged residues glutamic acid or lysine. We predicted that insertion of a charged residue would disrupt the hydrophobicity of the fusion peptide and therefore impair its ability to interact with membranes and initiate fusion. We chose the charged amino acids glutamic acid and lysine since they have side chain volumes similar to those of the replaced amino acids valine and isoleucine and display either a negative or a positive charge. We refer to the mutant Env proteins as Glu 1, Glu 2, Glu 3, Lys 1, Lys 2, and Lys 3, with the name designating the amino acid and position of the substitution (Fig. 1C). Note that although we have modeled this region as an alpha helix, it is quite possible that part or all of this region adopts alternate structures either before or during fusion (16).
FIG. 1.
ALSV putative fusion peptide region. (A) Overlay of the hydropathy plots of the TM subunits of Env glycoproteins of subgroups A (Schmidt-Ruppin), C (Prague), D (Schmidt-Ruppin), and E (RAV-0) ALSV. Arrow points to the candidate fusion peptide region. ∗, TM domain. (B) Amino acid sequence of the candidate fusion peptide region in four ALSV subgroups. •, identical amino acid at that position. (C) Putative fusion peptide region of ALSV-A modeled as an alpha helix. The hydrophobic face is outlined and has an average hydrophobicity index of 1.01. The position 1, 2, and 3 amino acids are circled. As discussed in the text, this region may adopt other structures either before or during fusion.
Expression, processing, and oligomerization of mutant Env proteins.
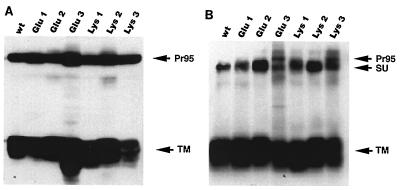
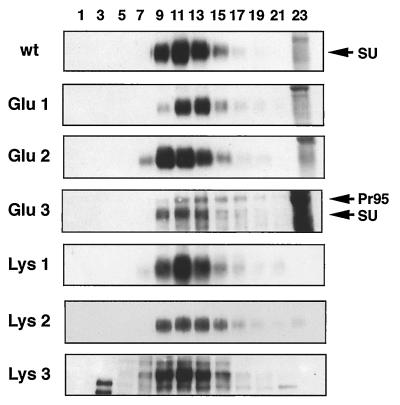
Because the designed mutations are within a highly conserved region that may be important for protein structure, we first characterized the expression, processing, and oligomerization of the mutant Envs. Stable NIH 3T3 cell lines expressing each of the mutant proteins were established. After induction with sodium butyrate for 16 to 18 h, cells were lysed and analyzed by Western blotting with an antibody against the cytoplasmic tail of Env A to assess total Env expression. Alternatively, cells were labeled with the membrane-impermeant reagent NHS-LC-biotin, lysed, and immunoprecipitated with anti-A tail antibody to assess cell surface forms of Env. As shown in Fig. 2A, a Pr95 polypeptide, similar in mobility to wt Pr95, was expressed in all of the mutant cell lines. The proteins were processed to the mature form since a TM subunit similar in mobility to the wt TM was detected in each mutant. Glu 1, Glu 2, Lys 1, and Lys 2 were processed and transported to the cell surface similar to wt Env since only mature SU and TM proteins were detected at the cell surface (Fig. 2B). The position 3 mutants were also expressed as mature protein at the cell surface. However, in these cases processing was less efficient, since some precursor (a band similar in size to Pr95) was transported to the surface. We next examined oligomerization of the mutant proteins. Cells were labeled with the membrane-impermeant biotinylating reagent, lysed, and subjected to sucrose density centrifugation (Fig. 3). All of the mutant proteins sedimented to a position similar to that of wt Env, consistent with trimer formation (5, 14). For all mutants except Glu 3 and Lys 3, only the mature protein was observed in the trimer fractions. For Glu 3 and Lys 3, some trimerized Pr95 was found as well.
FIG. 2.
Expression of mutant proteins in stable NIH 3T3 cell lines. (A) Lysates of Env-expressing NIH 3T3 cells were boiled and reduced in sample buffer, separated by SDS-PAGE (11.5% gel), transferred to nitrocellulose, and Western blotted with anti-A tail antibodies. (B) A parallel set of Env-expressing cells were biotinylated with the membrane-impermeant reagent NHS-LC-biotin, lysed, and immunoprecipitated with anti-A tail antibodies as described in Materials and Methods. Immunecomplexes were analyzed by SDS-PAGE (11% gel), transferred to nitrocellulose, probed with streptavidin-HRP, and detected by ECL. The SU, TM, and Pr95 proteins migrated as shown.
FIG. 3.
Sucrose density centrifugation analysis of cell surface Env proteins. Env-expressing NIH 3T3 cells were biotinylated as described in the legend to Fig. 2B, lysed, and subjected to sucrose gradient centrifugation as described in Materials and Methods. After centrifugation, fractions were collected, immunoprecipitated with an anti-A tail antibody, subjected to SDS-PAGE (9% gel), and transferred to nitrocellulose. Biotinylated proteins were detected as described in the legend to Fig. 2B. Fraction 1 indicates the top of the gradient.
In summary, all of the mutant Env proteins were processed, oligomerized, and expressed at the cell surface similarly to wt Env with the exception of the position 3 mutants, which were less efficiently processed and therefore expressed some precursor at the cell surface. Less efficient processing of the Glu 3 and Lys 3 mutants may be an indication of imprecise protein folding.
Ability of mutant Envs to interact with receptor and change conformation.
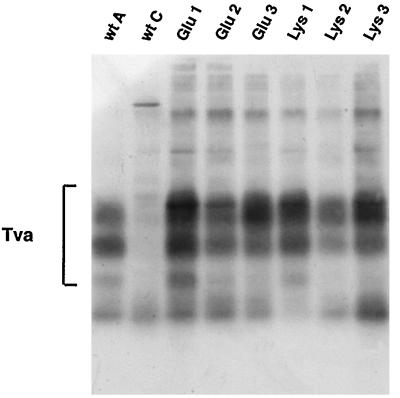
Tva is the receptor for ALSV-A (1, 35) and appears to be sufficient to mediate viral entry. We previously showed that Tva binds specifically to Env A but not to Env C as measured by a coimmunoprecipitation assay (12). We also showed that upon interaction with sTva, a soluble form of the Tva ectodomain, Env A undergoes a conformational change, demonstrated by the generation of a specific thermolysin digestion product of the SU subunit termed SU* (13). We tested whether the mutant Env proteins were able to bind Tva and, if so, whether they could undergo the receptor-induced conformational change.
To assess the ability of the mutant proteins to interact with Tva, we performed a coimmunoprecipitation assay. As seen in Fig. 4, Tva coimmunoprecipitated with all of the mutant Envs, demonstrating that the mutants retained the ability to bind receptor. Tva is a highly modified protein and runs as a broad smear on a gel with three prominent bands (1, 12). The relative amount of the lowest-molecular-weight Tva band precipitated varied from experiment to experiment in all samples. The mutants were next tested in the receptor-induced conformational change assay. The position 1 and 2 mutants behaved the same as wt Env; SU* was produced to significant levels only in the presence of sTva (Fig. 5). The position 3 mutants, however, were sensitive to thermolysin in both the absence and presence of sTva, and SU proteins were digested almost beyond detection. To increase sensitivity, we repeated the assays of Glu 3 and Lys 3 with three times more starting Env protein. Nonetheless, the Glu 3 and Lys 3 proteins were still digested by thermolysin to SU* in both the presence and absence of sTva (Fig. 5B). SU* generated from the position 3 mutants appeared to migrate at a slightly lower molecular weight than SU* from wt Env.
FIG. 4.
Receptor binding activity of mutant Env proteins. NIH 3T3 cells expressing Tva were biotinylated and lysed as described in the legend to Fig. 2B. Unlabeled lysates of cells expressing Env A, Env C, or mutant Env A proteins were mixed with the biotinylated Tva lysate and then immunoprecipitated with anti-A tail (or anti-C tail) antibodies. Samples were resolved on SDS-PAGE (12% gel) and blotted to nitrocellulose. Biotinylated proteins were detected as described in the legend to Fig. 2B.
FIG. 5.
Thermolysin digestion of envelope proteins in the absence and presence of sTva. (A) NIH 3T3 cells expressing Env proteins were biotinylated with the membrane-impermeant reagent NHS-LC-biotin, lysed, immunoprecipitated with anti-A or anti-C tail antibody, and washed. Immunecomplexes were then subjected to thermolysin digestion in the presence or absence of sTva as described previously (13). Samples were boiled, separated by SDS-PAGE (11% gel), transferred to nitrocellulose, probed with streptavidin-HRP, and visualized by ECL. The SU and TM subunits and SU* migrate as indicated. (B) The thermolysin digestion assay was repeated with the Glu 3 and Lys 3 mutants, using three times as much Env protein.
The findings presented in Fig. 5 suggest that the position 1 and 2 mutants change conformation in the presence of sTva like wt Env. As shown in Fig. 5A and B, the position 3 mutants are not folded the same way as wt Env. Certain mutations within the envelope glycoproteins of other viruses affect protein folding. In some cases, the defect can be overcome by incubating cells at a lower temperature (20). To test if the apparent misfolding of Glu 3 and Lys 3 was temperature sensitive, cells expressing Glu 3 and Lys 3 were incubated for 16 to 18 h at 28°C before being harvested for the thermolysin assay. Results similar to those presented in Fig. 5 were obtained (data not shown). The conformational change measured in the thermolysin assay may be related to activation of the fusion protein. It is interesting that SU* is seen in both the absence and presence of sTva with the position 3 mutants. This may be an indication that the position 3 Envs are “presprung,” or preactivated (3).
Infectivity of mutants.
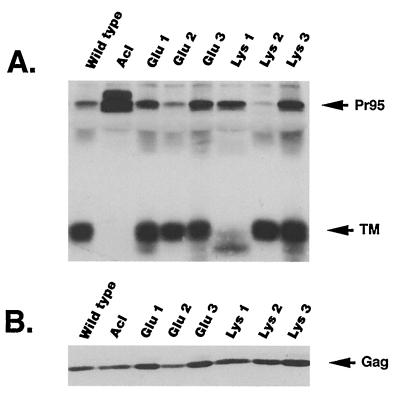
MLV particles efficiently incorporate ALSV Env into their envelopes (19). The resulting MLV(ALSV) pseudotypes are capable of infecting cells, but only if they express the appropriate ALSV receptor. We tested the ability of the mutant Envs to mediate entry of MLV(ALSV-A) pseudotypes into cells expressing Tva as a correlate of their fusion activity. MLV(ALSV-A) particles containing a lacZ reporter gene were made in 293T cells by using the three-plasmid transient transfection system (26). Briefly, the expression plasmids pHIT60 (containing MLV gag-pol), pHIT111 (containing lacZ), and pCB6-Env (either wt A, wt C, Acl, or a fusion peptide mutant) were cotransfected into 293T cells. Culture supernatants were collected 2 days posttransfection and assessed for particle production and envelope incorporation. Virus particles were pelleted from equal amounts of culture supernatants, resuspended in sample buffer, separated by SDS-PAGE, and Western blotted with the anti-A tail antibody (Fig. 6A). The same blot was then reprobed with the anti-MLV Gag antibody (Fig. 6B). All viral supernatants contained approximately the same number of viral particles since similar levels of Gag were detected. All mutant Env proteins were incorporated into virus at or near wt levels; equivalent amounts of TM protein (or Pr95 in the case of the Acl [13], a cleavage site mutant that is not processed to the mature SU and TM subunits) were detected in all of the samples.
FIG. 6.
Env incorporation into pseudotyped virus particles. Equal amounts of 293T cell viral supernatants were centrifuged through 20% sucrose as described in Materials and Methods. (A) Pellets were resuspended in sample buffer, boiled, analyzed on a Western blot probed with the anti-A tail antibody, and visualized by ECL. (B) The same blot was reprobed with the anti-Gag antibody and visualized by ECL.
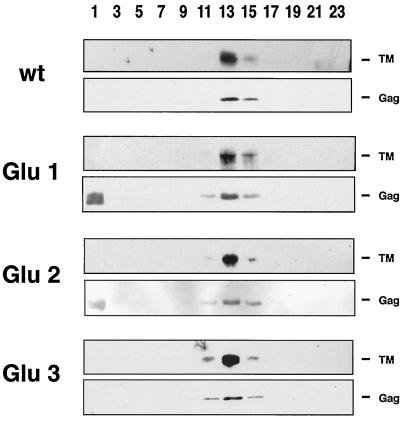
To confirm Env incorporation more rigorously, wt, Glu 1, Glu 2, and Glu 3 pseudotyped virions were also analyzed by sucrose density sedimentation. As shown in Fig. 7, wt Env as well as each of the mutant Envs comigrated with Gag at an approximate density of 1.15 g/ml, demonstrating that the Env proteins were, indeed, associated with intact virions (and not, for example, merely with plasma membrane vesicles). All of the mutant Envs appear to be incorporated into virions at levels approximately equal to each other and to that of wt Env.
FIG. 7.
Sedimentation analysis of pseudotyped virus particles. Equal amounts of 293T cell viral supernatants were pelleted through 20% sucrose as described in the text. Pellets were resuspended in 200 μl of TE buffer and subjected to centrifugation on a continuous 15 to 60% sucrose gradient as described in Materials and Methods. After centrifugation, fractions were chloroform-methanol precipitated, run on an SDS–12.5% gel, analyzed on a Western blot probed with the anti-A tail antibody, and visualized by ECL. Blots were then stripped as described in Materials and Methods, reprobed with the anti-Gag antibody, and visualized by ECL. 1 denotes the top of the gradient. A band is variably seen in fraction 1 of blots probed with the anti-Gag antibody. We do not think this is Gag because it migrates somewhat faster and always more broadly than Gag. No Pr95 was seen in these viral particles.
All mutant TM proteins from the pseudotyped virions migrated at the same apparent molecular weight as the wt TM except for the Lys 1 mutant, whose TM subunit migrated at a lower apparent molecular weight (Fig. 6A). Use of the terminal glycosylation inhibitor deoxymannojirimycin did not change its apparent molecular weight, and antibodies to both the N and C termini of TM recognized the protein, excluding the possibility of proteolysis at the termini (data not shown). Nevertheless, since all of the mutant Env proteins were efficiently incorporated into virus particles, if we detected any differences in pseudotype infectivity, they could not be attributed to differences in envelope incorporation or particle production.
It was somewhat surprising that the position 3 mutants were efficiently incorporated into virions given the results of the thermolysin assay, which showed that these proteins had altered folding. To test the possibility that these mutants were folded differently in the 293T cells or in the presence of Gag, the thermolysin assay was performed on the Glu 3 and Lys 3 Envs harvested from pelleted virions. The results paralleled the assay done with the stable 3T3 cell lines in that, for the position 3 mutants, SU* was produced in both the presence and absence of Tva (data not shown).
The pseudotyped viral particles (Fig. 6) were next used to infect NIH 3T3 cells stably expressing Tva. Infectivity was measured by in situ staining of infected cells for β-galactosidase 2 days postinfection. The results showed that all of the mutants were impaired in infectivity with titers ranging from 1 to 5 orders of magnitude lower than with wt Env (Table 1). The Acl mutant was 3 orders of magnitude lower in its ability to mediate infection. The position 3 mutants had titers 3 to 5 orders of magnitude lower than the wt Env titer. Given the behavior of the position 3 mutants in the thermolysin assay, however, it is possible that their loss of infectivity was due in part or in full to aberrant folding. The titers of the position 1 and position 2 mutants were consistently 1 to 2 and 2 to 3 orders of magnitude lower, respectively, than the wt Env titer. From these results, it is evident that the internal hydrophobic domain at residues 21 to 42, particularly residues 21 to 30, plays an important role in Env-mediated viral entry. A lysine substitution at each position appears to have a greater effect on fusion than a glutamic acid at the same position. The reason for this is not clear but may relate to the fact that lysine is more hydrophilic than glutamic acid (6) and would thereby disrupt the hydrophobicity of the region to a greater extent.
TABLE 1.
Infectivity of Env A fusion peptide mutants
| Mutant | Infectivitya | % of wt Env A level |
|---|---|---|
| wt Env A | 3.3 × 105 | 100 |
| wt Env C | 0 | 0 |
| Acl | 1.2 × 102 | 0.04 |
| Glu 1 | 2.3 × 104 | 6.9 |
| Glu 2 | 2.5 × 103 | 0.8 |
| Glu 3 | 2.2 × 102 | 0.07 |
| Lys 1 | 1.8 × 103 | 0.5 |
| Lys 2 | 9.0 × 102 | 0.3 |
| Lys 3 | 7.5 × 100 | 0.002 |
Expressed as infectious units per milliliter of viral stock.
Fusion activity of mutant Envs determined by cell-cell fusion assay.
The vaccinia virus/bacteriophage T7 system has been used previously to measure the fusion activity of viral envelope glycoproteins (7, 22). The assay measures the cytoplasmic activation of a reporter gene upon fusion of two different cell populations. We used a modified version of this system to measure fusion between cells expressing Env and cells expressing Tva. Briefly, Tva-expressing 3T3 cells were infected with a modified vaccinia virus Ankara (MVA), which encodes bacteriophage T7 RNA polymerase (34). MVA is a highly attenuated vaccinia virus strain that can assemble and replicate efficiently only in avian and BHK-21 cells (3a, 28). The infected cells were then overlaid with 293T cells which had been cotransfected with an Env expression plasmid and a plasmid with lacZ under the control of the T7 promoter (pOS8). Upon fusion, the lacZ gene is activated and the amount of β-galactosidase produced can be measured either by in situ staining or by an enzymatic assay of detergent cell lysates.
We first tested wt Env, Acl, and the position 1 and 2 mutants in the fusion assay (Fig. 8). The Glu 3 and Lys 3 mutants were tested separately since the assay conditions had to be modified to adjust for surface expression levels (see below). With wt Env, blue-stained cells were readily seen (Fig. 8A) and a β-galactosidase signal well above background was also measured (Fig. 8B). These findings indicated that fusion had occurred. For Acl and all of the position 1 and 2 mutants, no β-galactosidase activity above the mock-transfected (pOS8 alone) background was measured by either in situ staining or the enzymatic assay (Fig. 8A and B).
FIG. 8.
Cell-cell fusion assay of Glu 1, Glu 2, Lys 1, and Lys 2. 293T cells were cotransfected with pOS8 and pCB6 vectors expressing Env proteins; 48 h posttransfection, cells were lifted off the dish and split into three samples that were either checked for surface expression of envelope proteins or used in the cell-cell fusion assay. A portion of transfected 293T cells was overlaid onto Tva-expressing 3T3 cells which had been infected with MVA. After 12 h at 37°C, samples were either fixed and stained with X-Gal (A) or lysed and analyzed in the MUG β-galactosidase fluorometric assay (B). Results of the β-galactosidase fluorometric assay are expressed as fluorescence units and represent averages from three replicates; bars show standard deviations. All samples were read in the linear range of fluorescence emission. (C) A portion of transfected 293T cells were biotinylated with NHS-LC-biotin, lysed, and immunoprecipitated with anti-A tail antibody. Samples were analyzed as described in the legend to Fig. 2B. (D) Another subset of transfected 293T cells was incubated on ice with the anti-gp37 antibody in a total volume of 100 μl PBS–2% FCS for 30 min. Cells were washed twice in PBS and then incubated with a fluoresceinated goat anti-rabbit antibody (Jackson ImmunoResearch Laboratories) in 100 μl of PBS–2% FCS on ice. After 30 min, cells were washed twice with PBS, resuspended in PBS containing 2% paraformaldehyde, and analyzed on a Becton Dickinson flow cytometer.
Since fusion is dependent on protein expression at the cell surface, we analyzed the cells used in the fusion assay for cell surface expression of mutant Envs. 293T cells expressing Env were biotinylated, lysed, and immunoprecipitated with anti-A tail antibody (Fig. 8C). All of the position 1 and 2 mutant proteins were expressed at the cell surface at levels comparable to that of wt Env. As seen in Fig. 6, the TM form of Lys 1 appeared to migrate at a lower molecular weight than the others. Cell surface expression was further confirmed by FACS analysis using the anti-Ngp37 antibody, which recognizes the first 17 N-terminal residues of the TM domain. The results showed that all of the mutant proteins were expressed at the cell surface at levels comparable to that of wt Env (Fig. 8D). Thus, surface expression levels did not account for the low fusion activity of the Glu 1, Glu 2, Lys 1, and Lys 2 proteins.
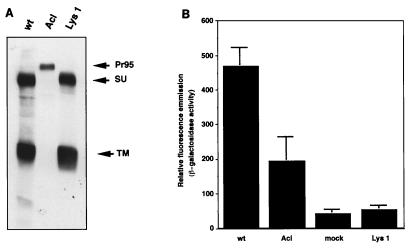
We next explored whether the low-molecular-weight form of the Lys 1 TM was responsible for its loss of fusion. The Lys 1 TM migrates at the same molecular weight as the wt TM when expressed in NIH 3T3 cells (Fig. 2) as opposed to when it is expressed in 293T cells (Fig. 6A and 8C). We therefore modified the fusion assay to use 3T3 cells. Stable 3T3 cells expressing wt, Acl, or Lys 1 mutant Env were infected with MVA and then overlaid with 293T cells transfected with pCB6-Tva and the β-galactosidase reporter plasmid. The results are seen in Fig. 9. The cell surface expression level of Lys 1 was comparable to that of wt Env as measured by immunoprecipitation of cell surface proteins (Fig. 9A). Despite this, Lys 1 expressed in 3T3 cells was again defective in fusion compared to wt Env (Fig. 9B). From this finding, we conclude that the fusion defect of Lys 1 was not due to the appearance of its TM subunit as a slightly lower molecular weight form in 293T cells. Lys 1 can therefore be considered a true fusion mutant.
FIG. 9.
Cell-cell fusion assay of Lys 1 mutant in 3T3 cells. NIH 3T3 cells expressing Env A, Env Acl, or Lys 1 were seeded in six-well dishes (105 cells per well) and incubated overnight. Cells were then infected with MVA at a multiplicity of infection of 1 as described in Materials and Methods and incubated overnight at 31°C in the presence of 100 μg of Ara-C per ml and sodium butyrate. Cells were then either analyzed for surface expression of Env proteins or used in the cell-cell fusion assay. (A) One well of a six-well dish of infected 3T3 cells was biotinylated with NHS-LC-biotin, lysed, and immunoprecipitated with anti-A tail antibody. Samples were analyzed as described in the legend to Fig. 2B. (B) Infected 3T3 cells were overlaid with 293T cells which had been cotransfected with pOS8 and pCB6-Tva 2 days earlier. After 24 h cells, were lifted off the dish, lysed and analyzed in the MUG β-galactosidase fluorometric assay. The data represent the averages of three replicates; bars show standard deviations.
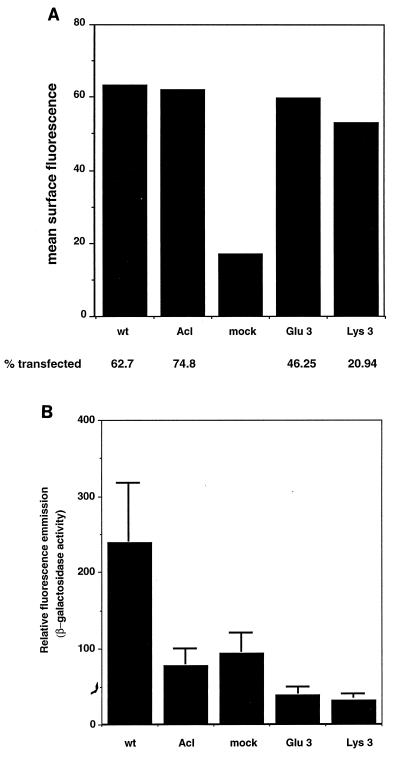
The position 3 mutants were tested in the cell-cell fusion assay to corroborate the results of the infectivity assay. While the Glu 3 and Lys 3 mutants were efficiently incorporated into virus particles, FACS analysis showed that the surface expression levels of both mutants were lower than the wt Env level when the same amount of input DNA was used (data not shown). Attempts to equalize surface expression were made by adjusting the amounts of transfected wt and mutant DNAs. Equal surface expression levels were achieved, but equal transfection efficiencies (percent Env-expressing cells) were never attained (Fig. 10A). This finding again suggests that the position 3 mutant proteins are not folded exactly as wt Env and are not transported efficiently to the cell surface in 293T cells. Even after normalizing for lower transfection efficiencies, we detected no fusion activity above background in the Glu 3 and Lys 3 mutants (Fig. 10B), consistent with the results from the infectivity assay. It was not expected that placement of a charged residue at position 3 would result in a misfolded protein since position 3 is flanked on both sides by charged residues (Fig. 1B and C). Since the position 3 mutants behave as if they are presprung (Fig. 5), the conservation of a hydrophobic residue at this position may be important for maintaining Env in a metastable prefusogenic conformation.
FIG. 10.
Cell-cell fusion assay of Glu 3 and Lys 3. 293T cells were cotransfected with pOS8 and either pCB6-Env A, Env Acl, Glu 3, Lys 3, or pCB6 alone; 48 h posttransfection, cells were lifted off the dish and either checked for cell surface expression or used in the cell-cell fusion assay. (A) FACS analysis of transfected 293T cells. (B) Transfected 293T cells were overlaid onto MVA-infected Tva-expressing 3T3 cells. After 24 h at 37°C, samples were lysed and analyzed in the MUG β-galactosidase fluorometric assay. The data represent the averages of three replicates; bars show standard deviations.
Summary.
The viral fusion proteins of orthomyxo-, paramyxo-, and retroviruses are arranged similarly, with a mature transmembrane and surface subunit being generated by proteolytic processing of a precursor protein. The newly revealed N terminus of the TM subunit contains the fusion peptide, a hydrophobic domain that interacts with the target membrane to initiate fusion (16). Internal fusion peptides are not as common but have been identified in the E1 and G proteins of SFV and VSV, respectively (4, 20, 32, 36). Mutations within these latter fusion peptides affect membrane fusion. Photolabeling experiments have shown that, like the hemagglutinin fusion peptide (15), the VSV fusion peptide interacts hydrophobically with target membranes (4).
Unlike most retroviruses, ALSV has a candidate fusion peptide internal to the N terminus of its TM subunit. Using site-directed mutagenesis, we have made six mutations in this region, each of which replaces a hydrophobic amino acid with a charged amino acid of approximately the same side chain volume. The mutant proteins all showed decreased fusion activity in both infectivity and cell-cell fusion assays. The mutations at positions 1 and 2 clearly impaired Env-mediated fusion without affecting its folding, oligomerization, cell surface expression, receptor binding, or ability to undergo a receptor-induced conformational change (Table 2). The behavior of the position 1 and 2 mutants therefore supports the contention that amino acids 21 to 42 of the TM subunit is the ALSV Env fusion peptide. Consistent with this is the fact that the Glu 2 mutant is significantly impaired in its ability to bind to liposomes when triggered by soluble receptor (16a). The mutants at position 3 had the greatest impairment in infectivity, but it is difficult to accurately assess their fusion phenotypes, as these proteins are not folded exactly as wt Env. In fact, our analysis of thermolysin susceptibility presented in Fig. 5 suggests that the position 3 mutants may be presprung into an inactive conformation much in the way that low pH pretreatment of influenza virus and SFV inactivates their fusion proteins (21, 27, 30).
TABLE 2.
Characterization of fusion peptide mutants
| Peptidea | Cell surface expression | Processing | Oligomer (trimer) | Receptor binding | SU* production
|
Infectivityb | Env viral incorporation | Fusion | |
|---|---|---|---|---|---|---|---|---|---|
| −Tva | +Tva | ||||||||
| wt | + | + | + | + | − | + | 3.3 × 105 | + | + |
| Mutants | + | + | |||||||
| Position 1 | |||||||||
| Glu | + | + | + | + | − | + | 2.3 × 104 | + | − |
| Lys | + | + | + | + | − | + | 1.8 × 103 | + | − |
| Position 2 | |||||||||
| Glu | + | + | + | + | − | + | 2.5 × 103 | + | − |
| Lys | + | + | + | + | − | + | 9.0 × 102 | + | − |
| Position 3 | |||||||||
| Glu | + | +/− | + | + | + | + | 2.2 × 102 | + | − |
| Lys | + | +/− | + | + | + | + | 7.5 × 100 | + | − |
Positions 1, 2, and 3 are underlined, in that order, in the sequence I F A S I L A P G V A A A Q A L R E I E R L.
Expressed as infectious units per milliliter of viral stock.
A structural model for the ALSV TM subunit is very similar to that for the carboxy-terminal 181 amino acids of the Ebola virus glycoprotein (9). Our results suggest that the analogous region in the Ebola virus glycoprotein (residues 608 to 623) may function as a fusion peptide.
ACKNOWLEDGMENTS
We thank Sue Delos for generating and characterizing the anti-Ngp37 antibody, Joanna Gilbert for help in designing the oligonucleotides used for mutagenesis, Paul Bates and John Balliet for supplying sTva, Bernard Moss and Linda Wyatt for supplying MVA and pOS8 and for advice on using MVA, Alan Kingsman for supplying the pHIT pseudotyping vectors, and William Ross at the UVA FACS Core Facility for performing the flow cytometry analysis.
This work was supported by grant A122470 from the National Institutes of Health to J.M.W. and a Howard Hughes predoctoral fellowship to L.D.H.
REFERENCES
- 1.Bates P, Young J A T, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 2.Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. [Google Scholar]
- 3.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 3a.Carroll M W, Moss B. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman cell line. Virology. 1997;238:198–211. doi: 10.1006/viro.1997.8845. [DOI] [PubMed] [Google Scholar]
- 4.Durrer P, Gaudin Y, Ruigrok R W H, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 5.Einfeld D, Hunter E. Oligomeric structure of a prototypic retrovirus glycoprotein. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 7.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 8.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 10.Gething M-J, Doms R W, York D, White J M. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert J, Mason D, White J. Fusion of Rous sarcoma virus with host cells does not require low pH. J Virol. 1990;64:5106–5113. doi: 10.1128/jvi.64.10.5106-5113.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert J M, Bates P, Varmus H E, White J M. The receptor for the subgroup A avian leukosis sarcoma virus binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;67:6889–6892. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert J M, Hernandez L D, Chernov-Rogan T, White J M. Generation of a water soluble oligomeric ectodomain of the Rous sarcoma virus envelope glycoprotein. J Virol. 1993;67:6889–6892. doi: 10.1128/jvi.67.11.6889-6892.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harter C, James P, Bächi T, Semenza G, Brunner J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the “fusion peptide.”. J Biol Chem. 1989;264:6459–6464. [PubMed] [Google Scholar]
- 16.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 16a.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 18.Lamb R A. Paramyxovirus fusion: a hypothesis for change. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 19.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh M, Helenius A. Virus entry into animal cells. Adv Virus Res. 1989;36:107–151. doi: 10.1016/S0065-3527(08)60583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoprotein analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J Virol. 1987;61:1609–1614. doi: 10.1128/jvi.61.5.1609-1614.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanes J R, Rubenstein J L, Nicolas J F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skehel J J, Bizebard T, Bullough P A, Hughson F M, Knossow M, Steinhauer D A, Wharton S A, Wiley D C. Membrane fusion by influenza hemagglutinin. Cold Spring Harbor Symp Quant Biol. 1995;60:573–580. doi: 10.1101/sqb.1995.060.01.061. [DOI] [PubMed] [Google Scholar]
- 26.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegmann T, Booy F P, Wilschut J. Effects of low pH on influenza virus. Activation and inactivation of the membrane fusion capacity of the hemagglutinin. J Biol Chem. 1987;262:17744–17749. [PubMed] [Google Scholar]
- 28.Sutter G, Moss B. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc Natl Acad Sci USA. 1992;89:10847–10851. doi: 10.1073/pnas.89.22.10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss R. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J, editor. The retroviruses. Vol. 2. New York, N.Y: Plenum Press; 1992. pp. 1–108. [Google Scholar]
- 30.White J, Kartenbeck J, Helenius A. Membrane fusion activity of influenza virus. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 32.Whitt M A, Zagouras P, Crise B, Rose J K. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J Virol. 1990;64:4907–4913. doi: 10.1128/jvi.64.10.4907-4913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wigler M, Silverstein S, Lee L-S, Pellicer A, Cheng Y-C, Axel R. Transfer of purified herpes virus thymidine kinase gene to cultured mouse cells. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 34.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 35.Young J A, Bates P, Varmus H E. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]